Abstract
Coat color and type are essential characteristics of domestic dog breeds. Although the genetic basis of coat color has been well characterized, relatively little is known about the genes influencing coat growth pattern, length, and curl. We performed genome-wide association studies of more than 1000 dogs from 80 domestic breeds to identify genes associated with canine fur phenotypes. Taking advantage of both inter- and intrabreed variability, we identified distinct mutations in three genes, RSPO2, FGF5, and KRT71 (encoding R-spondin–2, fibroblast growth factor–5, and keratin-71, respectively), that together account for most coat phenotypes in purebred dogs in the United States. Thus, an array of varied and seemingly complex phenotypes can be reduced to the combinatorial effects of only a few genes.
The tremendous phenotypic diversity of modern dog breeds represents the end point of a >15,000-year experiment in artificial and natural selection (1, 2). As has been demonstrated for traits such as body size (3) and coat color (4), marker-based associations with phenotypic traits can be explored within single breeds to initially identify regions of genetic association, and then expanded to multiple breeds for fine-mapping and mutation scanning (5, 6). Coat (pelage) phenotypes are particularly amenable to this strategy as they show a huge amount of variation across breeds but still allow for simple variation within single breeds (7). This offers a unique strategy for advancing the genetic understanding of a complex phenotype.
We used the structured pattern of fur variation in dogs to localize the genetic basis of three characteristics of the canine coat: (i) the presence or absence of “furnishings,” the growth pattern marked by a moustache and eyebrows typically observed in wire-haired dogs; (ii) hair length; and (iii) the presence or absence of curl. To accomplish this, we generated three genome-wide single-nucleotide polymorphism (SNP) data sets using the Affymetrix version 2.0 canine SNP chip (8, 9). The first data set consisted of 96 dachshunds segregating three coat varieties: wire-haired with furnishings, smooth, and long-haired without furnishings. The second data set comprised 76 Portuguese water dogs (PWDs), segregating the curl phenotype. The final data set, termed CanMap, included 903 dogs from 80 breeds representing a wide variety of phenotypes. An additional data set used to map furnishings included a panel of microsatellite markers (10), genotyped on a 96-dachshund pedigree segregating all three coat varieties.
The same strategy was used to map all three traits. First, a genome-wide association study (GWAS) within a breed segregating the phenotype was conducted to determine the most strongly associated locus. To rule out false-positives caused by population structure within the breeds (11), we did a second GWAS that used the CanMap data set divided into cases and controls based on the presence or absence of the phenotype in question. Fine-mapping of significant, concordant peaks was used to define the smallest shared haplotype, followed by sequencing to identify the putative causative mutations. Each mutation was validated in a large panel of at least 661 dogs from 108 breeds, including cases and controls for all phenotypes (table S1).
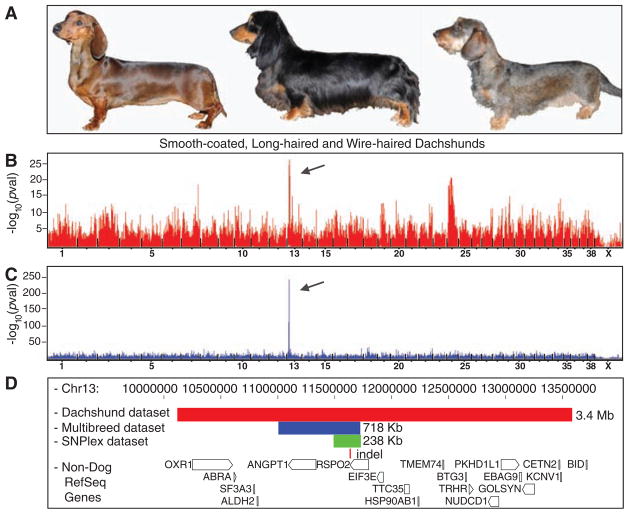
We initially mapped furnishings in the dachshund using smooth-coated and long-haired dogs as controls and wire-haired dogs as cases (Fig. 1A). Single-marker analysis of the dachshund GWAS data set and concurrent linkage analysis of the dachshund pedigree identified the same locus on canine chromosome 13 (CFA13) surrounding nucleotide 11,095,120 [P = 3.4 × 10−27, lod score (logarithm of the odds ratio for linkage) = 5.6; Fig. 1B]. We confirmed the association on CFA13 in the CanMap data set at nucleotide 11,659,792 (P = 10−241; Fig. 1C and table S2). A 718-kb homozygous haplotype in all dogs fixed with furnishings was located within both the original 3.4-Mb haplotype observed in the dachshund-only GWAS, and a 2.8-Mb haplotype identified in crossover analysis within the dachshund pedigree (Fig. 1D).
Fig. 1.
GWAS and fine-mapping identify RSPO2 as the associated gene for moustache and eyebrow growth pattern (furnishings). (A) Three types of coat segregate in dachshunds: (from left to right) smooth-coated, long-haired, and wire-haired with furnishings. (B) Results of the GWAS in the dachshund using wire-haired dogs as cases and smooth-coated and long-haired dogs as controls. The best P value (3.35 × 10−27), highlighted by the arrow, is located on CFA13 at position 11,095,120. (C) Results of the GWAS for furnishings in the CanMap data set. The arrow highlights the best association (P = 10−241) at CFA13 position 11,659,792. Both P values were obtained with single-marker χ2 analyses. (D) Homozygous regions identified in cases from GWAS and fine-mapping. The red rectangle represents the associated haplotype in the dachshund; the blue rectangle spans the homozygous region from 19 breeds fixed for furnishings based on the multibreed data set; the green rectangle indicates the region of homozygosity in 18 breeds fixed for the furnishings after fine-mapping. A 167-bp deletion, indicated by the small red rectangle, is located within all three haplotypes in the 3′UTR of the RSPO2 gene. The positions of genes in the region are represented by open boxes at the bottom of the figure, labeled to the left, with arrows indicating reading-frame direction (www.genome.ucsc.edu).
Fine-mapping allowed us to reduce the homozygous region to 238 kb spanning only the R-spondin–2 (RSPO2) gene, excluding the 5′ untranslated region (5′UTR) and the first exon (Fig. 1D, fig. S1, and table S3). RSPO2 is an excellent candidate for a hair-growth phenotype as it synergizes with Wnt to activate β-catenin (12), and Wnt signaling is required for the establishment of the hair follicles (13, 14). Moreover, the Wnt/β-catenin pathway is involved in the development of hair-follicle tumors, or pilo-matricomas (15), which occur most frequently in breeds that have furnishings (16). Recent studies have shown that a mutation in the EDAR gene, also involved in the Wnt pathway, is responsible for a coarse East-Asian hair type found in humans (17), with some similarity to canine wirehair.
All exons and conserved regions of RSPO2 were sequenced in dogs from seven breeds (table S4). Only an insertion of 167 base pairs (bp) within the 3′UTR at position 11,634,766 was perfectly associated with the furnishings trait in dogs from both the case/control study and the extended pedigree (table S5). The result was further confirmed in a set of 704 dogs of varying phenotypes. In total, 297 of 298 dogs with furnishings were either homozygous (268) or heterozygous (29) for the insertion, and all 406 dogs lacking the trait were homozygous for the ancestral state, as is consistent with a dominant mode of inheritance (table S1).
This mutation does not affect the protein-coding region of the RSPO2 gene. However, because the 3′UTR frequently encodes elements that influence mRNA stability [reviewed in (18)], we examined whether the insertion was associated with a change in the expression level of the RSPO2 gene. We found a threefold increase in RSPO2 transcripts in muzzle skin biopsies of dogs with furnishings, consistent with a transcript effect (fig. S2).
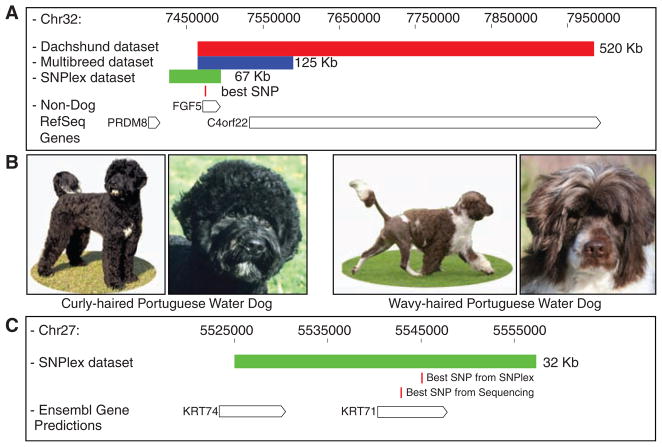
We applied the same mapping strategy to hair length. Previously, mutations in the FGF5 gene were identified in Welsh corgis segregating an atypical “fluffy” or long-haired phenotype (19) and associated with excess hair growth in mice and cats (20–22). Our study replicates these findings in an extended breed set. Indeed, association analyses in both the dachshund and CanMap data sets highlight the region on CFA32 containing FGF5 with P values of 3 × 10−27 and 9 × 10−44, respectively. After fine-mapping, a 67-kb homozygous region highlighted the FGF5 gene (Fig. 2A, fig. S3, and table S6). The strongest association was observed at position 7,473,337 (P = 1 × 10−157), in which a highly conserved Cys is changed to Phe (Cys95→Phe) in exon 1 of FGF5, consistent with the previous study (19). Sequencing within the homozygous haplotype revealed no SNPs with stronger association (table S7).
Fig. 2.
Regions of homozygosity identify genes for pelage length and curl. (A) Homozygous region found on CFA32 defining the length locus. The red bar indicates the 520-kb associated haplotype from 29 long-haired dachshunds; the blue bar spans the 125-kb homozygous region found in 319 dogs from 31 long-haired breeds; the green bar represents the 67-kb reduced homozygous region found after fine-mapping in 293 dogs from 39 long-haired breeds. The best-associated SNP, represented by the small red rectangle, is within these three homozygous regions in exon 1 of the FGF5 gene. (B) PWDs display two coat varieties: curly (two left panels) and wavy (two right panels). (C) Haplotype analysis at the curl locus on CFA27. The green bar represents the 32-kb homozygous haplotype found after fine-mapping in 65 dogs from five curly haired breeds. The best-associated SNP, represented by the small red rectangle, was found within the homozygous haplotype in exon 2 of the KRT71 gene. The positions of genes in both regions (A and C) are represented by open boxes at the bottom of each figure, labeled to the left, with arrows indicating reading-frame direction (www.genome.ucsc.edu). Curly PWD photos courtesy of M. Bloom (Copyright AKC).
This diagnostic SNP was typed in several hundred additional dogs of varying hair length. Within the dachshunds, all long-haired dogs had the TT genotype, whereas all short or wire-haired dogs had either the GT or GG genotypes, suggesting a recessive mode of inheritance, as predicted previously (23). Across all breeds, the T allele was found in 91% of the long-haired dogs, in only 3.9% of the short-haired dogs, and accounts for ~30% of genotypes found in medium-haired dogs. Three breeds with very long hair, including the Afghan hound, neither carry the Cys95→Phe variant nor show an association with CFA32, suggesting that additional loci exist that contribute to hair length in dogs (table S1).
To identify the gene that causes curly coat, we conducted a GWAS using PWDs (Fig. 2B) and identified a single associated SNP at position 5,444,030 on CFA27 (P = 4.5 × 10−7). A SNP in close proximity (5,466,995; P = 6.9 × 10−28) was associated with curly coat in the CanMap data set. Fine-mapping revealed a shared homozygous haplotype that included two keratin genes (Fig. 2C, fig. S4, and table S8). Sequence data covering 87% of the homozygous region identified one SNP at position 5,542,806 that segregated with the trait. Non–curly haired dogs carried the CC genotype; curly coated dogs had the TT genotype. In breeds where the trait segregates, such as PWDs, all three genotypes were observed. The relevant SNP is located in the KRT71 gene (previously called K6irs1, Kb34, and K71) and causes a nonsynonymous Arg151→Trp alteration (table S9). Genotyping an additional 661 samples at this SNP validated the association (P = 3 × 10−92) (table S1).
Keratins are obvious candidates for hair growth [reviewed in (24)], and mutations in KRT71 have been described in curly coated mice (25). The mutation described in our study is within the second exon of the gene and may affect either or both of two protein domains: a coiled-coil and a prefoldin domain (www.ensembl.org/Canis_familiaris/). Conceivably, sequence alterations in these domains could affect cellular targeting, receptor binding, or proper folding of the protein after translation [reviewed in (26)].
Notably, these three mutations in various combinations explain the observed pelage phenotype of 95% of dogs sampled, which include 108 of the ~160 American Kennel Club (AKC)–recognized breeds. A total of 622 dogs representing all identifiable coat phenotypes were genotyped at all three loci (table S10). By analyzing each of the three major traits both within and across multiple breeds, we show that combinations of these genotypes give rise to at least seven different coat types, encompassing most coat variation in modern domestic dogs (Fig. 3). Specifically, short-haired breeds display the ancestral state in all three genes. Wire-haired breeds, all of which have furnishings, carry the RSPO2 insertion. Dogs that carry both the RSPO2 and KRT71 mutations display “curly-wire” hair that is similar in texture to wire-hair but longer and curled or kinked rather than straight. Long-haired breeds carry the variant form of FGF5. Dogs carrying the FGF5 mutation, along with the RSPO2 insertion, have furnishings and long soft coats, rather than wiry ones. When dogs carry variants in both FGF5 and KRT71, the pelage is long and curly. Not surprisingly, coats must be of sufficient length to curl, and all curly haired dogs in our study were homozygous for the FGF5 mutation. Finally, if all three mutations are present, the phenotype is long and curly with furnishings.
Fig. 3.
Combinations of alleles at three genes create seven different coat phenotypes. Plus (+) and minus signs (−) indicate the presence or absence of variant (nonancestral) genotype. A characteristic breed is represented for each of the seven combinations observed in our data set: (A) short hair; (B) wire hair; (C) “curly-wire” hair; (D) long hair; (E) long, soft hair with furnishings; (F) long, curly hair; and (G) long, curly hair with furnishings. [Photos courtesy of M. Bloom (Copyright AKC)].
None of the mutations we observed were found in three gray wolves or the short-haired dogs, indicating that short-haired dogs carry the ancestral alleles (table S1). Our finding of identical haplotypes surrounding the variants in all dogs displaying the same coat type suggests that a single mutation occurred for each trait and was transferred multiple times to different breeds through hybridization. Because most breeds likely originated within the past 200 years (27), our results demonstrate how a remarkable diversity of phenotypes can quickly be generated from simple genetic underpinnings. Consequently, in domesticated species, the appearance of phenotypic complexity can be created through combinations of genes of major effect, providing a pathway for rapid evolution that is unparalleled in natural systems. We propose that in the wake of artificial selection, other complex phenotypes in the domestic dog will have similar tractable architectures that will provide a window through which we can view the evolution of mammalian form and function.
Supplementary Material
Acknowledgments
We gratefully acknowledge support from NSF grants 0733033 (R.K.W.) and 516310 (C.D.B.), NIH grants 1RO1GM83606 (C.D.B.) and GM063056 (K.G.L. and K.C.), the Nestlé Purina company, the AKC Canine Health Foundation, the University of California–Davis Veterinary Genetics Laboratory, and the Intramural Program of the National Human Genome Research Institute. We thank L. Warren and S. Stafford for providing pictures. Finally, we thank the many dog owners who generously provided us with samples from their pets.
Footnotes
www.sciencemag.org/cgi/content/full/1177808/DC1
Materials
Materials and Methods
Figs. S1 to S5
Tables S1 to S10
References
References and Notes
- 1.Vila C, et al. Science. 1997;276:1687. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- 2.Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Science. 2002;298:1610. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- 3.Sutter NB, et al. Science. 2007;316:112. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlsson EK, et al. Nat Genet. 2007;39:1321. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- 5.Lindblad-Toh K, et al. Nature. 2005;438:803. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 6.Wayne RK, Ostrander EA. Trends Genet. 2007;23:557. doi: 10.1016/j.tig.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 7.American Kennel Club. The Complete Dog Book. 19. Howell Book House; New York, NY: 1998. [Google Scholar]
- 8.Drogemuller C, et al. Science. 2008;321:1462. doi: 10.1126/science.1162525. [DOI] [PubMed] [Google Scholar]
- 9.Materials and methods can be found at Science Online.
- 10.Wong AK, Neff MW. Anim Genet. 2009;40:569. doi: 10.1111/j.1365-2052.2009.01875.x. [DOI] [PubMed] [Google Scholar]
- 11.Lander ES, Schork NJ. Science. 1994;265:2037. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 12.Kazanskaya O, et al. Dev Cell. 2004;7:525. doi: 10.1016/j.devcel.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Andl T, Reddy ST, Gaddapara T, Millar SE. Dev Cell. 2002;2:643. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- 14.Clevers H. Cell. 2006;127:469. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Chan EF, Gat U, McNiff JM, Fuchs E. Nat Genet. 1999;21:410. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 16.Meuten DJ, editor. Tumors in Domestic Animals. 4. Blackwell; Ames, Iowa: 2002. [Google Scholar]
- 17.Mou C, et al. Hum Mutat. 2008;29:1405. doi: 10.1002/humu.20795. [DOI] [PubMed] [Google Scholar]
- 18.Chatterjee S, Pal JK. Biol Cell. 2009;101:251. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 19.Housley DJ, Venta PJ. Anim Genet. 2006;37:309. doi: 10.1111/j.1365-2052.2006.01448.x. [DOI] [PubMed] [Google Scholar]
- 20.Sundberg JP, et al. Vet Pathol. 1997;34:171. doi: 10.1177/030098589703400301. [DOI] [PubMed] [Google Scholar]
- 21.Drogemuller C, Rufenacht S, Wichert B, Leeb T. Anim Genet. 2007;38:218. doi: 10.1111/j.1365-2052.2007.01590.x. [DOI] [PubMed] [Google Scholar]
- 22.Hebert JM, Rosenquist T, Gotz J, Martin GR. Cell. 1994;78:1017. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 23.Stockard CR. The Genetic and Endocrinic Basis for Differences in Form and Behavior. The Wistar Institute of Anatomy and Biology; Philadelphia: 1941. [Google Scholar]
- 24.Langbein L, Schweizer J. Int Rev Cytol. 2005;243:1. doi: 10.1016/S0074-7696(05)43001-6. [DOI] [PubMed] [Google Scholar]
- 25.Runkel F, et al. Mamm Genome. 2006;17:1172. doi: 10.1007/s00335-006-0084-9. [DOI] [PubMed] [Google Scholar]
- 26.Martin J, Gruber M, Lupas AN. Trends Biochem Sci. 2004;29:455. doi: 10.1016/j.tibs.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Ash EC. Dogs: Their History and Development. Benn; London: 1927. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.