Abstract
The history of fragment-based drug discovery, with an emphasis on crystallographic methods, is sketched, illuminating various contributions, including our own, which preceded the industrial development of the method. Subsequently, the creation of the BMSC fragment cocktails library is described. The BMSC collection currently comprises 68 cocktails of 10 compounds that are shape-wise diverse. The utility of these cocktails for initiating lead discovery in structure-based drug design has been explored by soaking numerous protein crystals obtained by our MSGPP (Medical Structural Genomics of Pathogenic Protozoa) consortium. Details of the fragment selection and cocktail design procedures, as well as examples of the successes obtained are given. The BMSC Fragment Cocktail recipes are available free of charge and are in use in over 20 academic labs.
Keywords: structural genomics, drug design, malaria, trypanosomiasis, leishmaniasis, history
HISTORICAL INTRODUCTION
Fragment-based drug discovery has matured to a powerful tool to arrive at drug leads over the course of the past decade. It has been the subject of many reviews [1–12], and refers to the screening of a drug target (almost always a protein) with a small number of small molecules (typically of the order of a thousand) with small molecular masses (often a Mr less than 250). The screening is primarily done by crystallography or NMR, though some groups use surface plasmon resonance or mass spectrometry analysis of covalent capture of thiolated fragments by strategically placed cysteine tags in the target. The name “fragment” has been coined to emphasize that the screened molecules could be considered substructures of the “normal” size small molecules utilized in high-throughput screening (HTS). In 2008 no fewer than 10 candidates derived from fragment-based screening were in clinical trials and another 6 in were the stage of preclinical research [12].
Many researchers pinpoint the start of fragment-based drug discovery to the appearance of a seminal paper by the Fesik group at Abbott Laboratories entitled “Discovering high-affinity ligands for proteins: SAR by NMR” [13]. This paper described how the binding of a weakly bound ligand out of a mixture of small molecules – called a cocktail – was mapped to shifts in the 15N-HSQC signals of binding site residues of the target, and it was further demonstrated that high affinity ligands could be obtained by linking the small ligands or “fragments”. Later this idea was transplanted to the world of crystallography, again by researchers at Abbott [14]. In brief, assuming one has crystals that allow for access to the binding site of interest, one can soak the crystal with a cocktail of small but shape-wise diverse “fragments” at high concentrations. With luck, one of the fragments will bind to the site of interest, and because of the shape diversity it should be possible to identify which fragment in the cocktail is fitting the electron density, avoiding lengthy deconvolution experiments. At the beginning of the millennium the method of fragment-based lead discovery using X-ray crystallography quickly became so appealing that specialized biotechnology companies were created that relied on it for their drug discovery efforts, such as SGX Pharmaceuticals (recently absorbed by Lilly), Plexxikon in California, Vertex Pharmaceuticals in Massachusetts, and Astex Therapeutics in the UK.
Fragment-based drug discovery did not emanate from a vacuum. The idea that leads can be elaborated from fragments that home in on hot spots in the targeted binding site was turned into a variety of tools by computational chemists. In 1985, Peter Goodford crafted the program GRID to predict such hot spots in the target for small molecular probes, most the size of just a functional group and hence the ultimate “fragment” [15]. The program MCSS which makes hot-spot predictions for fragment-sized molecules was reported in 1991 [16]. The first fragment libraries to be explored in a binding site were created by Boehm to “feed” his ligand design program LUDI in 1992 [17]. In the same year two of the current authors, then at the University of Groningen in The Netherlands, published an article with the title “In search of new lead compounds for African trypanosomiasis drug design: a linked-fragment approach” [18]. Their computational approach was guided by force-field calculations and entailed four stages: (1) a design pathway is defined based on the three-dimensional structure of a target protein; (2) this pathway is divided into subregions; (3) complementary building blocks, also called fragments, are designed in each subregion; complementarity is defined in terms of shape, hydrophobicity, hydrogen bond properties and electrostatics; and (4) fragments from different subregions are linked into potential lead compounds. Meanwhile experimentalists developed a crystallographic equivalent of GRID. Dagmar Ringe and colleagues replaced water in her crystals with pure organic solvent such as acetonitrile to determine the hot spots of the solvent molecules on the protein surface, a single-compound solvent per experiment [19].
The first crystallographic experiments using cocktails of fragments were carried out in the group of Wim Hol, at the University of Groningen, The Netherlands. Driven by a life-long interest in the use of structural biology for drug design [20] and inspired by an article in Scientific American about the revolution in RNA and peptide combinatorial chemistry for discovering high affinity ligands he conceived the idea of fragment cocktail crystallography on Oct 7 1990 (Fig. 1). He convinced Christophe Verlinde, a post-doc at the time, to test his approach with triose-phosphate isomerase of Ttypanosoma. brucei, the African sleeping sickness parasite. Soon thereafter compounds were selected, purchased and three cocktails of 128 compounds each were made by undergraduate student Gabby Rudenko (now Assistant professor at U. of Michigan at An Arbor) and technician Tjaard Pijning. Of the three cocktail soaks with 128 compounds each, one resulted in a convincing difference Fourier peak in the binding site. However, because of the limited resolution of 2.8 Å, the identification of the ligand from the shape of the electron density was not possible and we had to resort to deconvolution. Soaks with one of the sub-cocktails of 64, 32 and 16 different compounds revealed the same difference density but neither of the two sub-cocktails of 8 did – perhaps a chemical reaction in the 16 compounds cocktail produced the ligand. It deserves emphasis that all these experiments were carried out prior to the advent of fast data collection methods and equipment, hence requiring a substantial effort – up to several months for data collection and refinement, rather than the one day it would take in 2009. We were disappointed, but described these experiments in a book chapter in 1997 [21].
Fig. 1.
Excerpt from Wim Hol’s lab notebook dated Oct 7 1990 with transcription.
In hindsight, although the cocktail crystallography idea was novel there were several shortcomings in our approach. First, the selection of compounds was haphazard and mainly governed by budget constraints – we had no access to electronic compound catalogs. No effort was made to make sure that the compounds were sufficiently shape diverse for direct recognition from the electron density. Second, we underestimated the importance of solubility of the compounds, and therefore obtained at best a 1mM concentration per individual compound. Third, the number of compounds per cocktail was too high, causing solubility and compatibility issues, and problems with compound identification from the electron density. Each of these three practical problems was properly addressed nearly a decade later by researchers from Abbott Laboratories, undoubtedly benefiting from decades of experience in dealing with chemical screening in industry. In 2001 Vicki Nienaber, Jonathan Greer, Celerino Abad-Zapatero and Daniel Norbeck from Abbott obtained a patent “ligand screening and design by x-ray crystallography” [22].
Upon seeing around the year 2000 fragment-based ligand discovery blossoming in the biotech industry, we decided to revive the method at the Biomolecular Structure Center (BMSC) of the University of Washington in Seattle, within the framework of our Medical Structural Genomics for Protozoan Parasites (MSGPP) program project [23].
MSGPP COCKTAIL CRYSTALLOGRAPHY
The design and preparation of the BMSC fragment cocktails library can be briefly decribed as follows.
In silico cocktail creation
A starting list of 9,486 commercially available compounds was selected from the commercial ACD database version 2004.1 (Elsevier Molecular Design, San Leandro, CA) through use of the included ISIS program - the free ZINC database by the Shoichet group at UCSF [24] only became available a year later.
We used the following criteria:
Only allow [C, H, N, O, S, halogen] in the fragment, but allow [C, H, N, O, S, halogen, Li, Na, K, Ca, Mg] in counter ions or solvent molecules of solvate entities.
Number of halogens < 2.
Mr < 200.
clogP < 3.
Number of rotatable bonds < 3.
At least one ring, which may be part of a multi-cyclic system.
Eliminate reactive or undesirable groups: acryl amides, acyl halides, aldehydes, aziridines, carboxylic acid anhydrides and their chalcogen analogs, epoxides, hydrazines, hydrazones, isocyanates, isothiocyanates, maleimides, Michael acceptors, oximes, nitro compounds, peroxides, persulfides, semicarbazides, sulfonates and their esters, and sulfonyl halides.
Eliminate expensive (> $100 for 20 mg) or POA (Price On Application) compounds.
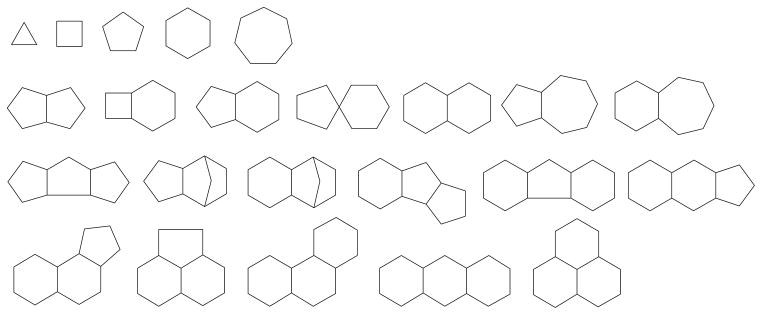
Subsequently, after removal of counter ions and solvent molecules the compounds were analyzed in terms of diversity of ring frameworks by fragmentation with the program MOE (CCG, Montreal, Canada). At the level of connectivity, 23 frameworks as shown in Fig. (2) could be distinguished, and representatives of each category, including saturated and aromatic rings, were further visually selected. Mutagens, poisons, and highly functionalized compounds were eliminated, though we disclaim responsibility for potential failure to recognize all mutagens and poisons. Deliberately, a substantial number of Br containing compounds were retained for the purpose of easier detection in the electron density. The final number of selected compounds was 680.
Fig. 2.
Diversity of the connectivity frameworks of the selected molecules.
In the next step the compounds were subdivided in ‘cocktails’ of 10 compounds by maximizing the shape diversity. Because the total number of ways to partition 680 compounds in 68 cocktails of 10 compounds is astronomical we refrained from carrying out the process manually. Instead for each compound a shape fingerprint with 4 descriptors was calculated with MOE consisting of the first kappa shape index of Hall and Kier (1K =Natoms(Natoms-1)/(Nbonds)2), where Natoms is the atom count and Nbonds is the bond count [25]), and the standard dimensions defined as the square root of each of the three largest eigenvalues of the covariance matrix of the atomic coordinates. The shape fingerprints were used to assess the similarity between compounds and 68 maximally diverse subsets of 10 compounds each were selected by non-parametric ranking in MOE.
Cocktail preparation and cocktail soaking
The 680 cocktail compounds were purchased from the following vendors: Acros, ABCR, Aldrich, Alfa Aesar, Bachem, Fluka, ICN, Maybridge, Lancaster, Oakwood, Pfaltz-Bauer, Sigma, Specs, TCI, and Toronto Research Chemicals. The cost of goods was about $30,000 in 2004, and the administrative effort of placing and tracking orders should not be underestimated. No guarantee can be given that all chemicals are still available from these vendors. The purity of all compounds was verified by LCMS and NMR. Stock solutions with a 100 mM final concentration of the cocktail compounds in 100 % DMSO. These stock solutions were diluted either into a specific cryo-buffer or into a fraction of the mother liquor, followed by crystal soaking. The final concentration of the cocktail chemicals was 5 to 10 mM, leading also to a residual 5 to 10% DMSO concentration. Soaking times as short as 10 s were shown to reveal bound ligands [26].
RESULTS
Over the last five years we have explored 26 MSGPP targets by cocktail crystallography. Seven targets gave hits while 19 targets did not yield results because of a variety of reasons. Some crystals were unable to tolerate the presence of even a low concentration of DMSO, other crystals cracked - presumably because of conformational changes in the protein induced by ligand binding. In other cases the diffraction resolution deteriorated beyond usefulness (at least about 2.8 Å is required for ligands that do not contain Br). Finally, some targets produced robust crystals but none of the cocktail soaks led to an interesting difference Fourier density. Here we summarize all of our successes except for the cocktail fragment hit MSGPP recently obtained for T. cruzi histidyl-tRNA synthetase (GeneDB identifier Tc00.1047053507019.40), solved at 1.7 Å resolution, as screening of all the cocktails has not been completed yet.
Plasmodium falciparum phosphoglycerate mutase (PGM)
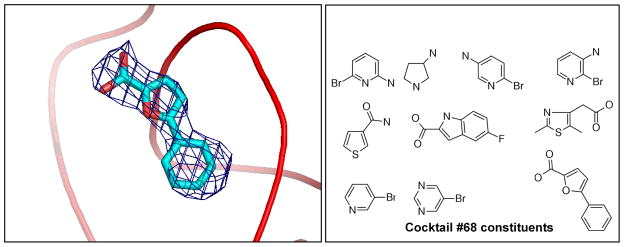
This glycolytic enzyme, with PlasmoDB [27] identifier PF11_0208, has so far not been shown to be essential for the parasite, but it was one of the first proteins we used for trying out our BMSC cocktails. The native structure was deposited in the PDB as 1XQ9, but is so far unpublished. We obtained a significant electron density in the difference Fourier after soaking in cocktail #68 (Fig. (3)). The observed density is consistent only with 5-phenyl-2-furoic acid.
Fig. 3.
Electron density for 5-Phenyl-2-furoic acid when bound to P.falciparum PGM (left); and cocktail 68 composition (right).
It must be noted that the PDB contains another unpublished phosphoglycerate mutase structure from P. falciparum with code 3EOZ. It corresponds to a different PlasmoDB identifier, PFD0660w. Neither of the two proteins has been characterized enzymatically to the best of our knowledge.
Trypanosoma brucei nucleoside 2-deoxyribosyltransferase (NDRT)
This target with GeneDB [28] identifier Tb927.5.1360 is critical for purine salvage and recycling in trypanosomatid parasites because of the absence of enzymes for de novo purine biosynthesis. Since there is no NDRT homologue in humans the T. brucei enzyme seems a particularly attractive target for drug development, however, due to overlapping salvage pathways in the parasite simultaneous blocking of more than one target may be necessary [35].
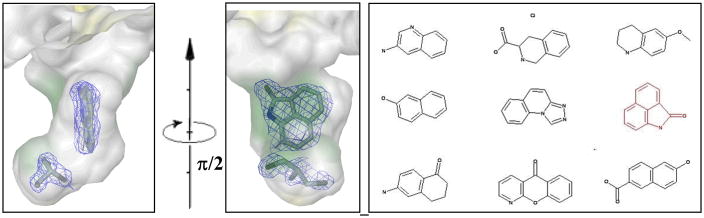
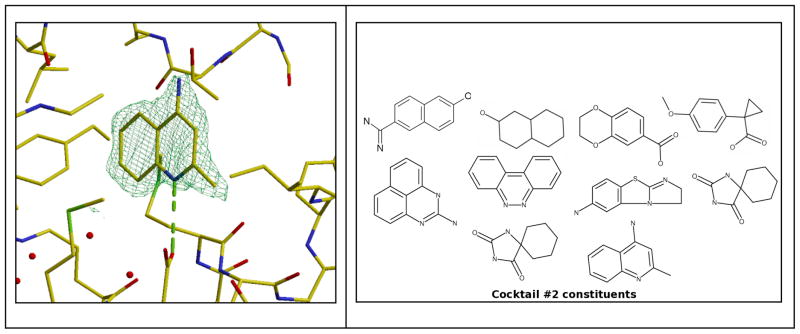
Initially, the crystal structure of the dimeric enzyme was solved at 1.8 Å resolution (PDB: 2A0K) [26]. The two active sites, lined by 9 hydrophobic, 4 charged, and 6 neutral side chains, were shown to be across the dimer interface and to possess a volume of around 1180 Å3 each. The crystals proved robust and were soaked with 31 of the 68 BMSC cocktails. From synchrotron data with a resolution slightly better than the native dataset ultimately four ligands were identified in the active site: benzo[cd]indol-2(1H)-one (PDB: 2F67); 5-aminoisoquinoline (PDB: 2F2T); 2-ethylbenzyl alcohol (PDB: 2F62); and 1-methylquinoline-2(1H)-one (PDB: 2F64) [26]. Remarkably in each case, in the immediate vicinity of the bound compound a glycerol molecule was observed whose oxygen atoms occupy positions that are equivalent to the O3′ and O5′ atoms of the substrate in the homologous Lactobacillus helveticus enzyme [29]. As an example the electron density for benzo[cd]indol-2(1H)-one is shown along with the cocktail composition in Fig. (4). It is clear that no other compound in the cocktail is compatible with the observations.
Fig. 4.
Electron density for benzo[cd]indol-2(1H)-one when bound to T.brucei NDRT (left); and cocktail 12 composition (right).
Leishmania major coproporphyrinogen III oxidase
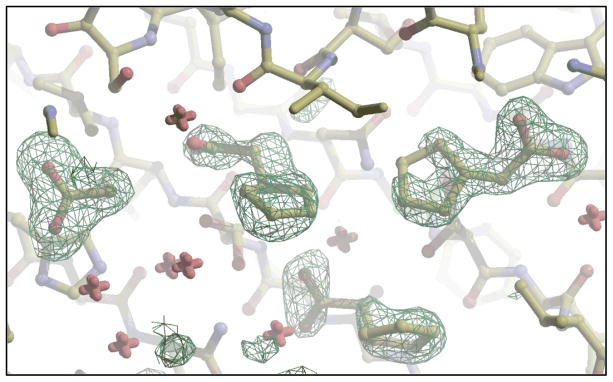
Leishmania major coproporphyrinogen III oxidase, with GeneDB identifier LmjF06.1270, is required for porphyrin biosynthesis, though there is no evidence so far that it is a valid drug target in trypanosomatids. In all, 66 different cocktails were soaked into 147 crystals, ultimately leading to 42 useful datasets. Fragments from two different cocktails were found to bind: 5-fluoroindole-2-carboxylic acid (PDB: 3DWS) and cyclopentyl acetic acid (PDB: 3DWR). Interestingly, as can be seen in Fig. (5), the dataset collected for the cocktail containing cyclopentyl acetic acid shows that three copies of cyclopentyl acetate occupy the active site as well as 1 acetate molecule, which is a “gift” from E.coli. We hypothesize that the 4 carboxylate groups of the fragments occupy the same positions as the 4 carboxylate moieties of the enzyme substrate coproporphyrinogen-III.
Fig. 5.
Electron density for 3 cyclopentyl acetate molecules plus 1 acetate molecule when bound to L. major CPO-III at 1.66 Å resolution.
Plasmodium falciparum D-ribulose 5-phosphate epimerase
This enzyme of the shikimate pathway, with PlasmoDB identifier PFL0960w, is of particular interest for therapeutic targeting because of its absence in mammals. MSGPP solved the apo structure at 2.0 Å resolution (PDB: 1TQX) [30]. Again, the crystals were used to hone our skills in cocktail crystallography. Soaking with cocktail 68 revealed electron density for 3-aminopyrrolidine at 2.0 Å (Fig. (6)).
Fig. 6.
Electron density for 3-amino-pyrrolidine when bound to P.falciparum D-ribulose- 5-phosphate epimerase; for cocktail 68 composition see fig. (2b).
Leishmania major putative adenylate kinase
GeneDB LmjF30.1890 is a putative L. major adenylate kinase. Its significance as a drug target is unknown, but we used the crystals to test our cocktails during the development phase. We found one hit (Fig. (7)).
Fig. 7.
Electron density for 4-amino-2-methylquinoline when bound to L.major adenylate kinase (left); and cocktail 2 composition (right).
Leishmania naiffi uracil-DNA glycosylase
This homolog of L. brasiliensis UDG with GeneDB identifier LbrM18_V2.0540 is responsible for excising uracil in DNA repair. The enzyme has been shown to be very important in the related parasite T. cruzi, whereas higher eukaryotes possess redundant pathways to repair the same DNA damage [31]. The 1.5 Å structure has been deposited in complex with 5-bromouracil with PDB accession code 3cxm. All 68 cocktails were screened, 42 of which led to 95 usable datasets, and no fewer than 4 fragments as well as DMSO were shown to bind to this target, as shown in Fig. (8). Subsequenlty, we designed a derivative of the hit 5-chloro-2-methoxybenzoic acid shown in Fig. (9), and thereby improved the IC50 from about 15 mM to 15±3 μM. The new inhibitor consists of 15 non-hydrogen atoms and exhibits a ligand efficiency (LE) of 0.44. This compares favorably with the 1 μM inhibitor of human UDG2 designed by Stivers [32], MA1, which possesses 25 atoms and an LE of only 0.34.
Fig. 8.
Fragments bound to L.naiffi uracil-DNA glycosylase: 4-morpholinopyridine (left upper), DMSO and 5-chloro-2-methoxybenzoic acid (right upper), DMSO and 8-hydroxyquinoline-2-carboxylic acid (left lower), and 1H-imidazole-4-carbothioamide (lower right).
Fig. 9.
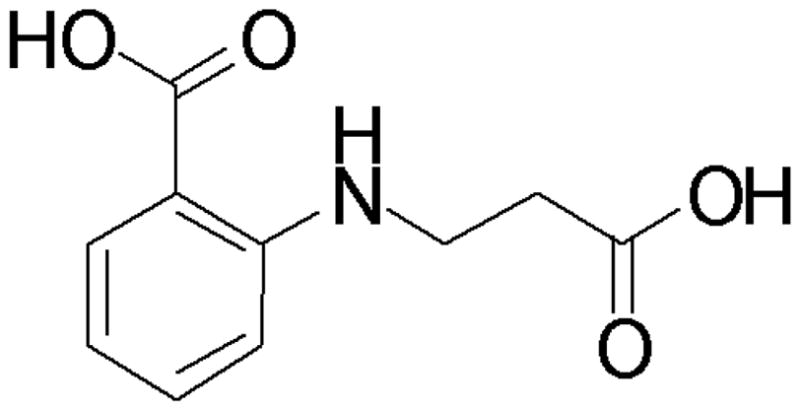
Designed inhibitor of L.naiffi uracil-DNA glycosylase.
DISCUSSION
The enormous advantage of screening smaller molecules than in HTS is that it allows for much better sampling of chemical space. The assay signal for a hit in HTS requires at least 1–10 μM affinity, which results in hits with an average molecular mass (Mr) of around 270, which is only slightly smaller than the average size of a drug [9]. This leaves little leeway for optimization by adding substituents because it has been established that compounds with an Mr above 500 have in general very poor bioavailability. In addition, in HTS up to several million compounds are screened against a target but the virtual space of drug-size molecules has been estimated to be of the order of 1060 to 10200, hence the sampling coverage is dismal. In contrast, the technologies used in fragment-based screening allow for the detection of hits with affinities as low as millimolar. There is a larger chance that a fragment is complementary to a binding site than is the case for a drug-size compound [33] and since the hits are small in size there is considerable margin for optimization within the bounds of the bioavailability molecular mass limit. Moreover, virtual chemical space for fragment molecules is much smaller than for drug-size molecules. A systematic study of chemical space below an Mr of 160, corresponding to 11 atoms, yielded an estimated 14 million possible compounds of which 36,000 are known (25% are acyclic) [34]. At least, meaningful sampling of the chemical space of the available fragment-size chemicals appears feasible, in contrast to HTS.
While fragments are small and consequently have poor affinity they provide for much better hit rates than HTS. Astex reports typical hit rates range from 0.5 to 10.0% depending on the protein, with cocktails covering over 300 compounds, and 4–8 compounds per cocktail [5]. We have similar results. Overall, seven out of 26 targets gave at least one hit which is 26%, and for datasets with useful resolution the hit rate defined as number of fragments seen in the difference maps divided by total number of soaked molecules is 2%. It should be mentioned that MSGPP did not yet try extensively to obtain robust crystal forms for the 19 targets that were not successful. Robust crystals with accessible binding sites that diffract to at least 2.8 Å when soaked with cocktails are key requirement for success with fragment-based cocktail crystallography, and could have increased significantly our success rate.
MSGPP hopes to transfer in the near future into a combination of projects focusing on ~ 10 to 20 well-validated drug targets from protozoan parasites causing global diseases such as malaria, African trypanosomiasis, Chagas disease, leishmaniasis and cryptosporidiosis. These projects will still be of limited scale and in an academic setting, but will contain a significant chemical synthesis component. However, the full power of all available fragment screening methods cannot be applied and, hence, fragment cocktail crystallography, supplemented by ultra-filtration methods [36] to find low-affinity binders in solution, will be the main method used for discovering initial binders and for finding opportunities for adding substituents to existing binders.
In a broader perspective, a lesson from the MSGPP efforts is that numerous crystals did not survive the fragment-cocktail soak procedure. As mentioned above, this is in certain cases most likely due to interference of certain fragments with crystal contacts. In other cases, conformational changes upon ligand binding destroy the crystalline order of pre-grown crystals, as Max Perutz already observed when adding oxygen to deoxyhemoglobin crystals more than half a century ago. Hence, for enhanced exploration of the intersection of chemical space with a potential drug target, it would be a major advantage if a substantial number of compounds could be set up in a large number of co-crystallization experiments. Since also numerous variants of a particular drug target need to be tried, one obtains quickly a very large number of crystallization trials needed. For instance, for ~ 100 different drug target variants [37–38], ~1000 crystallization conditions [39], and ~ 1000 different small molecule compounds, about 108 wells need to be explored. This number is quickly increased by a factor of 10 if different temperatures and protein-to-reservoir-solution volume ratios are also taken into account. Yet, with volumes of tens to a few hundreds of picoliters per well this is in principle possible since such small volumes with ~ 10 mgs/ml protein, can generate a small number of crystals of sufficient size, i.e. 5 to 10 μm dimensions, for detecting diffraction patterns or even solving protein structures [40]. This requires 10 mgs of each of the protein variants of the target under study, which is well possible with current techniques for many proteins. It is exciting that the following technological developments are currently taking place:
robotization has generated capacities reaching low nanoliter volumes in crystal screens [41–45];
automated imaging and crystal detection methods in crystallization drops have shown promise [46–48];
screening for crystal diffraction quality in the X-ray beam without mounting individual crystals has been reported [45,49];
cryo-protection of crystals in micromeshes has been shown to be possible [40,50], while further miniaturization of trays and other crystal screen tools for sub-nanoliter droplets may assist in high-throughput irradiation damage protection steps of many wells simultaneously.
By combining such technologies, and others, it might be possible to achieve the very high-throughput crystal growth screening (VHTXS) methods required for reaching the full power of fragment cocktail crystallography in future medical macromolecular crystallography.
Acknowledgments
We thank the staff of the SSRL and ALS synchrotron beam lines for their assistance. We also wish to thank contributions from other members of the initial SGPP consortium including Lori Anderson, Helen Neely, Margaret A. Holmes, Mark A. Robien, Lori W. Schoenfeld, Chris Mehlin, Mike Soltis, Thomas Earnest, George DeTitta and Joe Luft, to mention only a few. This project was supported in part by NIH grants P50GM64655 (SGPP) and P01AI067921 (MSGPP) to W.G.J.H.
References
- 1.Blundell TL, Jhoti H, Abell C. High-throughput crystallography for lead discovery in drug design. Nature Rev Drug Discov. 2002;1:45–54. doi: 10.1038/nrd706. [DOI] [PubMed] [Google Scholar]
- 2.Carr R, Jhoti H. Structure-based screening of low-affinity compounds. Drug Discov Today. 2002;7:522–527. doi: 10.1016/s1359-6446(02)02245-6. [DOI] [PubMed] [Google Scholar]
- 3.Erlanson DA, McDowell RS, O’Brien T. Fragment-based drug discovery. J Med Chem. 2004;47:3463–82. doi: 10.1021/jm040031v. [DOI] [PubMed] [Google Scholar]
- 4.Rees DC, Congreve M, Murray CW, Carr R. Fragment-based lead discovery. Nature Rev Drug Discov. 2004;3:660–672. doi: 10.1038/nrd1467. [DOI] [PubMed] [Google Scholar]
- 5.Hartshorn MJ, Murray CW, Cleasby A, Frederickson M, Tickle IJ, Jhoti H. Fragment-based lead discovery using X-ray crystallography. J Med Chem. 2005;48:403–413. doi: 10.1021/jm0495778. [DOI] [PubMed] [Google Scholar]
- 6.Zartler ER, Shapiro MJ. Fragonomics, fragment-based drug discovery. Curr Op Chem Biol. 2005;9:366–370. doi: 10.1016/j.cbpa.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Carr RA, Congreve M, Murray CW, Rees DC. Fragment-based lead discovery, leads by design. Drug Discov Today. 2005;10:987–992. doi: 10.1016/S1359-6446(05)03511-7. [DOI] [PubMed] [Google Scholar]
- 8.Jhoti H. A new school for screening. Nat Biotechnol. 2005;23:184–186. doi: 10.1038/nbt0205-184. [DOI] [PubMed] [Google Scholar]
- 9.Leach AR, Hann MM, Burrows JN, Griffen EJ. Fragment screening: an introduction. Mol BioSyst. 2006;2:429–446. doi: 10.1039/b610069b. [DOI] [PubMed] [Google Scholar]
- 10.Hajduk PJ, Greer J. A decade of fragment-based drug design, strategic advances and lessons learned. Nature Rev Drug Disc. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 11.Ciulli A, Abell C. Fragment-based approaches to enzyme inhibition. Curr Op Biotechnol. 2007;18:1–8. doi: 10.1016/j.copbio.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent developments in fragment-based drug discovery. J Med Chem. 2008;50:1116–1123. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- 13.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins, SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 14.Nienaber VL, Richardson PL, Klighofer V, Bouska JJ, Giranda VL, Greer J. Discovering novel ligands for macromolecules using X-ray crystallographic screening. Nature Biotechnol. 2000;18:1105–1108. doi: 10.1038/80319. [DOI] [PubMed] [Google Scholar]
- 15.Goodford P. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem. 1985;28:849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- 16.Miranker A, Karplus M. Functionality maps of binding sites, A multiple copy simultaneous search method. Proteins. 1991;11:29–34. doi: 10.1002/prot.340110104. [DOI] [PubMed] [Google Scholar]
- 17.Böhm HJ. LUDI, rule-based automatic design of new substituents for enzyme inhibitor leads. J Comput-Aided Mol Des. 1992;6:593–606. doi: 10.1007/BF00126217. [DOI] [PubMed] [Google Scholar]
- 18.Verlinde CLMJ, Rudenko G, Hol WGJ. In search of new lead compounds for African trypanosomiasis drug design, a linked-fragment approach. J Comput-Aided Mol Des. 1992;6:131–47. doi: 10.1007/BF00129424. [DOI] [PubMed] [Google Scholar]
- 19.Fitzpatrick PA, Steinmetz ACU, Ringe D, Klibanov A. Enzyme crystal in a neat organic solvent. Proc Natl Acad Sci USA. 1993;90:8653–8657. doi: 10.1073/pnas.90.18.8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hol WGJ. Protein crystallography and computer graphics - toward rational drug design. Angewandte Chemie (Int Ed) 1986;25:767–778. [Google Scholar]
- 21.Verlinde CLMJ, Kim H, Bernstein BE, Mande SC, Hol WGJ. Anti-trypanosomiasis drug development based on structures of glycolytic enzymes. In: Veerapandian P, editor. Structure-based drug design. Marcel Dekker; Inc; New York: 1997. pp. 365–394. [Google Scholar]
- 22.Nienaber VL, Greer J, Abad-Zapatero C, Norbeck DW. Ligand screening and design by x-ray crystallography. 6297021. US Pat. 2001 Sep 27;B1
- 23.Fan E, Baker D, Fields S, Gelb MH, Buckner FS, Van Voorhis WC, Phizicky E, Dumont M, Mehlin C, Grayhack E, Sullivan M, Verlinde C, DeTitta G, Meldrum DR, Merritt EA, Earnest T, Soltis M, Zucker F, Myler PJ, Schoenfeld L, Kim D, Worthey L, LaCount D, Vignali M, Li J, Mondal S, Massey A, Carroll B, Gulde S, Luft J, DeSoto L, Holl M, Caruthers J, Bosch J, Robien M, Arakaki T, Holmes M, Le Trong I, Hol WGJ. Structural Genomics of Pathogenic Protozoa: an overview. In: Kobe B, Guss M, Huber T, editors. Methods in Molecular Biology, Vol 426: Structural Proteomics. Humana Press; Totowa, NJ, USA: 2008. pp. 497–513. [DOI] [PubMed] [Google Scholar]
- 24.Irwin JJ, Shoichet BK. ZINC – a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall LH, Kier LB. The molecular connectivity chi indices and kappa shape indices in structure-property modeling. Rev Computational Chem. 1991;2:367–422. [Google Scholar]
- 26.Bosch J, Robien MA, Mehlin C, Boni E, Riechers A, Buckner FS, Van Voorhis WC, Myler PJ, Worthey EA, DeTitta G, Luft JR, Lauricella A, Gulde S, Anderson LA, Kalyuzhni O, Neely HM, Ross J, Earnest TN, Soltis M, Schoenfeld L, Zucker F, Merritt EA, Verlinde CLMJ, Hol WGJ. Identifying new lead compounds for inhibiting nucleoside 2-deoxyribose-transferase in Trypanosoma brucei by fragment cocktail crystallography. J Med Chem. 2006;49:5939–5946. doi: 10.1021/jm060429m. [DOI] [PubMed] [Google Scholar]
- 27.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ, Jr, Treatman C, Wang H. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2008;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, Parkhill J, Ivens AC, Rajandream MA, Barrell B. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anand R, Kaminski PA, Ealick SE. Structures of purine 2′-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 Å resolution. Biochemistry. 2004;43:2384–2393. doi: 10.1021/bi035723k. [DOI] [PubMed] [Google Scholar]
- 30.Caruthers J, Bosch J, Buckner F, Van Voorhis W, Myler P, Worthey E, Mehlin C, Boni E, DeTitta G, Luft J, Lauricella A, Kalyuzhniy O, Anderson L, Zucker F, Soltis M, Hol WG. Structure of a ribulose 5-phosphate 3-epimerase from Plasmodium falciparum. Proteins. 2006;62:338–342. doi: 10.1002/prot.20764. [DOI] [PubMed] [Google Scholar]
- 31.Peña-Diaz J, Akbari M, Sundheim O, Farez-Vidal ME, Andersen S, Sneve R, Gonzalez-Pacanowska D, Krokan HE, Slupphaug G. Trypanosoma cruzi contains a single detectable uracil-DNA glycosylase and repairs uracil exclusively via short patch base excision repair. J Mol Biol. 2004;342:787–799. doi: 10.1016/j.jmb.2004.07.043. [DOI] [PubMed] [Google Scholar]
- 32.Chung S, Parker JB, Bianchet M, Amzel LM, Stivers JT. Impact of linker strain and flexibility in the design of a fragment-based inhibitor. Nature Chem Biol. 2009;5:407–413. doi: 10.1038/nchembio.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hann MM, Leach AR, Harper G. Molecular complexity and its impact on the probability of finding leads for drug discovery. J Chem Inf Comput Sci. 2001;41:856–864. doi: 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- 34.Fink T, Bruggesser H, Reymond JL. Virtual exploration of the small-molecule chemical universe below 160 Daltons. Angew Chem. 2005;117:1528–1532. doi: 10.1002/anie.200462457. [DOI] [PubMed] [Google Scholar]
- 35.Craig SP, Eakin AE. Purine salvage enzymes of parasites as targets for structure-based inhibitor design. Parasitol Today. 1997;13:238–241. doi: 10.1016/s0169-4758(97)01042-9. [DOI] [PubMed] [Google Scholar]
- 36.Kamchonwongpaisan S, Vanichtanankul J, Tarnchompoo B, Yuvaniyama J, Taweechai S, Yuthavong Y. Stoichiometric selection of tight-binding inhibitors by wild-type and mutant forms of malarial (Plasmodium falciparum) dihydrofolate reductase. Anal Chem. 2005;77:1222–1225. doi: 10.1021/ac0487597. [DOI] [PubMed] [Google Scholar]
- 37.Choi KH, Groarke JM, Young DC, Rossmann MG, Pevear DC, et al. Design, expression, and purification of a Flaviviridae polymerase using a high-throughput approach to facilitate crystal structure determination. Protein Sci. 2004;13:2685–2692. doi: 10.1110/ps.04872204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graslund S, Sagemark J, Berglund H, Dahlgren LG, Flores A, et al. The use of systematic N and C-terminal deletions to promote production and structural studies of recombinant proteins. Protein Expr Purif. 2008;58:210–221. doi: 10.1016/j.pep.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, et al. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 40.Wagner A, Diez J, Schulze-Briese C, Schluckebier G. Crystal structure of ultralente - a microcrystalline insulin suspension. Proteins. 2009;74:1018–1027. doi: 10.1002/prot.22213. [DOI] [PubMed] [Google Scholar]
- 41.Santarsiero BD, Lee CC, Spraggon G, Gu J, Scheibe D, Uber EC, Cornell EW, Nordmeyer RA, Kolbe WF, Jin J, Jones AL, Jaklevic JM, Schultz PG, Stevens RC. An approach to rapid protein crystallization using nanodroplets. Journal of Applied Crystallography. 2002;35:278–281. [Google Scholar]
- 42.Hansen CL, Skordalakes E, Berger JM, Quake SR. A robust and scalable microfluidic metering method that allows protein crystal growth by free interface diffusion. Proc Natl Acad Sci U S A. 2002;99:16531–16536. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quake SR. Proc Natl Acad Sci USA. 2002;99:16531–16536. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng B, Roach LS, Ismagilov RF. Screening of protein crystallization conditions on a microfluidic chip using nanoliter-size droplets. J Am Chem Soc. 2003;125:11170–11171. doi: 10.1021/ja037166v. [DOI] [PubMed] [Google Scholar]
- 45.Gerdts CJ, Elliott M, Lovell S, Mixon MB, Napuli AJ, et al. The plug-based nanovolume Microcapillary Protein Crystallization System (MPCS) Acta Crystallogr D Biol Crystallogr. 2008;64:1116–1122. doi: 10.1107/S0907444908028060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson J. Towards the automated evaluation of crystallization trials. Acta Crystallogr D Boil Crystallogr. 2002;58:1907–1914. doi: 10.1107/s0907444902016633. [DOI] [PubMed] [Google Scholar]
- 47.Pan S, Shavit G, Penas-Centeno M, Xu DH, Shapiro L, et al. Automated classification of protein crystallization images using support vector machines with scale-invariant texture and Gabor features. Acta Crystallogr D Biol Crystallogr. 2006;62:271–279. doi: 10.1107/S0907444905041648. [DOI] [PubMed] [Google Scholar]
- 48.Liu R, Freund Y, Spraggon G. Image-based crystal detection: a machine-learning approach. Acta Crystallogr D Biol Crystallogr. 2008;64:1187–1195. doi: 10.1107/S090744490802982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacquamet L, et al. Automated Analysis of Vapor Diffusion Crystallization Drops with an XRay Beam. Structure. 2004;12:1219–1225. doi: 10.1016/j.str.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 50.Thorne RE, Stum Z, Kmetko J, O’Niell J, Gillilan R. Microfabricated mounts for high-throughput macromolecular cryocrystallography. J Applied Crystallography. 2003;36:1455–1460. [Google Scholar]