Abstract
The goal of this study was to identify downstream signaling molecules involved in mediating the IGF-independent effects of IGFBP-5 in osteoblasts. We identified RASSF1C, a member of the RASSF1 gene products, as a IGFBP-5 binding partner and as a potential mediator of IGFBP-5 effects on ERK phosphorylation and cell proliferation.
Introduction
It has been predicted that the intrinsic growth factor action of insulin-like growth factor binding protein (IGFBP)-5 involves either the binding of IGFBP-5 to a putative receptor to induce downstream signaling pathways and/or intracellular translocation of IGFBP-5 to bind to potential signaling molecules involved in osteoblast cell regulation. This study reports the characterization of isoform C of the Ras association family 1 (RASSF1C) gene as an interacting partner of IGFBP-5.
Materials and Methods
IGFBP-5 was used as bait in a yeast two-hybrid screen of a human osteosarcoma cDNA library. Expression levels of RASSF1C were measured by RT-PCR and/or Northern blot. IGFBP-5 effects on ERK phosphorylation were evaluated by immunoblot analysis. The effect of RASSF1C siRNA on cell proliferation was measured by the AlamarBlue assay.
Results
One of the clones that interacted strongly with the bait under high stringency conditions corresponded to RASSF1C. The interaction between RASSF1C and IGFBP-5 was confirmed by in vitro coimmunoprecipitation studies. Northern blot and RT-PCR analysis showed that RASSF1C was expressed in a variety of osteoblast cell types that produce IGFBP-5. Addition of synthetic RASSF1C-specific small interfering (si) RNA duplex or use of a RASSF1C-specific si-hairpin plasmid caused a decrease in cell number and abolished IGFBP-5–induced extracellular signal-regulated kinase (ERK)-1/2 phosphorylation but had no effect on IGFBP-5–induced increases in alkaline phosphatase (ALP) activity.
Conclusions
We have shown a novel interaction between IGFBP-5 and RASSF1C. Our findings that silencing of RASSF1C results in the reduction of osteoblast cell proliferation and that IGFBP-5 treatment increases phosphorylation of ERK-1/2 raise the possibility that RASSF1C, a Ras effector, could, in part, contribute to mediating the effects of IGFBP-5 on ERK phosphorylation and, consequently, cell proliferation.
Keywords: protein–protein interaction, yeast two-hybrid screen, small interfering RNA, extracellular signal-regulated kinase-1/2 phosphorylation, insulin-like growth factor–independent mechanism
INTRODUCTION
INSULIN-LIKE GROWTH FACTORS (IGFs) regulate proliferation, differentiation, and apoptosis of bone cells through their cognate receptors, which are subject to regulation by IGF binding proteins (IGFBPs) and their proteases.(1–3) IGFBPs are unique in that they not only regulate the activity of IGFs but can also act independent of ligands.(4–7) Although a great deal is known about the mechanisms by which IGFBPs modulate IGF actions, little is known about the molecular mechanism(s) behind the IGF-independent action of IGFBPs.
IGFBP-5 has been shown to regulate both osteoblast cell proliferation and activity in vitro.(8–11) Recent findings showed that IGFBP-5 itself is a growth factor with cellular effects that are independent of IGFs.(12) Consistent with this, IGFBP-5 treatment increases bone formation parameters in vitro and in vivo in osteoblasts derived from IGF-I knockout mice. Furthermore, we and others have shown that transgenic overexpression of IGFBP-5 inhibits bone formation, in part, through an IGF-independent pathway.(13,14) IGFBP-5 binds to a putative receptor on the osteoblast cell surface, which may induce downstream signaling pathways.(1,15,16) IGFBP-5 also contains a nuclear localization sequence that mediates transport of IGFBP-5 to the cell nucleus,(16,17) where it may affect gene transcription. More recently, it has been shown that IGFBP-5 has IGF-receptor–independent effects in normal human intestinal smooth muscle cells, through activation of the P38 MAP kinase and extracellular signal-regulated kinase (ERK)-1/2 pathways, leading to stimulation of cell proliferation and secretion of IGF-I.(18)
To understand the molecular mechanism(s) by which IGFBP-5 stimulates bone formation through an IGF-independent pathway, it is essential to identify the cellular proteins that interact with IGFBP-5. These IGFBP-5 interacting proteins could be IGFBP-5 receptors, nuclear proteins, and signaling proteins that mediate IGF-independent actions of IGFBP-5. Therefore, we used a yeast two-hybrid assay screen(19) to identify proteins that bind to IGFBP-5 using human IGFBP-5 as bait for screening a human osteosarcoma U2 cDNA library. We have recently reported on IGFBP-5 interaction with the Four and One-half LIM domain protein 2 (FHL2) and we have shown that FHL2 binds IGFBP-5, but not IGFBP-4 or IGFBP-6.(20) In this article, we report on the characterization of isoform C of the Ras association family 1 (RASSF1) gene as an interacting partner of IGFBP-5. The RASSF1 gene is predicted to generate four isoforms (RASSF1A, 1B, 1C, and 1F) as a result of alternative splicing and alternative promoter use.(21,22) RASSF1A is the most extensively studied isoform of the RASSF1 gene. Inactivation of RASSF1A is involved in tumor growth progression of various human cancers, whereas overexpression of RASSF1A inhibits tumor growth.(23)
Because IGFBP-5 regulates bone formation parameters both in vitro and in vivo, RASSF1C's interaction with IGFBP-5 provided a hypothesis that RASSF1C may play a role in regulating osteoblast cell proliferation and/or activity and could thereby mediate the action of IGFBP-5. To define the function of RASSF1C in osteoblast cells in vitro, the small interfering RNA (siRNA) technique was used.(24–27) Reduction in RASSF1C expression reduced cell growth in multiple osteoblast cell lines, showing for the first time that RASSF1C, one of four isoforms of the RASSF1 gene, could promote cell proliferation in contrast to the tumor suppressing function predicted for RASSF1A.
MATERIALS AND METHODS
Yeast two-hybrid screen
IGFBP-5 was used as bait to screen a human osteosarcoma cell cDNA library as previously described.(20)
Osteoblast cell culture
Normal human osteoblasts were isolated, as previously described,(28) from calvaria and rib bone specimens obtained from the Cooperative Human Tissue Network, which is supported by the National Cancer Institute. Cell culture work was carried out as previously described.(20)
RNA isolation and Northern analysis
Total RNA from untransformed normal human osteoblasts derived from calvaria and rib and human osteosarcoma cell lines (SaOs-2 and U2) was isolated using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) or the RNAeasy kit (Qiagen, Valencia, CA, USA). Northern blot analysis was carried out as previously described.(20)
Expression analysis of RASSF1C by RT-PCR
To determine the expression of the RASSF1C gene in different osteosarcoma cell lines, the following PCR primers were used: forward, 5′-CTGCAGCCAAGAGGACTCGG-3′; reverse, 5′-GGGTGGCTTCTTGCTGGAGGG-3′. Total RNA (200 ng) was used for the RT reaction using the Omniscript kit (Qiagen). One microliter of the RT reaction was used for PCR using the HotStart master mix (Qiagen). The PCR reactions were run at the following conditions: 95°C for 15 minutes, 95°C for 1 minute, 60°C for 30 s, and 72°C for 30 s for 35 cycles.
Preparation of pYGFP-RASSF1C plasmid construct
The RASSFIC cDNA was amplified by PCR using the following primers: the forward primer contains SalI site 5′-ACGCGTCGACATGGGCGAGGCTGAAACA-3′ and the reverse primer contains BamHI 5′-CGCGGATCCAAGGTCACCCCAAAGGACA-3′. The restriction sites for SalI and BamHI are shown in bold. The PCR reaction was run at the following conditions: 2 minutes at 95°C, 1 minute at 95°C, 30 s at 55°C, 1 minute at 72°C, and 10 minutes at 72°C for 35 cycles. The amplified PCR product was run on 2% agarose gel, purified, and digested with SalI/BamHI. The PCR product was subsequently ligated to SalI/BamHI digested pYGFP plasmid using a standard method.(29) The ligated product was used to transform TOP10 competent E. coli cells using the heat shock method,(29) transformed cells were spread on complete medium containing ampicillin at 200 μg/ml, and plates were incubated at 37°C overnight.
Single colonies were used to inoculate liquid E. coli culture containing ampicillin and were grown overnight with shaking at 37°C. The E. coli culture was used to prepare plasmid DNA using a miniprep kit from Qiagen. The plasmid DNA was subsequently digested with SalI/BamHI and run on a 0.8% agarose gel. Plasmids containing RASSF1C inserts were sequenced to confirm that the RASSF1C cDNA was in frame with the YGFP sequence.
Preparation of phCMV2-RASSF1C expression plasmid
The phCMV2Xi-clone PCR cloning kit was used to construct the mammalian RASSF1C expression vector (Gene Therapy Systems, San Diego, CA, USA). The phCMV2RASSF1C forward primer: 5′-TAACAATG-TACCCATACGATGTTCCGGATTACGCTGGCGAGGCTGAAACACCTT-3′ (underlined sequence corresponds to the HA tag and the italicized sequence corresponds to RASSF1C coding sequence; the start codon is in bold) and the phCMV2RASSF1C reverse primer 5′-CCCGGGCCCGCGGTACCGTCGACTGCAGAA-TTACCCCAAAGGACAGGCGTGCA-3′ (the stop codon is underlined and the italicized sequence corresponds to the 3′ UTR end of the RASSF1C cDNA) were used to amplify the RASSF1C cDNA sequence from our clone identified from the yeast two-hybrid screen. All cloning and transformation procedures were carried out as recommended by the GTS user manual. The structures of recombinant clones were confirmed using DNA sequencing and PCR.
Transfection of U2 cell line with phCMV2-RASSF1C plasmids
Transfection of U2 human osteosarcoma cells with the phCMV2-RASSF1C plasmid construct were carried out either in 6-well plates or in 100-cm2 tissue culture dishes. Cells were plated at 20,000 cells/cm2 in DMEM supplemented with 10% calf serum. After 24 h, the cells were transfected with 1 μg plasmid DNA using Effectene reagent (Qiagen). Cells were collected 24 h after transfection and were lysed in a 50 mM Tris HCl, pH 7.4 buffer containing 150 mM NaCl and 1% tritron ×100.
Co-immunoprecipitation
To confirm the IGFBP-5/RASSF1C interaction observed in our yeast two-hybrid assay, co-immunoprecipitation studies were carried out as previously described(20) using HA (Covance, Berkley, CA, USA) and IGFBP-5 antisera.
siRNA duplex design
The siRNA duplexes for the RASSF1C and alkaline phosphatase (ALP) genes were designed using the Oligoengine software program (www.oligoengine.com). The sequence of RASSFIC siRNA-258 (5′-GGACUACAAUGGCCAGAUC-3′ dTdT) was based on the cDNA sequence obtained from our clone 21. The sequence for the ALP siRNA is 5′-CCCGGACUUCUGGAACCGCdTdT-3′.
We also designed siRNA corresponding to the RASSF1C mRNA sequence (accession AF132676.1): siRNA-186; 5′-GCUGAGAUUGAGCAGAAGAdTdT-3′; siRNA-252: sense 5′-CAAGGACGGUUCUUACACAdTdT-3′. The siRNA oligos were ordered from Qiagen.
Construction of the RASSF1C silencer plasmid
We also cloned the sequence encoding the RASSFIC siRNA-258 duplex in the pSilencer 1.0-U6 siRNA expression vector (Ambion, Austin, TX, USA). The pSilencer plasmid 1.0-U6 was digested with ApaI and EcoRI as recommended by the manufacturer's instructions. After the digestion, the linearized plasmid was gel purified using the QiaXII gel purification kit (Qiagen). The siRNA oligos were ordered from Invitrogen as gel-purified and in lyophilized form. The oligos were annealed and ligated to the pSilencer plasmid as recommended in the user manual. Plasmids containing siRNA inserts were later checked by DNA sequencing using the T3 primer and were used to transfect cells. Plasmids that did not contain siRNA inserts were used as a negative control in transfection studies.
Treatment of cells with siRNA molecules
Cells were seeded at 4000 cells/well in 96-well plates or 35,000 cells/well in 6-well plates in DMEM supplemented with 10% calf serum a day before treatment with siRNA molecules. On the treatment day, the medium was replaced, and cells were transfected with siRNA (25–200 nM) using a Gene Silencer siRNA transfection reagent as recommended by the GeneSilencer reagent manual (Gene Therapy Systems, San Diego, CA, USA). The cells treated with siRNA molecules were incubated for 48 h before cell proliferation assay using alamarBlue.
To evaluate the effect of blockage of RASSF1C expression on IGFBP-5–induced ERK phosphorylation, MC3T3-E1 cells were plated at a density of 150,000 cells/well in α-MEM containing 10% calf serum in 12-well plates. After overnight incubation, medium was removed and replaced with transfection medium containing 200 nM siRNA or vehicle, as described above. After overnight incubation, the medium was removed and replaced with fresh serum-free α-MEM for 24 h before the addition of IGFBP-5 or vehicle. Ten minutes after the addition of IGFBP-5, cells were lysed with electrophoresis sample buffer and used for immunoblot analysis.
AlamarBlue assay
AlamarBlue is reduced by reactions innate to cellular metabolism, and therefore it provides an indirect measure of viable cell number (AccuMed International, Westlake, OH, USA). Cells were rinsed 48–72 h after siRNA treatment with PBS, and the medium was replaced 100 μl of 1× AlamarBlue diluted in phenol red-free DMEM. Direct light was avoided while using the dye, and the plates were incubated for 4 h in 37°C. Fluorescence was determined using a fluorescent plate reader (Fluorolite 1000; Dynex Technologies, Chantilly, VA, USA).
Transfection of MG63 and MC3T3-E1 cells with pSilencer and RT-PCR analysis
MG63 and MC3T3-E1 cells were transfected with 2 μg/ml of pSilencer plasmid or pSilencer-si258 DNA using Effectene (Qiagen). Seventy-two hours after transfection, RNA was extracted, and 200 ng was used to prepare cDNA with Omniscript kit (Qiagen). One microliter of the RT reaction was used for PCR using the HotStart master mix (Qiagen). The PCR reactions were run at the following conditions: 95°C for 15 minutes, 95°C for 1 minute, 60°C for 30 s, and 72°C for 30 s for 35 cycles. The RASSFIC- and actin-specific primers were used for PCR.
Apoptosis
For trypan blue staining, MC3T3-E1 and MG63 cells were treated with siRNA or vehicle, rinsed with PBS, and stained with 0.5% trypan blue for 5 minutes. Subsequently, cells were washed with PBS and examined for dye uptake using a light microscope.
For caspase assay, MC3T3-E1 and MG63 cells were cultured in medium containing 10% calf serum as described previously. Media were removed and rinsed with PBS, and serum-free α-MEM was added before the addition of effectors. Twenty-four hours later, apoptosis was measured using the Apo-ONE homogeneous caspase-3/7 assay kit (Promega, Madison, WI, USA).
ALP assay
For the ALP assay, TE85 and MC3T3-E1 cells were plated in α-MEM containing 10% calf serum, 50 μg/ml ascorbic acid, and 100 mm β-glycerophosphate in 96-well plates. After 24 h, cells were rinsed with serum-free α-MEM containing ascorbic acid and β-glycerophosphate, and effectors were added and incubated for 72 h before determination of ALP activity as previously described.(30) Protein concentration in cell extract was measured by the Bradford Method using a commercial kit (Bio-Rad Laboratories, Richmond, CA, USA). ALP activity was standardized based on cellular protein content and expressed as mu/mg protein.
Effects of IGFBP-5 on ERK-1/2 phosphorylation in MC3T3 cells
The phosphorylation of ERK-1/2 was measured by Western blot analysis using standard methods, as previously described.(20) MC3T3-E1 cells were plated at 200,000 cells/well in α-MEM containing 10% calf serum in a 6-well plate. Twenty-four hours later, serum-free α-MEM was added after rinsing cells with PBS to remove serum. After a 24-h incubation in serum-free α-MEM, cells were treated with recombinant human IGFBP-5 (100 ng/ml), IGF-1 (30 ng/ml), or PBS for various periods of time. After treatment, cells were rapidly extracted with 1× sample loading buffer, boiled for 5 minutes, and subjected to Western blot analysis. The Western blots in duplicate were analyzed using phospho-p44/42 MAP kinase (Thr202/Tyr204; Cell Signaling Technology, Beverly, MA, USA) and pan ERK (BD Biosciences, Palo Alto, CA, USA) antibodies to detect levels of phosphorylated and unphosphorylated ERK-1/2, respectively. Bands of interest corresponding to the phosphorylated and unphosphorylated ERK-1/2 were visualized by chemiluminescence and quantified with densitometry.
RESULTS
IGFBP-5 interacts with RASSF1C
We used the yeast two-hybrid system 3 (THS), in which IGFBP-5, fused to a GAL binding domain, was used as bait to identify candidate proteins that interact with IGFBP-5. We screened a U2 human osteosarcoma cDNA library fused to the GAL activation domain (AD) in the expression vector pACT2. We picked several positive AD-cDNA clones that activated all of the three reporter genes in the AH109 strain. The AD-cDNA plasmids were isolated and were sequenced, as previously described.(20) One of the positive clones (clone 21) matched the isoform C sequence of the RASSF1 gene. The clone contained the full-length cDNA sequence encoding 270 amino acids and interacted with IGFBP-5 as shown by the two-hybrid assay. RASSF1C cDNA obtained from clone 21 shows a 100% identity with the mouse RASSFIC sequence, whereas it shares 93.3% identity with the human RASSF1C sequence deposited in the GenBank at the amino acid level (Fig. 1).
FIG. 1.
Alignment of 270 amino acid sequence encoded by clone 21 (c21) with mouse (m1C) and the human RASSF1C (h1C) amino acid sequence. The RASSF1C cDNA obtained from clone 21 shows 100% identity with the mouse RASSF1C sequence, whereas it shows a 93.3% identity with the human RASSF1C sequence deposited in the GenBank.
The RASSF1 gene encodes four isoforms, specifically RASSF1A, RASSF1C, RASSF1B, and RASSF1F.(21,22) The RASSF1A encodes 340 amino acids, RASSF1C encodes 270 amino acids, RASSF1B encodes 189 amino acids, and RASSF1F encodes a truncated protein of 92 amino acids. After identifying the RASSF1C AD-cDNA clone by DNA sequencing, we transformed AH109 yeast cells with the BD-IGFBP-5 and AD-cDNA plasmids to reconfirm the IGFBP-5/RASSF1C interaction. The growth of the AH109 reporter strain on high stringency medium points toward a strong interaction between RASSF1C and IGFBP-5. No growth was observed when either the BD-IGFBP-5 or the AD-RASSF1C plasmids were tested with control plasmids pGADT7-T and pGBKT7-53 to transform the reporter strain, respectively (data not shown). Therefore, the IGFBP-5/RASSF1C interaction was specific.
RASSF1C gene is expressed in bone cells
To determine if RASSF1C is expressed in other bone cell types in addition to U2 human osteosarcoma cells, total RNA from untransformed normal human bone cells derived from calvaria (HBC) and rib (HBR), human osteosarcoma cell lines (U2, MG63, TE85), and mouse calvaria cell line MC3T3-E1 was used to carry out either Northern blot or RT-PCR. With the Northern blot analyses, RASSFIC probe detected signals in all cell types tested (Fig. 2A). RT-PCR using RASSF1C-specific primers confirmed expression of RASSF1C in multiple osteoblast cell types (Fig. 2B).
FIG. 2.
(A) Northern blot analysis of total RNA extracted from various human osteoblast cell preparations using the RASSF1C cDNA as a probe. RNA was extracted from 70–80% confluent cultures of normal human osteoblasts derived from calvaria (HBC) and rib (HBR) and from SaOs-2 and U2 osteosarcoma cells. Twenty micrograms of total RNA was loaded per lane, and the blot was probed with [32P]labeled RASSF1C cDNA. The probe detected a 1.7-kb band in all cell lines tested. (B) RT-PCR analysis of total RNA extracted from various osteoblast cell types using RASSF1C and actin specific primers. One microgram of total RNA was RT reacted using oligodT. One microliter of the RT reaction was used for PCR reactions. The RASSF1C-specific primers amplified a 280-bp PCR product from all the cell lines tested. Actin-specific primers amplified a 200-bp product, as expected. (C) Co-immunoprecipitation of RASSF1C/IGFBP-5 in U2 cells overexpressing HA-RASSF1C. Lane 1, 100 μl of U2 cell lysate overexpressing HA-RASSFIC incubated with 50 μl protein A-Sepharose coupled to IGFBP-5 antibody-recombinant IGFBP-5 complex; lane 2, the same as lane 1, but recombinant IGFBP-5 was not included in the complex; lane 3, the same as lane 1, but control cell lysate from untransfected U2 cells was used. After 4 h of incubation at 4°C, the protein complex was washed, and bound proteins were extracted with SDS sample buffer and subjected to SDS-PAGE and immunoblotting using the HA-antibody. RASSF1C was co-immunoprecipitated in lane 1 but not in lane 2 or 3.
IGFBP-5/RASSF1C interaction determined by co-immunoprecipitation
To further confirm the RASSF1C/IGFBP-5 interaction observed in the yeast two-hybrid assay, cell lysates from U2 cells overexpressing RASSF1C as a fusion with the HA tag were immunoprecipitated with IGFBP-5 protein using IGFBP-5 antiserum and probed with HA-tag monoclonal antibody in Western immunoblot analysis. The RASSF1C/ IGFBP-5 complex was co-immunoprecipitated by IGFBP-5 antibody (Fig. 2C). This interaction was specific because no RASSF1C was detected in the immunoprecipitate when either RASSF1C or IGFBP-5 was omitted from the reaction mixture. These data, together with the yeast two-hybrid data, provide evidence that IGFBP-5 and RASSF1C do interact. However, it remains to be determined whether the interaction between IGFBP-5 and RASSF1C occurs at normal physiological levels in osteoblasts.
Silencing of RASSF1C expression by siRNA
We synthesized the siRNA duplex specific for RASSF1C mRNA using our clone 21 cDNA sequence (si258) as well as human RASSF1C mRNA sequences deposited in the GenBank (si186 and si252) and tested their effects on cell number in multiple osteoblast cell types. Figure 3A shows that various synthetic siRNA molecules decreased cell number by 20–40% in various osteoblast cell types. Furthermore, the magnitude of inhibition caused by si258 on cell number was dose-dependent in TE85 and MG63 human osteosarcoma cells (data not shown). The inhibitory effect of RASSF1C siRNA on osteoblast cell number seems to be specific, because a control siRNA duplex, specifically ALP siRNA, did not produce a similar effect on osteoblast cell number (data not shown). In addition to using synthetic siRNA molecules, the RASSF1C siRNA-258 duplex was cloned into the pSilencer 1.0 U6 plasmid and used for evaluation of cell number. The cell number was significantly reduced in cells treated with pSilencer-RASSF1C plasmid compared with cells transfected with pSilencer plasmid alone (Fig. 3A). To confirm that the inhibitory effect of RASSF1C siRNA on cell number is caused by reduced expression of RASSF1C, we measured RASSF1C mRNA levels in cultures treated with psi258 or control plasmid. Figure 3B shows that psi258 treatment reduced RASSF1C mRNA levels by 70% and 60%, respectively, in MG63 and MC3T3-E1 cells. To determine if the inhibitory effect of RASSF1C siRNA was caused by increased apoptosis, we evaluated apoptosis by trypan blue staining and by caspase assay in siRNA-treated and vehicle-treated control cultures. We found that apoptosis was not affected by IGFBP-5 or RASSF1C siRNA treatment by either trypan blue assay (data not shown) or by caspase assay (Fig. 3C) in serum-free cultures of MC3T3-E1 and MG63 cells. In contrast, treatment with TNF-α, a known stimulator of apoptosis, increased caspase activity significantly in both MC3T3-E1 and MG63 cells.
FIG. 3.
(A) Effects of RASSF1C siRNA on cell proliferation. For cell proliferation experiments involving RASSF1C-specific siRNA duplexes, cells were plated at 2000 cells/well in media containing 10% cell serum (CS). After a 24-h treatment, cells were transfected with 0.3 ug of siRNA per well using the Transmessenger reagent kit (Qiagen). For treatment with pSilencer-RASSF1C-258 or pSilencer plasmids (control), cells were plated at 15,000–20,000 cells per well in 6-well plates. After 24 h, cells were transfected with 2 μg of plasmid DNA using the effectene. Forty-eight or 72 h after addition of siRNA duplex or pSilencer plasmid, cell number was determined by alamarBlue assay. Values are mean ± SE of six to eight replicates per treatment. Ap < 0.01 vs. corresponding controls. Data shown in the figure have been reproduced in at least one independent experiment. RASSF1C siRNA decreased cell number in various osteoblast cell types. (B) RT-PCR analysis of RASSF1C expression from MG63 and MC3T3-E1 cells transfected with pSilencer-RASSF1C-258 or pSilencer plasmid. Two hundred nanograms of total RNA was used for RT, and 1 μl of RT was used for PCR reactions using RASSF1C- and actin-specific primers. Lane 1, RASSF1C amplification from control cells; lane 2, RASSF1C amplification from treated cells; lanes 3 and 4, actin amplification from control and treated cells, respectively. RASSF1C siRNA decreased expression of RASSF1C by 60–70%. (C) Effects of IGFBP-5, TNF-α, and RASSF1C siRNA on caspase activity in MC3T3-E1 and MG63 cells. To evaluate the effects of IGFBP-5, cells were treated for 48 h with 100 ng/ml IGFBP-5 or vehicle before caspase assay. TNF-α (10 ng/ml) was used as a positive control. To evaluate the effects of RASSF1C siRNA, MG63 and MC3T3-E1 cells were transfected with psi258 or control plasmid and incubated with or without IGFBP-5 for 48 h before caspase assay. Values are expressed as percentage of vehicle-treated control and are mean ± SE of eight replicates per treatment. The fluorescence units (mean ± SE) for vehicle-treated control MC3T3-E1 and MG63 cultures were 359 ± 8 and 1079 ± 45, respectively. Ap < 0.01 vs. corresponding controls. Neither IGFBP-5 nor RASSF1C siRNA had any significant effect on caspase activity. (D) Effects of RASSF1C siRNA on ALP activity in MC3T3-E1 and TE85 cells. Cells were plated and treated with siRNA or plasmid and effectors. Seventy-two hours after treatment with effectors, ALP activity and protein content were measured in cell extracts. ALP activity was standardized on the basis of milligrams of protein and compared with vehicle-treated control cultures. ALP activity in vehicle-treated control cultures were 127 mu/mg protein and 2347 mu/mg protein, respectively, in MC3T3-E1 and TE85 cells. Values are percentage of vehicle-treated control and are mean ± SE of eight replicate wells. Ap < 0.05 vs. vehicle-treated control. Bp < 0.05 vs. si258-treated control cultures. ALP activity was significantly increased in the presence of IGFBP-5 in both control and siRNA-treated cultures of MC3T3-E1 and TE85 cells.
In previous studies, we and others have found evidence that IGFBP-5 treatment increased ALP activity in bone cells.(11,31) We therefore tested the effects of RASSF1C siRNA on ALP activity in TE85 and MC3T3-E1 cells. We found that RASSF1C siRNA did not block IGFBP-5–induced increase in ALP activity (Fig. 3D).
IGFBP-5 stimulates the phosphorylation of ERK-1/2 isoforms in osteoblasts
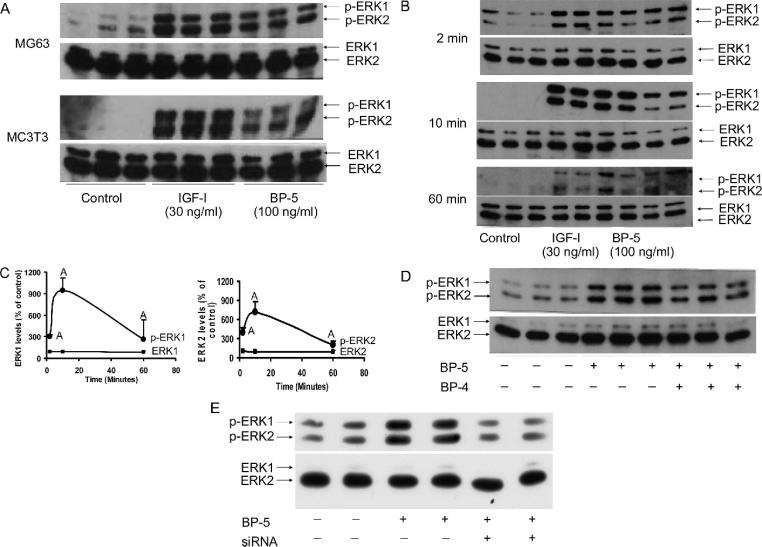
Because RASSF1C contains the Ras effector domain, we predicted that the interaction between IGFBP-5 and RASSF1C could lead to activation of MAPK/ERK pathways. Therefore, we first examined the effect of IGFBP-5 on the phosphorylation of ERK-1/2 isoforms in MG63 human osteosarcoma cells and MC3T3-E1 mouse osteoblasts. Figure 4A shows that IGFBP-5 treatment increased phosphorylation of ERK-1/2 in both cell types. The magnitude of IGFBP-5–induced increase in ERK-1/2 phosphorylation, however, was less compared with an equivalent dose of IGF-1 at 10 minutes. Figure 4B shows that neither IGFBP-5 nor IGF-1 treatment caused a time-dependent increase in phosphorylation of ERK-1 and -2. At 2 minutes, IGFBP-5 increased phosphorylation of ERK-1/2 by 3- to 4-fold and by 7- to 9-fold at 10 minutes. The level of phosphorylation was decreased to ~3-fold after 60 minutes of IGFBP-5 treatment, suggesting that the changes in phosphorylation induced by IGFBP-5 treatment are transient (Fig. 4C). Although the acute effect of IGFBP-5 on ERK phosphorylation is consistent with the direct effect of IGFBP-5, it is possible that endogenously produced IGF-1 contributed to this effect. To confirm that the IGFBP-5–induced increase in ERK phosphorylation was IGF-independent, we evaluated ERK phosphorylation in the presence of excess IGFBP-4 that binds IGF and neutralizes its action. Figure 4D shows that IGFBP-5–induced phosphorylation was not abolished by exogenous addition of IGFBP-4, providing evidence that the effect of IGFBP-5 on ERK phosphorylation is indeed partially IGF-independent. The data obtained in this study is consistent with what has been found in quiescent muscle cells.(18)
FIG. 4.
(A) MG63 human osteosarcoma and MC3T3-E1 mouse osteoblasts were treated with 30 ng/ml IGF-1, 100 ng/ml IGFBP-5, or vehicle for 10 minutes. Cell lysate was used for determination of total and phosphorylated ERK-1/2 using specific antibodies. Three replicates were used for each treatment. Both IGF-1 and IGFBP-5 increased ERK-1/2 phosphorylation. (B) MC3T3-E1 mouse osteoblasts were treated with 30 ng/ml IGF-I, 100 ng/ml IGFBP-5, or vehicle under serum-free conditions. Two, 10, and 60 minutes after treatment, cells were lysed, and cell lysate was used for determination of total and phosphorylated ERK-1/2 levels. Three replicates were used for each treatment. IGFBP-5, like IGF-1, increased phosphorylation levels of ERK-1/2 in a time-dependent manner. (C) Levels of total and phosphorylated ERK-1/2 in IGFBP-5–treated cultures were quantitated by densitometric scanning of bands and are expressed as percentage of vehicle-treated control for data shown in B. Values are mean ± SD of three replicates per group. Ap < 0.05 vs. control. (D) Effects of IGFBP-4 pretreatment on IGFBP-5–induced increase in ERK phosphorylation. MC3T3-E1 cells were pretreated for 60 minutes with 300 ng/ml IGFBP-4 or vehicle before the addition of IGFBP-5. Ten minutes later, total and phosphorylated ERK-1/2 levels were determined by Western immunoblot analysis. IGFBP-5 increased ERK-1/2 phosphorylation in the presence of IGFBP-4. (E) Effects of RASSF1C siRNA on IGFBP-5–induced ERK-1/2 phosphorylation. MC3T3-E1 cells were incubated with transfection medium containing vehicle or siRNA258. Twenty-four hours after transfection, the media were changed to serum-free and incubated for 24 h before the addition of the vehicle or 100 ng/ml IGFBP-5. Ten minutes later, phosphorylated and total levels of ERK-1/2 were determined by Western immunoblot using specific antibodies. RASSF1C siRNA blocked IGFBP-5–induced increase in ERK-1/2 phosphorylation.
RASSF1C siRNA blocks IGFBP-5–induced ERK-1/2 phosphorylation
Recently, it has been shown that RASSF1 gene products bind to CNK, a multidomain scaffold protein that has been identified as a positive regulator of the Ras/Raf/MAPK pathway in Drosophila.(32) If RASSFIC interaction with IGFBP-5 is involved in activation of the MAPK/ERK pathway, it should be possible to block IGFBP-5-induced ERK-1/2 phosphorylation by inhibiting RASSFIC expression. Accordingly, Fig. 4E shows that pretreatment of MC3T3-E1 cells with RASSFIC siRNA blocked IGFBP-5–induced ERK-1/2 phosphorylation. RASSF1C siRNA treatment did not, however, alter the total level of ERK proteins.
Subcellular localization of RASSF1C in osteoblasts
If RASSF1C acts to regulate IGFBP-5 actions through modulating Ras/Raf/ERK pathways, we would predict that RASSF1C would be localized in the subcellular compartment where Ras proteins are localized. To evaluate RASSF1C's subcellular localization, RASSF1C was expressed as a fusion with yellow fluorescence protein (YFP) in U2 cells (Fig. 5). The YFP-RASSF1C fusion protein seems to principally localize on the endoplasmic reticulum (ER) and the Golgi. Although we have found that localization of neither IGFBP-5 nor FHL2 in osteoblasts is altered by addition of GFP (data not shown), additional studies are needed to rule out the possibility that YFP modifications of RASSF1C did not alter the subcellular distribution of RASSF1C. Recently, it has been shown that a fusion of the Ras association binding domain (RBD) of Raf1 to GFP bound activated Ras proteins on both the ER and Golgi membranes,(33,34) indicating that Ras signaling is not confined to the plasma membrane. Although, the localization data reported here are consistent with subcellular localization patterns seen for Ras proteins, further studies showing co-localization of Ras and RASSF1C are needed to prove that these two proteins are co-localized in the same subcellular compartments.
FIG. 5.
Subcellular localization of YFPRASSF1C fusion protein in U2 cells. The entire open reading frame of RASSF1C cDNA was fused to the YFP sequence. One microgram of plasmid DNA was used to transfect U2 cells in 6-well plates using the effectene reagent. Twenty-four hours after transfection, cells were analyzed using a fluorescent microscope with the appropriate filters. The YFP-RASSF1C fusion protein appears to be mainly localized in the endoplasmic reticulum and Golgi. N, nucleus.
DISCUSSION
In this study, we characterized a 1.4-kb clone identified under high stringency conditions using IGFBP-5 as bait and a U2 osteosarcoma cell cDNA library as prey in a yeast two-hybrid screen. Sequence analysis of the 1.4-kb clone revealed a 100% and 93.3% sequence identity with mouse and human RASSF1C sequences at the amino acid level, respectively. Because we used the human osteosarcoma library to fish out the IGFBP-5 interacting clones, we expected the RASSF1C clone to exhibit a 100% sequence identity with the human RASSF1C gene. In this regard, we previously identified a clone using the same library that exhibited 100% sequence identity with the human FHL2.(20) One potential explanation for this discrepancy is that the yeast plasmid PCAT2 library that we purchased from Clonetech may have been made using RNA from both U2 cells and mouse osteoblasts. Consistent with this interpretation, other independent yeast two-hybrid screens using the PCAT2 library with different baits yielded cDNA sequences that exhibited 100% sequence identity with mouse, but not human, genes (unpublished observations). Future studies are needed to sequence random cDNA clones from the PCAT2 library to confirm the assumption that this library is contaminated with mouse cDNA.
In previous studies, it has been shown that the RASSF1A gene is inactivated in several tumors and that overexpression of RASSF1A inhibits tumor growth.(23) Based on these and other findings, it has been predicted that RASSF1A functions as a tumor suppressor. In contrast to RASSF1A, little is known about the role of RASSF1C in any cell type. If RASSF1C functions in a manner similar to that of RASSF1A, we would predict RASSF1C to inhibit osteoblast cell proliferation. On the other hand, if RASSF1C functions, in part, to mediate the stimulatory effects of IGFBP-5 on osteoblast cell proliferation, RASSF1C should act as a positive regulator of cell proliferation. Therefore, we tested the consequence of silencing the expression of RASSF1C using siRNA in multiple osteoblast cell types. Our findings that osteoblast cell numbers were reduced by 20–40% by different siRNA complexes reveal that RASSF1C is a promoter of cell proliferation in contrast to what was predicted based on the established role of RASSF1A.
To determine if RASSF1C regulates apoptosis, we next evaluated the effect of RASSF1C siRNA on caspase activity in MC3T3-E1 and MG63 cells. We found that neither IGFBP-5 treatment nor RASSF1C siRNA treatment had any significant effect on MC3T3-E1 or MG63 cell apoptosis, suggesting that the reduced cell number, as determined by AlamarBlue assay in cultures treated with RASSF1C siRNA, could not be explained by increased cell death. In contrast to our finding, other studies have shown that IGFBP-5 treatment both promoted and inhibited apoptosis in different cell types.(35,36) Thus, the biological effects of IGFBP-5 on proliferation, differentiation, and apoptosis may vary depending on the cell type studied, cell culture, and treatment conditions.
In terms of the molecular pathways for IGF-independent actions of IGFBP-5, we have shown that IGFBP-5 caused a time-dependent increase in ERK-1/2 phosphorylation that was not abolished by inhibitory IGFBP-4. IGFBP-5–induced ERK-1/2 phosphorylation is not unique to osteoblasts, because Kuemmerle and Zhou(18) have reported that IGFBP-5 stimulated Ras-dependent activation of P38 MAP kinase and ERK-1/2 pathways. Based on the findings that that RASSF1C interacts with IGFBP-5 and that RASSF1C contains a Ras association domain, we postulated that the interaction between IGFBP-5 and RASSF1C may be involved in regulation of IGFBP-5–induced ERK phosphorylation. Accordingly, we found that treatment of MC3T3-E1 mouse osteoblasts with RASSF1C siRNA reduced IGFBP-5 effects on ERK phosphorylation. The mechanism by which IGFBP-5 binding to RASSF1C leads to activation of the ERK pathway can only speculated at this time. In this regard, a number of nonenzymatic accessory proteins have recently been identified in the regulation of Ras-Raf-MAP kinase cascade.(37) These include molecules such as KSR1, Sur-8, CNK, and MP-1, which have been suggested to play a role as scaffolding and/or adaptor proteins.(37–40) CNK1 has been shown to directly bind to both RASSF1A and RASSF1C proteins.(40) Furthermore, Praskova et al.(41) have recently shown that RASSF1A and RASSF1C bind to KRS2, also known as MST1, and modulate ATP-mediated autoactivation of KRS2. Based on these data, together with our findings that RASSF1C binds to IGFBP-5 and that inhibition of RASSF1C expression in osteoblasts abolishes IGFBP-5–induced ERK phosphorylation, it can be speculated that IGFBP-5 and RASSF1C may serve as components of adapter/scaffold protein complex for Raf/MAP kinase cascade and may function to localize or stabilize one or more kinases in the Ras-Raf-MAP kinase pathway. Future studies are needed to address if the binding of IGFBP-5 to RASSF1C is involved in the recruitment or activation of the Raf/MAP kinase.
The involvement of the MAP kinase/ERK pathway in regulating osteoblast cell proliferation has been well established.(42) It is therefore tempting to speculate that the IGFBP-5–induced increase in ERK phosphorylation could be involved in mediating the IGFBP-5 effects on cell proliferation. In this regard, RASSF1C siRNA decreased the IGFBP-5 effect on ERK phosphorylation and decreased cell number, but had no effect on IGFBP-5–induced increases in ALP activity. Future studies are needed to establish the cause and effect association between the IGFBP-5 interaction with RASSF1C, ERK phosphorylation, and cell proliferation to provide experimental data that the interaction between IGFBP-5 and RASSF1C leads to an increase in ERK phosphorylation, which mediates the IGFBP-5 effects on cell proliferation. In addition to interacting with RASSF1C, IGFBP-5 has also been shown to interact with FHL2, a potential transcription modulator.(20) The questions of whether RAASF1C and/or FHL2 interacts with IGFBP-5 under normal physiological conditions and whether such interactions are involved in mediating the biological effects of IGFBP-5 in vivo remain to be established.
ACKNOWLEDGMENTS
The authors acknowledge the technical assistance provided by Joe Rung-Aroon and Sharanya Bala and the secretarial assistance provided by Sean Belcher and Shelley Levtzow. This work was supported by National Institute of Health Grants AR31062 and AR07543. All work was performed at facilities provided by the Jerry L Pettis VA Medical Center (Loma Linda, CA, USA).
Footnotes
The authors have no conflict of interest.
REFERENCES
- 1.Mohan S, Baylink DJ. IGF system components and their role in bone metabolism. In: Rosenfeld RG, Roberst C, editors. IGFs in Health and Diseases. Humana Press; Totowa, NJ, USA: 1999. pp. 457–496. [Google Scholar]
- 2.Mohan S, Baylink DJ. Insulin-like growth factor system components and the coupling of bone formation to resorption. Horm Res. 1996;45(Suppl 1):59–62. doi: 10.1159/000184833. [DOI] [PubMed] [Google Scholar]
- 3.Rosen CJ, Donahue LR. Insulin-like growth factors and bone: The osteoporosis connection revisited. Proc Soc Exp Biol Med. 1998;219:1–7. doi: 10.3181/00379727-219-44310. [DOI] [PubMed] [Google Scholar]
- 4.Fanayan S, Firth SM, Butt AJ, Baxter RC. Growth inhibition by insulin-like growth factor-binding protein-3 in T47D breast cancer cells requires transforming growth factor-beta (TGF-beta) and the type II TGF-beta receptor. J Biol Chem. 2000;275:39146–39151. doi: 10.1074/jbc.M006964200. [DOI] [PubMed] [Google Scholar]
- 5.Oh Y, Muller HL, Lamson G, Rosenfeld RG. Insulin-like growth factor (IGF)-independent action of IGF-binding protein-3 in Hs578T human breast cancer cells. Cell surface binding and growth inhibition. J Biol Chem. 1993;268:14964–14971. [PubMed] [Google Scholar]
- 6.Rajah R, Valentinis B, Cohen P. Insulin-like growth factor (IGF)-binding protein-3 induces apoptosis and mediates the effects of transforming growth factor-beta1 on programmed cell death through a p53- and IGF-independent mechanism. J Biol Chem. 1997;272:12181–12188. doi: 10.1074/jbc.272.18.12181. [DOI] [PubMed] [Google Scholar]
- 7.Spagnoli A, Hwa V, Horton WA, Lunstrum GP, Roberts CT, Jr, Chiarelli F, Torello M, Rosenfeld RG. Antiproliferative effects of insulin-like growth factor-binding protein-3 in mesenchymal chondrogenic cell line RCJ3.1C5.18. relationship to differentiation stage. J Biol Chem. 2001;276:5533–5540. doi: 10.1074/jbc.M005088200. [DOI] [PubMed] [Google Scholar]
- 8.Andress DL, Loop SM, Zapf J, Kiefer MC. Carboxy-truncated insulin-like growth factor binding protein-5 stimulates mitogenesis in osteoblast-like cells. Biochem Biophys Res Commun. 1993;195:25–30. doi: 10.1006/bbrc.1993.2004. [DOI] [PubMed] [Google Scholar]
- 9.Bauss F, Lang K, Dony C, Kling L. The complex of recombinant human insulin-like growth factor-I (rhIGF-I) and its binding protein-5 (IGFBP-5) induces local bone formation in murine calvariae and in rat cortical bone after local or systemic administration. Growth Horm IGF Res. 2001;11:1–9. doi: 10.1054/ghir.2000.0181. [DOI] [PubMed] [Google Scholar]
- 10.Miyakoshi N, Richman C, Kasukawa Y, Linkhart TA, Baylink DJ, Mohan S. Evidence that IGF-binding protein-5 functions as a growth factor. J Clin Invest. 2001;107:73–81. doi: 10.1172/JCI10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richman C, Baylink DJ, Lang K, Dony C, Mohan S. Recombinant human insulin-like growth factor-binding protein-5 stimulates bone formation parameters in vitro and in vivo. Endocrinology. 1999;140:4699–4705. doi: 10.1210/endo.140.10.7081. [DOI] [PubMed] [Google Scholar]
- 12.Mohan S, Nakao Y, Honda Y, Landale E, Leser U, Dony C, Lang K, Baylink DJ. Studies on the mechanisms by which insulin-like growth factor (IGF) binding protein-4 (IGFBP-4) and IGFBP-5 modulate IGF actions in bone cells. J Biol Chem. 1995;270:20424–20431. doi: 10.1074/jbc.270.35.20424. [DOI] [PubMed] [Google Scholar]
- 13.Devlin RD, Du Z, Buccilli V, Jorgetti V, Canalis E. Transgenic mice overexpressing insulin-like growth factor binding protein-5 display transiently decreased osteoblastic function and osteopenia. Endocrinology. 2002;143:3955–3962. doi: 10.1210/en.2002-220129. [DOI] [PubMed] [Google Scholar]
- 14.Salih DA, Mohan S, Kasukawa Y, Tripathi G, Lovett FA, Anderson NF, Carter EJ, Wergedal JE, Baylink DJ, Pell JM. Insulin-like growth factor binding protein-5 (IGFBP-5) induces a gender-related decrease in bone mineral density (BMD) in transgenic mice. Endocrinology. 2005;146:931–940. doi: 10.1210/en.2004-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andress DL. Insulin-like growth factor-binding protein-5 (IGFBP-5) stimulates phosphorylation of the IGFBP-5 receptor. Am J Physiol. 1998;274:E744–E750. doi: 10.1152/ajpendo.1998.274.4.E744. [DOI] [PubMed] [Google Scholar]
- 16.Schedlich LJ, Le Page SL, Firth SM, Briggs LJ, Jans DA, Baxter RC. Nuclear import of insulin-like growth factor-binding protein-3 and -5 is mediated by the importin beta subunit. J Biol Chem. 2000;275:23462–23470. doi: 10.1074/jbc.M002208200. [DOI] [PubMed] [Google Scholar]
- 17.Schedlich LJ, Young TF, Firth SM, Baxter RC. Insulin-like growth factor-binding protein (IGFBP)-3 and IGFBP-5 share a common nuclear transport pathway in T47D human breast carcinoma cells. J Biol Chem. 1998;273:18347–18352. doi: 10.1074/jbc.273.29.18347. [DOI] [PubMed] [Google Scholar]
- 18.Kuemmerle JF, Zhou H. Insulin-like growth factor-binding protein-5 (IGFBP-5) stimulates growth and IGF-I secretion in human intestinal smooth muscle by Ras-dependent activation of p38 MAP kinase and Erk1/2 pathways. J Biol Chem. 2002;277:20563–20571. doi: 10.1074/jbc.M200885200. [DOI] [PubMed] [Google Scholar]
- 19.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 20.Amaar YG, Thompson GR, Linkhart TA, Chen ST, Baylink DJ, Mohan S. Insulin-like growth factor-binding protein 5 (IGFBP-5) interacts with a four and a half LIM protein 2 (FHL2) J Biol Chem. 2002;277:12053–12060. doi: 10.1074/jbc.M110872200. [DOI] [PubMed] [Google Scholar]
- 21.Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, Sekido Y, Latif F, Milchgrub S, Toyooka S, Gazdar AF, Lerman MI, Zabarovsky E, White M, Minna JD. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, Yoon JH, Dammann R, Pfeifer GP. Frequent hypermethylation of the RASSF1A gene in prostate cancer. Oncogene. 2002;21:6835–6840. doi: 10.1038/sj.onc.1205814. [DOI] [PubMed] [Google Scholar]
- 24.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 25.Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 26.Leirdal M, Sioud M. Gene silencing in mammalian cells by preformed small RNA duplexes. Biochem Biophys Res Commun. 2002;295:744–748. doi: 10.1016/s0006-291x(02)00736-2. [DOI] [PubMed] [Google Scholar]
- 27.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 28.Linkhart TA, Linkhart SG, MacCharles DC, Long DL, Strong DD. Interleukin-6 messenger RNA expression and interleukin-6 protein secretion in cells isolated from normal human bone: Regulation by interleukin-1. J Bone Miner Res. 1991;6:1285–1294. doi: 10.1002/jbmr.5650061204. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Kaboratory Manual. 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor; NY, USA: 1989. [Google Scholar]
- 30.Farley JR, Hall SL, Tanner MA, Wergedal JE. Specific activity of skeletal alkaline phosphatase in human osteoblast-line cells regulated by phosphate, phosphate esters, and phosphate analogs and release of alkaline phosphatase activity inversely regulated by calcium. J Bone Miner Res. 1994;9:497–508. doi: 10.1002/jbmr.5650090409. [DOI] [PubMed] [Google Scholar]
- 31.Yin P, Xu Q, Duan C. Paradoxical actions of endogenous and exogenous insulin-like growth factor-binding protein-5 revealed by RNA interference analysis. J Biol Chem. 2004;279:32660–32666. doi: 10.1074/jbc.M401378200. [DOI] [PubMed] [Google Scholar]
- 32.Douziech M, Roy F, Laberge G, Lefrancois M, Armengod AV, Therrien M. Bimodal regulation of RAF by CNK in Drosophila. EMBO J. 2003;22:5068–5078. doi: 10.1093/emboj/cdg506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bivona TG, Philips MR. Ras pathway signaling on endo-membranes. Curr Opin Cell Biol. 2003;15:136–142. doi: 10.1016/s0955-0674(03)00016-4. [DOI] [PubMed] [Google Scholar]
- 34.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, II, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 35.Cobb LJ, Salih DA, Gonzalez I, Tripathi G, Carter EJ, Lovett F, Holding C, Pell JM. Partitioning of IGFBP-5 actions in myogenesis: IGF-independent anti-apoptotic function. J Cell Sci. 2004;117:1737–1746. doi: 10.1242/jcs.01028. [DOI] [PubMed] [Google Scholar]
- 36.Marshman E, Green KA, Flint DJ, White A, Streuli CH, Westwood M. Insulin-like growth factor binding protein 5 and apoptosis in mammary epithelial cells. J Cell Sci. 2003;116:675–682. doi: 10.1242/jcs.00263. [DOI] [PubMed] [Google Scholar]
- 37.Sternberg PW, Alberola-Ila J. Conspiracy theory: RAS and RAF do not act alone. Cell. 1998;95:447–450. doi: 10.1016/s0092-8674(00)81612-8. [DOI] [PubMed] [Google Scholar]
- 38.Anselmo AN, Bumeister R, Thomas JM, White MA. Critical contribution of linker proteins to Raf kinase activation. J Biol Chem. 2002;277:5940–5943. doi: 10.1074/jbc.M110498200. [DOI] [PubMed] [Google Scholar]
- 39.Lanigan TM, Liu A, Huang YZ, Mei L, Margolis B, Guan KL. Human homologue of Drosophila CNK interacts with Ras effector proteins Raf and Rlf. FASEB J. 2003;17:2048–2060. doi: 10.1096/fj.02-1096com. [DOI] [PubMed] [Google Scholar]
- 40.Rabizadeh S, Xavier RJ, Ishiguro K, Bernabeortiz J, Lopez-Ilasaca M, Khokhlatchev A, Mollahan P, Pfeifer GP, Avruch J, Seed B. The scaffold protein CNK1 interacts with the tumor suppressor RASSF1A and augments RASSF1A-induced cell death. J Biol Chem. 2004;279:29247–29254. doi: 10.1074/jbc.M401699200. [DOI] [PubMed] [Google Scholar]
- 41.Praskova M, Khoklatchev A, Ortiz-Vega S, Avruch J. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem J. 2004;381:453–462. doi: 10.1042/BJ20040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau KH, Baylink DJ. Molecular mechanism of action of fluoride on bone cells. J Bone Miner Res. 1998;13:1660–1667. doi: 10.1359/jbmr.1998.13.11.1660. [DOI] [PubMed] [Google Scholar]