Abstract
Dysregulation of metabolism is a common phenomenon in cancer cells. The NADP+-dependent isocitrate dehydrogenases 1 and 2 (IDH1 and IDH2) function at a crossroads of cellular metabolism in lipid synthesis, cellular defense against oxidative stress, oxidative respiration, and oxygen-sensing signal transduction. We review the normal functions of the encoded enzymes, frequent mutations of IDH1 and IDH2 recently found in human cancers, and possible roles for the mutated enzymes in human disease. IDH1 and IDH2 mutations occur frequently in some types of World Health Organization grades 2–4 gliomas and in acute myeloid leukemias with normal karyotype. IDH1 and IDH2 mutations are remarkably specific to codons that encode conserved functionally important arginines in the active site of each enzyme. To date, all IDH1 mutations have been identified at the Arg132 codon. Mutations in IDH2 have been identified at the Arg140 codon, as well as at Arg172, which is aligned with IDH1 Arg132. IDH1 and IDH2 mutations are usually heterozygous in cancer, and they appear to confer a neomorphic enzyme activity for the enzymes to catalyze the production of D-2-hydroxyglutarate. Study of alterations in these metabolic enzymes may provide insights into the metabolism of cancer cells and uncover novel avenues for development of anticancer therapeutics.
Normal Functions of NADP+-Dependent Isocitrate Dehydrogenases 1 and 2
Isocitrate dehydrogenases (IDHs) catalyze the oxidative decarboxylation of isocitrate to α-ketoglutarate and reduce NAD(P)+ to NAD(P)H. This process involves oxidation of isocitrate to oxalosuccinate, with NAD(P)H as the electron acceptor, followed by decarboxylation of oxalosuccinate to form α-ketoglutarate. Humans and other eukaryotes have both NAD+- and NADP+-dependent IDHs. NAD+-dependent IDH, or IDH3, is a multisubunit enzyme that is localized to the mitochondrial matrix and is classically thought to play a central role in aerobic energy production in the tricarboxylic acid (TCA) cycle. IDH1 and IDH2 are NADP+ dependent, share considerable sequence similarity (70% identity in humans), and are unrelated to IDH3. Importantly, IDH1 and IDH2 catalyze reversible reactions and have no known allosteric modifiers, whereas the reaction catalyzed by IDH3 is irreversible and allosterically regulated by a variety of positive (calcium, ADP, and citrate) and negative (ATP, NADH, and NADPH) effectors (1). In this review, we use IDH to refer to both IDH1 and IDH2 but not IDH3.
IDH1 is highly expressed in the mammalian liver and moderately expressed in other tissues (2). It contains a C-terminal tripeptide peroxisome targeting signal 1 sequence (3) and localizes to varying extents to the cytoplasm and peroxisome of yeast and mammalian cells (4–6). IDH2 contains an N-terminal mitochondrial signal peptide and localizes to the mitochondria (3). It is highly expressed in mammalian heart, muscle, and activated lymphocytes and moderately expressed elsewhere (2,7).
Structure and Mechanism of Regulation
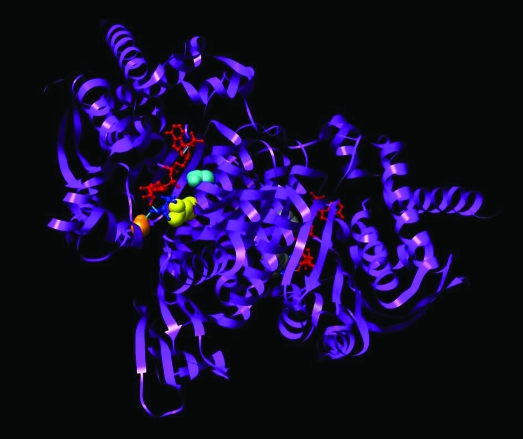
Early studies of IDH structure were performed on bacterial homologs of human IDH (8–13). Ceccarelli et al. (14) solved the crystal structure of porcine IDH2. Xu et al. (15) reported the crystal structure of human IDH1. Bacterial homologs of IDH, as well as porcine IDH2 and human IDH1, function as homodimers (13–15). Each homolog comprises a large domain, a clasp domain, and a small domain. In human IDH1, the large domain is made up of residues 1–103 and 286–414, the clasp encompasses residues 137–185, and the small domain consists of residues 104–136 and 186–285. IDH homodimers contain two asymmetric active sites, with each active site made up of a cleft formed by the large and small domains of one IDH1 molecule and the small domain of the other IDH1 molecule in the dimer. The active sites are exposed to solvent and are accessible to the substrate and cofactor. The clasp functions to hold the two subunits together to form this active site (15). Human IDH1 transitions between an inactive open, an inactive semi-open, and a catalytically active closed conformation. In the inactive open conformation, Asp279 occupies the position where the isocitrate substrate normally forms hydrogen bonds with Ser94 (Figure 1). This steric hindrance by Asp279 to isocitrate binding is relieved in the active closed conformation.
Figure 1.
Isocitrate dehydrogenase 1 (IDH1) dimer in closed conformation. Structure of both molecules of the IDH1 dimer in the active closed conformation. The crystal structure of IDH1 is shown in ribbon format (PDBID:1T0L) (15). The dimer contains two active sites, each of which contains a NADP+-binding site and a metal ion–binding site. One active site is shown in the closed conformation, with the substrate isocitrate in dark blue and the cofactor NADP+ in red. Mutations that alter Arg132 (yellow) to histidine, cysteine, or other amino acids are associated with human gliomas and other cancers. This residue forms three hydrogen bonds with the isocitrate substrate (dark blue). Ser94 (orange) also forms one hydrogen bond with the isocitrate substrate. In the inactive open conformation (data not shown), Asp279 (cyan) contacts Ser94 and sterically hinders isocitrate binding. To transition to the active closed conformation shown here, Asp279 must swing away from Ser94 to relieve this steric hindrance. During this transition, Asp279 contacts Arg132 (15), suggesting that Arg132 plays a role in the transition between inactive and active enzyme conformations. Displayed image was created with University of California San Francisco Chimera software version 1.3 (San Francisco, CA) (16).
Roles in Normal Cellular Metabolism
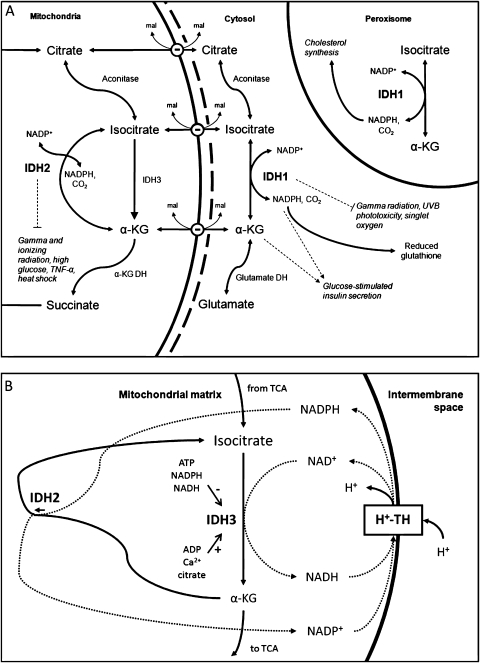
IDH1 and IDH2 play prominent yet distinctive roles in cellular metabolism (Figure 2, A), with IDH1 involved in lipid metabolism and glucose sensing and IDH2 involved in the regulation of oxidative respiration. In yeast, IDH1 aids in the beta oxidation of polyunsaturated fatty acids within the peroxisome by providing NADPH, a cofactor for 2,4-dienoyl CoA reductase (30). In higher eukaryotes, polyunsaturated fatty acid oxidation also occurs in mitochondria, and, because of this localization, likely uses NADPH supplied by IDH2. NADPH generated by IDH1 also contributes to the cellular defense against reactive oxygen species (ROS) generated during lipid oxidation and other processes (31). In mammalian hepatocytes, IDH1 provides NADPH for peroxisomal fat and cholesterol synthesis, and its expression is regulated and activated by sterol regulatory element-binding proteins 1a and 2 (32).
Figure 2.
Functions of isocitrate dehydrogenase 1 (IDH1) and IDH2 in the normal cell. A) IDH1 and IDH2 catalyze the reversible conversion of isocitrate to α-ketoglutarate (α-KG) and NADP+ to NADPH. IDH1 is located in the cytosol and the peroxisome (4–6). In the cytosol, IDH1 produces NADPH to contribute to the reduction of glutathione, the major cellular antioxidant (17). In the peroxisome, NADPH contributes to cholesterol synthesis (18). IDH1 has been shown to protect cells against gamma radiation (19), singlet oxygen (20), and UVB radiation (21) (light dashed line). IDH1 facilitates glucose-stimulated insulin secretion in pancreatic islets (22) (dashed arrows), which may be mediated by NADPH interactions with voltage-gated potassium channels or α-ketoglutarate interactions with α-ketoglutarate hydroxylases (data not shown). Conversion of α-ketoglutarate may be important for glia-specific glutamate and glutamine metabolism. IDH2 is located in the mitochondria and may function as the major catalyst of the isocitrate to α-ketoglutarate reaction in the tricarboxylic acid (TCA) cycle in some tissues (23). IDH3 is also located in the mitochondria and catalyzes the irreversible conversion of isocitrate to α-ketoglutarate and NAD+ to NADH. IDH2 has been shown to protect cells against gamma and ionizing radiation (19,24), high glucose (25), tumor necrosis factor-α (TNF-α) (26), and heat shock (27) (light dashed line). Carriers exchange malate (mal) for isocitrate, citrate, or α-ketoglutarate at the inner mitochondrial membrane. Cofactors for enzymes other than IDH1 and IDH2 are not indicated. B) A mitochondrial isocitrate/α-ketoglutarate cycle has been proposed by Sazanov and Jackson (28). In some tissues, flux through IDH2 proceeds in reverse, with α-ketoglutarate converted to isocitrate and NADPH converted to NADP+ (29). IDH3 converts isocitrate back to α-ketoglutarate and NAD+ to NADH. H+-transhydrogenase (H+-TH) completes this cycle by transferring electrons from NADH to NADPH (28). The transhydrogenase reaction is coupled to the transport of protons down their electrochemical potential gradient into the mitochondrial matrix (28). This cycle has the net effect of dissipating the electrochemical potential gradient across the inner mitochondrial membrane and producing heat. IDH2 also reduces the net flux from isocitrate to α-ketoglutarate (28). Because of this effect, allosteric modifiers of IDH3 (ATP, Ca2+, citrate, ATP, NADH, and NADPH) have a larger relative impact on net flux from isocitrate to α-ketoglutarate for the TCA cycle. DH = dehydrogenase; UVB = ultraviolet B.
IDH1 plays a crucial role in cellular glucose sensing. In mammalian pancreatic islets, knockdown of IDH1 results in impairment of glucose-stimulated insulin secretion and an increase in lactate production (33). In addition, knockdown or inhibition of the mitochondrial citrate/isocitrate carrier impairs glucose-stimulated insulin secretion and promotes glucose incorporation into fatty acids and glucose-induced increases in NADPH/NADP+ (22). Together, these results point to a central role for IDH1 in glucose sensing, and IDH1 has been proposed to function in a novel anaplerotic pyruvate cycle that mediates this process (22). In this cycle, glucose-derived pyruvate enters the TCA cycle through pyruvate carboxylase, is converted to isocitrate, and exits the mitochondria via the citrate/isocitrate carrier. IDH1 converts this isocitrate to α-ketoglutarate, producing NADPH. α-Ketoglutarate and/or NADPH promote insulin secretion, possibly by modulating α-ketoglutarate hydroxylases or voltage-gated potassium channels, respectively; α-ketoglutarate may be cycled back to pyruvate through a pathway that remains to be elucidated. The role for IDH1 in cellular metabolism is evidenced by the phenotype of transgenic mice overexpressing IDH1, which exhibit fatty liver, hyperlipidemia, and obesity (34). The IDH1 overexpression phenotype is especially consistent with overactive glucose-stimulated insulin secretion, as expected for an enzyme with a role in this pathway.
IDH2 plays a key role in TCA cycle regulation in multiple tissues. Analysis of families with retinitis pigmentosa has determined that some of these families are homozygous for a defective IDH3 subunit allele (23). Because these individuals have no obvious pathology outside the retina, it has been proposed that IDH1 or IDH2, or both, may function to convert isocitrate to α-ketoglutarate for the TCA cycle in nonretinal tissues. In contrast to this idea, the IDH2 reaction has been shown to proceed in reverse in mammalian heart and liver, catalyzing the conversion of α-ketoglutarate and NADPH to isocitrate and NADP+ (29). A recent study in glioma cells supports this role for IDH2 by showing that small interfering RNA knockdown of IDH2, but not IDH3, results in lowered conversion of glutamine to citrate, a process that would require this reverse flux (35). An isocitrate/α-ketoglutarate cycle has been proposed in which IDH2 proceeds in this reverse direction, and IDH3 converts isocitrate back to α-ketoglutarate. This cycle is completed by the transfer of electrons from NADH to NADPH by H+-transhydrogenase, which is driven by the proton electrochemical gradient of the inner mitochondrial membrane (Figure 2, B). The isocitrate/α-ketoglutarate cycle dissipates this gradient, generating heat, which provides tight control of flux through the TCA cycle by the allosteric regulators of IDH3 (Ca2+, ADP/ATP, citrate, and NAD(P)H) and by the state of the inner mitochondrial membrane (28). It is difficult to reconcile a reverse flux through IDH2 with the idea that IDH2 can rescue the forward flux of IDH3. Perhaps in retinitis pigmentosa, flux through IDH2 in tissues like the heart and liver is forced into the forward direction, but this unnatural metabolic state results in apparent pathology only in the retina.
The evidence points to a central role for the isocitrate to α-ketoglutarate reaction in the cell. At this crossroads, allosteric regulators of IDH3 and the mitochondrial membrane electrochemical gradient modulate overall TCA cycle flux; metabolites may exit the mitochondria and provide cellular machinery with information on glucose status; and metabolites may continue through the TCA cycle or cycle back to pyruvate through an anaplerotic pathway.
Response to Cellular Insults
In mammalian cells, IDH activity increases in response to a variety of oxidative insults, but activity of other TCA cycle enzymes, including α-ketoglutarate dehydrogenase and IDH3, decreases (36), suggesting that these enzymes play a role in the cellular response to such insults, in addition to their roles in normal cellular metabolism. Both enzymes can produce NADPH, a reducing equivalent essential for the reduction of glutathione by glutathione reductase and for the activity of the thioredoxin system [for a review, see (37)], both of which confer cellular protection against oxidative damage. Additionally, α-ketoglutarate serves as a potent antioxidant, and both IDH1 and IDH2 activity can modulate the availability of this compound, which can be exchanged for malate across the inner mitochondrial membrane. Although glucose-6-phosphate dehydrogenase is classically thought to provide NADPH for reductive processes in cells, the IDHs have increasingly been implicated as the major source for this reduced compound (17). Because the inner mitochondrial membrane is impermeable to NADPH, IDH2 may serve as the source for this antioxidant to protect against mitochondrial-specific stressors, such as ROS produced by the respiratory electron transport chain. This process would require flux through IDH2 to proceed in the forward direction, which may occur in tissues other than the liver and heart, or generally under conditions of stress.
Park and colleagues (38) have confirmed the role of IDH1 and IDH2 as protectors against various insults. They have shown that IDH1 or IDH2 deficiency leads to increased lipid peroxidation, oxidative DNA damage, intracellular peroxide generation, and decreased survival after oxidant exposure and that overexpression of either IDH confers protection from these effects (38). These findings suggest that IDH2 can modulate processes originally associated with IDH1 activity and does so via production of α-ketoglutarate in the mitochondria, followed by localization to the cytosol. Cellular IDH1 levels are associated with protection from apoptosis after exposure to ROS (39) or singlet oxygen species (20) and with protection from cell death following ultraviolet B-induced phototoxicity (21). IDH2 protects against apoptosis following heat shock (27), treatment with tumor necrosis factor-α (26), exposure to high glucose (25), or exposure to ionizing radiation (24). Both IDH1 and IDH2 have been shown to be induced by and to protect against gamma irradiation (19). It has been reported that IDH1 and IDH2 activity varies according to age in several tissues from ad libitum-fed rats but not in tissues from diet-restricted rats (40) and that IDH1 expression decreases over time in the brains of aging mice (41).
Although IDH1 and IDH2 play a crucial role in the defense against oxidative stress, they are inactivated by oxidation. Lipid peroxidation products (42), singlet oxygen (43), hypochlorous acid (44), ROS (45), nitric oxide (46), and peroxynitrite (47) all inactivate the enzymes. This process is likely mediated by modifications including glutathionylation in the presence of high levels of oxidized glutathione (48), S-nitrosylation in the presence of reactive nitrogen species (46,49), and nonenzymatic glycation in the presence of high glucose levels, as demonstrated in diabetic human and rat tissue (50). It has been suggested that other antioxidant enzymes may protect the IDHs from oxidative inactivation and that cells respond to such inactivation with de novo IDH protein synthesis (51).
Mutations in Human Cancer
IDH1 Arg132 mutations were discovered in a genome-wide mutation analysis of 22 human World Health Organization grade 4 glioblastomas (52). Two genetically distinct classes of glioblastomas exist: primary glioblastomas, which arise de novo, and secondary glioblastomas, which progress from the less malignant grade 2 diffuse astrocytomas and grade 3 anaplastic astrocytomas. In addition, grade 2 well-differentiated oligodendrogliomas are gliomas that can progress to grade 3 anaplastic oligodendrogliomas, and the mixed grade 2 oligoastrocytomas can progress to grade 3 anaplastic oligoastrocytomas and grade 4 secondary glioblastomas [for a review, see (53)]. Subsequent analyses revealed that mutations in IDH1 Arg132 are in fact common (50%–94%) in grades 2 and 3 gliomas and secondary glioblastomas and also occur less frequently in primary glioblastomas and other cancers (Table 1). In addition, IDH1 Arg132 mutations have been identified in acute myeloid leukemia (AML), and rare cases have been reported in B-acute lymphoid leukemia, prostate cancer, and colorectal cancer (Table 1). IDH1 mutations were identified in 7% of AML patients in one large study (Table 1) (63) but not in two others (Table 1) (61,54). Most AMLs with IDH1 mutations are cytogenetically normal (63). The cytogenetic status of patients in the two AML studies that did not find IDH1 mutations (61,54) was not reported, but a difference in the AML cytogenetic subtype distribution between studies could explain the different findings. In cancer, the vast majority of IDH1 mutations are heterozygous with a wild-type allele. In gliomas, most IDH1 mutations (89.3%, Table 2) were G395A (R132H), whereas a slight majority of AMLs contain C394T (R132C) mutations (Table 2) (54–59,61,63,66). Mutations in IDH2 at Arg172, the exact analog of Arg132 in IDH1, have also been found in grades 2 and 3 gliomas and in AMLs (Tables 1 and 2) (35,54,58,64,65). Also, mutations in Arg140 of IDH2 have been found in AMLs (Table 2) (35,64) but not in gliomas (54). The analogous residue to IDH2 Arg140 in IDH1, Arg100, has not been reported to be mutated in cancer at this time, and recurring somatic mutations in other residues of IDH1 or IDH2, or in IDH3, have not been identified in cancer. Because few cell lines have been derived from grades 2 and 3 gliomas and karyotypically normal AMLs, it is not surprising that no cell lines have been reported to contain IDH mutations at this time.
Table 1.
Frequency of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) mutations in human cancers
| Cancer | IDH1 mutated* | % IDH1 mutated† | IDH2 mutated* | % IDH2 mutated† | Reference |
| Gliomas | |||||
| Diffuse astrocytoma (grade 2) | 34/46, 25/30, 60/68, 13/22, 165/227, 10/12 | 59–88 | 2/5‡, 2/227 | 0.9 | (54–59) |
| Anaplastic astrocytoma (grade 3) | 29/47, 36/52, 21/27, 32/62, 0/2, 146/228, 9/18 | 50–78 | 2/16‡, 2/228 | 0.9 | (54–59,60) |
| Secondary glioblastoma (grade 4) | 5/6, 7/8, 11/13, 28/34, 5/10, 11/15,10/13 | 73–88 | 0/2 | (52,54–57,59,60) | |
| Primary glioblastoma (grade 4) | 7/99, 7/99, 6/123, 3/59, 6/173, 11/94, 4/25, 11/183 | 3–16 | 0/117‡, 0/17 | (52,54–59,61,62) | |
| Pediatric glioblastoma (grade 4) | 1/14, 0/15 | 0–7 | 0/15 | (54,55) | |
| Well-differentiated oligodendroglioma (grade 2) | 36/51, 41/51, 31/39, 23/34, 105/128, 41/54 | 68–82 | 2/10‡, 3/128 | 2.3 | (54–59) |
| Anaplastic oligodendroglioma (grade 3) | 36/54, 31/36, 6/8, 12/20, 1/2, 121/174, 24/49 | 49–86 | 3/5‡, 6/174 | 3.4 | (54–59,60) |
| Oligoastrocytoma (grade 2) | 36/46, 3/3, 16/17, 10/20, 62/76, 26/34 | 50–94 | 1/76 | 1.3 | (54–59) |
| Anaplastic oligoastrocytoma (grade 3) | 29/37, 7/7, 6/8, 18/23, 76/177, 34/54 | 43–78 | 6/177 | 3.4 | (54–59) |
| Other cancers | |||||
| Primitive neuroectodermal tumor (grade 4) | 3/9, 3/31, 0/8 | 10 | (55–57) | ||
| Acute myeloid leukemia | 0/45, 0/100, 16/188, 3/16, 11/145, 6/78 | 0–8 | 0/45, 0/188, 2/16, 2/145, 12/78 | 0-15 | (35,54,61,63–65) |
| B-acute lymphoblastic leukemia | 1/60 | 2 | (61) | ||
| Prostate cancer | 0/7, 0/4, 2/75 | 3 | 0/123 | (54,60–62) | |
| Colorectal cancer | 1/11§, 0/114, 0/128, 0/97 | 0/83 | (54,60–62,66) | ||
Number of mutated samples/total number of samples of given type analyzed for each report of the given sample type.
Percentages or ranges of percentages are given if any analysis of more than 10 samples found a mutation and reflect only studies that include 10 or more samples.
IDH2 mutations were analyzed only in tumors that did not contain IDH1 mutations and are not included in any range of percentages for IDH2-mutated tumors.
Identified in a genomic analysis of colorectal cancer (66). Percentage not reported.
Table 2.
Relative frequency of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) mutations in gliomas and number of reported cases of IDH1 and IDH2 mutations in acute myeloid leukemia (AML) patients
| Gene | Codon | Amino acid substitution | % in gliomas* | No. of AML patients† |
| IDH1 | CGT>CAT | R132H | 89.3 | 12 |
| IDH1 | CGT>TGT | R132C | 3.9 | 17 |
| IDH1 | CGT>AGT | R132S | 1.5 | 1 |
| IDH1 | CGT>GGT | R132G | 1.3 | 4 |
| IDH1 | CGT>CTT | R132L | 0.3 | |
| IDH2 | CGA>CAA | R140Q‡ | n.r.‡ | 7 |
| IDH2 | AGG>AAG | R172K | 2.7 | 2 |
| IDH2 | AGG>ATG | R172M | 0.8 | |
| IDH2 | AGG>TGG | R172W | 0.7 |
Epidemiology and Association With Other Genetic Changes
IDH-mutated cancers are associated with younger age at diagnosis in most glioma tumor types (54–57) but not in AML (63). However, they are rare in glioma patients aged 18 years or younger (58,67,68). Glioma patients with IDH mutations survive longer than patients with wild-type IDH (52,54,56), and multivariable analyses have shown IDH1 mutation status to be an independent positive prognostic factor for glioblastomas (59,69). Initial studies did not reveal a significant association between survival and IDH status in AML (35, 63, 65). Studies including larger numbers of AML patients may identify any association between IDH status and clinical outcomes in this disease.
Tumor protein p53 (TP53) mutations are common in grades 2 and 3 astrocytomas and in secondary glioblastomas. In some studies, IDH mutations have been found to associate with TP53 mutations (54,56,57), although other studies did not find a statistically significant association (55,59). Biopsies taken at different times from astrocytoma patients demonstrate that IDH mutation occurs before TP53 mutation (57). In addition, patients with germline TP53 mutations, which predispose to grade 2–4 astrocytomas, had tumors that contained somatic IDH1R132C mutations (70). Loss of chromosome arms 1p and 19q is commonly observed in oligodendroglial tumors and is frequently observed in IDH-mutated but not in IDH wild-type oligodendroglial tumors (54,56). IDH mutations are inversely associated with many of the hallmark genetic changes of primary glioblastomas, such as epidermal growth factor receptor amplification, cyclin-dependent kinase inhibitor 2A or 2B deletion, and phosphatase and tensin homolog mutations (54,56).
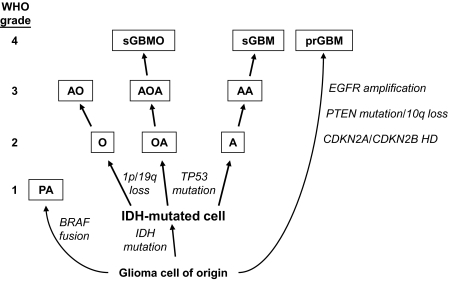
IDH mutations do not increase in frequency in the progression to higher-grade gliomas (Table 1) and occur before other genetic changes, leading to the suggestion that these mutations arise at some point in the transition from a normal cell to a clinically evident tumor (54–57,71). This suggestion has led to refinement of the genetic model for the formation and progression of different glioma subtypes (56,71). Common genetic changes associated with different glioma subtypes and the hypothesis that grade 2 gliomas arise from an IDH-mutated population of cells are integrated into the model shown in Figure 3.
Figure 3.
Common genetic alterations in glioma tumorigenesis and progression. Grade 2 gliomas include well-differentiated oligodendrogliomas (O), oligoastrocytomas (OA), and diffuse astrocytomas (A) (72). All three types of tumors undergo two sequential genetic alterations for tumorigenesis: first, a mutation in isocitrate dehydrogenase 1 (IDH1) or IDH2, and second, homozygous deletion of chromosome arms 1p and 19q or tumor protein p53 (TP53) mutation. The IDH mutation event occurs at some point in the transformation from the glioma cell of origin to a glioma cell. The second genetic event contributes to the histopathological and clinical phenotype of the resulting tumor. For example, oligodendrogliomas usually contain 1p and 19q loss, whereas astrocytomas have TP53 mutations. The oligoastrocytomas can contain either genetic alteration. Grade 2 tumors can progress to grade 3 anaplastic oligodendrogliomas (AO), anaplastic oligoastrocytomas (AOA), and anaplastic astrocytomas (AA), as well as to grade 4 secondary glioblastomas (sGBM) and secondary glioblastomas with oligodendroglial component (sGBMO) (72). Grade 1 pilocytic astrocytomas (PA) and grade 4 primary GBMs (prGBM) arise de novo (72), do not frequently contain IDH mutations, and contain other alterations that are rare in the IDH mutation–containing tumors (54). It is not known whether these tumors arise from the same cell type. CDKN2A and 2B = cyclin-dependent kinase inhibitor 2A and 2B; EGFR = epidermal growth factor receptor; HD = homozygous deletion; PTEN = phosphatase and tensin homolog; WHO = World Health Organization.
Functional Properties of IDH Mutants
Arg132 of IDH1 is aligned with Arg172 of IDH2, and this arginine is conserved in all known homologs of IDH in other species (3). Uniquely, among all residues involved in isocitrate binding, Arg132 forms three hydrogen bonds with the α- and β-carboxyl of the isocitrate substrate (73). Arg132, as well as Gln277, contact Asp279 in the transitional semi-open state of IDH1 (15). Arg132 may therefore be important in the transition from the open state, in which Asp279 sterically hinders isocitrate binding by contacting Ser94, to the active closed state (Figure 1). Consistent with the importance of Arg132 in this transition, the x-ray structure of IDH1R132H appears to favor the closed state compared with the structure of IDH1wt, indicating that alteration of Arg132 changes the equilibrium between the open and closed states (74). Extrapolating from IDH1, one can expect Arg172 of IDH2 to also play a role in the transition between conformations of this enzyme (14,15). Notably, Arg140 of IDH2, which is mutated in AML, also forms hydrogen bonds with the β-carboxyl of the isocitrate substrate (35). In addition, Arg140 is adjacent to Arg172 in the active site of IDH2 (35), and these two residues may both function in the transition between the open and closed states of IDH2. Early research showed that purified rat IDH1R132E is essentially inactive (75). In another study, a porcine mutant analogous to human IDH2R172Q displayed twofold reduced specific activity and a 100-fold increased Km for isocitrate (76). Following their discovery in gliomas, the enzymatic activities, as measured by production of NADPH, of IDH1R132H, IDH1R132S, IDH1R132C, IDH1R132G, IDH2R172G, IDH2R172K, and IDH2R172M were found to be greatly reduced compared with wild type in lysates of cells overexpressing these enzymes (54,56). Furthermore, the Km for isocitrate of purified IDH1R132H, IDH1R132C, and IDH1R132S was increased by more than 50-fold (73).
Oncogenes or Tumor Suppressors?
Since the discovery of IDH mutations, their function in cancer has puzzled cancer geneticists. The inactivation of proteins that protect the cell is at first reminiscent of a tumor suppressor, such as TP53. However, no homozygous deletions or other inactivating alterations of IDH1 or IDH2 have been reported in cancer, as is observed for classical tumor suppressors. Furthermore, the heterozygous nature and specificity of the IDH mutations evokes activating “hotspots” in oncogenes, such as v-raf murine sarcoma viral oncogene homolog B1 (BRAF), v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), and phosphoinositide-3-kinase, catalytic, alpha polypeptide (PIK3CA) and has led to the proposal that such mutations result in gain of function (52). A gain of similar function for IDH1 and IDH2 would also help to explain why mutations in either gene appear to provide cancer cells with a similar selective advantage. For example, mutations could alter yet undescribed normal IDH interactions with other molecules in the cell, disrupt normal covalent modifications of the enzymes, or lead to depletion or replenishment of other compounds by acquisition of new enzymatic activity.
Dominant Negative Activity
One possible function of IDH mutations is dominant inhibition of the wild-type copy of the enzyme. Zhao et al. showed that IDH1wt and IDH1R132H can form a heterodimer that exhibits 4% of the activity of the IDH1wt homodimer (69). The wild-type homodimer displays a sigmoidal curve of cooperative binding to isocitrate, whereas the heterodimer exhibits a hyperbolic curve with a higher Km for isocitrate, indicating that the heterodimer loses both affinity and cooperativity for the substrate (69). Although the purified wild-type:mutant heterodimer has reduced activity in vitro, the extent to which this heterodimer forms in vivo is unclear. To exert dominant negative activity in vivo, a substantial fraction of IDH1 molecules would need to exist as heterodimers. For this to happen, the binding between IDH1R132H and IDH1wt molecules would have to outcompete the binding between IDH1wt and other IDH1wt molecules, as well as the binding between IDH1R132H and other IDH1R132H molecules. This has not been demonstrated nor has it been shown that IDH1R132H molecules can lower the IDH activity of cells containing IDH1wt. Also, dominant negative activity may be harmful to cancer cells because knockdown of IDH1 or IDH2 slows the growth of glioma cells (35). Given the protective role for IDH1 and IDH2 in the cell, the lack of other inactivating genetic alterations observed for IDH1 or IDH2 in cancer, and the lack of evidence that dominant negative activity occurs in vivo, it seems unlikely that dominant negative activity is a major function of the IDH mutations in cancer.
Neomorphic Enzyme Activity
Recently, unbiased metabolite profiling revealed that expression of IDH1R132H in glioma cells leads to production of 2-hydroxyglutarate (74). Further study revealed that IDH1 Arg132 and IDH2 Arg172 mutants reduce α-ketoglutarate to D-2-hydroxyglutarate while converting NADPH to NADP+ (35,65,74). In addition, cancer tissue samples containing IDH1 Arg132, IDH2 Arg140, or IDH2 Arg172 mutations have more than 100-fold higher concentrations of this metabolite than cancers with wild-type copies of these genes (35,65,74). Mutation of IDH1 Arg132 appears to favor the active closed state of the enzyme, increasing its affinity for NADPH, which may promote reduction of α-ketoglutarate to D-2-hydroxyglutarate under low concentrations of NADPH (74). The mutation also results in reorganization of the enzyme active site in a way that apparently favors reduction of α-ketoglutarate to D-2-hydroxyglutarate rather than oxidative decarboxylation of isocitrate to α-ketoglutarate or the reverse reductive carboxylation of α-ketoglutarate to isocitrate (74).
The fact that neomorphic enzyme activity is a shared feature of the IDH1 and IDH2 mutations points to the importance of this activity in cancer, which has led to speculation that D-2-hydroxyglutarate acts as an oncometabolite (74). Individuals with L-2-hydroxyglutaric aciduria, a genetic metabolic defect leading to accumulation of L-2-hydroxyglutarate, the opposite enantiomer of D-2-hydroxyglutarate, appear to have a higher risk of developing malignant brain tumors (77). However, patients with D-2-hydroxyglutaric aciduria, in which D-2-hydroxyglutarate accumulates because of a genetic defect (78), are not known to have an increased risk for developing brain tumors. Although D- and L-2-hydroxyglutarate are mirror images of each other, they may have quite different biological functions because patients with D- and L-2-hydroxyglutaric aciduria have distinct clinical symptoms and because different enzymes function on each enantiomer (78,79). It is also important to consider that this neomorphic enzyme activity may alter flux through α-ketoglutarate, NADP+, or NADPH in ways that benefit cancer cells (80).
Aberrant Glucose Sensing
What might a change in IDH activity achieve for cancer cells? A possible selective advantage for cancer cells could stem from the role of IDH1 in glucose sensing. IDH1 has been shown to participate in a glucose-sensing pathway in pancreatic islets (33) and may communicate the presence of high glucose to downstream members of this pathway by raising NADPH levels. A similar pathway may be intact in other tissue types. Rather than stimulating insulin secretion, as in islet cells, this pathway may regulate nutrient satiety or cellular differentiation based on nutrient availability in other cell types. For example, cells that detect high levels of glucose would signal, via high NADPH levels, to stop taking up high levels of nutrients or to differentiate. IDH1 and IDH2 mutants consume NADPH as they convert α-ketoglutarate to 2-hydroxyglutarate (35,65,74), and IDH1-mutated tissues have lower NADP+-dependent IDH activity (81). In cells with IDH1 mutations, this may lead to low cytosolic NADPH levels, which would aberrantly signal a low nutrient status to downstream players in the glucose-sensing pathway. The cell may then compensate for perceived low nutrient status by increasing cellular nutrient consumption or by blocking cellular differentiation. Increased nutrient consumption is a hallmark of cancer and may give cancer cells a selective growth advantage. Both glioma and AML cells have been characterized as relatively undifferentiated (35), and blocking differentiation may benefit cancer cells by allowing them to continue to self-renew and accumulate advantageous genetic alterations. However, whether a glucose-sensing pathway involving IDH1 and NADPH functions in the cell lineages that give rise to gliomas and AMLs, which downstream players function in this pathway, and what effect IDH mutations have on cellular NADPH levels have yet to be elucidated. Moreover, the inner mitochondrial membrane is impermeable to NADPH, and it is unknown whether a putative depletion of NADPH in the mitochondria by IDH2 mutants would lead to depletion of cytosolic NADPH.
Hypoxia Signal Transduction
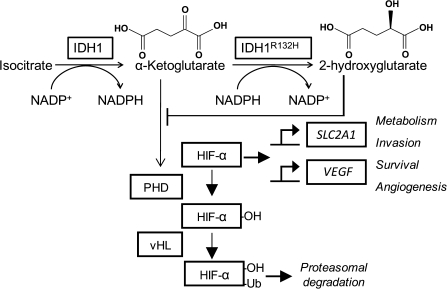
Overexpression of IDH1R132H in mammalian cells increases the stability of hypoxia-inducible factor 1α (HIF-1α, Figure 4) (73). HIF-1α is a transcription factor with targets that modulate apoptosis, cell survival, and angiogenesis, and its increased expression has been implicated in other cancer types with frequent loss-of-function mutations in TCA cycle enzymes [for a review, see (82)]. One proposed mechanism for IDH mutant–mediated stabilization of HIF-1α is by inhibition of prolyl hydroxylases. Prolyl hydroxylases use α-ketoglutarate as a substrate for a reaction that normally targets HIF-1α for degradation. IDH mutants could lead to lower cellular α-ketoglutarate levels by consuming this compound, which may lead to prolyl hydroxylase inactivation. Alternatively, 2-hydroxyglutarate produced by IDH mutants has been thought to competitively inhibit prolyl hydroxylases by occupying the α-ketoglutarate-binding site on these enzymes (Figure 4) (83). However, grades 2 and 3 gliomas do not demonstrate angiogenesis, as would be expected for tumors that activate this hypoxia signaling pathway (82). Finally, increased expression of HIF-1 target genes is not found in AMLs (63), which calls into question the idea that HIF-1α stabilization is a major function of the IDH mutations.
Figure 4.
Model for activation of hypoxia-inducible factor 1 (HIF-1) by isocitrate dehydrogenase 1 (IDH1) mutations. HIF-1α is hydroxylated by HIF prolyl hydroxylase (PHD), which targets HIF-1α for ubiquitylation by von Hippel Lindau protein (vHL) and subsequent proteasomal degradation. PHDs require O2 and α-ketoglutarate (α-KG) as substrates (82). When stabilized, HIF-1α dimerizes with HIF-1β and activates transcription of targets such as solute carrier family 2 member 1 (SLC2A1) and vascular endothelial growth factor (VEGF) (82). IDH1wt converts isocitrate to α-ketoglutarate, and IDH1R132H converts α-ketoglutarate to 2-hydroxyglutarate. By consuming α-ketoglutarate, IDH1R132H may lower the availability of this substrate, which would decrease PHD activity and lead to HIF-1α stabilization. Also, based on its structural similarity to α-ketoglutarate, 2-hydroxyglutarate has been hypothesized to competitively inhibit PHD activity by occupying PHD α-ketoglutarate binding sites (83). Ub = ubiquitin.
Mutagenic Program
IDH1 and IDH2 mutations could also contribute to tumorigenesis and cancer progression through increased mutagenesis (56). Loss of a wild-type IDH1 or IDH2 allele, combined with a new enzyme activity that consumes NADPH and α-ketoglutarate, could lead to depletion of these two compounds that normally help to defend the cell against oxidative stress. Unchecked oxidative stress could lead to mutagenesis as ROS interact with the genome. Alternatively, 2-hydroxyglutarate itself may act as a mutagen through an as-yet unknown mechanism. Grades 2 and 3 astrocytomas and secondary glioblastomas frequently contain IDH mutations and later develop missense mutations in TP53 and other genes (52,54,55,57–59), supporting the idea that early IDH mutations could promote later advantageous mutations that underlie the formation and progression of these cancers.
Future Directions
The functional significance of IDH mutations in human cancer remains, to a large extent, a mystery. IDH mutations generally associate with specific gene expression signatures (84), and determination of the gene expression environment in mutated and nonmutated tumors of specific tumor types may provide insight into the mechanism of IDH-mutated gliomagenesis. Such studies may clarify the functional role of the mutations and possibly offer information to help guide glioma management in the clinic. Determining whether IDH status is an independent prognostic factor for any of the tumor types that contain these mutations may guide clinical management of a lethal group of cancers. One study has already taken advantage of the remarkable sensitivity and specificity of IDH1 mutations for diffuse astrocytomas to distinguish these lesions from pilocytic astrocytomas, which do not frequently contain IDH1 mutations (85). This could be useful in cases for which scant material is available for histopathological analysis. Analysis of 2-hydroxyglutarate levels may also simplify the diagnosis and management of glioma and AML patients, especially if this compound is also elevated in cerebrospinal fluid, serum, or urine of patients (80). Furthermore, the IDHs operate at a metabolic crossroads. It will be crucial to understand whether and how IDH mutations alter tumor cell metabolic profiles, lipid biosynthesis, the defense against oxidative stress, oxidative respiration, and hypoxia signal transduction in cancer.
Long-term research will require the use of mammalian tissue and model organisms and may uncover the basis for tumor-type specificity of IDH mutations and the specific timing of mutation acquisition during the progression from a cell of origin to a cancer cell. As prevalent and specific alterations, IDH mutants are possible targets for molecular therapies. Given the tumor specificity of metabolic enzyme mutations and the striking difference between the cellular metabolism of cancer and normal cells, metabolic enzymes may prove to be fruitful targets for anticancer therapies (86). In addition, a clear understanding of the mechanism of IDH-mutated carcinogenesis may reveal exciting new targets for cancer treatment.
Funding
National Institutes of Health (NIH) Medical Scientist Training Program training (2T32GM007171 to Z.J.R.); Pediatric Brain Tumor Foundation, the National Brain Tumor Society, and NIH (R01CA118822 to H.Y.).
Footnotes
The funders did not have any involvement in the design of the study; the collection, analysis, and interpretation of the data; the writing of the article; or the decision to submit the article for publication.
We thank Rachel Karchin and Parminder Mankoo for contributions of rendered IDH1 structures in University of California San Francisco Chimera. We thank Giselle Y. Lopez and Thy N. Huynh for editorial feedback.
References
- 1.Gabriel JL, Zervos PR, Plaut GW. Activity of purified NAD-specific isocitrate dehydrogenase at modulator and substrate concentrations approximating conditions in mitochondria. Metabolism. 1986;35(7):661–667. doi: 10.1016/0026-0495(86)90175-7. [DOI] [PubMed] [Google Scholar]
- 2.Jennings GT, Sechi S, Stevenson PM, Tuckey RC, Parmelee D, McAlister-Henn L. Cytosolic NADP(+)-dependent isocitrate dehydrogenase. Isolation of rat cDNA and study of tissue-specific and developmental expression of mRNA. J Biol Chem. 1994;269(37):23128–23134. [PubMed] [Google Scholar]
- 3.Nekrutenko A, Hillis DM, Patton JC, Bradley RD, Baker RJ. Cytosolic isocitrate dehydrogenase in humans, mice, and voles and phylogenetic analysis of the enzyme family. Mol Biol Evol. 1998;15(12):1674–1684. doi: 10.1093/oxfordjournals.molbev.a025894. [DOI] [PubMed] [Google Scholar]
- 4.Geisbrecht BV, Gould SJ. The human PICD gene encodes a cytoplasmic and peroxisomal NADP(+)-dependent isocitrate dehydrogenase. J Biol Chem. 1999;274(43):30527–30533. doi: 10.1074/jbc.274.43.30527. [DOI] [PubMed] [Google Scholar]
- 5.Henke B, Girzalsky W, Berteaux-Lecellier V, Erdmann R. IDP3 encodes a peroxisomal NADP-dependent isocitrate dehydrogenase required for the beta-oxidation of unsaturated fatty acids. J Biol Chem. 1998;273(6):3702–3711. doi: 10.1074/jbc.273.6.3702. [DOI] [PubMed] [Google Scholar]
- 6.Yoshihara T, Hamamoto T, Munakata R, Tajiri R, Ohsumi M, Yokota S. Localization of cytosolic NADP-dependent isocitrate dehydrogenase in the peroxisomes of rat liver cells: biochemical and immunocytochemical studies. J Histochem Cytochem. 2001;49(9):1123–1131. doi: 10.1177/002215540104900906. [DOI] [PubMed] [Google Scholar]
- 7.Luo H, Shan X, Wu J. Expression of human mitochondrial NADP-dependent isocitrate dehydrogenase during lymphocyte activation. J Cell Biochem. 1996;60(4):495–507. doi: 10.1002/(sici)1097-4644(19960315)60:4<495::aid-jcb6>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 8.Dean AM, Lee MH, Koshland DE., Jr. Phosphorylation inactivates Escherichia coli isocitrate dehydrogenase by preventing isocitrate binding. J Biol Chem. 1989;264(34):20482–20486. [PubMed] [Google Scholar]
- 9.Hurley JH, Thorsness PE, Ramalingam V, Helmers NH, Koshland DE, Jr, Stroud RM. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc Natl Acad Sci U S A. 1989;86(22):8635–8639. doi: 10.1073/pnas.86.22.8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean AM, Koshland DE., Jr Electrostatic and steric contributions to regulation at the active site of isocitrate dehydrogenase. Science. 1990;249(4972):1044–1046. doi: 10.1126/science.2204110. [DOI] [PubMed] [Google Scholar]
- 11.Hurley JH, Dean AM, Sohl JL, Koshland DE, Jr, Stroud RM. Regulation of an enzyme by phosphorylation at the active site. Science. 1990;249(4972):1012–1016. doi: 10.1126/science.2204109. [DOI] [PubMed] [Google Scholar]
- 12.Hurley JH, Dean AM, Thorsness PE, Koshland DE, Jr, Stroud RM. Regulation of isocitrate dehydrogenase by phosphorylation involves no long-range conformational change in the free enzyme. J Biol Chem. 1990;265(7):3599–3602. doi: 10.2210/pdb4icd/pdb. [DOI] [PubMed] [Google Scholar]
- 13.Dean AM, Koshland DE., Jr Kinetic mechanism of Escherichia coli isocitrate dehydrogenase. Biochemistry. 1993;32(36):9302–9309. doi: 10.1021/bi00087a007. [DOI] [PubMed] [Google Scholar]
- 14.Ceccarelli C, Grodsky NB, Ariyaratne N, Colman RF, Bahnson BJ. Crystal structure of porcine mitochondrial NADP+-dependent isocitrate dehydrogenase complexed with Mn2+ and isocitrate. Insights into the enzyme mechanism. J Biol Chem. 2002;277(45):43454–43462. doi: 10.1074/jbc.M207306200. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Zhao J, Xu Z, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279(32):33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 16.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 17.Winkler BS, DeSantis N, Solomon F. Multiple NADPH-producing pathways control glutathione (GSH) content in retina. Exp Eye Res. 1986;43(5):829–847. doi: 10.1016/s0014-4835(86)80013-6. [DOI] [PubMed] [Google Scholar]
- 18.Haselbeck RJ, McAlister-Henn L. Function and expression of yeast mitochondrial NAD- and NADP-specific isocitrate dehydrogenases. J Biol Chem. 1993;268(16):12116–12122. [PubMed] [Google Scholar]
- 19.Lee SH, Jo SH, Lee SM, et al. Role of NADP+-dependent isocitrate dehydrogenase (NADP+-ICDH) on cellular defence against oxidative injury by gamma-rays. Int J Radiat Biol. 2004;80(9):635–642. doi: 10.1080/09553000400007680. [DOI] [PubMed] [Google Scholar]
- 20.Kim SY, Lee SM, Tak JK, Choi KS, Kwon TK, Park JW. Regulation of singlet oxygen-induced apoptosis by cytosolic NADP+-dependent isocitrate dehydrogenase. Mol Cell Biochem. 2007;302(1–2):27–34. doi: 10.1007/s11010-007-9421-x. [DOI] [PubMed] [Google Scholar]
- 21.Jo SH, Lee SH, Chun HS, et al. Cellular defense against UVB-induced phototoxicity by cytosolic NADP(+)-dependent isocitrate dehydrogenase. Biochem Biophys Res Commun. 2002;292(2):542–549. doi: 10.1006/bbrc.2002.6667. [DOI] [PubMed] [Google Scholar]
- 22.Joseph JW, Jensen MV, Ilkayeva O, et al. The mitochondrial citrate/isocitrate carrier plays a regulatory role in glucose-stimulated insulin secretion. J Biol Chem. 2006;281(47):35624–35632. doi: 10.1074/jbc.M602606200. [DOI] [PubMed] [Google Scholar]
- 23.Hartong DT, Dange M, McGee TL, Berson EL, Dryja TP, Colman RF. Insights from retinitis pigmentosa into the roles of isocitrate dehydrogenases in the Krebs cycle. Nat Genet. 2008;40(10):1230–1234. doi: 10.1038/ng.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JH, Kim SY, Kil IS, Park JW. Regulation of ionizing radiation-induced apoptosis by mitochondrial NADP+-dependent isocitrate dehydrogenase. J Biol Chem. 2007;282(18):13385–13394. doi: 10.1074/jbc.M700303200. [DOI] [PubMed] [Google Scholar]
- 25.Shin AH, Kil IS, Yang ES, Huh TL, Yang CH, Park JW. Regulation of high glucose-induced apoptosis by mitochondrial NADP+-dependent isocitrate dehydrogenase. Biochem Biophys Res Commun. 2004;325(1):32–38. doi: 10.1016/j.bbrc.2004.09.218. [DOI] [PubMed] [Google Scholar]
- 26.Kil IS, Kim SY, Lee SJ, Park JW. Small interfering RNA-mediated silencing of mitochondrial NADP+-dependent isocitrate dehydrogenase enhances the sensitivity of HeLa cells toward tumor necrosis factor-alpha and anticancer drugs. Free Radic Biol Med. 2007;43(8):1197–1207. doi: 10.1016/j.freeradbiomed.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Shin SW, Kil IS, Park JW. Silencing of mitochondrial NADP+-dependent isocitrate dehydrogenase by small interfering RNA enhances heat shock-induced apoptosis. Biochem Biophys Res Commun. 2008;366(4):1012–1018. doi: 10.1016/j.bbrc.2007.12.067. [DOI] [PubMed] [Google Scholar]
- 28.Sazanov LA, Jackson JB. Proton-translocating transhydrogenase and NAD- and NADP-linked isocitrate dehydrogenases operate in a substrate cycle which contributes to fine regulation of the tricarboxylic acid cycle activity in mitochondria. FEBS Lett. 1994;344(2–3):109–116. doi: 10.1016/0014-5793(94)00370-x. [DOI] [PubMed] [Google Scholar]
- 29.Comte B, Vincent G, Bouchard B, Benderdour M, Des Rosiers C. Reverse flux through cardiac NADP(+)-isocitrate dehydrogenase under normoxia and ischemia. Am J Physiol Heart Circ Physiol. 2002;283(4):H1505–H1514. doi: 10.1152/ajpheart.00287.2002. [DOI] [PubMed] [Google Scholar]
- 30.van Roermund CW, Hettema EH, Kal AJ, van den Berg M, Tabak HF, Wanders RJ. Peroxisomal beta-oxidation of polyunsaturated fatty acids in Saccharomyces cerevisiae: isocitrate dehydrogenase provides NADPH for reduction of double bonds at even positions. EMBO J. 1998;17(3):677–687. doi: 10.1093/emboj/17.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minard KI, McAlister-Henn L. Dependence of peroxisomal beta-oxidation on cytosolic sources of NADPH. J Biol Chem. 1999;274(6):3402–3406. doi: 10.1074/jbc.274.6.3402. [DOI] [PubMed] [Google Scholar]
- 32.Shechter I, Dai P, Huo L, Guan G. IDH1 gene transcription is sterol regulated and activated by SREBP-1a and SREBP-2 in human hepatoma HepG2 cells: evidence that IDH1 may regulate lipogenesis in hepatic cells. J Lipid Res. 2003;44(11):2169–2180. doi: 10.1194/jlr.M300285-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Ronnebaum SM, Ilkayeva O, Burgess SC, et al. A pyruvate cycling pathway involving cytosolic NADP-dependent isocitrate dehydrogenase regulates glucose-stimulated insulin secretion. J Biol Chem. 2006;281(41):30593–30602. doi: 10.1074/jbc.M511908200. [DOI] [PubMed] [Google Scholar]
- 34.Koh HJ, Lee SM, Son BG, et al. Cytosolic NADP+-dependent isocitrate dehydrogenase plays a key role in lipid metabolism. J Biol Chem. 2004;279(38):39968–39974. doi: 10.1074/jbc.M402260200. [DOI] [PubMed] [Google Scholar]
- 35.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mailloux RJ, Beriault R, Lemire J, et al. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS ONE. 2007;2(1)) doi: 10.1371/journal.pone.0000690. e690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura H. Thioredoxin and its related molecules: update 2005. Antioxid Redox Signal. 2005;7(5–6):823–828. doi: 10.1089/ars.2005.7.823. [DOI] [PubMed] [Google Scholar]
- 38.Lee SM, Koh HJ, Park DC, Song BJ, Huh TL, Park JW. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med. 2002;32(11):1185–1196. doi: 10.1016/s0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 39.Lee SM, Park SY, Shin SW, Kil IS, Yang ES, Park JW. Silencing of cytosolic NADP(+)-dependent isocitrate dehydrogenase by small interfering RNA enhances the sensitivity of HeLa cells toward staurosporine. Free Radic Res. 2009;43(2):165–173. doi: 10.1080/10715760802653661. [DOI] [PubMed] [Google Scholar]
- 40.Kil IS, Lee YS, Bae YS, Huh TL, Park JW. Modulation of NADP(+)-dependent isocitrate dehydrogenase in aging. Redox Rep. 2004;9(5):271–277. doi: 10.1179/135100004225006056. [DOI] [PubMed] [Google Scholar]
- 41.Yang S, Liu T, Li S, et al. Comparative proteomic analysis of brains of naturally aging mice. Neuroscience. 2008;154(3):1107–1120. doi: 10.1016/j.neuroscience.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 42.Yang JH, Yang ES, Park JW. Inactivation of NADP+-dependent isocitrate dehydrogenase by lipid peroxidation products. Free Radic Res. 2004;38(3):241–249. doi: 10.1080/10715760310001657712. [DOI] [PubMed] [Google Scholar]
- 43.Kim SY, Tak JK, Park JW. Inactivation of NADP(+)-dependent isocitrate dehydrogenase by singlet oxygen derived from photoactivated rose bengal. Biochimie. 2004;86(8):501–507. doi: 10.1016/j.biochi.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Park SY, Lee SM, Shin SW, Park JW. Inactivation of mitochondrial NADP+-dependent isocitrate dehydrogenase by hypochlorous acid. Free Radic Res. 2008;42(5):467–473. doi: 10.1080/10715760802098834. [DOI] [PubMed] [Google Scholar]
- 45.Lee SM, Huh TL, Park JW. Inactivation of NADP(+)-dependent isocitrate dehydrogenase by reactive oxygen species. Biochimie. 2001;83(11–12):1057–1065. doi: 10.1016/s0300-9084(01)01351-7. [DOI] [PubMed] [Google Scholar]
- 46.Yang ES, Richter C, Chun JS, Huh TL, Kang SS, Park JW. Inactivation of NADP(+)-dependent isocitrate dehydrogenase by nitric oxide. Free Radic Biol Med. 2002;33(7):927–937. doi: 10.1016/s0891-5849(02)00981-4. [DOI] [PubMed] [Google Scholar]
- 47.Yang ES, Lee JH, Park JW. Ethanol induces peroxynitrite-mediated toxicity through inactivation of NADP+-dependent isocitrate dehydrogenase and superoxide dismutase. Biochimie. 2008;90(9):1316–1324. doi: 10.1016/j.biochi.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Shin SW, Oh CJ, Kil IS, Park JW. Glutathionylation regulates cytosolic NADP+-dependent isocitrate dehydrogenase activity. Free Radic Res. 2009;43(4):409–416. doi: 10.1080/10715760902801525. [DOI] [PubMed] [Google Scholar]
- 49.Lee JH, Yang ES, Park JW. Inactivation of NADP+-dependent isocitrate dehydrogenase by peroxynitrite. Implications for cytotoxicity and alcohol-induced liver injury. J Biol Chem. 2003;278(51):51360–51371. doi: 10.1074/jbc.M302332200. [DOI] [PubMed] [Google Scholar]
- 50.Kil IS, Lee JH, Shin AH, Park JW. Glycation-induced inactivation of NADP(+)-dependent isocitrate dehydrogenase: implications for diabetes and aging. Free Radic Biol Med. 2004;37(11):1765–1778. doi: 10.1016/j.freeradbiomed.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 51.Batinic-Haberle I, Benov LT. An SOD mimic protects NADP+-dependent isocitrate dehydrogenase against oxidative inactivation. Free Radic Res. 2008;42(7):618–624. doi: 10.1080/10715760802209639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 54.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 56.Ichimura K, Pearson DM, Kocialkowski S, et al. IDH1 mutations are present in the majority of common adult gliomas but are rare in primary glioblastomas. Neuro Oncol. 2009;11(4):341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 59.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 60.Bleeker FE, Lamba S, Leenstra S, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30(1):7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 61.Kang MR, Kim MS, Oh JE, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125(2):353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 62.Park SW, Chung NG, Han JY, et al. Absence of IDH2 codon 172 mutation in common human cancers. Int J Cancer. 2009;125(10):2485–2486. doi: 10.1002/ijc.24647. [DOI] [PubMed] [Google Scholar]
- 63.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Green A, Beer P. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med. 2010;362(4):369–370. doi: 10.1056/NEJMc0910063. [DOI] [PubMed] [Google Scholar]
- 65.Gross S, Cairns RA, Minden MD. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207(2):339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 67.De Carli E, Wang X, Puget S. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(21):2248;. doi: 10.1056/NEJMc090593. author reply 2249. [DOI] [PubMed] [Google Scholar]
- 68.Reitman Z, Yan H. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(21):2249;. doi: 10.1056/NEJMc090593. author reply 2249. [DOI] [PubMed] [Google Scholar]
- 69.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15(19):6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 70.Watanabe T, Vital A, Nobusawa S, Kleihues P, Ohgaki H. Selective acquisition of IDH1 R132C mutations in astrocytomas associated with Li-Fraumeni syndrome. Acta Neuropathol. 2009;117(6):653–656. doi: 10.1007/s00401-009-0528-x. [DOI] [PubMed] [Google Scholar]
- 71.Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci. 2009;15(19):6002–6007. doi: 10.1111/j.1349-7006.2009.01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324(5924):261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jennings GT, Minard KI, McAlister-Henn L. Expression and mutagenesis of mammalian cytosolic NADP+-specific isocitrate dehydrogenase. Biochemistry. 1997;36(44):13743–13747. doi: 10.1021/bi970916r. [DOI] [PubMed] [Google Scholar]
- 76.Soundar S, Danek BL, Colman RF. Identification by mutagenesis of arginines in the substrate binding site of the porcine NADP-dependent isocitrate dehydrogenase. J Biol Chem. 2000;275(8):5606–5612. doi: 10.1074/jbc.275.8.5606. [DOI] [PubMed] [Google Scholar]
- 77.Haliloglu G, Jobard F, Oguz KK, et al. L-2-hydroxyglutaric aciduria and brain tumors in children with mutations in the L2HGDH gene: neuroimaging findings. Neuropediatrics. 2008;39(2):119–122. doi: 10.1055/s-2008-1081217. [DOI] [PubMed] [Google Scholar]
- 78.Kranendijk M, Struys EA, Gibson KM. Evidence for genetic heterogeneity in D-2-hydroxyglutaric aciduria. Hum Mutat. 2009;31(3):279–283. doi: 10.1002/humu.21186. [DOI] [PubMed] [Google Scholar]
- 79.Struys EA, Gibson KM, Jakobs C. Novel insights into L-2-hydroxyglutaric aciduria: mass isotopomer studies reveal 2-oxoglutaric acid as the metabolic precursor of L-2-hydroxyglutaric acid. J Inherit Metab Dis. 2007;30(5):690–693. doi: 10.1007/s10545-007-0697-5. [DOI] [PubMed] [Google Scholar]
- 80.Reitman ZJ, Parsons DW, Yan H. IDH1 and IDH2: not your typical oncogenes. Cancer Cell. 2010;17(3):215–216. doi: 10.1016/j.ccr.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bleeker FE, Atai NA, Lamba S. The prognostic IDH1 (R132) mutation is associated with reduced NADP (+)-dependent IDH activity in glioblastoma. Acta Neuropathol. 2010;119(4):487–494. doi: 10.1007/s00401-010-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25(34):4675–4682. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- 83.Frezza C, Tennant DA, Gottlieb E. IDH1 mutations in gliomas: when an enzyme loses its grip. Cancer Cell. 2010;17(1):7–9. doi: 10.1016/j.ccr.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 84.Ducray F, Marie Y, Sanson M. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(21):2248;. author reply 2249. [PubMed] [Google Scholar]
- 85.Korshunov A, Meyer J, Capper D, et al. Combined molecular analysis of BRAF and IDH1 distinguishes pilocytic astrocytoma from diffuse astrocytoma. Acta Neuropathol. 2009;118(3):401–405. doi: 10.1007/s00401-009-0550-z. [DOI] [PubMed] [Google Scholar]
- 86.Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. N Engl J Med. 2009;360(8):813–815. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]