See “An inactivating mutation in HDAC2 leads to dysregulation of apoptosis mediated by APAF1,“ by Hanigan CL, Van Engeland M, De Bruine P, et al on page 1654.
The function of the class I histone deacetylase, HDAC2, in drug response and progression of colorectal tumorigenesis has been the subject of intense investigation, although contrasting effects have recently been reported.1-3 HDAC2 is a transcriptional co-repressor that interacts with a number of transcription factors. Upon recruitment to promoter regions, HDAC2 mediates transcriptional repression by inducing local histone deacetylation and remodelling chromatin into a confirmation less permissive for transcription. Additionally, HDAC2 can catalyze the deacetylation of transcription factors themselves, modifying their transcriptional activity.4
In the current issue of GASTROENTEROLOGY, Hanigan et al5 confirm and extend a previous study by Ropero et al2 describing the presence of a frameshift mutation within a poly A tract (9 adenines) in exon 1 of the HDAC2 gene. This mutation, which is largely confined to colon tumors with microsatellite instability (MSI), produces a premature stop codon that results in loss of HDAC2 protein expression. Consistent with previous findings,2 the authors also demonstrate that the HDAC2-deficient RKO colon cancer cell line is highly refractory to apoptosis induced by histone-deacetylase inhibitors (HDACi), an emerging class of therapeutics presently approved for the treatment of cutaneous T-cell lymphoma, and also in trials for treatment of different tumors, including colon cancer.
To understand the basis for this resistance, the effect of HDACi treatment on expression of a series of pro- and antiapoptotic proteins was examined in HDACi-sensitive and -resistant cell lines. HDACi failed to induce apoptotic protease-activating factor 1 (APAF1), a proapoptotic protein, specifically in HDAC2-deficient RKO cells, establishing a link between APAF1 induction and HDACi-induced apoptosis. In HCT116 cells, HDAC2 was shown to localize to the APAF1 promoter, and siRNA-mediated down-regulation of HDAC2 induced APAF1 expression, suggesting HDAC2 directly represses APAF1 expression. Consistent with this model, basal levels of APAF1 were elevated in HDAC2 mutant RKO cells. Based on these findings, the authors propose that HDACi treatment results in inhibition of HDAC2 activity and de-repression of APAF1, which subsequently promotes apoptosis. The authors also raise the intriguing possibility that HDAC2 deficient cells may have clonally evolved to up-regulate other cell survival pathway components to compensate for the elevated APAF1 levels, providing the basis for resistance to HDACi.
However, the hypothesis that HDAC2 mutation status is a critical determinant of HDACi response was recently questioned by Ree et al,3 and is currently an issue of debate.3 Ree et al3 report that the RKO cell line cultured in their laboratory does express HDAC2, as do RKO cells obtained directly from the American Type Culture Collection. Notably, radiotoxicity was similarly amplified by HDACi in both HDAC2 expressing and nonexpressing RKO lines.3 Nevertheless, additional studies should be performed to further resolve this issue. For example, the effect of stable or transient HDAC2 down-regulation on HDACi-induced apoptosis should be determined. Likewise, the effect on HDACi-induced apoptosis, of reexpression of HDAC2 in HDAC2-deficicent cells could be examined.
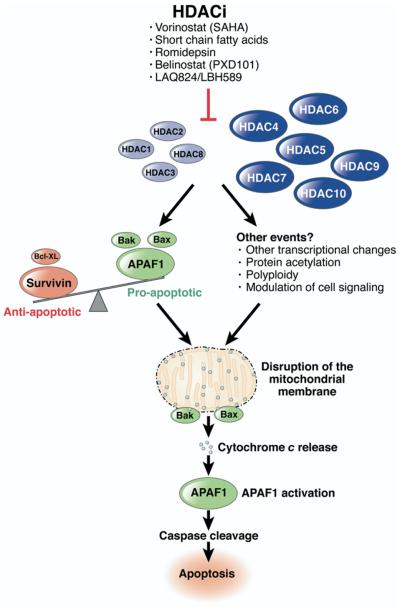
The role of HDAC2 mutation aside, the present study provides important insights into the mechanism of apoptosis induction by HDACi. This has been the subject of intense investigation over several years, with both the intrinsic/mitochondrial and extrinsic/death receptor pathways implicated.4 In colon cancer cells, inhibitors of caspase 3 and 9, but not 8, attenuate HDACi-induced apoptosis, indicating that the mitochondrial pathway is utilized primarily.6 The authors now demonstrate that siRNA-mediated inhibition of APAF1 attenuates HDACi-induced apoptosis, providing additional and independent evidence for the utilization of the mitochondrial pathway in HDACi-induced apoptosis in colon cancer cells (Figure).
Figure.
Proposed mechanisms of HDACi-induced apoptosis in colon cancer cells involving the mitochondrial pathway. Class I HDACs are indicated in light blue, and class II HDACs in dark blue.
APAF1 induction alone, however, is unlikely to completely explain HDACi-induced apoptosis as APAF1 requires cytochrome c to be released from the mitochondria for activation.7 Additional initiating events that lead to permeabilization of the mitochondrial membrane and the release of cytochrome c would therefore also be required for HDACi-induced apoptosis. In colon cancer cells, disruption of the balance in expression of pro- and antiapoptotic proteins in favor of proapoptotic factors has been suggested as a possible mechanism by which this may occur. This includes increased expression of the proapoptotic Bak protein, and decreased expression of the antiapoptotic proteins, Bcl-XL and survivin.4 HDACi also induce mitochondrial localization of the proapoptotic Bax and Bak proteins.4 However, as indicated in the present study, consistency in these effects across multiple sensitive cell lines is not observed. Identification of the early events induced by HDACi, which lead to cytochrome c release and APAF1 activation, will provide a more comprehensive understanding of the basis of sensitivity and resistance to HDACi.
HDAC2 Mutations in MSI Colon Cancer: Driver or Bystander?
A separate issue raised by the current findings, and one of importance and interest for understanding the mechanisms of tumor onset, is whether inactivating HDAC2 mutations facilitate tumorigenesis. Colon tumors with MSI, where HDAC2 mutations are primarily observed, comprise 15%–20% of colon tumors, and are driven by mutation or epigenetic inactivation in 1 of several DNA mismatch repair genes including Mlh1, Msh2, Msh6, PMS1, or PMS2. The inability to repair DNA mismatches results in a high mutation rate in these tumors (called “mutator phenotype”), with mono-, di- or trinucleotide repeat elements particularly prone to replication error. Distinguishing driver from bystander mutations in MSI cancers therefore presents a significant challenge, although this can be approached in several ways. For instance, driver mutations would be characterized by a mutation rate above that of background, homozygous inactivation (in the case of a tumor suppressor gene), and the presence of mutations within the same pathway in microsatellite stable (MSS) colon tumors.8
Assessment of these criteria in relation to the data available for HDAC2 mutations suggests this likely reflects a bystander event in tumorigenesis. First, the frequency of mutations in HDAC2 observed in MSI cancers was 43% by Hanigan et al,5 and 20% by Ropero et al (mean, 31.5%), which is within the range reported for A9 repeat elements in intergenic regions (3%–53%; mean, 22%).8 In comparison, an A10 repeat in the transforming growth factor-βRII gene, a bone fide target in tumor development, is mutated in 82% in MSI tumors, above the background mutation rate for A10 repeats (7%–76%; mean, 40%). Second, in relation to biallelic inactivation of HDAC2, whereas the RKO cell line was found to be homozygous mutant, the HCT116 and SW48 lines were heterozygous. HCT116 and SW48 cells also retained strong expression of HDAC2 mRNA and protein, indicating loss of heterozygosity by a second independent event, such as promoter methylation had not occurred. Biallelic inactivation of HDAC2 in vitro is therefore not consistently present. To address the frequency of biallelic inactivation of HDAC2 in MSI tumors in vivo, the authors examined HDAC2 protein expression by immunohisto-chemistry in tumors harboring HDAC2 mutations. Most tumors with HDAC2 mutations had either widespread or regional specific loss of HDAC2 protein. Although the distribution of tumors with these staining patterns is not described, the heterogenous staining raises the possibility that HDAC2 may be undergoing constant mutation, giving rise to portions of the tumor which are wild type (WT), heterozygous of mutant for HDAC2. In support of this, Hanigan et al5 noted that the MSI HCT116 cell line cultured in their laboratory was heterozygous for the HDAC2 mutation, whereas newly passaged HCT116 cells are WT, suggesting this mutation arose during in vitro culture. As described, Ree et al3 have also pointed out the existence of variations of the RKO cell line which have intact HDAC2 expression, indicating that RKO sublines may have arisen in different laboratories that are either WT, mutant, or heterozygous for HDAC2. Finally, the frequency of HDAC2 mutations in MSS tumors reported in both studies is extremely low, and no mutations in other HDAC enzymes or components of the HDAC2 transcriptional complex have yet been described in MSS colon cancers. Collectively, suggesting that this mutation is not actively selected for during colon cancer progression.
With regard to functional studies, Ropero et al9 directly addressed the significance of HDAC2 mutations by reintroducing HDAC2 into the HDAC2-deficient RKO cell line. HDAC2 reexpression reduced RKO cell growth, suggesting an antiproliferative/tumor-suppressor-like function for this protein.2 A follow-up gene profiling study identified a number of HDAC2 target genes, which may facilitate this process.9 However, expansion of these findings to additional cell lines, and utilization of approaches other than overexpression, are needed to demonstrate a tumor suppressor role for HDAC2.
In particular this is necessary because several studies have demonstrated a protumorigenic role for HDAC2 in colon cancer. First, multiple studies have demonstrated HDAC2 overexpression in colorectal tumors,1,10,11 and high tumoral HDAC2 expression is a prognostic indicator for reduced patient survival.11 Second, crossing HDAC2-deficient mice with ApcMin mice decreases tumor incidence in vivo.12 Finally, reduced expression of HDAC2 by siRNA has been shown by several groups to inhibit cell growth and to induce apoptosis of colon cancer cell lines.1,10,11 Similar protumorigenic effects for HDAC2 have also been suggested in other tumor types.13 Collectively, therefore, the majority of the data generated to date suggests a protumorigenic role for HDAC2.
Nevertheless, the demonstration in the present study of the existence of homozygous HDAC2-deficient colon cancer cell lines and tumors indicates that colon cancer cells are able to survive and grow in the absence of this protein. In this regard, it is also noted that the growth-inhibitory and apoptotic effects induced upon HDAC2 down-regulation in colon cancer cell lines are significantly less than that induced by HDACi. One explanation may be the redundant function HDAC2 shares with other class I HDACs, particularly the highly homologous HDAC1 protein. In support of this, transient siRNA-mediated repression of HDAC2 induces expression of HDAC1 and vice versa.10 Redundancy between these two proteins is also observed in vivo, where cardiac-specific deletion of HDAC1 or HDAC2 alone failed to evoke a phenotype, whereas simultaneous deletion of both genes induced a range of cardiac defects.14
In addition to providing a potential explanation for how colon cancer cells may survive in the absence of HDAC2, these findings may also have important implications for the further development of HDACi. Although the design of class or isoform-specific HDACi has been proposed as a means of reducing side effects to these agents, the redundancy that exists among HDAC family members suggests agents that simultaneously inhibit multiple family members may elicit greater therapeutic benefit.
Interpreting the role of HDAC2 and inactivating HDAC2 mutations in tumorigenesis and drug response is therefore complicated by several factors. In particular, the mutator phenotype of the tumor and the potentially redundant role played by other HDAC family members should be considered in ongoing studies to dissect the biological role of HDAC2 and other HDACs in tumorigenesis.
Footnotes
The author discloses no conflicts.
References
- 1.Zhu P, Martin E, Mengwasser J, et al. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell. 2004;5:455–463. doi: 10.1016/s1535-6108(04)00114-x. [DOI] [PubMed] [Google Scholar]
- 2.Ropero S, Fraga MF, Ballestar E, et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet. 2006;38:566–569. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]
- 3.Ree AH, Folkvord S, Flatmark K. HDAC2 deficiency and histone acetylation. Nat Genet. 2008;40:812–813. doi: 10.1038/ng0708-812. [DOI] [PubMed] [Google Scholar]
- 4.Mariadason JM. HDACs and HDAC inhibitors in colon cancer. Epigenetics. 2008;3:28–37. doi: 10.4161/epi.3.1.5736. [DOI] [PubMed] [Google Scholar]
- 5.Hanigan CL, Van Engeland M, De Bruine P, et al. An activating mutation in HDAC2 leads to dysregulation of apoptosis mediated APAF1. Gastroenterology. 2008;135:1654–1664. doi: 10.1053/j.gastro.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 6.Ruemmele FM, Schwartz S, Seidman EG, et al. Butyrate induced Caco-2 cell apoptosis is mediated via the mitochondrial pathway. Gut. 2003;52:94–100. doi: 10.1136/gut.52.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Amelio M, Tino E, Cecconi F. The apoptosome: emerging insights and new potential targets for drug design. Pharm Res. 2008;25:740–751. doi: 10.1007/s11095-007-9396-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sammalkorpi H, Alhopuro P, Lehtonen R, et al. Background mutation frequency in microsatellite-unstable colorectal cancer. Cancer Res. 2007;67:5691–5698. doi: 10.1158/0008-5472.CAN-06-4314. [DOI] [PubMed] [Google Scholar]
- 9.Ropero S, Ballestar E, Alaminos M, et al. Transforming pathways unleashed by a HDAC2 mutation in human cancer. Oncogene. 2008;27:4008–4012. doi: 10.1038/onc.2008.31. [DOI] [PubMed] [Google Scholar]
- 10.Wilson AJ, Byun DS, Popova N, et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 11.Weichert W, Roske A, Niesporek S, et al. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008;14:1669–1677. doi: 10.1158/1078-0432.CCR-07-0990. [DOI] [PubMed] [Google Scholar]
- 12.Zimmermann S, Kiefer F, Prudenziati M, et al. Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res. 2007;67:9047–9054. doi: 10.1158/0008-5472.CAN-07-0312. [DOI] [PubMed] [Google Scholar]
- 13.Weichert W, Roske A, Gekeler V, et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montgomery RL, Davis CA, Potthoff MJ, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]