Abstract
Different cell types can form patterns within fungal communities; for example, colonies of Saccharomyces cerevisiae form two sharply defined layers of sporulating cells separated by an intervening layer of unsporulated cells. Because colony sporulation patterns have only been investigated in a single laboratory strain background (W303), in this report we examined these patterns in other strain backgrounds. Two other laboratory strain backgrounds (SK1 and Σ1278b) that differ from W303 with respect to colony morphology, invasive growth, and sporulation efficiency nevertheless displayed the same colony sporulation pattern as W303. This pattern was also observed in colonies of wild isolates of S. cerevisiae and S. paradoxus. The wild yeast colonies sporulated on a much wider range of carbon sources than did the lab yeast and displayed a similar layered sporulation pattern when grown on either acetate or glucose medium and on either rich or synthetic medium. SK1 and Σ1278b and wild yeast colonies invaded the agar surface. The region of invasion varied between strains with respect to the organization and appearance of cells, but this invasion was always accompanied by sporulation. Thus, sporulation patterns are a general property of S. cerevisiae, and sporulation in colonies can be coordinated with invasive growth.
Keywords: Saccharomyces cerevisiae, Saccharomyces paradoxus, Colony, Sporulation, Patterning, Pseudohyphae, Invasive growth, Meiosis, Organization
1. Introduction
Cell patterning is the lynchpin of development in metazoans, but its role in microbial function is still poorly understood. Nevertheless, patterns of different cell types have been observed in microbial colonies and biofilms of many species, including eukaryotes (reviewed in (Kimmel and Firtel, 2004, Shapiro, 1998)). Recently, we reported that sporulation occurs in a sharply defined pattern within colonies of the budding yeast Saccharomyces cerevisiae (Piccirillo et al., 2010). Specifically, once colony growth is complete, sporulation initiates in two distinct horizontal layers within the colony-- a narrow layer at the bottom of the colony and an interior layer near the center of the colony. Over time, this central band of sporulation expands to eventually include the top of the colony. Even after extended incubation, colonies remain divided into sharply divided layers of sporulated and unsporulated regions.
Sporulation in S. cerevisiae is comprised of meiosis, whereby a diploid cell is converted into four haploid gametes, and spore formation, whereby each gamete is packaged within a spore wall. Three main nutrient criteria must be met for sporulation to occur: 1) fermentable carbon sources such as glucose must be absent, 2) non-fermentable carbon sources such as acetate must be present, and 3) nitrogen or another essential nutrient must be absent (reviewed in (Honigberg and Purnapatre, 2003, Zaman et al., 2008)). Notably, these three conditions are expected during late stages of colony growth under natural conditions. In the wild, S. cerevisiae and closely related yeast are found primarily on decaying fruit or tree exudates, both of which contain primarily fermentable carbon sources; however, during late stages of growth, these fermentable carbon sources will have been converted via glycolysis to non-fermentable carbon sources, primarily ethanol. Non-fermentable carbon sources stimulate sporulation through multiple pathways (Jambhekar and Amon, 2008), including pathways that lead to bicarbonate production and export, which in turn stimulates sporulation by raising the pH of the medium (Hayashi et al., 1998, Ohkuni et al., 1998). In colonies, alkali signals between cells, sensed through the Rim101p pathway are required for the gradual expansion of the region of sporulation from a narrow band in the interior of the colony to a much broader region including the top of the colony (Piccirillo et al., 2010).
It is sometimes useful to compare biological processes such as colony patterning in different laboratory strains of yeast. Laboratory strains of yeast can differ at hundreds of polymorphic loci, including both deletions and duplications (Schacherer et al., 2007). Related to these genetic differences, laboratory yeast strains differ dramatically in a number of related and easily observable phenotypes such as colony morphology (Granek and Magwene, 2010, Vopalenska et al., 2005), ability to form pseudohyphae (Liu et al., 1996) and efficiency of sporulation (Esposito and Klapholz, 1981).
Differences between laboratory strain backgrounds in colony morphology, invasive growth and ability to form pseudohyphae can be attributed in part to allelic difference in the flocculin family of genes, which encode cell surface proteins that mediate cell-cell adhesion (reviewed in (Gancedo, 2001). In particular, some flocculin alleles are not expressed or are nonfunctional in some laboratory strains. For example, the S288C strain background lacks a functional allele of FLO8, which encodes a transcriptional activator of the FLO11 and FLO1 flocculin genes (Liu et al., 1996). As a result, S288C fails to express most flocculin genes and is incapable of pseudohyphal differentiation or invasive growth, but restoring a functional FLO8 allele to this strain allows both pseudohyphal differentiation and invasive growth (Kobayashi et al., 1999). Similarly, differences in sporulation efficiency between strains are attributable in part to allelic differences in signal transduction components and transcriptional regulation (Ben-Ari et al., 2006, Deutschbauer and Davis, 2005, Gerke et al., 2009).
Some traits of laboratory yeast, such as the ability to disperse as single cells in liquid culture, may have been selected during domestication in the laboratory (Mortimer and Johnston, 1986). In addition, because most lab strains are derived from vineyard strains, other traits may have been selected during vineyard cultivation (Liti et al., 2009, Mortimer, 2000). As a result it can be useful to investigate biological processes in wild isolates. For example, S. cerevisiae isolated from tree exudates (Naumov et al., 1998) have probably evolved separately from domesticated strains (Aa et al., 2006, Fay and Benavides, 2005). Of course, for any wild strain of S. cerevisiae, the possibility of genetic exchange with domesticated yeast, e.g. from vineyards, cannot be entirely ruled out (Ruderfer et al., 2006). For this reason, it can also be useful to examine these traits in a closely related yeast species, Saccharomyces paradoxus (Johnson et al., 2004, Naumov et al., 1998), that has not been domesticated (Koufopanou et al., 2006).
To determine whether the colony sporulation pattern we previously observed in the W303 background (Piccirillo et al., 2010) was a general property of yeast or reflected properties acquired during domestication or unique to specific laboratory strains, we examined sporulation patterns in a variety of laboratory and wild strains and in a range of growth conditions. Because colonies of different strain backgrounds differ with respect to colony morphology, sporulation efficiency, invasive growth and pseudohyphal differentiation, we investigated the relationship between these different processes in colony development.
2. Materials and methods
2.1. Strains and media
Yeast strains used in this study are shown in Supplementary Table 1. SH1020 is a derivative of W303 (Lee and Honigberg, 1996), SH2533 is a derivative of Σ1278b (Palecek et al., 2000), and SH561 is a derivative of SK1 (Smith et al., 1990). The flo11Δ mutant was constructed as described (Gray and Honigberg, 2001). S. cerevisiae and S. paradoxus strains isolated from tree exudates have been described (Sniegowski et al., 2002). Except as noted below, colonies were grown on YNA medium (2% potassium acetate (pH 7.0), 0.25% yeast extract, 2% agar, supplemented with the minimal amino acids necessary to balance auxotrophies) at 30° C. For the experiment reported in Table 3, in some cultures the acetate in YNA was replaced with 2% of ethanol, raffinose, galactose or glucose, or 0.5% glucose. 0.5% SC is synthetic complete medium (Kaiser et al., 1994) containing 0.5% glucose.
Table 3.
Effect of carbon source on frequency of sporulation in coloniesa
| Strain Bkgb | Carbon source |
|||||
|---|---|---|---|---|---|---|
| OAc | EtOH | Raf | Galac | 2% Gluc | 0.5% Gluc | |
| W303 | 48±4 | <1 | <1 | <1 | <1 | <1 |
| SKI | 79±1 | 3±1 | 2±1 | <1 | <1 | <1 |
| Σ1278b | 30±1 | <1 | <1 | <1 | <1 | <1 |
| wild S. cerevisiae | 72±4 | 5±1 | 23±6 | 4±1 | 5±2 | 16±2 |
| wild S. paradoxus | 42±4 | 22±3 | 28±5 | 10±2 | 14±1 | 27±5 |
Percentage of cells in colony that formed spores after 8 days (mean±SEM, n=4)
Strain numbers shown in Supplementary Table 1
2.2. Colony embedding and section
Sectioning colonies was performed as described (Piccirillo et al., 2010). In brief, colonies were grown on agar medium and then encased in melted agar. The encased colony was perfused first with fixative, then with ethanol and finally with Spurr’s resin. Sections were stained with toluidine blue. Distribution of asci within colonies was quantified by superimposing a vertical stack of nine equal-sized rectangles on the colony image. For each rectangle, all sporulated and non-sporulated cells were counted (50–100 cells), and the percentage of total cells that had formed asci was calculated.
To measure the percentage of sporulated cells in a section that were dyads, the number of asci with two spores visible in the section was multiplied by the estimated fraction of such asci that were authentic dyads (rather than tetrads with only two spores visible. This latter fraction was estimated by examining serial sections of these two-spored asci. For each two-spored asci, if additional spores were visible in at least one other serial sections, then the ascus was considered to be a tetrad; in contrast, if only one spore of the ascus was visible in at least one other serial sections, then the dyad was considered an “authentic” dyad. (Fig. S2 in supplementary material). The estimated fraction of authentic dyads was determined separately for asci in the invasive region and in the center of the colony. These fractions were based on examining 50–80 classifiable two-spored asci from at least three independent colonies.
2.3. Measuring efficiency of sporulation and agar invasion
To measure overall sporulation efficiency in colonies, the entire colony was scraped from the agar and resuspended in water. This suspension was examined under a light microscope, and 250–300 cells were classified as sporulated or unsporulated. Percentage sporulation is the fraction of the total cells counted that had formed tetrad or dyad asci. To quantify the efficiency of agar invasion, colony sections with more than five cells present below the agar surface were considered invasive. As an additional measure of invasion, plate washing assays were performed as described (Guldal and Broach, 2006). Plates were photographed before and after washing and the two images compared to determine the fraction of colonies retained after washing.
For experiments comparing sporulation efficiency in colonies growing on different carbon sources, “spot” colonies were analyzed. These colonies were inoculated by spotting 0.5 μl of culture containing 1.0 ×105 cells on to plates. To ensure an equivalent environment for each colony, 20 equally spaced colonies were spotted on each plate in a ring approximately 5 mm from the edge of plate.
2.4. Image acquisition and processing
All microscope images were captured using a Colorview II camera and AnalySIS software (SIS). Images were adjusted for brightness in Canvas 8.
3. Results
3.1. Patterns of sporulation are conserved among yeast with different colony morphologies
Because we previously observed a layered pattern of sporulation in colonies using the S. cerevisiae laboratory strain background W303 (Piccirillo et al., 2010), we examined two other common laboratory strains of yeast (SK1 and Σ1278b), which differ dramatically from one another with respect to colony morphology (Granek and Magwene, 2010). As a preliminary step, we compared the timing and efficiency of sporulation in colonies of all three strains (Fig. 1). When grown on rich acetate medium, SK1 and W303 colonies formed spores with similar timing, though the final efficiency of spore formation in SK1 was somewhat higher. In contrast, Σ1278b colonies formed spores significantly less rapidly and efficiently than the other two strains.
Fig. 1. Timing of spore formation in colonies of three common laboratory yeast strains.
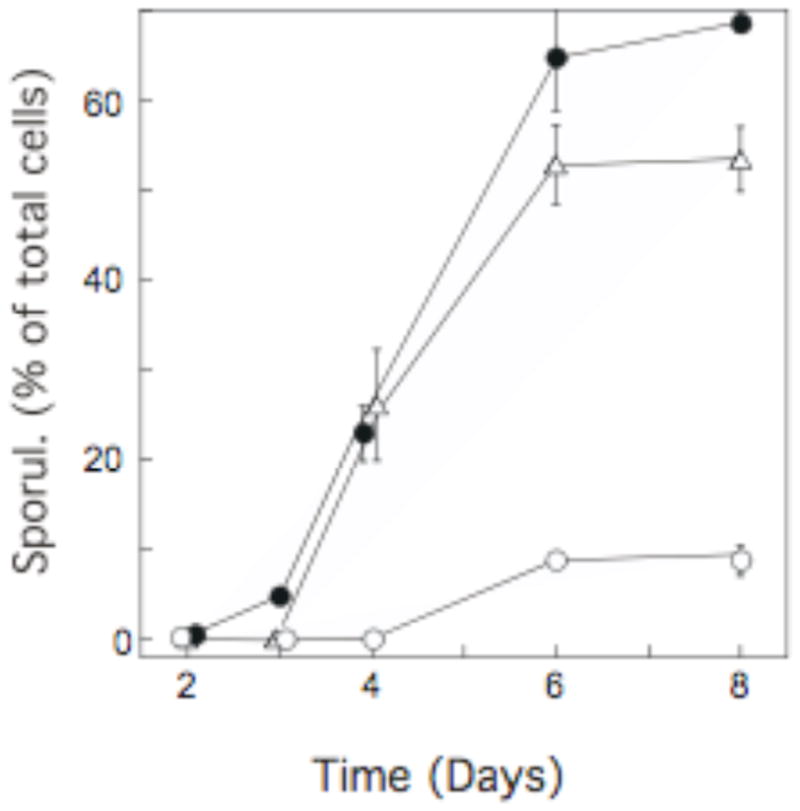
Colonies of SK1 SH561 (SK1 background, ●), SH1020 (W303 background, △), or SH2533 (Σ1278b background, ○) were plated on YNA medium. At the indicated times, the frequency of asci among the total cells in the colony was determined for each of three colonies from a single experiment. The mean frequency of asci in these three colonies is represented by the data points, and the error bars represent the SEM. Where error bars are not visible, their length is less than that of the symbol.
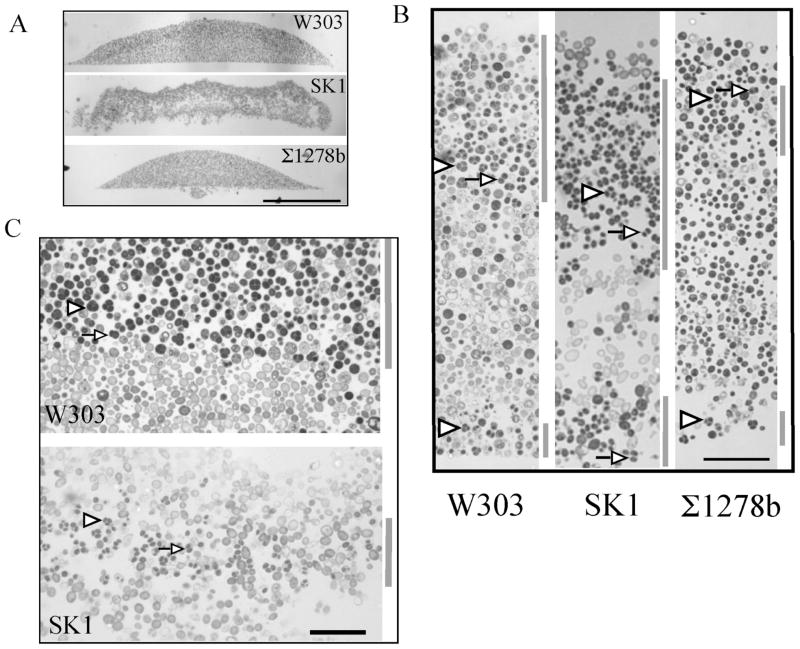
We next examined the colony morphology and distribution of sporulated cells within colonies for all three laboratory-strain backgrounds. When grown on the above medium, SK1 colonies were lacey or flocculent, so that sections of these colonies often contained regions with few or no cells. In contrast, W303 and Σ1278b colonies were smooth and sections of these colonies revealed a compact structure (Fig. 2A–2B). Despite differences in colony morphology and the timing and efficiency of sporulation, the overall pattern of sporulation within mature colonies was very similar for all three strains. As reported previously for W303 (Piccirillo et al., 2010), sporulated cells in all three strains were observed in a broad layer near the top of the colony and in a narrow layer at the bottom of the colony (Fig. 2B, regions with high frequencies of asci indicated by gray bar to right of each image).
Fig. 2. Pattern of sporulation in colonies of three common laboratory yeast strains.
Colonies from the indicated strain backgrounds were grown as in Fig. 1, embedded and sectioned. A) Cross-section after 8 days of incubation at 30° C, scale bar = 0.5 mm. B) Section from top to bottom of colony after 8 days of incubation, scale bar = 40 μm. C) Boundary between the upper region of sporulation in cells and the underlying region after 10 days incubation (W303) or 4 days incubation (SK1), scale bar = 25 μm. Gray bars to the left of images in B) and C) indicate region of colonies with high frequencies of asci. Arrowheads in the figures indicate representative tetrad asci (three or four spores visible), whereas arrows indicate representative dyad asci (two spores). For each of the above strains, at least three independent colonies were examined; all colonies from the same strain displayed the same sporulation pattern.
For SK1, the development of colony sporulation patterns over time (Fig. 3) was similar to the pattern previously reported for W303 (Piccirillo et al., 2010). Specifically, spores were initially observed in an internal layer within SK1 colonies (Fig. 2C) as well as at the bottom of the colony. Over time, this internal layer expanded to eventually include the top of the colony (Fig. 3). We previously demonstrated that in W303, this changing pattern of sporulation is not attributable to colony growth, since growth, as measured either by colony diameter or by cell number, is largely complete before sporulation initiates (Piccirillo et al., 2010). Importantly, SK1 colonies, like W303 colonies, developed sharp boundaries between regions of sporulation and regions of non-sporulation; indeed, an almost straight line can be drawn between the two regions (Fig. 2C). Although the colony sporulation pattern for Σ1278b is similar to the other two strains, because Σ1278b colonies sporulate with relatively low efficiency, it was not possible in this strain to monitor the development of sporulation patterns over time or to observe sharp boundaries.
Fig. 3. Distribution of sporulated cells in SK1 colonies.
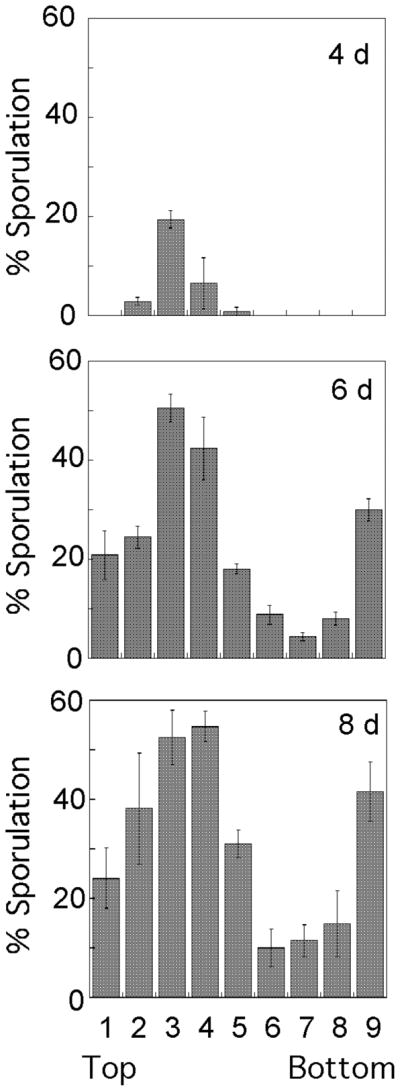
Colonies were incubated for the times indicated on each graph (4, 6, or 8 days) and then sectioned. The frequency of asci in each region of the colony is shown in the bar graphs; the leftmost bar represents the top of the colony and the rightmost bar represents the bottom of the colony. The data is the average of three independent colonies, and error bars represent the SEM.
3.2. High frequency of pseudohyphae is not required for invasive growth
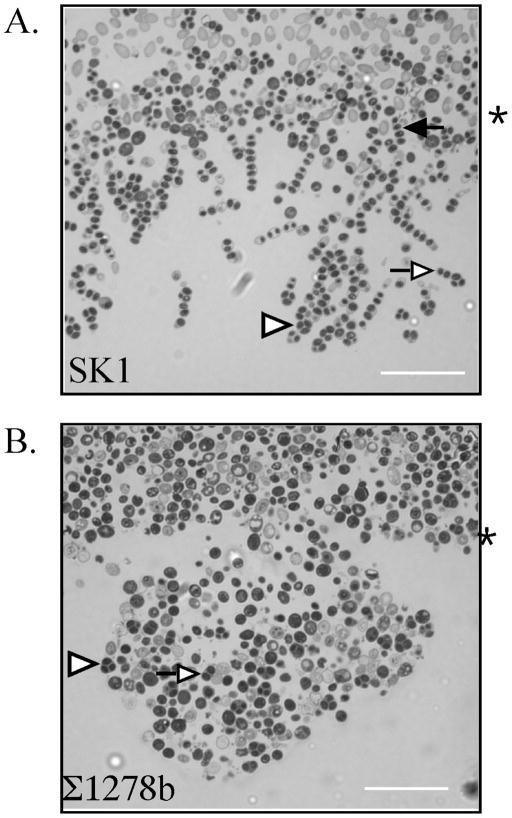
W303 colonies did not invade the agar surface; in contrast, greater than half of SK1 and Σ1278b colonies penetrated the agar (Table 1). The structure of the region of invasion can be clearly visualized in sectioned colonies (Fig. 2A and Fig. 4). Interestingly, SK1 formed a different structure in this region than does Σ1278b. In SK1, chains of cells invaded the agar at multiple sites through the central third of the colony; whereas, in Σ1278b there was a single site of invasion near the center of the colony that expanded outward into a pocket or bubble of cells beneath the surface. A second difference between the region of invasion in SK1 and Σ1278b was the shape of cells in this region (Fig. 4). SK1 colonies contained many elongated cells (pseudohyphae) at the region of invasion, most of which had also sporulated (discussed below). In contrast, Σ1278b colonies grown under the same conditions did not contain detectable pseudohyphae. These results indicate that invasion of the agar can be accompanied by pseudohyphal growth, but that extensive pseudohyphal growth is not a prerequisite for cells to invade the agar.
Table 1.
Efficiency of Invading Agar
| Strain Bkga | Medium | Time (days) | % invb | nc |
|---|---|---|---|---|
| SKI | YNA | 8 | 94 | 18 |
| Σ1278b | YNA | 8 | 62 | 13 |
| Wild S. c. | YNA | 6 | 100 | 9 |
| 0.5% SC | 8 | 0 | 10 | |
| Wild S. p. | YNA | 6 | 83 | 6 |
| 0.5% SC | 8 | 0 | 6 |
Strains numbers shown in Supplementary Table 1
(colonies invading agar) × 100%/(total colonies)
number of colonies examined
Fig. 4. Structure of region of SK1 and Σ1278b colonies that invade the agar surface.
A) A portion of the region of an SK1 colony which has invaded the agar, B) Region of an Σ1278b colony which has invaded the agar. Colonies incubated for eight days prior to sectioning. Asterisk to right of images indicates surface of agar, arrowheads in the figures indicate representative tetrad asci (three or four spores visible), open arrows indicate representative dyad asci (two spores), and the filled arrow indicates a linear four-spored ascus. Scale bar = 25 μm.
As an independent measure of the efficiency that the three laboratory strain backgrounds invade the agar under the standard conditions of this study, we also performed “wash assays” in which all but the invasive region of colonies are washed away under a stream of water. After extensive washing, more than 90% of either SK1 or Σ1278b colonies retained enough cells attached to the agar to be easily visible by eye (Fig. S1 in supplementary material). In contrast, no visible cells were retained when W303 colonies, or an SK1 strain deleted for the FLO11 adhesin were washed under identical conditions. Consistent with the differences between SK1 and Σ1278b colony sections in the appearance of the invasive region, in SK1 colonies most of the agar surface contacting the colony retained cells after washing, whereas in Σ1278b colonies only a small central portion of the agar surface contacting the colony retained cells.
3.3. Asci present in the region of invasive growth
In both SK1 and Σ1278b colonies, asci were frequently observed in the region of invasion (Fig. 4). Indeed, analysis of serial sections revealed that the frequency of sporulation in the invasive region was comparable to the frequency of sporulation in the top layer of the colony for both SK1 colonies and Σ1278b colonies (Table 2, column 3). However, for SK1 colonies, the appearance of asci was different in the invasive region than in the top layer of sporulation. In the top layer, the majority of asci were tetrahedral (tetrads), whereas in the invasive region, significantly more of the asci contained only two spores (dyads) (P<0.001), and sometimes contained three spores in a linear arrangement (compare Fig. 2B to Fig. 4, and Table 2, column 4). We reasoned that these dyads and linear asci likely derived from pseudohyphae that invade the agar and subsequently sporulate.
Table 2.
Dyad asci in regions of colonya
| Strain | Region | spor. (%)b | dyads/asci (%)c |
|---|---|---|---|
| SK1 | Top Layer | 38±5 (3) | 43±2 (3) |
| Invasive | 39±4 (3) | 72±1 (3) | |
| Σ1278b | Top Layer | 17±2 (4) | 33±2 (4) |
| Invasive | 13±2 (4) | 26±1 (4) |
Mean ± SEM (number of colonies)
(asci ×100%)/total cells
(dyad asci ×100%)/total asci
In contrast to SK1 colonies, asci in Σ1278b colonies were mostly tetrads rather than dyads (Table 2, rows 3–4), even though the overall frequency of sporulation in the invasive region of Σ1278b colonies was approximately the same as in the top region. As a result, the frequency of dyads/total asci within the invasive region was significantly lower in Σ1278b colonies than in SK1 colonies (P<0.001). Thus, pseudohyphal differentiation could accompany and perhaps stimulate efficient invasive growth and sporulation, as in SK1; however, both invasive growth and sporulation could also occur in the absence of efficient pseudohyphal differentiation, as in Σ1278b.
3.4. Sporulation patterns and invasive structures in wild yeast colonies are similar to laboratory yeast
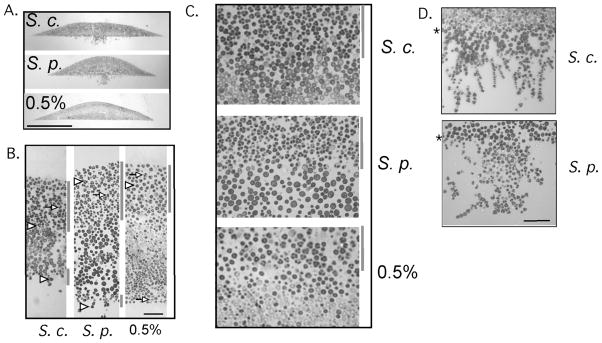
We next asked whether colonies formed from wild strains of yeast would display sporulation patterns and invasive growth similarly to lab yeast. For this purpose, two strains each of S. cerevisiae and Saccharomyces paradoxus were tested. All four of these wild strains were isolated from tree exudates and are unrelated to lab strains (Naumov et al., 1992, Naumov et al., 1998). Colonies of these strains were grown as colonies for 6 days and sectioned as above (Fig. 5A). We found that all four wild strains yielded sporulation patterns similar to laboratory strains, with a narrow layer of sporulation along the bottom surface of the colony and a broader layer of sporulation through approximately one-third of the colony (Fig. 5B). Because almost all cells in the upper layer formed asci, the boundary between this layer and the underlying unsporulated layer was especially distinct (Fig. 5C).
Fig. 5. Pattern of sporulation in wild isolates of S. cerevisiae and S. paradoxus.
Colonies of wild S. cerevisiae (S. c.) and S. paradoxus (S. p.) were grown on YNA medium for six days, or colonies of wild S. cerevisiae were grown on 0.5% synthetic glucose medium (0.5%) for 8 days, then embedded and sectioned as in Fig. 1. Arrowheads indicate representative tetrad asci (three or four spores visible), whereas arrows indicate representative dyad asci (two spores). A) Section through center of entire colony, scale bar = 0.5 mm, B) Vertical slice of section, from top to bottom of colony, scale bar = 30 μm. C) Portion of colony containing boundary between high sporulating region at top and low sporulating region. D) Portion of colonies grown on YNA medium that invades the agar. Asterisks to left of images indicate the agar surface, scale bar = 30 um. Gray bars to right of images in (B) and (C) indicate regions containing a high frequency of asci. For the images in B–D, at least three independent colonies from each of two wild S. cerevisiae and two S. paradoxus strains were examined and yielded similar patterns.
Colonies from both species of wild yeast invaded the agar efficiently when grown on standard medium (Table 1, rows 3 and 5; Fig. S1). Interestingly, the region of invasion of wild S. cerevisiae colonies resembled those of SK1 colonies, whereas the region of invasion of the wild S. paradoxus colonies was more similar to that of Σ1278b colonies (Fig. 5D).
3.5. Effect of carbon and nitrogen source on colony sporulation
To investigate the effect of carbon source on colony sporulation, we first determined the total frequency of sporulated cells in colonies grown on either fermentable or non-fermentable carbon sources (Table 3). The three laboratory strain backgrounds tested (W303, SK1, and Σ1278b) formed spores efficiently only on acetate; neither a different non-fermentable carbon source (ethanol) nor any of three fermentable carbon sources (glucose, galactose or raffinose) allowed appreciable sporulation in these strains. In contrast, wild S. cerevisiae and S. paradoxus colonies formed spores on a wide range of carbon sources, including moderate concentrations of glucose, though with higher efficiency on some carbon sources than on others. Wild S. cerevisiae and S. paradoxus differed in the effects of different carbon sources on sporulation; both isolates of the wild S. cerevisiae sporulated almost twice as efficiently as the S. paradoxus isolates on acetate medium, but 2–4 times less efficiently than S. paradoxus on most other carbon sources.
Because wild yeast colonies can sporulate on media containing a range of carbon sources, we next asked whether sporulation and invasive growth would also occur in colonies grown on synthetic rather than rich medium. Nitrogen and other nutrients are supplied by yeast extract and peptone in rich medium, whereas these nutrients are supplied in synthetic (i.e. defined) medium as ammonium sulfate and yeast nitrogen base. When wild-type colonies were incubated for seven days on synthetic medium containing 0.5% glucose,, 22 ± 3% of cells in S. cerevisiae colonies and 28 ± 3% of cells in S. paradoxus colonies formed asci. Unlike colonies grown on rich acetate medium, colonies grown on the synthetic glucose medium completely failed to invade the agar (Table 1, rows 4 and 6; Fig. 5A).
3.6. Colony sporulation patterns are similar on synthetic glucose medium and rich acetate medium
Because the wild S. cerevisiae strains formed colonies with relatively high frequencies of asci on synthetic medium containing 0.5% glucose, we examined the pattern of sporulation in these colonies after 8 days of incubation. We found that these colonies displayed a layered pattern of sporulation (Fig. 5B and 5C) that was indistinguishable from the pattern formed on rich acetate medium. Thus, the pattern of sporulation characteristic of yeast colonies was not limited to a particular carbon or nitrogen source.
4. Discussion
We compared the properties of colonies from three laboratory yeast strains and four wild yeast strains. Colonies from these strains varied dramatically with respect to their overall morphology, their ability to form pseudohyphae and their ability to invade the agar surface. In addition, nutrient requirements for sporulation in colonies varied between these strains. Despite these differences, all seven strains tested displayed the same characteristic colony sporulation pattern, with two layers of sporulated cells and an intervening layer of unsporulated cells. Furthermore, under all conditions tested in which sporulation occurred efficiently in colonies, these colonies exhibited sharply defined boundaries between regions containing asci and regions lacking asci. These results indicate that patterns of sporulation within colonies are likely to be a general property of S. cerevisiae.
The wild yeast strains tested in this study formed spores on a much wider range of carbon sources than did laboratory yeast. Indeed, wild yeast colonies sporulated relatively efficiently even on medium containing glucose in moderate concentrations (0.5%), perhaps indicating that wild yeast are more efficient than lab yeast at converting glucose to non-fermentable carbon sources. In any case, wild yeast formed colony sporulation patterns on synthetic glucose media that were indistinguishable from those formed on rich acetate medium. Thus, the layered colony sporulation pattern probably does not depend on a specific nutrient combination.
Pseudohyphal differentiation and invasive growth are closely coupled processes, and a number of genes are required for both, notably the flocculins FL011 and FLO10 (Fichtner et al., 2007, Van Mulders et al., 2009). As indicated previously (Guo et al., 2000), standard assays for invasive growth (e.g. cells remaining after washing plates) may not fully distinguish invasive growth from cell affinity to the agar surface. Nevertheless, there have been several reports of mutants that affect these two processes differently (Birkaya et al., 2009, Palecek et al., 2000). These earlier studies suggest that pseudohyphae may contribute to invasive growth without being necessary for this invasion. Our results support this view; Σ1278b can form pseudohyphae under a variety of conditions, but under the conditions used in this study, Σ1278b colonies invade the agar without forming detectable pseudohyphae. Invasive growth may require only a few pseudohyphae at the initial site of invasion, but clearly a high level of pseudohyphal differentiation is not required for invasion.
Although pseudohyphal differentiation (the dimorphic switch) is not required to invade the agar, this switch may stimulate invasion. Two distinct types of structures were observed at the site of invasion, in different strains. In Σ1278b (and wild S. paradoxus) colonies, invasion occurred at a single central point in the colony, forming a bubble-like pocket of cells beneath the surface. In contrast, in SK1 (and wild S. cerevisiae) colonies, pseudohyphae invaded the agar at multiple points through the central approximately one-third of the colony. Thus pseudohyphae may be more efficient at invading the agar surface than are ovoid cells.
The region of invasion in SK1, Σ1278b and wild yeast colonies contain relatively high levels of sporulated cells. Interestingly, the asci in these regions of invasion were mostly two-spore dyads for SK1 and mostly tetrads for Σ1278b. This difference is consistent with earlier studies demonstrating that that elongated yeast cells, such as zygotes formed after mating and some S. cerevisiae hybrid species, result in linear rather than tetrahedral asci (Hawthorne, 1955, Thomas and Botstein, 1987). Thus, either typical ovoid or elongated (pseudohyphal) cells are capable of sporulating once they have invaded the agar, but the arrangement of spores within the asci may conform to the shape of the pre-existing cell.
The conserved pattern of sporulation in yeast colonies described in this study raises the question of what (if any) selective advantage colony patterning may confer on yeast. While meiosis in higher organisms is considered important in maintaining genetic diversity in populations, sequence analysis indicates that wild yeast strains are probably completely homozygous (Diezmann and Dietrich, 2009, Koufopanou et al., 2006, Sniegowski et al., 2002). This homozygosity results from the ability of haploids to efficiently switch mating-types followed by mating. Of course, sporulation in a clonal homozygous population would not generate genotype diversity. However, sequence comparisons between diverse natural isolates in yeast suggest that mating between unrelated strains of wild yeast (outcrossing), though rare, is nevertheless important for maintaining genotype diversity in yeast populations (Johnson et al., 2004, Liti et al., 2009, Ruderfer et al., 2006). Outcrossing in nature likely occurs in the context of a yeast community containing a mixture of strains. If these natural communities, like clonal colonies, sequester sporulation in one region of the community, this would tend to increase the frequency of these inter-strain crosses.
In addition to increasing the possibility of outcrossing, sporulation patterning may help to conserve nutrients in colonies. Although sporulation is a response to starvation, the combined programs of meiosis and spore formation require continued respiration in order to induce the hundreds of genes required for meiotic recombination, chromosome segregation and spore wall formation. Limiting sporulation to only a portion of the colony may ensure that scarce nutrients are utilized by only a subset of the colony’s population. Another possibility is that the non-sporulating cells in colonies die rapidly, and these dead cells supply nutrients to other cells in the colony. Finally, positioning asci at the upper surface of colonies may promote their dispersion to new environments. This dispersion occurs primarily when they are ingested to insects such as Drosophila or attach to the appendages of these insects (Begon, 1986). Spores are much more viable within the gut of Drosophila than are vegetative (growing) cells (Coluccio et al., 2008). Thus, localization of sporulation to the top surface of colonies might increase dispersal to new locations as well as increase the efficiency of interstrain mating and nutrient utilization.
Supplementary Material
Fig. S1 Invasive growth of laboratory and wild strains of budding yeast as measured by the agar wash assay. Colonies of SK1 (SH561), Σ1278b, W303, a flo11Δ derivative of SK1 (SH4325) and an isolate of wild S. cerevisiae and wild S. paradoxus were grown for 8 days on YNA medium, the plates photographed (left column), washed extensively, and then rephotographed (see Methods). The percentages indicated to the right of the images represent the percent retention calculated as (colonies retaining cells after washing) × 100%/(total colonies). Percentages are the average of at least three independent plates ± SEM.
Fig. S2 Serial sections of invasive region of SK1 Adjacent serial sections of the same colony were identified because of similar patterns of cells in the section. (A–D) Four serial sections from the invasive region of an SK1 colony. Arrows indicate asci in pwhich exactly two spores are visible in at least one serial section and only one spore is visible in at least one other section (or both spores are much weaker). Arrowhead indicates an ascus in which two spores are visible in one section (C) and three spores are visible in two other sections (A & B).
Acknowledgments
We are grateful to Drs. Aaron Mitchell (Carnegie-Mellon University), Stephen Kron (Univ. of Chicago) and Paul Sniegowski (Univ. of Pennsylvania) for strains. Colony sectioning was performed by Jennifer Baumler (School of Biological Sciences EM facility, UMKC). Funding was from NIH grant R15GM80710.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aa E, Townsend JP, Adams RI, Nielsen KM, Taylor JW. Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:702–715. doi: 10.1111/j.1567-1364.2006.00059.x. [DOI] [PubMed] [Google Scholar]
- Begon M. Yeasts and Drosophila. In: Ashburner M, Carson H, Thompson JN, editors. The Genetics and Biology of Drosophila. Academic Press; London: 1986. pp. 345–384. [Google Scholar]
- Ben-Ari G, et al. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet. 2006;2:e195. doi: 10.1371/journal.pgen.0020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkaya B, Maddi A, Joshi J, Free SJ, Cullen PJ. Role of the cell wall integrity and filamentous growth mitogen-activated protein kinase pathways in cell wall remodeling during filamentous growth. Eukaryot Cell. 2009;8:1118–1133. doi: 10.1128/EC.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio AE, Rodriguez RK, Kernan MJ, Neiman AM. The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS ONE. 2008;3:e2873. doi: 10.1371/journal.pone.0002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer AM, Davis RW. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat Genet. 2005;37:1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- Diezmann S, Dietrich FS. Saccharomyces cerevisiae: population divergence and resistance to oxidative stress in clinical, domesticated and wild isolates. PLoS One. 2009;4:e5317. doi: 10.1371/journal.pone.0005317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito RE, Klapholz S. Meiosis and ascospore development. In: Strathern JN, Jones EW, Broach JR, editors. Molecular biology of the yeast Saccharomyces: Life cycle and inheritance. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1981. pp. 211–287. [Google Scholar]
- Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner L, Schulze F, Braus GH. Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell-cell and cell-substrate adherence of S. cerevisiae S288C. Mol. Microbiol. 2007;66:1276–1289. doi: 10.1111/j.1365-2958.2007.06014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:107–123. doi: 10.1111/j.1574-6976.2001.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Gerke J, Lorenz K, Cohen B. Genetic interactions between transcription factors cause natural variation in yeast. Science. 2009;323:498–501. doi: 10.1126/science.1166426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granek JA, Magwene PM. Environmental and genetic determinants of colony morphology in yeast. PLoS Genet. 2010;6:e1000823. doi: 10.1371/journal.pgen.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M, Honigberg SM. Effect of chromosomal locus, GC content and length of homology on PCR- mediated targeted gene replacement in Saccharomyces. Nucleic Acids Res. 2001;29:5156–5162. doi: 10.1093/nar/29.24.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldal CG, Broach J. Assay for adhesion and agar invasion in S. cerevisiae. J Vis Exp. 2006:64. doi: 10.3791/64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Styles CA, Feng Q, Fink GR. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci USA. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne DC. The Use of Linear Asci for Chromosome Mapping in Saccharomyces. Genetics. 1955;40:511–518. doi: 10.1093/genetics/40.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Ohkuni K, Yamashita I. Control of division arrest and entry into meiosis by extracellular alkalization in Saccharomyces cerevisiae. Yeast. 1998;14:905–913. doi: 10.1002/(SICI)1097-0061(199807)14:10<905::AID-YEA290>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Honigberg SM, Purnapatre K. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J Cell Sci. 2003;116:2137–2147. doi: 10.1242/jcs.00460. [DOI] [PubMed] [Google Scholar]
- Jambhekar A, Amon A. Control of meiosis by respiration. Curr Biol. 2008;18:969–975. doi: 10.1016/j.cub.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LJ, Koufopanou V, Goddard MR, Hetherington R, Schafer SM, Burt A. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics. 2004;166:43–52. doi: 10.1534/genetics.166.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics: A Cold Spring Harbor laboratory course manual. 1994. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1994. [Google Scholar]
- Kimmel AR, Firtel RA. Breaking symmetries: regulation of Dictyostelium development through chemoattractant and morphogen signal-response. Curr Opin Genet Dev. 2004;14:540–549. doi: 10.1016/j.gde.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kobayashi O, Yoshimoto H, Sone H. Analysis of the genes activated by the FLO8 gene in Saccharomyces cerevisiae. Curr Genet. 1999;36:256–261. doi: 10.1007/s002940050498. [DOI] [PubMed] [Google Scholar]
- Koufopanou V, Hughes J, Bell G, Burt A. The spatial scale of genetic differentiation in a model organism: the wild yeast Saccharomyces paradoxus. Philos Trans R Soc Lond B Biol Sci. 2006;361:1941–1946. doi: 10.1098/rstb.2006.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RH, Honigberg SM. Nutritional regulation of late meiotic events in Saccharomyces cerevisiae through a pathway distinct from initiation. Mol Cell Biol. 1996;16:3222–3232. doi: 10.1128/mcb.16.6.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 2000;10:403–409. doi: 10.1101/gr.10.4.403. [DOI] [PubMed] [Google Scholar]
- Naumov GI, Naumova E, Korhola M. Genetic identification of natural Saccharomyces sensu stricto yeasts from Finland, Holland and Slovakia. Antonie Van Leeuwenhoek. 1992;61:237–243. doi: 10.1007/BF00584230. [DOI] [PubMed] [Google Scholar]
- Naumov GI, Naumova ES, Sniegowski PD. Saccharomyces paradoxus and Saccharomyces cerevisiae are associated with exudates of North American oaks. Can J Microbiol. 1998;44:1045–1050. [PubMed] [Google Scholar]
- Ohkuni K, Hayashi M, Yamashita I. Bicarbonate-mediated social communication stimulates meiosis and sporulation of Saccharomyces cerevisiae. Yeast. 1998;14:623–631. doi: 10.1002/(SICI)1097-0061(199805)14:7<623::AID-YEA264>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Palecek SP, Parikh AS, Kron SJ. Genetic analysis reveals that FLO11 upregulation and cell polarization independently regulate invasive growth in Saccharomyces cerevisiae. Genetics. 2000;156:1005–1023. doi: 10.1093/genetics/156.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S, White MG, Murphy JC, Law DJ, Honigberg SM. The Rim101p/PacC pathway and alkaline pH regulate pattern formation in yeast colonies. Genetics. 2010;184:707–716. doi: 10.1534/genetics.109.113480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer DM, Pratt SC, Seidel HS, Kruglyak L. Population genomic analysis of outcrossing and recombination in yeast. Nat Genet. 2006;38:1077–1081. doi: 10.1038/ng1859. [DOI] [PubMed] [Google Scholar]
- Schacherer J, Ruderfer DM, Gresham D, Dolinski K, Botstein D, Kruglyak L. Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS ONE. 2007;2:e322. doi: 10.1371/journal.pone.0000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- Smith HE, Su SS, Neigeborn L, Driscoll SE, Mitchell AP. Role of IME1 expression in regulation of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6103–6113. doi: 10.1128/mcb.10.12.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Dombrowski PG, Fingerman E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Botstein D. Ordered Linear Tetrads Are Produced by the Sporulation of Newly Formed Zygotes of Saccharomyces cerevisiae. Genetics. 1987;115:229–232. doi: 10.1093/genetics/115.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mulders SE, et al. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 2009;9:178–190. doi: 10.1111/j.1567-1364.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- Vopalenska I, Hulkova M, Janderova B, Palkova Z. The morphology of Saccharomyces cerevisiae colonies is affected by cell adhesion and the budding pattern. Res Microbiol. 2005;156:921–931. doi: 10.1016/j.resmic.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Invasive growth of laboratory and wild strains of budding yeast as measured by the agar wash assay. Colonies of SK1 (SH561), Σ1278b, W303, a flo11Δ derivative of SK1 (SH4325) and an isolate of wild S. cerevisiae and wild S. paradoxus were grown for 8 days on YNA medium, the plates photographed (left column), washed extensively, and then rephotographed (see Methods). The percentages indicated to the right of the images represent the percent retention calculated as (colonies retaining cells after washing) × 100%/(total colonies). Percentages are the average of at least three independent plates ± SEM.
Fig. S2 Serial sections of invasive region of SK1 Adjacent serial sections of the same colony were identified because of similar patterns of cells in the section. (A–D) Four serial sections from the invasive region of an SK1 colony. Arrows indicate asci in pwhich exactly two spores are visible in at least one serial section and only one spore is visible in at least one other section (or both spores are much weaker). Arrowhead indicates an ascus in which two spores are visible in one section (C) and three spores are visible in two other sections (A & B).