SUMMARY
Type I interferons (IFNs) are secreted cytokines that orchestrate diverse immune responses to infection. Although typically considered to be most important in the response to viruses, type I IFNs are also induced by most, if not all, bacterial pathogens. Although diverse mechanisms have been described, bacterial induction of type I IFNs occurs upon stimulation of two main pathways: (1) Toll-like receptor (TLR) recognition of bacterial molecules such as lipopolysaccharide (LPS); (2) TLR-independent recognition of molecules delivered to the host cell cytosol. Cytosolic responses can be activated by two general mechanisms. First, viable bacteria can secrete stimulatory ligands into the cytosol via specialized bacterial secretion systems. Second, ligands can be released from bacteria that lyse or are degraded. The bacterial ligands that induce the cytosolic pathways remains uncertain in many cases, but appear to include various nucleic acids. In this review, we discuss recent advances in our understanding of how bacteria induce type I interferons and the roles type I IFNs play in host immunity.
INTRODUCTION
Type I interferons (IFNs) are secreted cytokines that include a single IFNβ protein, as well as numerous IFNα and other IFN family members (Decker et al., 2005). All type I IFNs signal via a heterodimeric receptor (IFNAR) and act locally and systemically to coordinate diverse responses to infection. An important local effect of type I IFN is the induction of the “anti-viral state”, which involves expression of host genes that interfere with viral replication (Zuniga et al., 2007). Some genes induced by type I IFN also exhibit anti-bacterial activity, such as the p47 GTPases (Taylor et al., 2004). Type I IFN can also sensitize host cells to apoptosis, which is thought to counteract the ability of viruses or bacteria to utilize the host’s intracellular niche for replication. In addition to local responses, type I IFN functions systemically, for example to activate Natural Killer and CD8+ T cell cytotoxicity, or to induce the upregulation of genes required for antigen presentation and activation of adaptive immunity.
The ability to produce type I IFN appears to be a universal property of all cells in the body, but the proximal pathogen-sensing receptors and signaling mechanisms leading to type I IFN induction differ significantly depending on the stimulatory ligand and responding cell type. Despite their diversity, the signaling pathways leading to induction of type I IFN do converge upon some common downstream elements, including the ubiquitin ligase TRAF3 and transcription factors such as IRF3 and IRF7. Once activated by phosphorylation in the cytosol, the IRFs enter the nucleus and assemble with NF-κB and other transcription factors on the IFNβ promoter in a complex (Panne et al., 2007) that activates extremely robust (e.g., 1000-fold) transcriptional induction of the Ifnb gene. In this review, we discuss recent advances in our understanding of the type I IFN host response to bacteria.
INDUCTION OF TYPE I INTERFERONS BY BACTERIA
TLR-dependent pathways
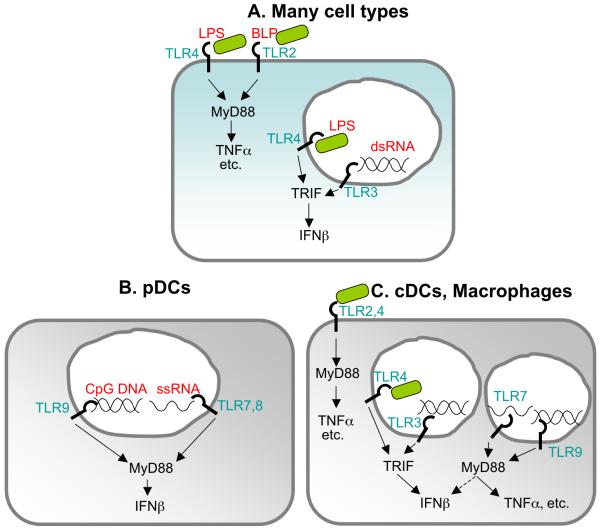
The Toll-like receptors (TLRs) are a family of cell surface or endosome localized receptors that recognize a variety of conserved microbial molecules (Kumar et al., 2009). TLR2, 3, 4, 5, 7, 8 and 9 are the primary TLRs that are potentially able to recognize bacterial products, and with the exception of TLR5, all have been linked to the induction of type I IFN. Interestingly, the mechanism of IFN induction by these TLRs varies considerably, and in many cases, TLR signaling only results in IFN induction in dedicated cell types (Figure 1).
Figure 1. TLR Receptors and Ligands.
A. TLR2 and 4 recognize bacterial lipoprotein (BLP) and lipopolysaccharide (LPS), respectively, and signal from the cell surface via the adaptor MyD88 to activate proinflammatory cytokines including TNFα and IL-6. TLR3 recognizes double stranded RNA (dsRNA) and signals via the adaptor TRIF from an intracellular compartment to induce IFNβ. Upon endocytosis, TLR4 can also signal via TRIF to induce type I IFN. B. Plasmacytoid dendritic cells primarily express nucleic acid sensing TLRs, which are localized to intracellular compartments. pDCs produce vast amounts of IFNβ upon stimulation via a MyD88-dependent pathway. C. Conventional dendritic cells and macrophages express many TLRs. However, only cDCs have been reported to induce IFNβ via TLR7 and 9. In macrophages, TLR7 and 9 signaling induces proinflammatory cytokines such as TNFα.
Type I IFN induction by TLR4
The best-characterized mechanism by which bacteria induce type I IFN is via TLR4, a cell-surface localized receptor that recognizes the lipid A moiety of lipopolysaccharide (LPS) from the outer membrane of gram-negative bacteria. TLR4 may recognize other bacterial ligands (Ashkar et al., 2008, Mossman et al., 2008, Thanawastien et al., 2009). TLR4 signals via two cytosolic adaptor proteins, MyD88 and TRIF, which are recruited sequentially to the cytoplasmic tail of TLR4 (Kagan et al., 2008). TRIF, but not MyD88, is required for induction of type I IFN by TLR4. TLR4 signaling induces type I IFN in many cell types and this broad capacity to induce type I IFN is shared by TLR3, which recognizes double-stranded RNA and is the only other TLR that utilizes TRIF for its downstream signaling (Kumar et al., 2009). However, there are few examples of TLR3-dependent recognition of bacteria. The other TLRs that induce type I IFN do so only in specialized cell types such plasmacytoid dendritic cells (pDCs) and conventional dendritic cells (cDCs) in a MyD88-dependent pathway (Figure 1). It is not clear why TLR4 would have evolved the unique capacity to stimulate type I IFN in many cell types in response to LPS. As discussed below, it does not appear that type I IFN is particularly critical for defense against gram-negative bacteria.
The microbes that are best recognized by TLR4 tend to be gram-negative commensals that reside on mucosal surfaces, such as E. coli in the gut, or closely related pathogenic genera, such as Salmonella. These microbes tend to produce hexaacylated lipid A that is the optimal ligand for TLR4. There is speculation that some mucosal bacteria may benefit by producing LPS that is recognized by TLR4 (Munford et al., 2006), but how they might benefit is not yet clear. Despite a widespread portrayal of TLR ligands as highly conserved across diverse bacterial species, gram-negative bacteria produce a tremendous variety of LPS molecules, many of which are poor ligands for TLR4. For example, the LPS of many human pathogens, including Legionella pneumophila, Helicobacter pylori, Francisella tularensis, Coxiella burnetii, and Brucella abortus, is poorly detected by TLR4 (Munford et al., 2006). In the case of Yersinia pestis, production of a specific LPS that evades TLR4 recognition is essential for virulence (Montminy et al., 2006). In other cases, it remains unclear if evasion of TLR4 is critical for virulence. Indeed, as discussed below, most TLR4-evasive gram-negative bacteria still induce type I IFNs via TLR-independent pathways.
Type I IFN induction by other TLRs
In contrast to TLR4, which is localized to the cell surface, the other TLRs that stimulate type I IFN (i.e., TLR3, 7, 8 and 9) localize to intracellular compartments (Figure 1). TLR2 was recently reported to induce type I IFN, but only in inflammatory monocytes and selectively in response to viral, not bacterial, ligands (Barbalat et al., 2009). TLR7, and its paralog TLR8, recognize single-stranded RNA ligands, whereas TLR9 recognizes DNA containing unmethylated CpG motifs. These TLRs signal exclusively via MyD88 and can stimulate the production of inflammatory cytokines such as TNFα in many cell types, but exhibit the ability to induce type I IFNs only in specialized cell types, most notably pDCs. Although pDCs are not numerous, they are capable of producing vast amounts of type I IFN on a per cell basis, and are important for inducing systemic levels of type I IFN in the response to viruses. However, the role of pDCs in bacterial infections has not been extensively investigated. In fact, although DNA from many bacterial species contains the unmethylated CpG motifs that can be recognized by TLR9, there is remarkably little evidence that TLR9 participates significantly in the response to bacterial infections in any cell type. A recent study provided surprising evidence for cell-type specific TLR-dependent IFN responses to bacteria. In this study, cDCs, but not macrophages or pDCs, were shown to produce type I IFN in a TLR7-MyD88-IRF1-dependent manner via phagolysosomal degradation of group A and B Streptococcus (GAS, GBS) (Mancuso et al., 2009). Previously, in response to viruses, TLR7 was thought to induce type I IFN primarily in pDCs, not cDCs. Thus, the results of Mancuso et al. may describe a bacterial-specific TLR-dependent pathway for induction of type I IFN.
Cytosolic pathways that induce type I IFN
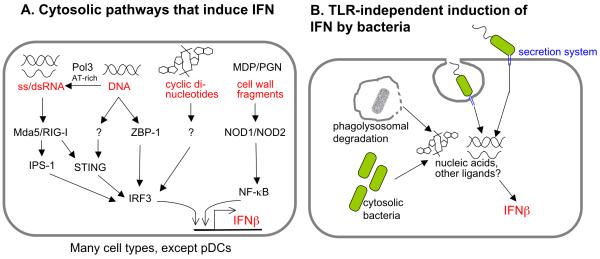
In addition to surface- or endosome-localized TLRs, host cells also express several cytosolic sensors that induce type I IFN in response to nucleic acid ligands, such as RNA, DNA, and cyclic-di-GMP (Figure 2A). The mechanisms by which bacteria stimulate cytosolic sensors are under investigation. As discussed below, one current model is that nucleic acids can be released from lysed bacteria. Additionally, bacterial secretion systems may leak or secrete nucleic acid ligands during infection (Figure 2B).
Figure 2. Bacterial Induction of Type I IFN via Cytosolic Receptors and Ligands.
A. In many cell types, except pDCs, cytosolic IFN-inducing receptors are expressed that sense nucleic acids, including RNA, DNA, and cyclic-di-nucleotides. AT-rich DNA can be transcribed by RNA polymerase III to generate ligands for the RNA-sensing pathway. Other sensors for DNA and c-di-nucleotides appear to exist, but remain to be identified. Cytosolic pathways that recognize bacterial cell wall fragments can synergize with nucleic acid sensing pathways to induce IFNβ. B. Extracellular or phagosomal bacteria utilizing secretion systems can leak or secrete nucleic acids that are sensed via cytosolic pathways outlined in A. Bacteria replicating in the cytosol activate type I IFN either by transport or lysis that releases IFN-inducing ligands. Degradation of phagocytosed bacteria can lead to IFN induction in many ways, and in some cases ligands generated in the phagolysosome access the cytosol via a pathway that remains to be elucidated.
Cytosolic RNA Sensing
RIG-I, MDA5, and LPG2 (collectively called RIG-I-like receptors or RLRs) are cytosolic receptors that bind directly to RNA and induce a type I IFN response to many RNA viruses (Wilkins et al., 2010) (Figure 2A). All three RLRs bind RNA via a DExD/H box-containing RNA helicase domain. RIG-I and MDA5 contain caspase-recruitment domains (CARDs), which are required for signaling through a downstream signaling adaptor, IPS-1 (also called MAVS, Cardif, VISA). IPS-1 appears to localize to the mitochondria where it serves as an essential adaptor for coordinating activation of IRF3/7, NF-κB, and MAP kinases (Yoneyama et al., 2009).
The precise role of LGP2 remains to be fully clarified, but it is clear that RIG-I and MDA5 function non-redundantly in viral recognition due to distinct specificities for different RNA structures. RIG-I preferentially recognizes 5′-triphosphate RNA, a motif modified in self RNAs by capping (mRNA) or removal (tRNA, rRNA) (Pichlmair et al., 2006, Hornung et al., 2006). RIG-I can also recognize short double-stranded synthetic RNAs, which may or may not contain a 5′ triphosphate (Kato et al., 2008, Takahasi et al., 2008, Schmidt et al., 2009, Schlee et al., 2009). In contrast, MDA5 ligands are less well characterized, but appear to include double-stranded RNAs greater than one kilobase in length that lack a 5′ triphosphate (Kato et al., 2008). While studies with synthetic or purified ligands have been informative as to what ligands can stimulate RLRs, not much is known about the physiological ligands that do activate RLRs during infection. One exception is a recent study showing that during influenza or Sendai virus infection, ssRNA viral genomes with 5′-triphosphates serve as the dominant RIG-I ligand, and RNAs from viral transcripts, replication-derived dsRNA intermediates, or processed self RNAs do not contribute (Rehwinkel et al., 2010). Since bacterial mRNAs are not capped and can contain 5′ triphosphates (Bieger et al., 1989), bacteria are potentially able to generate RNA ligands that can be recognized by RLRs, though there are few specific examples (see below).
Two recent reports suggest an unusual mechanism by which DNA could stimulate a cytosolic RNA sensor. In this mechanism, RNA polymerase III transcribes cytosolic DNA thereby generating RNA ligands for RIG-I (Ablasser et al., 2009, Chiu et al., 2009). Only highly AT-rich DNA appears to be a suitable template for Pol III. Epstein-Barr Virus, which produces Pol III-transcribed EBER RNAs, is an example of a pathogen sensed via the Pol III pathway (Ablasser et al., 2009, Chiu et al., 2009).
Cytosolic DNA Sensing
In addition to the Pol III-dependent pathway, both mouse and human cells express at least one additional cytosolic DNA-sensing pathway (Ablasser et al., 2009, Stetson et al., 2006). The identity of this DNA sensor remains uncertain, but it appears to be able to sense dsDNA from many sources without a requirement for specific sequence motifs. A candidate sensor, ZBP-1 (DLM-1/DAI), was reported to bind DNA and activate IRF3 to induce type I IFN (Takaoka et al., 2007, Wang et al., 2008). ZBP-1 appears to be important for the type I IFN response to human cytomegalovirus (HCMV) (DeFilippis et al., 2010). However, Zpb1 deficiency shows no discernable defect in IFN induction in response to other viral or bacterial infections (Monroe et al., 2009, Ishii et al., 2008, Lippmann et al., 2008) possibly because of redundancy with other DNA sensors.
STING (stimulator of interferon genes, also known as MITA, ERIS) was recently identified as a downstream signaling adaptor required for IFN induction in response to cytosolic DNA (Ishikawa et al., 2008, Zhong et al., 2008, Sun et al., 2009). In addition, STING appears to function as a signaling adaptor downstream of RIG-I, but not MDA5 (Figure 2A). Listeria monocytogenes and Chlamydia muridarum both require STING for type I IFN induction in vitro (Ishikawa et al., 2008, Prantner et al., 2010). Many other bacteria probably require STING for type I IFN induction since it appears to function in multiple cytosolic nucleic acid sensing pathways.
Cytosolic cyclic-di-GMP sensing
Cyclic-di-GMP is a bacterial second messenger produced by nearly all bacterial species, but not by mammalian cells, and therefore, could be a more specific target for innate immune recognition of bacteria than other nucleic acids. In fact, cyclic-di-GMP has been shown to exhibit immunostimulatory properties (Karaolis et al., 2007), including robust induction of a cytosolic pathway leading to type I IFN production (McWhirter et al., 2009). Induction of type I IFN by c-di-GMP requires IRF3, but is independent of known cytosolic sensors or TLRs (McWhirter et al., 2009). At present, the cytosolic DNA-sensing and c-di-GMP-sensing pathways are genetically indistinguishable, and both pathways may utilize the same unknown sensor, though this seems unlikely given the significant structural differences between c-di-GMP (a cyclic diribonucleotide) and IFN-inducing DNA (double-stranded deoxyribonucleotide polymer of ~40bp or more). There is no evidence that induction of type I IFN by any bacterial species requires c-di-GMP, though it is doubtful that innate sensing of c-di-GMP evolved by chance. One key question is how c-di-GMP would reach the host cell cytosol. C-di-GMP may be small enough to be transported (or leak) into host cells via specialized secretion systems that are essential for type I IFN induction by diverse bacterial species (see below, Figure 2B), though this remains to be established. Recent identification of diadenylate cyclase activity in a bacterium (Witte et al., 2008) led to speculation that c-di-AMP may also elicit a type I IFN response (McWhirter et al., 2009).
Other cytosolic pathways that affect IFN induction
There are few examples of non-nucleic acid molecules contributing to induction of type I IFN via cytosolic pathways. One example is the recognition of bacterial cell wall fragments, such as muramyl dipeptide (MDP), by the cytosolic sensors NOD1 and NOD2. NOD1 and NOD2 signal through the kinase RIP2, which leads to NF-κB activation (Park et al., 2007). Reports vary as to whether NOD signaling is sufficient for IFN-β induction. Although stimulation of NOD2 by MDP appears insufficient to induce type I IFN, stimulation with an N-glycolyl-modified form of MDP made by Mycobacterium tuberculosis was sufficient to induce significant type I IFN via NOD2 and the transcription factor IRF5 (Pandey et al., 2009). In response to viruses, ssRNA has been reported to induce IFNβ via NOD2 (Sabbah et al., 2009). However, in response to bacteria, NOD signaling most often appears to contribute to induction of type I IFNs primarily via NF-κB, which synergizes with other transcription factors, but alone is insufficient to induce IFNβ (Leber et al., 2008).
Induction of type I IFN via Bacterial Secretion Systems
Bacteria can stimulate cytosolic signaling pathways by various mechanisms. As outlined in the following examples, one common mechanism appears to involve delivery of bacterial ligands to the host cell cytosol via a variety of specialized secretion systems. Secretion systems are commonly employed by bacterial pathogens to deliver effector proteins to the cell cytosol from a phagosome or extracellular location. Although translocated effectors allow pathogens to manipulate their hosts, molecules delivered to the host cell cytosol can become targets of innate immune recognition.
Legionella
Few bacteria have been shown to induce type I IFN via the RNA sensing pathway involving MDA5, RIGI or IPS-1. One exception is Legionella pneumophila, a gram-negative pathogen that replicates in macrophages by employing a type IV secretion system (T4SS) to translocate effectors into the macrophage cytosol and orchestrate the creation of its replicative vacuole (Isberg et al., 2009). Interestingly, induction of type I IFN by L. pneumophila requires its type IV secretion system but not TLRs (Stetson et al., 2006), suggesting that IFN is induced upon cytosolic recognition of a translocated L. pneumophila molecule. In human epithelial-like A549 cells, knockdown of IPS-1, the signaling adaptor for RIG-I and MDA5, reduced the induction of type I IFN by L. pneumophila (Opitz et al., 2006). Two other studies demonstrated that mouse bone marrow macrophages carrying a targeted deletion or an shRNA to knockdown Ips-1, Rig-i or Mda5 were partially defective in IFN induction in response to L. pneumophila (Chiu et al., 2009, Monroe et al., 2009). Despite agreement that an RNA-sensing pathway can respond to L. pneumophila, there is uncertainty over the underlying molecular mechanism. Chiu et al. favor a model in which L. pneumophila translocates DNA into host cells, leading to production of RNA ligands via Pol III transcription. This model is consistent with the ability of the L. pneumophila T4SS to conjugate DNA plasmids to recipient bacteria (Vogel et al., 1998), but no there is no direct evidence DNA translocation occurs during infection. Monroe et al. demonstrated that transfection of macrophages with L. pneumophila RNA, but not DNA, induced Rig-i-dependent type I IFN in macrophages. It remains unclear whether L. pneumophila RNA, DNA, or perhaps another IFN-inducing molecule, is the physiological ligand translocated through the T4SS. Helicobacter pylori is another gram-negative pathogen with a T4SS that may stimulate an IFN response in host cells via a mechanism similar to that of L. pneumophila (Rad et al., 2009).
Listeria
The gram-positive bacterium Listeria monocytogenes employs a pore-forming toxin, listeriolysin O (LLO), to disrupt the phagosomal membrane and escape into the cell cytosol where it replicates (Portnoy et al., 1988). LLO-deficient Listeria are trapped in a vacuole and induce a MyD88-dependent response, but do not induce type I IFN, whereas wildtype Listeria that reach the cytosol induce a distinct, non-overlapping IRF3-dependent type I IFN response (Leber et al., 2008, O’Riordan et al., 2002). Induction of type I IFN by Listeria requires IRF3 but is independent of TLRs and the cytosolic RNA-sensing pathway. An unbiased genetic screen identified a role for multidrug resistant transporters in the induction of the cytosolic IFN response to Listeria (Crimmins et al., 2008). MDRs selectively transport small molecules rather than large RNA/DNA polymers, so these data suggest a model in which MDRs transport a small molecule that is sensed by a cytosolic immunosurveillance pathway.
Francisella
Francisella tularensis is a gram-negative bacterium that is the causative agent of tularemia. F. tularensis utilizes a type VI secretion system, encoded within the Francisella pathogencitiy island (FPI), to escape into the macrophage cytosol where it replicates. The FPI is also required for induction of type I IFN, via a pathway that requires IRF3, but is independent of TLRs or the cytosolic RNA sensors (RIG-I, MDA5) (Henry et al., 2007). It is possible that Francisella either secretes an IFN-inducing ligand, or leaks immunostimulatory DNA after lysis in the cytosol. It has also been suggested that nucleic acids from phagosomally degraded Francisella are released into the cytosol upon disruption of the phagosomal membrane via the T6SS (Fernandes-Alnemri et al., 2010). Francisella DNA that reaches the cytosol activates IRF3-dependent type I IFN signaling, which is critical for activation of the DNA-sensing AIM2 inflammasome (Fernandes-Alnemri et al., 2010, Rathinam et al., 2010).
Yersinia
LPS from the gram-negative genus Yersinia can serve as a potent ligand for TLR4, but in addition, a recent report identified a TLR-independent type I IFN response to extracellular Yersinia expressing a functional T3SS (Auerbuch et al., 2009). The TLR-independent response to Yersinia occurs in the absence of known translocated effectors, yet requires the pore-forming proteins YopB or YopD (Auerbuch et al., 2009). These data are consistent with a model in which a bacterial molecule reaches the host cytosol in a T3SS-dependent manner and stimulates a cytosolic pathway leading to IFN induction. Neither the stimulatory bacterial molecule nor the cytosolic sensor or host factors mediating this response have been identified.
Mycobacterium
Mycobacterium tuberculosis resides in a membrane bound compartment within infected host cells and gains access to the cytosol via a type VII secretion system (T7SS, formerly known as ESX-1). Like other pathogens discussed above, M. tuberculosis relies on its secretion system for virulence, and in addition, the secretion system is required for type I IFN induction in vitro and in vivo (Stanley et al., 2007). Despite conflicting reports as to which host pathways are required, there is agreement that TLR signaling is not required (Stanley et al., 2007, Pandey et al., 2009). Leber et al. and Pandey et al. found a partial requirement for NOD2, whereas Stanley et al. found no requirement for RIP2 (a kinase downstream of NOD2) in IFNβ induction. As previously mentioned, NOD2 has been proposed to induce type I IFN in response to N-glycolyl-MDP from M. tubercuolosis (Pandey et al., 2009). Taken together, it appears that M. tuberculosis induces type I IFN by delivery of nucleic acids and/or cell wall fragments to the cytosol, but it remains unclear whether the T7SS translocates these molecules, or simply permeabilizes the phagosomal membrane, allowing for leakage of bacterial molecules to the cytosol.
Induction of type I IFN by ligands released by bacteria degraded in the phagosome
As discussed above, it is thought that viable bacteria induce type I IFNs by secretion of molecules into host cells. However, there are several reports of bacteria that induce type I IFN upon degradation by innate immune cells. Degraded or lysed bacteria that remain confined in a phagosome can activate TLRs, as exemplified by TLR7 recognition of Streptococcus in cDCs (see above; (Mancuso et al., 2009)). Different cell types appear to vary in their degradative capacity. For example, it was found that cDCs and macrophages, but not pDCs, generate nucleic acid ligands for TLR7 and 9 upon bacterial infection, alternatively, pDCs may have a reduced ability to phagocytose bacteria (Mancuso et al., 2009). In addition, there are several examples of bacteria that activate cytosolic IFN-inducing pathways once degraded in a phagosome. In macrophages, Group B Streptococcus was shown to induce TLR-independent type I IFN in manner requiring degradation of phagolysosomal bacteria and disruption of the phagosomal membrane by pore-forming toxins (Charrel-Dennis et al., 2008). Cytosolic recognition of GBS required IRF3, but not IPS-1, RIP2 or ZBP-1. The data presented were consistent with a model in which liberated bacterial genomic DNA activates an unknown cytosolic DNA sensor (Charrel-Dennis et al., 2008). Studies have also suggested that live Borrelia burgdorferi induces type I IFN by a TLR-independent mechanism likely involving degradation of bacteria in the phagosome (Miller et al., 2008, Salazar et al., 2009).
A similar mechanism was previously found to be relevant in type II IFN (IFNγ)-activated macrophages infected with Listeria (Herskovits et al., 2007). IFNγ pretreatment of macrophages mimics conditions expected to exist in vivo after the innate immune response has already been initiated. In contrast to naïve macrophages, IFN-γ-activated macrophages are able to produce type I IFN during infection with Listeria deficient in hemolysin (LLO), a pore-forming toxin required for bacterial entry into the cytosol. Induction of type I IFN seemed to result from rapid phagosomal degradation of LLO-deficient Listeria and subsequent release of ligands into the cytosol that signal, in part, through NOD2 and IRF3 (Herskovits et al., 2007). The involvement of NOD2 suggested PGN fragments were being delivered to the cytosol, potentiating IFN induction by activating NF-κB (Leber et al., 2008). In this case, the primary IFN-inducing signal could be nucleic acid or another ligand, released from degraded bacteria. Lysozyme-sensitive mutants of Listeria that were rapidly degraded in naïve macrophages were found to induce type I IFNs, but this induction was unexpectedly found to be entirely TLR2-dependent and only partially MyD88-dependent (Boneca et al., 2007), which is difficult to reconcile with the existing literature. An additional unresolved issue is how ligands generated by phagosomally degraded Listeria reach the host cell cytosol. Nevertheless, it has become clear that many extracellular and intracellular vacuolar bacteria induce a cytosolic type I IFN response. Type I IFN induction by phagosomal degradation of bacteria may or may not be independent of bacterial secretion systems, and leads to the release of ligands capable of activating cytosolic IFN-inducing pathways.
THE FUNCTION OF TYPE I INTERFERONS IN THE HOST RESPONSE TO BACTERIA
Although type I IFNs are well known to induce a robust antiviral host response, the role of type I IFNs in response to bacterial infection is variable, and is even sometimes detrimental to the host. For example, type I IFN plays an important role in mediating the pathology of LPS-induced toxic shock (Karaghiosoff et al., 2003). A bigger surprise has been several studies demonstrating that type I IFN can actually impair bacterial clearance. For example, Ifnar-deficient mice exhibit lower Listeria monocytogenes burdens in the liver and spleen, as compared to wild type mice (Auerbuch et al., 2004, O’Connell et al., 2004, Carrero et al., 2004). Type I IFN signaling is also detrimental to the clearance of Mycobacterium tubercuolosis from the spleen (Stanley et al., 2007) and the lung during infection with various Mtb strains (Ordway et al., 2007). Furthermore, type I IFN impairs clearance of Chlamydia from the genital tract and lungs (Nagarajan et al., 2008, Qiu et al., 2008), and is detrimental to host survival during infection with Francisella tularensis (Henry et al., 2010). The in vivo mechanisms by which type I IFN signaling increases host susceptibility to bacterial infection remain uncertain. One suggestion is that abundant type I IFN predisposes lymphocytes to apoptosis, resulting in suppression of innate responses via increased IL-10 (Carrero et al., 2006). The observation that type I IFN stimulates production of IL-27, a cytokine that strongly suppresses IL-17A production (Guo et al., 2008), hints at another mechanism (Henry et al., 2010). IL-17A is a cytokine produced by γδT cells that appears to play an important role in restricting Listeria replication by orchestrating neutrophil responses (Hamada et al., 2008, Meeks et al., 2009). In fact, Ifnar-deficient mice induce more IL-17A in response to Francisella and Listeria (Henry et al., 2010). Therefore, one way type I IFN signaling could result in increased host susceptibility is by suppressing IL-17 responses, which are necessary for neutrophil-mediated bacterial clearance. Another report shows that crosstalk between cytokine signaling pathways can reduce the host’s ability to mount an appropriate innate immune response. Induction of type I IFN by Listeria was shown to suppress macrophage activation by reducing the ability to respond to IFNγ, a critical cytokine for resistance to Listeria (Rayamajhi et al., 2010).
Given that type I IFN appears to be a universal host response to bacterial infection, it would be surprising if type I IFN never played a role in host protection. In fact, type I IFN is crucial for host resistance to some bacterial infections. For example, Ifnar-deficient mice exhibit decreased survival and increased bacterial burdens upon infection with Group B Streptococcus, Streptococcus pneumoniae, and E. coli (Mancuso et al., 2007). The susceptibility of Ifnar-deficient mice to these infections correlated with reduced cytokine production such as TNFα and IFNγ. Type I IFN also plays a role in restricting L. pneumophila replication in macrophages (Coers et al., 2007), but Ifnar−/− mice do not appear to exhibit increased susceptibility in vivo (Monroe et al., 2009), potentially due to redundancy. In response to F. tularensis, type I IFN signaling has also been observed to induce the expression of inflammasome components, a molecular signaling complex involved in interleukin-1β and IL-18 production (Henry et al., 2007). Thus, in the context of bacterial infection, type I IFN appears to modulate a broad range of pro- and anti-inflammatory effects. The mechanisms by which the immunomodulatory effects of type I IFNs are regulated are only beginning to be understood (Rothlin et al., 2007).
The large number of recent studies on the induction and function of type I IFNs in response to bacterial infections has led to an increasing appreciation for the complexity of this family of cytokines. Given that most, if not all, bacteria induce type I IFNs, via multiple pathways, it is clearly too simplistic to fall back on the old notion that type I IFNs are primarily ‘antiviral’ cytokines. On the other hand, in the context of bacterial infections, it is difficult to provide a simple statement of the function of type I IFNs. It appears, instead, that in response to bacteria, types I IFNs serve a variety of beneficial and detrimental immune functions, many of which remain to be fully understood.
Acknowledgements
K.M.M. is supported by an NSF Graduate Student Fellowship. R.E.V. is a recipient of fellowships from the Cancer Research Institute and the Burroughs Wellcome Fund. We also acknowledge the support of NIH grants AI080749 and AI082357.
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009 doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkar AA, Mossman KL, Coombes BK, Gyles CL, Mackenzie R. FimH adhesin of type 1 fimbriae is a potent inducer of innate antimicrobial responses which requires TLR4 and type 1 interferon signalling. PLoS Pathog. 2008;4:e1000233. doi: 10.1371/journal.ppat.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbuch V, Golenbock DT, Isberg RR. Innate immune recognition of Yersinia pseudotuberculosis type III secretion. PLoS Pathog. 2009;5:e1000686. doi: 10.1371/journal.ppat.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol. 2009;10:1200–1207. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieger CD, Nierlich DP. Distribution of 5′-triphosphate termini on the mRNA of Escherichia coli. J Bacteriol. 1989;171:141–147. doi: 10.1128/jb.171.1.141-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci U S A. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J Exp Med. 2004;200:535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero JA, Unanue ER. Lymphocyte apoptosis as an immune subversion strategy of microbial pathogens. Trends Immunol. 2006;27:497–503. doi: 10.1016/j.it.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Charrel-Dennis M, Latz E, Halmen KA, Trieu-Cuot P, Fitzgerald KA, Kasper DL, Golenbock DT. TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell Host Microbe. 2008;4:543–554. doi: 10.1016/j.chom.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA Polymerase III Detects Cytosolic DNA and Induces Type I Interferons through the RIG-I Pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J, Vance RE, Fontana MF, Dietrich WF. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol. 2007;9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- Crimmins GT, Herskovits AA, Rehder K, Sivick KE, Lauer P, Dubensky TW, Jr., Portnoy DA. Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc Natl Acad Sci U S A. 2008;105:10191–10196. doi: 10.1073/pnas.0804170105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T, Muller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- DeFilippis VR, Alvarado D, Sali T, Rothenburg S, Fruh K. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J Virol. 2010;84:585–598. doi: 10.1128/JVI.01748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010 doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Chang EY, Cheng G. The type I IFN induction pathway constrains Th17-mediated autoimmune inflammation in mice. J Clin Invest. 2008;118:1680–1690. doi: 10.1172/JCI33342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Umemura M, Shiono T, Tanaka K, Yahagi A, Begum MD, et al. IL-17A produced by gammadelta T cells plays a critical role in innate immunity against listeria monocytogenes infection in the liver. J Immunol. 2008;181:3456–3463. doi: 10.4049/jimmunol.181.5.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Kirimanjeswara GS, Ruby T, Jones JW, Peng K, Perret M, et al. Type I IFN Signaling Constrains IL-17A/F Secretion by {gamma}{delta} T cells during Bacterial Infections. J Immunol. 2010 doi: 10.4049/jimmunol.0902065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits AA, Auerbuch V, Portnoy DA. Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog. 2007;3:e51. doi: 10.1371/journal.ppat.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaghiosoff M, Steinborn R, Kovarik P, Kriegshauser G, Baccarini M, Donabauer B, et al. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- Karaolis DK, Means TK, Yang D, Takahashi M, Yoshimura T, Muraille E, et al. Bacterial c-di-GMP is an immunostimulatory molecule. J Immunol. 2007;178:2171–2181. doi: 10.4049/jimmunol.178.4.2171. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- Leber JH, Crimmins GT, Raghavan S, Meyer-Morse N, Cox JS, Portnoy DA. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann J, Rothenburg S, Deigendesch N, Eitel J, Meixenberger K, van Laak V, et al. IFNbeta responses induced by intracellular bacteria or cytosolic DNA in different human cells do not require ZBP1 (DLM-1/DAI) Cell Microbiol. 2008;10:2579–2588. doi: 10.1111/j.1462-5822.2008.01232.x. [DOI] [PubMed] [Google Scholar]
- Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, Akira S, et al. Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat Immunol. 2009;10:587–594. doi: 10.1038/ni.1733. [DOI] [PubMed] [Google Scholar]
- Mancuso G, Midiri A, Biondo C, Beninati C, Zummo S, Galbo R, et al. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J Immunol. 2007;178:3126–3133. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med. 2009;206:1899–1911. doi: 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks KD, Sieve AN, Kolls JK, Ghilardi N, Berg RE. IL-23 is required for protection against systemic infection with Listeria monocytogenes. J Immunol. 2009;183:8026–8034. doi: 10.4049/jimmunol.0901588. [DOI] [PubMed] [Google Scholar]
- Miller JC, Ma Y, Bian J, Sheehan KC, Zachary JF, Weis JH, et al. A critical role for type I IFN in arthritis development following Borrelia burgdorferi infection of mice. J Immunol. 2008;181:8492–8503. doi: 10.4049/jimmunol.181.12.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe KM, McWhirter SM, Vance RE. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog. 2009;5:e1000665. doi: 10.1371/journal.ppat.1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montminy SW, Khan N, McGrath S, Walkowicz MJ, Sharp F, Conlon JE, et al. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat Immunol. 2006;7:1066–1073. doi: 10.1038/ni1386. [DOI] [PubMed] [Google Scholar]
- Mossman KL, Mian MF, Lauzon NM, Gyles CL, Lichty B, Mackenzie R, et al. Cutting edge: FimH adhesin of type 1 fimbriae is a novel TLR4 ligand. J Immunol. 2008;181:6702–6706. doi: 10.4049/jimmunol.181.10.6702. [DOI] [PubMed] [Google Scholar]
- Munford RS, Varley AW. Shield as signal: lipopolysaccharides and the evolution of immunity to gram-negative bacteria. PLoS Pathog. 2006;2:e67. doi: 10.1371/journal.ppat.0020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan UM, Prantner D, Sikes JD, Andrews CW, Jr., Goodwin AM, Nagarajan S, Darville T. Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect Immun. 2008;76:4642–4648. doi: 10.1128/IAI.00629-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan M, Yi CH, Gonzales R, Lee KD, Portnoy DA. Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc Natl Acad Sci U S A. 2002;99:13861–13866. doi: 10.1073/pnas.202476699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz B, Vinzing M, van Laak V, Schmeck B, Heine G, Gunther S, et al. Legionella pneumophila induced IFNbeta in lung epithelial cells via IPS-1 and IRF3 which also control bacterial replication. J Biol Chem. 2006;281:36173–36179. doi: 10.1074/jbc.M604638200. [DOI] [PubMed] [Google Scholar]
- Ordway D, Henao-Tamayo M, Harton M, Palanisamy G, Troudt J, Shanley C, et al. The hypervirulent Mycobacterium tuberculosis strain HN878 induces a potent TH1 response followed by rapid down-regulation. J Immunol. 2007;179:522–531. doi: 10.4049/jimmunol.179.1.522. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Yang Y, Jiang Z, Fortune SM, Coulombe F, Behr MA, et al. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Portnoy DA, Jacks PS, Hinrichs DJ. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prantner D, Darville T, Nagarajan UM. Stimulator of IFN Gene Is Critical for Induction of IFN-{beta} during Chlamydia muridarum Infection. J Immunol. 2010;184:2551–2560. doi: 10.4049/jimmunol.0903704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Fan Y, Joyee AG, Wang S, Han X, Bai H, et al. Type I IFNs enhance susceptibility to Chlamydia muridarum lung infection by enhancing apoptosis of local macrophages. J Immunol. 2008;181:2092–2102. doi: 10.4049/jimmunol.181.3.2092. [DOI] [PubMed] [Google Scholar]
- Rad R, Ballhorn W, Voland P, Eisenacher K, Mages J, Rad L, et al. Extracellular and intracellular pattern recognition receptors cooperate in the recognition of Helicobacter pylori. Gastroenterology. 2009;136:2247–2257. doi: 10.1053/j.gastro.2009.02.066. [DOI] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010 doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M, Humann J, Penheiter K, Andreasen K, Lenz LL. Induction of IFN-alphabeta enables Listeria monocytogenes to suppress macrophage activation by IFN-gamma. J Exp Med. 2010;207:327–337. doi: 10.1084/jem.20091746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB, Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131:1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, et al. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar JC, Duhnam-Ems S, La Vake C, Cruz AR, Moore MW, Caimano MJ, et al. Activation of human monocytes by live Borrelia burgdorferi generates TLR2-dependent and -independent responses which include induction of IFN-beta. PLoS Pathog. 2009;5:e1000444. doi: 10.1371/journal.ppat.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Taylor GA, Feng CG, Sher A. p47 GTPases: regulators of immunity to intracellular pathogens. Nat Rev Immunol. 2004;4:100–109. doi: 10.1038/nri1270. [DOI] [PubMed] [Google Scholar]
- Thanawastien A, Montor WR, Labaer J, Mekalanos JJ, Yoon SS. Vibrio cholerae proteome-wide screen for immunostimulatory proteins identifies phosphatidylserine decarboxylase as a novel Toll-like receptor 4 agonist. PLoS Pathog. 2009;5:e1000556. doi: 10.1371/journal.ppat.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, et al. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci U S A. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C, Gale M., Jr. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010 doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Zuniga EI, Hahm B, Oldstone MB. Type I interferon during viral infections: multiple triggers for a multifunctional mediator. Curr Top Microbiol Immunol. 2007;316:337–357. doi: 10.1007/978-3-540-71329-6_16. [DOI] [PubMed] [Google Scholar]