Abstract
Immunotherapy of colorectal carcinoma (CRC) has great promise as the presence of T lymphocytes in CRC tissues in situ is correlated with reduced recurrence and increased survival. Thus, identification of the antigens recognized by T cells of CRC patients may permit development of vaccines with potential benefit for these patients. Using expression cloning, we identified the antigen, nucleophosmin (Npm), recognized by an HLA-A1 restricted cytotoxic T lymphocyte (CTL) line derived from the peripheral blood mononuclear cells (PBMC) of a rectal cancer patient. A decamer peptide derived from the Npm sequence sensitized peptide-pulsed HLA-A1 positive cells to lysis by the CTL line. The peptide also induced proliferative and cytotoxic T lymphocytes in the PBMC of 4 of 6 CRC patients which lysed HLA-A1 positive peptide-pulsed target cells and CRC cells endogenously expressing Npm. Overexpression of Npm by tumors of various histological types, recognition of the antigen by T cells derived from different CRC patients, and association of the antigen with poor prognostic outcome make it a promising target for immunotherapeutic intervention in cancer patients.
Keywords: Colorectal carcinoma patients, Cytotoxic T cells, antigens, tumor immunity
Introduction
Despite advances in screening and treatment, colorectal cancer (CRC) remains the second leading cause of cancer-related deaths in the USA (American Cancer Society. Cancer Facts & Figures 2008. Atlanta: American Cancer Society; 2008). Therefore new improved treatments are needed. Immunotherapy of CRC has great promise as the presence of T lymphocytes in CRC tissues in situ is correlated with reduced recurrence and increased survival 1–3, suggesting a role of T lymphocytes in tumor rejection. Thus, identification of the antigens recognized by T cells of CRC patients may permit vaccine development. We have previously described an HLA-A1 restricted CTL (CTL007) derived from the peripheral blood mononuclear cells (PBMC) of a rectal carcinoma patient. The CTL recognized exclusively HLA-A1-positive CRC cell lines 4. In the present study, the antigen recognized by the CTL was identified as nucleophosmin (Npm). We show here that Npm is recognized by T cells derived from 4 of 6 CRC patients and expressed by all CRC, melanoma and breast carcinoma cell lines tested. Npm is overexpressed in CRC tissues as compared to normal colon. Npm is a nucleolar phosphoprotein, described as a multifunctional protein shuttling between the nucleus and cytoplasm (reviewed in 5, 6). The role of Npm in oncogenesis is controversial as both oncogenic and tumor suppressive functions have been attributed to Npm 5, 6. In some haematological malignancies translocation of Npm occurs 5, 7–9, and in several other solid tumors Npm is overexpressed. Expression of Npm by bladder cancer patients’ tumors has been associated with poor prognosis increasing the risk of tumor recurrence and progression 10. Thus, overexpression of Npm by tumors of various histological types, recognition of the antigen by T cells derived from different CRC patients and association of the antigen with poor prognostic outcome render it a promising target for immunotherapeutic intervention in cancer patients.
Materials and Methods
Cell lines, tissues and PBMC
CRC cell lines WC:007, 008, 013, and 020 were established in CRC medium 11. EBV–B 007 has been described 11. HT29 (CRC), K562 (erythroleukemia) and Daudi (Burkitt lymphoma) cell lines were obtained from American Type Culture Collection (Manassas, VA). Established breast cancer (ZR75-1, MDA-MB453) and melanoma (WM35, WM793) cell lines were described and maintained in RPMI 1640 medium containing 10% FBS, DMEM medium containing 10% FBS and MCDB153-L15 medium containing 2% FBS, respectively 12–14. Fetal fibroblast cell line FF2475 was maintained in DMEM medium with 10% FBS. HEK293 cells were obtained from Invitrogen (Carlsbad, CA) and maintained in DMEM with 10% FBS. CTL007 has been described and was grown in T cell medium 11. CRC, normal colon and breast tissues were obtained from cancer patients during surgery. PBMC were obtained from patients’ peripheral blood on the day of surgery or as late as 9 months after surgery (273649). All tissues and PBMC were obtained under informed consent and a protocol approved by the Institutional Review Boards of The Wistar Institute, Fox Chase Cancer Center and Virtua Memorial Hospital.
Antibodies
MAb 289HA-1 (IgM; anti-HLA-A1) was obtained from One Lambda (Canoga Park, CA), anti-HLA class II antibody (B33.1) from B. Perussia (Thomas Jefferson University, Philadelphia, PA), anti-interferon-γ antibodies from Endogen (Pierce Biotechnology, Rockford, IL), anti-CD4 and anti-CD8 antibodies from BD Pharmingen (San Diego, CA), anti-Npm mAb FC82291 from GeneTex (San Antonio, TX), anti-β-1 and anti-β-3 antibodies from BD Pharmingen, anti-actin antibody from Sigma (St. Louis, MO), normal mouse IgG from Jackson ImmunoResearch (West Grove, PA) and HRP-labeled anti-mouse-IgG antibody from MP Biomedicals (Irvine, CA). Anti-CRC mAb GA733 (IgG2a) has been described 15.
Cloning of HLA-A1 cDNA from WC007 cells
mRNA was isolated from WC007 cells using the FastTrack® 2.0 mRNA isolation kit (Invitrogen). HLA-A1 cDNA was cloned using specific primers (5’ primer: 5’-AAAACTCGAGATGGCCGTCATGGCGC-3’, 3’ primer: 5’-AAAAGAATTCACACTTTACAAGCTGTGAGAGA-3’).
Identification of the cDNA clone recognized by CTL007
The expression gene cloning method 16 was used to clone the antigen recognized by CTL007. In brief, a tumor cDNA library was synthesized using the Lambda ZAP-CMV XR library construction kit (Stratagene, La Jolla, CA) and five µg of polyA+ WC007 RNA. The resulting plasmid library was divided into pools of 100 clones per well. For library screening, 3×104 HEK293 cells per well were co-transfected with 100 ng HLA-A1 cDNA and 200 ng library pool cDNA (100 clones per well) using lipofectamine 2000 (Invitrogen). Twelve to 16 hours after transfection, 1×104 T cells were added per well. Supernatants were harvested after an additional 24–30 h to determine IFN-γ levels using an ELISA kit (Endogen).
Down-regulation of NPM RNA by shRNA
Npm protein levels were down-regulated using the MISSION® shRNA lentiviral transduction target set for nucleophosmin (NM_002520, Sigma-Aldrich, St. Louis, MI). WC007 cells were transduced in the presence of polybrene (Sigma-Aldrich) with lentiviral particles. Eleven days after transduction, cells were used as targets for CTL007. At the same time Npm protein levels were determined by standard western blot analysis using anti-Npm antibody (Gene-Tex). Anti-β-actin antibody (Sigma) was used as control.
DNA sequencing
DNA sequencing was performed by The Wistar Institute’s DNA sequencing facility. The amino acid sequence was deduced from the DNA sequence using ExPASy (http://us.expasy.org/tools/dna.html). DNA and deduced amino acid sequence comparisons were performed with the BLAST program (http://www.ncbi.nlm.nih.gov/blast ).
Peptide design
Potential HLA-A*01 binding epitopes were determined from the deduced amino acid sequence of the isolated cDNA clone using the Rammensee epitope prediction model 17 and were limited to epitopes with an HLA-A1 binding score ≥20. Selected peptides were synthesized and HPLC-purified by Invitrogen. The following peptides were used: IVEAEAMNY, with a binding score of 23 (peptide 1, Npm position 59–67); GCELKADKDY, with an HLA-A1 binding score of 24 (peptide 2, Npm position 20–29); KVEAKFINY, with a binding score of 25 (peptide 3, Npm position 263–271), and an Npm-unrelated HLA-A1-binding control peptide, SSTDSTFLY, with a binding score of 21.
T cell proliferation assay
The assay was done as previously described 18. In brief, PBMC were isolated by Ficoll-Hypaque density gradient centrifugation of heparinized blood. Adherent monocytes (5×104 per well) were pulsed for 8 hours with Poly (DL-lactide-co-glycolide)-peptide microspheres 19, excess peptides were removed and the peptide-pulsed monocytes were cocultured with CTL007 or PBMC (105 cells/well) in T cell medium. Proliferative lymphocyte responses were determined by 3H-thymidine incorporation assay on day 5. CD4 and CD8 expression by the proliferating lymphocytes was determined by FACS analysis. All determinations were done in triplicate.
Cytotoxicity Assays
CTL activity was tested in standard 4–18-h 51Cr release assays. In brief, tumor targets were labeled with 51Cr as described earlier 11, 20 , pulsed with peptide (25 µM) in the presence of β2-microglobulin (1µg/ml, Sigma) for 8h or left unpulsed and then co-cultured with T cells. At the end of incubation, supernatants were harvested and counted for 51Cr release. For target cells treated with Npm specific shRNA, a live/dead viability/cytotoxicity kit (Molecular Probes, Invitrogen) was used. In some experiments, CD4+ and CD8+ T cells were isolated using anti-CD4 or anti-CD8 Ab-coated magnetic beads (Invitrogen) before using them as effector cells in cytotoxity assays.
Determination of Npm protein levels
Cells were lysed in lysis buffer (300 mM NaCl, 50 mM Na-phosphate pH 8.0, 0.5% Triton X-100, 1 mM EDTA, and 1 mM PMSF) and protein concentration determined with a BCA protein assay kit (Pierce, Rockford, IL). Equal amounts of protein were loaded onto an 8–16% SDS-polyacrylamide gel (Life-Gels, Clarkston, GA) and after separation transferred onto a PVDF membrane (Bio-Rad, Hercules, CA). Western blots were probed using anti-Npm or anti-β-actin antibodies, followed by incubation with HRP-coupled goat anti-mouse IgG antibody. Signals were detected using chemoluminescence (Millipore, Billerica, MA). The NE-PERR nuclear and cytoplasmic extraction reagents (Thermo Scientific, Rockford, Il) were used to isolate nuclear and cytoplasmic proteins following the manufacturer's protocol.
Statistical analyses
Differences between experimental and control values were analyzed for significance by 2-sided Student’s t-test.
Results
Recognition of Npm by CTL007
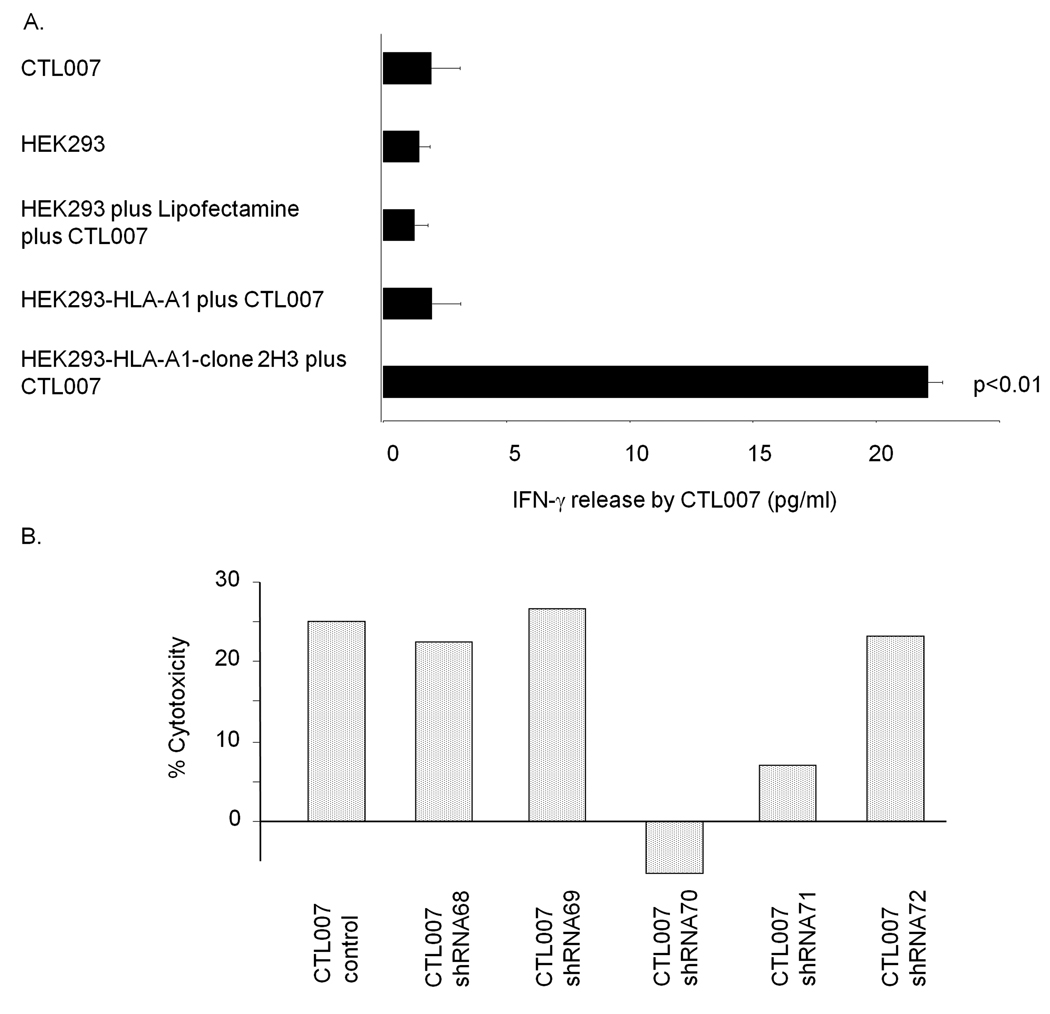
The HLA-A1 cDNA clone isolated from WC007 cells was 100% identical to the sequence published in Genbank (data not shown). This clone and a cDNA expression library from WC007 cells were transfected into HEK293 cells for initial screening of pools of 100 cDNA clones each for reactivity with CTL007. Positive pools were subdivided into smaller pools until a single clone (2H3) recognized by CTL007 was isolated. This clone had an insert size of 1260bp and the sequence was 100% identical to the published Npm sequence (Genbank gi:33876659, data not shown). Following transfection of the clone into HEK293 cells, it significantly (p<0.01) stimulated IFN-γ release in CTL007 (Fig.1A). Of the five available Npm-specific MISSION® shRNA lentiviral transduction particles, one (clone 70) reduced Npm protein levels in WC007 cells by >90% compared to mock transduced control cells (data not shown). Treatment of WC007 cells with this Npm shRNA completely abolished lysis of the cells by CTL007 (Fig. 1B). Thus, CTL007 recognizes Npm on CRC cells.
Fig. 1.
A. Stimulation of CTL007 by cDNA clone 2H3. HEK293 cells were treated as indicated and CTL007 added after 24 hours. IFN-γ release by HEK293 cells transfected with both HLA-A1 and 2H3 cDNA is statistically significantly higher (p<0.01) than the release by each of the control cells. Bars indicate mean IFN-γ release +/− S.D. of triplicate determinations. B. WC007 cells were stably transduced with Npm-specific shRNA lentiviral particles (#68–72) or mock transduced. Lysis of the target cells was determined in live/dead viability/cytotoxicity assay.
Epitope determination
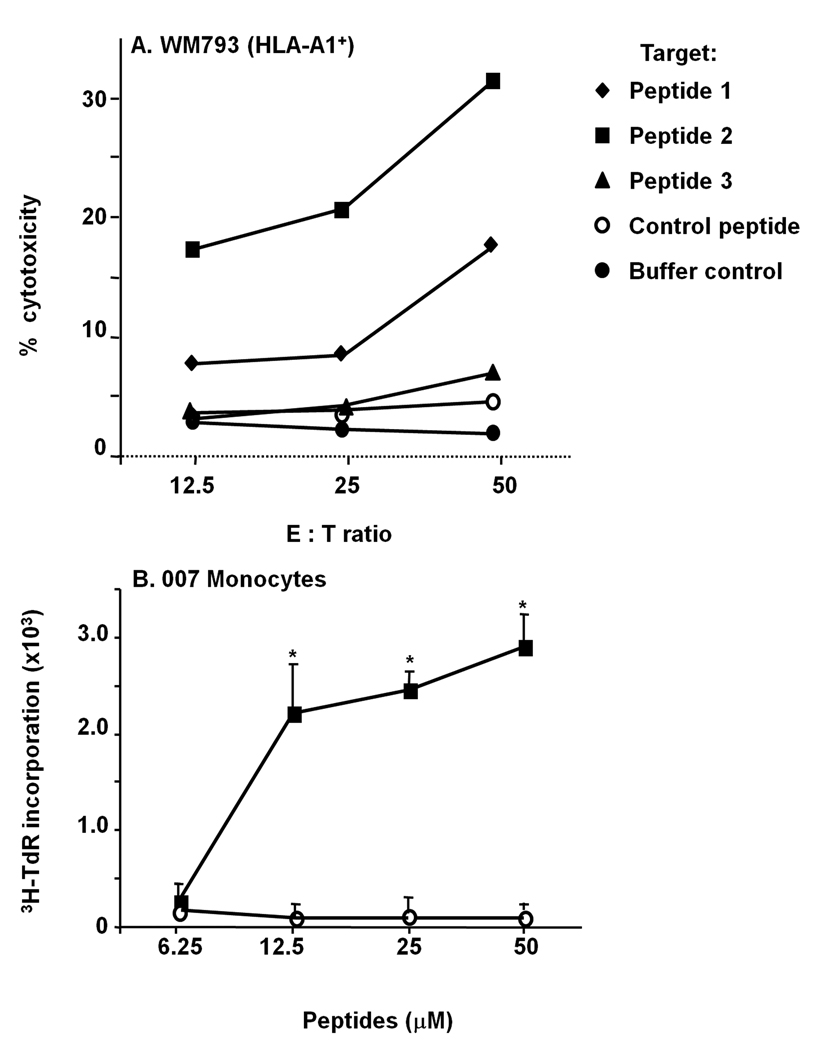
To determine the epitope recognized by CTL007, HLA-A1 positive WM793 melanoma cells, which were not lysed by the CTL, were pulsed with 3 Npm peptides predicted to bind to HLA-A1 and pulsed target cells were tested for lysis by the CTL. Peptide 2 (GCELKADKDY, position 20–29) showed the highest reactivity in this assay, whereas low lysis was seen with peptide 1. CTL007 did not significantly lyse WM793 cells pulsed with control peptide, peptide 3 or left unpulsed (Fig. 2A). Monocytes derived from patient 007 and pulsed with peptide 2 significantly and specifically stimulated CTL007 (Fig. 2B).
Fig. 2.
Recognition of peptide-pulsed WM793 cells or 007 monocytes by CTL007
A. HLA-A1-positive WM793 cells were pulsed with the different Npm or control peptides and incubated with CTL007. CTL activity of CTL007 was measured in standard 51Cr release assay. B. HLA-A1 positive 007 monocytes were pulsed with different concentrations of peptide 2 or control peptide and the proliferative response of CTL007 was measured. The proliferative response to stimulation with peptide 2-pulsed monocytes was statistically significant (p<0.01).
Recognition of Npm peptide 2 by PBMC of CRC patients
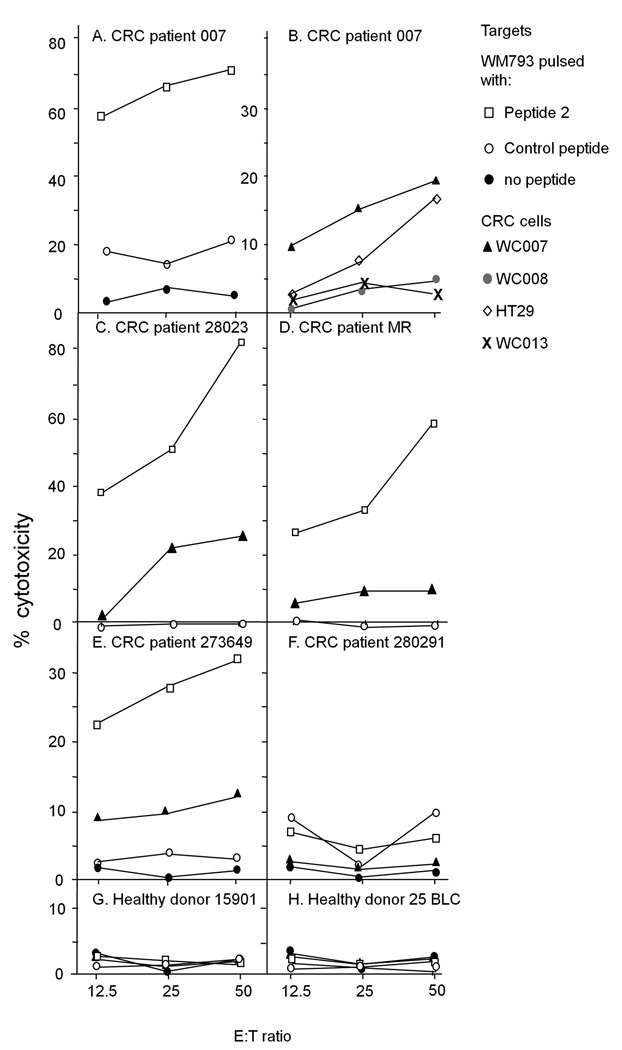
To determine whether Npm peptide 2 is recognized by CRC patients’ lymphocytes, PBMC from patient 007, five additional HLA-A1 positive CRC patients and two HLA-A1 positive healthy donors were stimulated twice with peptide 2 (25µM) or control peptide-pulsed autologous monocytes in vitro. Peptide 2-stimulated 007PBMC significantly lysed WM793 melanoma cells pulsed with peptide 2 as compared to control targets (Fig. 3A). Peptide 2-stimulated 007PBMC showed low, but reproducible (in 3 independent experiments) lysis of HLA-A1 positive WC007 and HT29 CRC cells and negligible lysis of HLA-A1 positive WC008 and WC013 CRC cells (Fig. 3B), replicating the behavior of CTL007 4. Control peptide-stimulated 007PBMC did not proliferate in culture after peptide stimulation excluding cytotoxicity assays with these cells.
Fig. 3.
Cytotoxic activity of peptide 2-stimulated PBMC of HLA-A1 positive CRC patients and healthy donors against different target cells. PBMC were stimulated twice with peptide 2-pulsed monocytes in vitro and tested for cytotoxic activity against different target cells in 51Cr release assay.
Of the five other HLA-A1 positive CRC patients, three patient’s lymphocytes (28023, MR and 273649) recognized specifically peptide 2-pulsed WM793 cells (Fig 3 C to E) and, two of them to a very low degree, also WC007 CRC cells, after stimulation of the PBMC with peptide 2, as demonstrated by specific lysis of the target cells (Fig. 3C and E). PBCMs of the HLA-A1 positive CRC patient 28029 did not react with peptide 2-pulsed WM793 or unpulsed WC007 cells (Fig 3 F) and PBMC of the last patient (NM) did not grow after in vitro stimulation with peptide-pulsed autologous monocytes and therefore were not available for the CTL assay. Control peptide-stimulated PBMC of all patients reacted poorly with peptide 2-pulsed WM793 target cells and WC007 CRC cells (not shown). PBMC of two HLA-A1-positive healthy donors did not significantly lyse peptide 2-pulsed WM793 or WC007 target cells after repeated in vitro stimulation with peptide 2 (Fig. 3G and H).
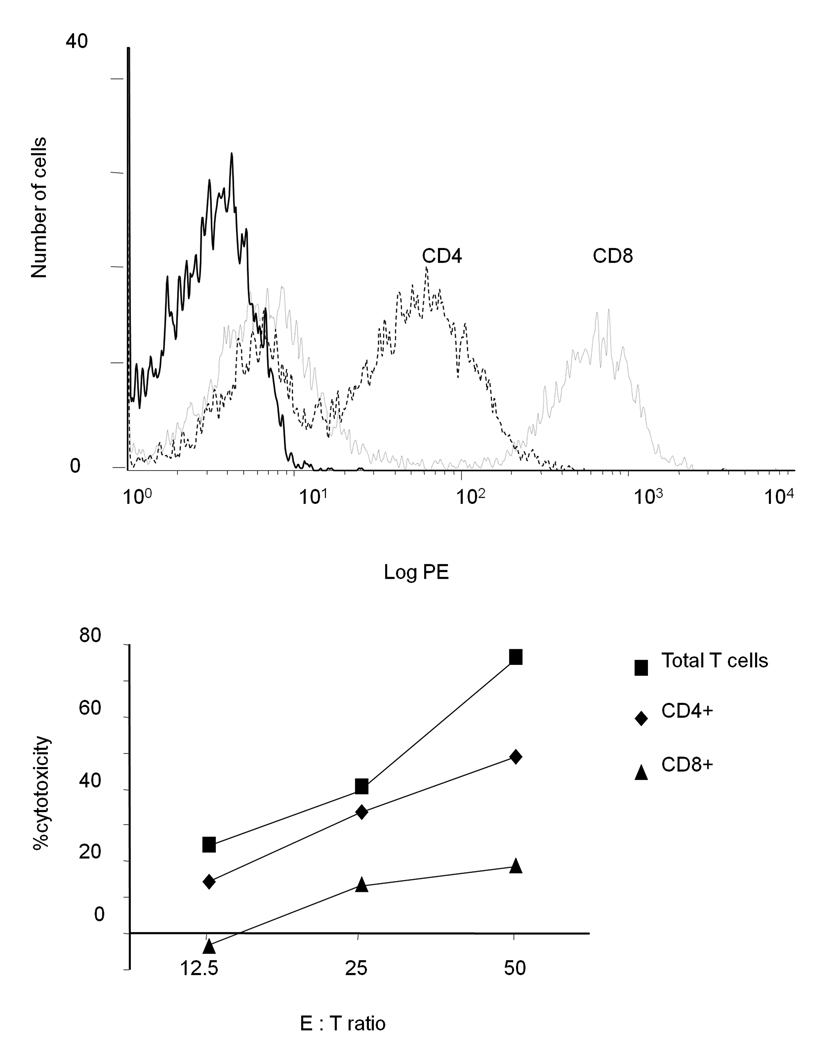
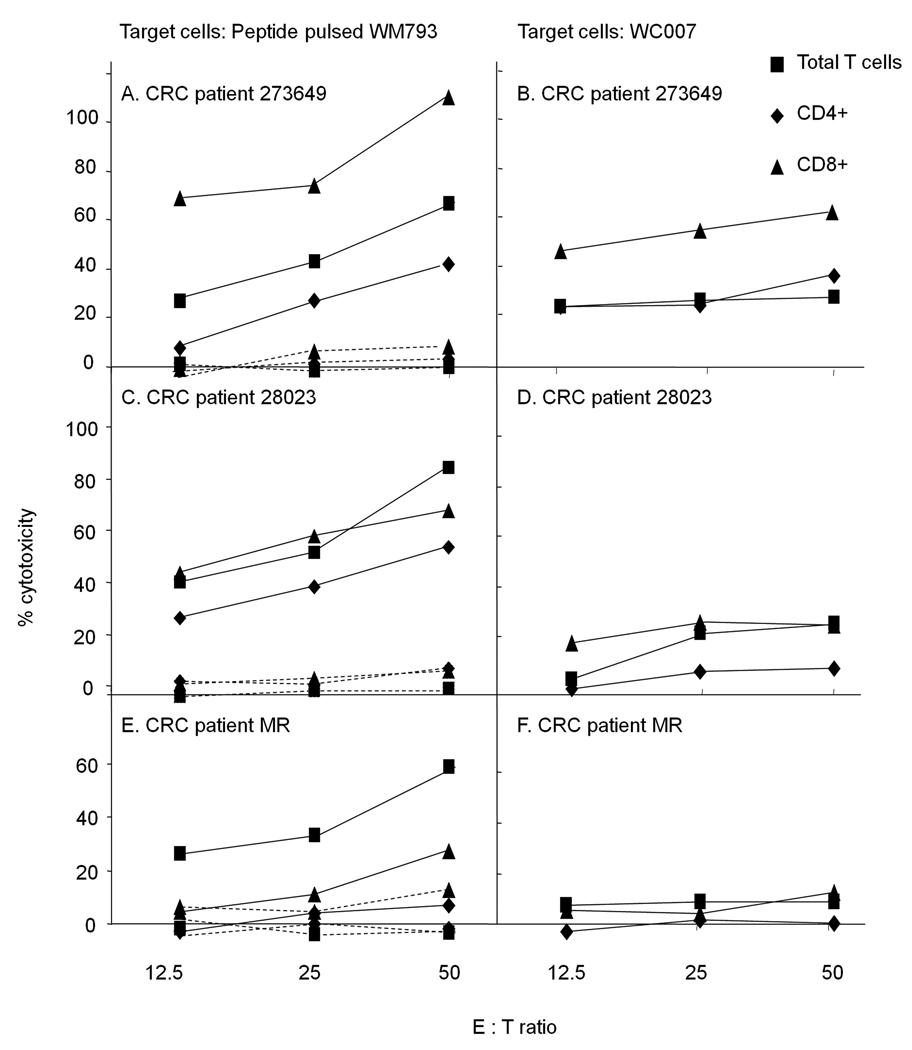
PBMC of patient 007 proliferating after stimulation with peptide 2 were of CD4 and CD8 phenotype (Fig. 4A). When the two subpopulations were sorted using magnetic beads and tested for cytotoxic reactivity, CD4+ cells lysed WC007 target cells more effectively than CD8+ cells, albeit only at E:T ratio of 50:1 (Fig. 4B).In contrast, the CTL induced in the three other patients (Fig. 3 C–E) were predominantly of CD8 phenotype (Fig. 5).
Fig. 4.
A. Phenotype of 007PBMC stimulated with peptide 2. PBMC of patient 007 were stimulated twice with peptide 2 and after 3 weeks the cells were analyzed for binding to anti-CD4 (dashed line), anti-CD8 (dotted line) or isotype - matched control (solid line) antibodies by FACS analysis. B. PBMC were sorted into CD4+ and CD8+ cells using magnetic beads. The two subpopulations as well as the unsorted PBMC were used in a CTL assay using WC007 as target cells
Fig. 5.
Phenotype of CRC patients' PBMC stimulated with peptide 2. PBMC were sorted into CD4+ and CD8+ cells using magnetic beads. The two subpopulations as well as the unsorted PBMC were tested for cytotoxic activity in a CTL assay using peptide 2-loaded (solid line) and control-peptide loaded (dashed lines) WM793 cells or WC007 cells as target cells.
Npm protein in lysates of tumor and normal cells
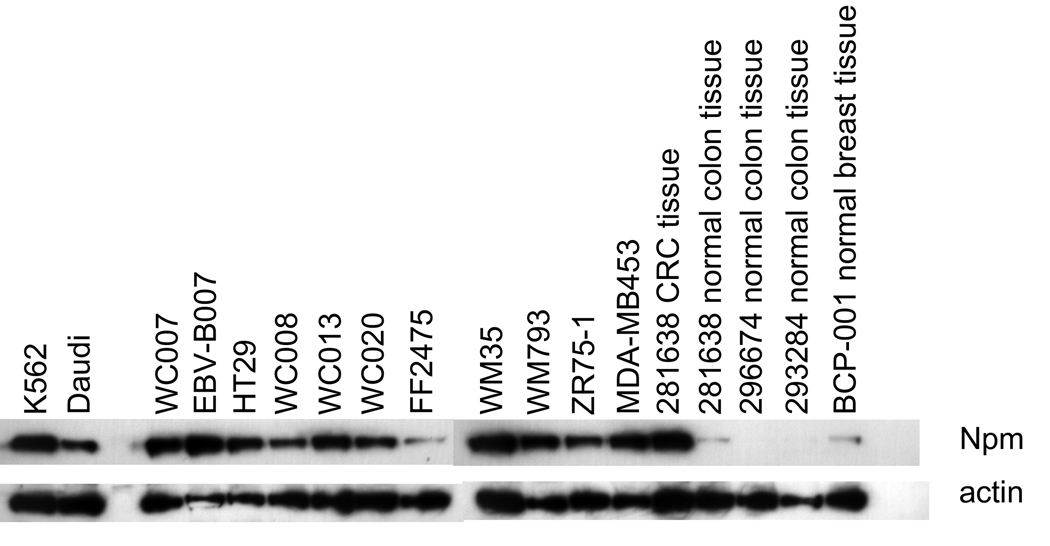
Npm was expressed by all tumor cell lines tested. It was also expressed by 007EBV-B cells, but minimally by cultured FF2475 fibroblasts. Uncultured CRC tissue expressed high amounts of Npm, whereas normal colonic and breast tissues expressed much lower amounts of the protein (Fig. 6), which is in line with previous publications reporting overexpression of Npm in tumor versus normal tissues 8, 21–23.
Fig. 6.
Npm expression by tumor and normal cells. Lysates of tumor cell lines and normal tissues were tested for Npm levels with anti-Npm antibody by Western blot analysis.
Discussion
Using gene expression cloning, we have identified Npm as the antigen recognized by CTL007 on CRC cells. This is the first identification of Npm as a T cell antigen. The cDNA clone we have isolated from CRC cells encodes full-length non-mutated Npm. In contrast, genomic translocation of Npm and expression of a fusion protein (Npm –ALK) are common in several haematological malignancies 24, 25. Npm mRNA and protein analysis in WC007 CRC cells recognized by CTL007 excluded the presence of an Npm fusion protein in these cells. Npm protein levels were increased in CRC cells compared to normal colon tissue (Fig. 6), in agreement with the demonstrated increase in Npm mRNA levels in CRC 8, 22. Increased tumor expression of Npm, which has been related to decreased cell doubling time 26, was correlated with unfavorable prognosis in bladder cancer patients 10. A possible role of Npm as an oncogene has been suggested (reviewed in 5, 6). Antigens encoded by oncogenes that drive proliferation of tumor cells are unlikely to be lost from the cells and hence may make more stable and consistent target antigens. Furthermore, immunological targeting of such antigens may be beneficial for patients as their expression by tumors may be associated with poor prognostic outcome of the disease. A growth stimulatory role has been suggested for the antigens ABCE1 27 , TYMS and PGK1 28 which are recognized by CRC patients’ CTL, and therefore these antigens, in addition to Npm, have potential as immunotherapeutic targets for CRC patients.
CTL007 recognized 2 of 3 Npm peptides tested (Fig. 2). Recognition of two peptides with quite distinct amino acid sequences by the CTL may be explained by the polyclonality of the CTL. However, Npm may be the only antigen recognized by the CTL line as treatment of the target cells with shRNA specific for Npm totally abolished target cell lysis by the CTL (Fig. 1B). Peptide 2, which induced highest sensitization of target cells for lysis by CTL007 (Fig. 2), not only induced CTL in PBMC of patient 007, but also in the PBMC of three of five additional HLA-A1 positive CRC patients available to us for study (Fig. 3). The CTL induced by the peptide in three patients not only lysed peptide-coated target cells, but also, albeit to a lesser extent, CRC cells endogenously expressing Npm (Fig. 3 A, C, E). Unfortunately autologous tumor cells were not available in any of the patients. It needs to be emphasized that CTL007 which was induced by stimulating 007PBMC with autologous WC007 CRC cells showed significantly higher lysis [up to 54%; 4] of the CRC cells than the CTL induced in the same PBMC by peptide 2 (up to 20% lysis in various experiments; Fig. 3 shows one such experiment). Tumor cells presenting multiple Npm epitopes, including both CTL and T helper epitopes, may be superior T cell stimulators than a peptide representing only a single CTL epitope. High peptide concentrations (12.5µM) were needed to generate CTLs from PBMC (data not shown), suggesting that the CTLs are of low avidity.
CTL007 lysed 2 (WC007, HT29) of 4 HLA-A1 expressing CRC cell lines tested 4 (Table 1). Target cell lysis was not associated with Npm total protein expression levels, as all 4 CRC cell lines tested and one non-lysed HLA-A1 positive melanoma cell line (WM793) expressed the protein by Western blot analysis (Fig. 6 and Table 1). These data suggest that differences in the susceptibility of target cells to lysis by CTL007 cannot be explained by differences in Npm protein expression levels by the target cells. Npm gene sequence analysis of a few cell lines (WC007, WC013) indictated no difference in the Npm gene sequence from cells lysed by CTL007 and cells not lysed (data not shown). There was also no difference in the amount or size of nuclear and cytoplasmic Npm protein in cells lysed or not lysed by CTL007 (data not shown). The reason for differences in lysis of different target cells is unknown and needs further investigation. Npm may provide a vaccine for HLA-A1 positive CRC patients and patients with tumors of other tissue origins expressing Npm, such as melanoma (29 and Fig. 6), breast carcinoma (Fig. 6), hepatocellular carcinoma 9, and squamous cell carcinoma 21. Furthermore, in addition to HLA-A1 binding epitopes, Npm expresses multiple other epitopes potentially binding to each of the HLA listed at http://www.syfpeithi.de and therefore also has vaccine potential for patients with these HLA types.
Table 1.
Absence of correlation of Npm protein expression and lysis of cells by CTL007
In conclusion, overexpression of Npm by tumors of various histological types, recognition of the antigen by T cells derived from different CRC patients, expression of anchors for all known HLA types on the protein, and association of the antigen with poor prognostic outcome make it a promising target for immunotherapeutic intervention in cancer patients.
Acknowledgements
We thank J. Faust of the flow cytometry facility of the Wistar Institute for FACS analysis. This work was supported by NIH grants CA74294 and CA10815, and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health (D.H). D.T was supported in part by grants from the National Cancer Institute (CA98166) and the Kentucky Science and Technology Corporation.
Abbreviations
- CRC
colorectal carcinoma
- CTL
cytotoxic T lymphocyte
- HLA
human leokocyte antigen
- MHA
mixed hemadsorption assay
- Npm
nucleophosmin
- PBMC
peripheral blood mononuclear cell
- PVDF
polyvinylidene fluoride
- SRBC
sheep red blood cell
Footnotes
In this paper we describe the identification by expression cloning of an antigen, nucleophosmin (Npm), recognized by an HLA–A1 restricted cytotoxic T lymphocyte line derived from the peripheral blood mononuclear cells (PBMC) of a rectal cancer patient. This is the first time the protein encoded by the potential oncogene Npm is described as a T cell antigen. A decamer peptide derived from the Npm sequence sensitized peptide-pulsed HLA-A1 positive cells to lysis by the CTL line. The peptide also induced proliferative and cytotoxic T lymphocytes in the PBMC of 4 of 6 CRC patients which lysed HLA-A1 positive peptide-pulsed target cells and CRC cells endogenously expressing Npm. Overexpression of Npm by tumors of various histological types, recognition of the antigen by T cells derived from different CRC patients, and association of the antigen with poor prognostic outcome make it a promising target for immunotherapeutic intervention in cancer patients.
Competing interest statement
The authors declare that they have no competing financial interests.
References
- 1.Clark WH, Jr, Elder DE, Guerry Dt, Braitman LE, Trock BJ, Schultz D, Synnestvedt M, Halpern AC. Model predicting survival in stage I melanoma based on tumor progression. J Natl Cancer Inst. 1989;81:1893–1904. doi: 10.1093/jnci/81.24.1893. [DOI] [PubMed] [Google Scholar]
- 2.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 3.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, Meatchi T, Bruneval P, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 4.Somasundaram R, Jacob L, Swoboda R, Caputo L, Song H, Basak S, Monos D, Peritt D, Marincola F, Cai D, Birebent B, Bloome E, et al. Inhibition of cytolytic T lymphocyte proliferation by autologous CD4+/CD25+ regulatory T cells in a colorectal carcinoma patient is mediated by transforming growth factor-beta. Cancer Res. 2002;62:5267–5272. [PubMed] [Google Scholar]
- 5.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 6.Lim MJ, Wang XW. Nucleophosmin and human cancer. Cancer Detect Prev. 2006;30:481–490. doi: 10.1016/j.cdp.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naoe T, Suzuki T, Kiyoi H, Urano T. Nucleophosmin: a versatile molecule associated with hematological malignancies. Cancer Sci. 2006;97:963–969. doi: 10.1111/j.1349-7006.2006.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nozawa Y, Van Belzen N, Van der Made AC, Dinjens WN, Bosman FT. Expression of nucleophosmin/B23 in normal and neoplastic colorectal mucosa. J Pathol. 1996;178:48–52. doi: 10.1002/(SICI)1096-9896(199601)178:1<48::AID-PATH432>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 9.Yun JP, Miao J, Chen GG, Tian QH, Zhang CQ, Xiang J, Fu J, Lai PB. Increased expression of nucleophosmin/B23 in hepatocellular carcinoma and correlation with clinicopathological parameters. Br J Cancer. 2007;96:477–484. doi: 10.1038/sj.bjc.6603574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsui KH, Cheng AJ, Chang PL, Pan TL, Yung BY. Association of nucleophosmin/B23 mRNA expression with clinical outcome in patients with bladder carcinoma. Urology. 2004;64:839–844. doi: 10.1016/j.urology.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 11.Jacob L, Somasundaram R, Smith W, Monos D, Basak S, Marincola F, Pereira S, Herlyn D. Cytotoxic T-cell clone against rectal carcinoma induced by stimulation of a patient's peripheral blood mononuclear cells with autologous cultured tumor cells. Int J Cancer. 1997;71:325–332. doi: 10.1002/(sici)1097-0215(19970502)71:3<325::aid-ijc3>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 12.Engel LW, Young NA, Tralka TS, Lippman ME, O'Brien SJ, Joyce MJ. Establishment and characterization of three new continuous cell lines derived from human breast carcinomas. Cancer Res. 1978;38:3352–3364. [PubMed] [Google Scholar]
- 13.Soker S, Fidder H, Neufeld G, Klagsbrun M. Characterization of novel vascular endothelial growth factor (VEGF) receptors on tumor cells that bind VEGF165 via its exon 7-encoded domain. J Biol Chem. 1996;271:5761–5767. doi: 10.1074/jbc.271.10.5761. [DOI] [PubMed] [Google Scholar]
- 14.Somasundaram R, Swoboda R, Caputo L, Lee A, Jackson N, Marincola FM, Guerry D, Herlyn D. A CD4+, HLA-DR7-restricted T-helper lymphocyte clone recognizes an antigen shared by human malignant melanoma and glioma. Int J Cancer. 2003;104:362–368. doi: 10.1002/ijc.10964. [DOI] [PubMed] [Google Scholar]
- 15.Herlyn D, Herlyn M, Ross AH, Ernst C, Atkinson B, Koprowski H. Efficient selection of human tumor growth-inhibiting monoclonal antibodies. J Immunol Methods. 1984;73:157–167. doi: 10.1016/0022-1759(84)90041-3. [DOI] [PubMed] [Google Scholar]
- 16.Brichard V, Van Pel A, Wolfel T, Wolfel C, De Plaen E, Lethe B, Coulie P, Boon T. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1993;178:489–455. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 18.Somasundaram R, Zaloudik J, Jacob L, Benden A, Sperlagh M, Hart E, Marks G, Kane M, Mastrangelo M, Herlyn D. Induction of antigen-specific T and B cell immunity in colon carcinoma patients by anti-idiotypic antibody. J Immunol. 1995;155:3253–3261. [PubMed] [Google Scholar]
- 19.Somasundaram R, Swoboda R, Caputo L, Otvos L, Weber B, Volpe P, van Belle P, Hotz S, Elder DE, Marincola FM, Schuchter L, Guerry D, et al. Human leukocyte antigen-A2-restricted CTL responses to mutated BRAF peptides in melanoma patients. Cancer Res. 2006;66:3287–3293. doi: 10.1158/0008-5472.CAN-05-1932. [DOI] [PubMed] [Google Scholar]
- 20.Somasundaram R, Robbins P, Moonka D, Loh E, Marincola F, Patel A, Guerry D, Herlyn D. CD4(+), HLA class I-restricted, cytolytic T-lymphocyte clone against primary malignant melanoma cells. Int J Cancer. 2000;85:253–259. doi: 10.1002/(sici)1097-0215(20000115)85:2<253::aid-ijc17>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 21.Chang JT, Wang HM, Chang KW, Chen WH, Wen MC, Hsu YM, Yung BY, Chen IH, Liao CT, Hsieh LL, Cheng AJ. Identification of differentially expressed genes in oral squamous cell carcinoma (OSCC): overexpression of NPM, CDK1 and NDRG1 and underexpression of CHES1. Int J Cancer. 2005;114:942–949. doi: 10.1002/ijc.20663. [DOI] [PubMed] [Google Scholar]
- 22.van Belzen N, Dinjens WN, Eussen BH, Bosman FT. Expression of differentiation-related genes in colorectal cancer: possible implications for prognosis. Histol Histopathol. 1998;13:1233–1242. doi: 10.14670/HH-13.1233. [DOI] [PubMed] [Google Scholar]
- 23.Yokota T, Kouno J, Adachi K, Takahashi H, Teramoto A, Matsumoto K, Sugisaki Y, Onda M, Tsunoda T. Identification of histological markers for malignant glioma by genome-wide expression analysis: dynein, alpha-PIX and sorcin. Acta Neuropathol (Berl) 2006;111:29–38. doi: 10.1007/s00401-005-1085-6. [DOI] [PubMed] [Google Scholar]
- 24.Drexler HG, Gignac SM, von Wasielewski R, Werner M, Dirks WG. Pathobiology of NPM-ALK and variant fusion genes in anaplastic large cell lymphoma and other lymphomas. Leukemia. 2000;14:1533–1559. doi: 10.1038/sj.leu.2401878. [DOI] [PubMed] [Google Scholar]
- 25.Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 26.Derenzini M, Sirri V, Trere D, Ochs RL. The quantity of nucleolar proteins nucleolin and protein B23 is related to cell doubling time in human cancer cells. Lab Invest. 1995;73:497–502. [PubMed] [Google Scholar]
- 27.Shichijo S, Ishihara Y, Azuma K, Komatsu N, Higashimoto N, Ito M, Nakamura T, Ueno T, Harada M, Itoh K. ABCE1, a member of ATP-binding cassette transporter gene, encodes peptides capable of inducing HLA-A2-restricted and tumor-reactive cytotoxic T lymphocytes in colon cancer patients. Oncol Rep. 2005;13:907–913. [PubMed] [Google Scholar]
- 28.Shichijo S, Azuma K, Komatsu N, Ito M, Maeda Y, Ishihara Y, Itoh K. Two proliferation-related proteins, TYMS and PGK1, could be new cytotoxic T lymphocyte-directed tumor-associated antigens of HLA-A2+ colon cancer. Clin Cancer Res. 2004;10:5828–5836. doi: 10.1158/1078-0432.CCR-04-0350. [DOI] [PubMed] [Google Scholar]
- 29.Bernard K, Litman E, Fitzpatrick JL, Shellman YG, Argast G, Polvinen K, Everett AD, Fukasawa K, Norris DA, Ahn NG, Resing KA. Functional proteomic analysis of melanoma progression. Cancer Res. 2003;63:6716–6725. [PubMed] [Google Scholar]