Abstract
Neuropeptides are produced from larger precursors by limited proteolysis, first by endopeptidases and then by carboxypeptidases. Major endopeptidases required for these cleavages include prohormone convertase (PC) 1/3 and PC2. In the present study, quantitative peptidomics analysis was used to characterize the specific role PC1/3 plays in this process. Peptides isolated from hypothalamus, amygdala, and striatum of PC1/3 null mice were compared to those from heterozygous and wild-type mice. Extracts were labeled with stable isotopic tags and fractionated by HPLC, after which relative peptide levels were determined using tandem mass spectrometry. In total, 92 peptides were found, of which 35 were known neuropeptides or related peptides derived from 15 distinct secretory pathway proteins: 7B2, chromogranin A and B, cocaine- and amphetamine-regulated transcript, procholecystokinin, proenkephalin, promelanin concentrating hormone, proneurotensin, propituitary adenylate cyclase-activating peptide, proSAAS, prosomatosatin, provasoactive intestinal peptide, provasopressin, secretogranin III, and VGF. Among the peptides derived from these proteins, ~1/3 were decreased in the PC1/3 null mice relative to wild-type mice, ~1/3 showed no change, and ~1/3 increased in PC1/3 null. Cleavage sites were analyzed in peptides that showed no change or that decreased in PC1/3 mice, and these results were compared with peptides that showed no change or decreased in previous peptidomic studies with PC2 null mice. Analysis of these sites showed that while PC1/3 and PC2 have overlapping substrate preferences, there are particular cleavage site residues that distinguish peptides preferred by each PC.
Keywords: peptide processing, proprotein convertase, protease, peptidase, proSAAS, neuropeptide
INTRODUCTION
Neuropeptides are produced from the selective cleavage of precursor proteins at specific sites. Post-translational processing includes cleavage by endopeptidases such as the prohormone/proprotein convertases (PCs), followed by removal of remaining basic residues from carboxy termini by carboxypeptidases (CPs) (Zhou et al. 1999;Seidah and Chretien 1997;Reznik and Fricker 2001). This can be followed by further steps such as amidation of carboxy-terminal glycine of the peptide (Prigge et al. 2000). Secretory pathway endopeptidases include PC1/3, PC2, PC4, PC5/6, PC7, furin, and PACE4 which overlap in their expression and subcellular location. All are serine proteases with subtilisin-like catalytic domains (Steiner 1998;Seidah and Chretien 2004b;Seidah and Chretien 2004a). PC1/3 and PC2 are the major PCs found within mature secretory vesicles and are thought to function in the production of many neuroendocrine peptides (Steiner 1998;Seidah and Chretien 2004b;Seidah and Chretien 2004a). Other PCs such as furin and PC7 function in proprotein processing in the trans Golgi network (Steiner 1998). In addition to PCs, other endopeptidases may be involved in peptide biosynthesis. For example, it has been proposed that cathepsin L is present in secretory vesicles and contributes to the biosynthesis of neuropeptides in a pathway distinct from the subtilisin-like prohormone convertases (Hook 2006a). An important question in the field is the contribution each enzyme makes to peptide processing in vivo.
The substrate specificities of PC1/3 and PC2 have been studied using several approaches. Early in vitro studies involved incubating purified enzyme with candidate substrates and analysis of the products (Benjannet et al. 1991;Zhou and Lindberg 1993;Dupuy et al. 1994;Johanning et al. 1998;Day et al. 1998). Other studies have co-expressed the PCs with candidate substrates in neuroendocrine cell lines, and then examined the products secreted into the media and/or present inside the cells (Mathis and Lindberg 1992;Breslin et al. 1993;Zhou and Mains 1994;Paquet et al. 1996). Analysis of processed substrates allowed for the determination of consensus sites for PC cleavage. Precursors are usually cleaved at the general motif (K/R)-(X)n -(K/R)↓, where n = 0, 2, 4 or 6 and X is any amino acid other than Cys (Seidah and Chretien 2004b;Seidah and Chretien 2004a). To study the role of the PCs in vivo, mice with disruptions of the genes encoding PC1/3 and PC2 have been created (Zhu et al. 2002b;Zhu et al. 2002a;Furuta et al. 1997). Mice that lack either PC1/3 or PC2 alone are viable, but double knock-out mice are embryonic lethal (X.Zhu, unpublished). PC1/3 null mice display a syndrome of severe postnatal growth impairment and are 60% the size of their wild-type counterparts at 10 weeks (Zhu et al. 2002b;Zhu et al. 2002a). These mice also exhibit hyperproinsulinemia, but with normal glucose tolerance. In contrast, PC2 null mice are normal at birth and exhibit only a small decrease in growth rate compared to wild-type mice (Furuta et al. 1997). The knockout (KO) mouse models made it possible to evaluate the physiological role of PC1/3 and PC2 in mouse tissues. Examination of the processing of a number of neuropeptide precursors in the KO mouse models has been conducted and a number of neuropeptide and peptide hormone processing defects have been found (Furuta et al. 1997;Furuta et al. 2001;Furuta et al. 1998;Johanning et al. 1998;Berman et al. 2000;Allen et al. 2001;Zhu et al. 2002a;Dey et al. 2003). These previous studies used radioimmunoassays to neuropeptides. Radioimmunoassays have a reasonably high sensitivity but are restricted to known peptides for which specific antisera are available.
Recent studies have used a quantitative peptidomics technique to examine the pituitary of PC1/3 null mice (Pan et al. 2005), and the hypothalamus and other brain regions of PC2 null mice (Pan et al. 2006;Zhang et al. 2010). The quantitative peptidomics technique involves separately labeling peptides extracted from brain regions from different groups of mice with stable isotopic tags (Che and Fricker 2005;Che et al. 2005a;Fricker et al. 2006). The differentially-labeled peptides are pooled and analyzed using liquid chromatography and electrospray ionization mass spectrometry (LC/MS). This analysis allows the detection, quantification, and identification of many of the known neuropeptides as well as novel peptides (Baggerman et al. 2004;Svensson et al. 2007;Hatcher et al. 2008).
To gain a better understanding of the role of PC1/3 in vivo, we applied the quantitative peptidomics technique to examine peptides in several brain regions of the PC1/3 null mice. A number of peptides are substantially decreased in the PC1/3 null mice, relative to wild-type (WT) mice, indicating an important role for this enzyme in the production of these peptides. Taken together with results from analysis of PC2 null mice, both PC1/3 and PC2 appear to have complementary roles although with some redundancy.
MATERIALS AND METHODS
PC1/3-null mice were generated by deletion of exon 1 and several upstream transcriptional control elements (CRE, ICS, AP-1 et Sp1) from the PCSK1 gene, by inserting a neomycin cassette in C57Bl/6 mice background. The mice used in the present experiments were backcrossed for 11 generations, where PC1/3-heterozygous (Het) mice were mated with CD1 mice in order to transfer PC1/3-Het mice into the CD1 background. To obtain PC1/3-null mice, male and female PC1/3-Het mice were mated. Genotyping of the offspring revealed prenatal lethality of the nulls because the Mendelian ratio was not exact. Only 16% of PC1/3-null mice were generated instead of 25% and of these, only 33% survived. This yielded an overall yield of ~5% for the PC1/3-null mice. Genotyping was carried out by PCR and the primer used was GT-S2 :5’-GCC TCT GAA AGA TCA AAA CAC GAG-3’; GT-W2: 5’-CTC CAT TCT TCA AAG AGA ACC GC-3’ and GT-N2: 5’-CGC CTG TGC TCT AGT AGC TTT ACG-3’. A wild-type amplicon was obtained with GT-S2 and GT-W2, whereas the neomycin gene was amplified with GT-S2 and GT-N2. The PCR profile was 1 cycle of 5 min at 94°C, followed by 35 cycles of 30 sec at 94°C, 30 sec at 55°C, 1 min at 72°C and finally 1 cycle of 3 min at 72°C.
All mice used for the analysis were males, and those for the groups to be compared (WT, Hets, and KOs) were 4–5 months of age. Mice were sacrificed by decapitation and the head was immediately irradiated in a conventional microwave oven for 8 s at full power; this has previously been shown to raise the brain temperature to 80°C (Che et al. 2005b). After cooling, the brain was removed and cut with a razor blade into coronal sections for subsequent dissection. Coronal cuts were made on either side of the hypothalamus at bregma 0.00 and −3.00 (Paxinos and Franklin 2001). The striatum was dissected from the 1.94 to 0.00 section by removing the cortex. This 'striatum' sample includes the caudate putamen, nucleus accumbens, septum, and ventral palladium. The coronal section of bregma 0.00 to −3.00 was dissected into the amygdala, and hypothalamus. Tissue was frozen in dry ice and stored at −70°C until analysis.
Extraction of tissue
Tissue was extracted as previously described (Che et al. 2007). In brief, the dissected brain regions were sonicated twice for 20s at 1 pulse/s in 200 µL ice-cold water using an ultrasonic processor (W-380; Ultrasonic Inc., Farmingdale, NY, USA). The homogenates were incubated in a 70°C water bath for 20 min and then cooled on ice and acidified with 0.1 M HCl to a final concentration of 10 mM HCl. The homogenates were centrifuged at 13 000 g for 40 min at 4°C and the supernatant was then transferred to a low-retention tube. The pH of the peptide extracts was adjusted to 9.5 by the addition of 0.4 M phosphate buffer. In addition to the brain regions from individual WT, Het, and PC1/3 KO mice, two pools of brain regions from Het mice were also included (pools A and B); these pools consisted of tissue from 3 mice, and each pool was extracted in 600 µl of water. Following extraction, these pools were divided into three aliquots for labeling with isotopic tags, as described below.
Isotopic labeling
Labeling was performed as previously described using trimethylammoniumbutyryl-N-hydroxysuccinimide (TMAB) containing either no deuteriums (D0-TMAB), 3 deuteriums (D3-TMAB), 6 deuteriums (D6-TMAB), 9 deuteriums (D9-TMAB) or one 13C and 9 deuteriums (referred to as D12-TMAB because the mass is equivalent to the D0-TMAB plus 12 daltons); labels were synthesized as described (Morano et al. 2008). The TMAB reagents were dissolved in DMSO at 350 µg/ µL and used to label the samples, as described in Supplemental Figure S1. Briefly, 2.2 µL of labeling reagent solutions were added to each tissue extract and incubated for 10 min at 23–25°C. Then an appropriate volume of 1.0 M NaOH was added to adjust the pH back to 9.5 and the sample extract solution was incubated another 10 min at 23–25°C. These two steps were repeated seven times to ensure all the peptides were completely labeled and then the mixture was incubated at 23–25°C for 40 min, followed by the addition of 10 µL of 2.5 M glycine to quench the remaining TMAB reagents. After labeling, samples were pooled as indicated (Figure S1) and filtered through an Amicon Ultra-4 Ultracel unit (Millipore) to remove proteins > 10 kDa. To remove any TMAB labels from Tyr residues in the peptides, the filtrate was adjusted to pH 9.0 and 2.0 M hydroxylamine (in DMSO) was added in three aliquots of 3.3 µL each, with an interval of 10 min between each addition. After desalting with a PepClean C-18 spin column (Pierce), peptides were eluted with 80 µL of 70% acetonitrile and 0.1% trifluoroacetic acid in water, frozen, and concentrated to 20 µL in a vacuum centrifuge. Aliquots of the peptide samples were stored at −70°C until further MS analysis.
Mass spectrometry and data analysis
To detect and quantify the labeled peptides from PC1/3 null and WT mouse tissues, liquid chromatography and tandem mass spectrometery (LC/MS/MS) was performed on a Waters Q-TOF - Ultima Mass Spectrometer (Micromass, Manchester, UK) as previously described (Berti et al. 2009). The most intense ion in each MS survey scan was fragmented to generate MS/MS spectra.
Peptides were identified based on a combination of approaches. Mascot (http://www.matrix-science/com) was used to search the MS/MS data for matches to the mouse NCBI database. In Mascot searching, modifications were selected as either GIST-Quat (i.e. the Mascot name for TMAB) or one of the heavy forms of the TMAB tags (such as GIST-Quat:2(H)3, 6, and 9) at the N-terminus and on all Lys residues. To reduce the number of false-positives, all Mascot hits were manually interpreted as previously described (Berti et al. 2009;Morano et al. 2008;Zhang et al. 2008). The criteria for accepting a peptide as identified have been previously published; in brief (i) the peptide mass had to be within 40 ppm of the theoretical mass; (ii) the observed number of TMAB tags on the peptide matched the predicted number of free amines available (i.e. lysine residue and N-terminus); (iii) the observed charge state(s) of the peptide was consistent with the expected number of positive charges; (iv) 80% or more of the major fragments observed in MS/MS matched (within 40 ppm) to predicted fragmentation ions; and (v) at least five fragmentation ions matched (within 40 ppm) to either b- and/or y-series ions. Some peptides detected in MS mode were not selected for fragmentation in MS/MS mode. If these peptides eluted from the LC column at the same relative time as a previously sequenced peptide, and also satisfied the above criteria regarding mass, tag number, and charge state, these peptides were considered identified.
Quantitation was performed as previously described (Morano et al. 2008;Zhang et al. 2008). In brief, the peak intensity for the monoisotopic peak and the peak containing one 13C atom were averaged. In spectra with overlapping peaks in which the signals with multiple 13C atoms extended into the range of the next TMAB tag, the isotopic distribution of the peptide was calculated and the contribution from the 13C-containing peaks subtracted, as described (Morano et al. 2008). Each LC/MS run contained tissue extract from one WT mouse, one Het mouse, and one mouse homozygous for the mutation (KO), as described above and in Figure S1. The relative peak intensity for each of these groups was determined for each of the three runs, and these ratios were averaged. Statistical testing was performed using Student’s t-test, comparing the ratio of the KO/WT peaks to the Het/WT peaks for each run. In some cases, this yielded statistically significant changes (described below). However, in other cases, both the KO and the Het levels appeared to be altered, and there was no statistical difference between these two groups. For these peptides, previously published data comparing the level of the same peptides between WT groups was used for the statistical testing (Zhang et al. 2008).
RESULTS
A total of 92 distinct peptides were detected in the 9 LC/MS runs, of which approximately half were identified from MS/MS sequence analysis. Of these, 35 were known neuropeptides or related peptides derived from a total of 15 different secretory pathway proteins. The remainder of the identified peptides was from proteins localized to the cytosolic and/or other intracellular locations (mitochondria, nucleus); these intracellular peptides were not further analyzed in the present study.
Relative peptide levels were quantified in each of the LC/MS runs by measurement of the peak intensities for each isotopic form detected in the spectra. For this analysis, the intensity of the monoisotopic peak and the peak containing one atom of 13C were measured. Typical data are shown in Supplemental Figures S2 and S3. Some peptides were found to be present at comparable levels in WT, Het, and KO mice, such as the peptide named Big LEN in the striatum (Figure S3B). Some peptides are elevated in the KO mice, relative to levels in the Het and WT mice (Figure S3A), while others are decreased in the KO mice, relative to Het and WT mice (Figure S3C and D). For this analysis, peptides were counted separately if found in multiple brain regions, and so the total number of peptides considered was 57 (i.e. each line in Table 1 was counted). Altogether, 18 peptides were found to be elevated in the PC1/3 KO mice (PC1/3 KO to WT ratio≥1.30); these may represent substrates of the enzyme, or peptides otherwise elevated by the absence of PC1/3. However, many of these showed large variation among the replicates, reflected in a large standard error of the mean in Table 1. Twenty one peptides did not show large changes in the PC1/3 KO mice, with ratios between 0.70 and 1.30 (Table 1). Eighteen peptides decreased in the PC1/3 KO mice (ratio ≤0.70), and may represent products of this enzyme.
Table 1.
Peptidomics results for PC1/3 null mice
| Precursor | Peptide name | Upstream | Sequence | Downstream | Theor mass |
Obs Mass |
ppm | z | #T | Reg. | PC1 Het/WT |
PC1 KO/WT | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Avg | SEM | Avg | SEM | Sig | n | |||||||||||
| 7B2 | 200–212 | .LDNVVAKK | SVPHFSEEEKEAE | * | 1516.67 | 1516.71 | 25 | 3 | 2 | Amy | 1.12 | 0.20 | 1.40 | 0.12 | 3 | |
| 7B2 | 200–212 | .LDNVVAKK | (SVPHFpSEEEKEAE) | * | 1596.63 | 1596.65 | 12 | 3 | 2 | Amy | 1.19 | 0.13 | 1.30 | 0.08 | 3 | |
| 7B2 | 200–212 | .LDNVVAKK | (SVPHFpSEEEKEAE) | * | 1596.63 | 1596.62 | −2 | 3 | 2 | Stri | 0.88 | 0.16 | 0.91 | 0.20 | 3 | |
| CART | 82–86 | .LKKLKSKR | (IPIYE) | KKYGQVPM. | 633.33 | 633.32 | 16 | 1 | 1 | Hyp | 2.21 | 0.22 | 1.33 | 0.07 | 3 | |
| Chromogranin A | 392–402 | .MKLSFRTR | AYGFRDPGPQL | RRGWRPSS. | 1219.60 | 1219.62 | 20 | 2 | 1 | Hyp | 1.67 | 0.07 | 1.48 | 0.13 | 3 | |
| Chromogranin B | 438–446 | .LDTREEKR | LLDEGHYPV | RESPIDTA. | 1041.52 | 1041.53 | 10 | 2 | 1 | Hyp | 1.01 | 0.11 | 0.23 | 0.07 | **a | 3 |
| Chromogranin B | 438–446 | .LDTREEKR | LLDEGHYPV | RESPIDTA. | 1041.52 | 1041.50 | −16 | 2 | 1 | Stri | 0.87 | 0.31 | 0.37 | 0.05 | nsa, **b | 3 |
| Chromogranin B | 588–597 | .VNWGYEKR | SFARAPQLDL | KRQYDGVA. | 1116.59 | 1116.60 | 11 | 2 | 1 | Amy | 1.05 | 0.10 | 1.25 | 0.09 | 3 | |
| Chromogranin B | 588–597 | .VNWGYEKR | SFARAPQLDL | KRQYDGVA. | 1116.59 | 1116.62 | 26 | 2 | 1 | Hyp | 1.45 | 0.23 | 1.19 | 0.04 | 3 | |
| Chromogranin B | 588–597 | .VNWGYEKR | SFARAPQLDL | KRQYDGVA. | 1116.59 | 1116.58 | −6 | 2 | 1 | Stri | 0.95 | 0.23 | 0.90 | 0.08 | 3 | |
| Chromogranin B | 438–454 | .LDTREEKR | LLDEGHYPVRESPIDTA | KRYPQSKW. | 1910.93 | 1910.96 | 14 | 3 | 1 | Amy | 1.10 | 0.13 | 1.48 | 0.23 | 3 | |
| Chromogranin B | 438–454 | .LDTREEKR | LLDEGHYPVRESPIDTA | KRYPQSKW. | 1910.93 | 1911.02 | 48 | 3 | 1 | Hyp | 1.01 | 0.26 | 1.38 | 0.41 | 3 | |
| Chromogranin B | 438–454 | .LDTREEKR | LLDEGHYPVRESPIDTA | KRYPQSKW. | 1910.93 | 1910.94 | 7 | 3 | 1 | Stri | 1.09 | 0.27 | 0.99 | 0.15 | 3 | |
| Chromogranin B | 313–330 | .HPLSEERR | PSPKESKEADVATVRLGE | KRSHHLAH. | 1911.99 | 1911.97 | −12 | 4 | 3 | Amy | 1.26 | 0.03 | 1.41 | 0.14 | 3 | |
| Chromogranin B | Phosphorylated 357–373 |
.GEESRSYR | GLQYRGRGpSEEDRAPRP | RSEESQER. | 2022.97 | 2022.94 | −15 | 4 | 1 | Amy | 0.86 | 0.01 | 0.36 | 0.09 | **a | 3 |
| Chromogranin B | Phosphorylated 357–374 |
.GEESRSYR | GLQYRGRGpSEEDRAPRPR | SEESQERE. | 2179.07 | 2179.05 | −10 | 5 | 1 | Amy | 0.81 | 0.15 | 0.57 | 0.10 | nsa, **b | 3 |
| Procholecystokinin | 36–44 | .ATDPVEQR | (AQEAPRRQL) | RAVLRTDG. | 1067.58 | 1067.59 | 4 | 3 | 1 | Stri | 1.38 | 0.23 | 1.31 | 0.31 | 2 | |
| Proenkephalin | Leu-Enkephalin | .WWMDYQKR | YGGFL | KRFAESLP. | 555.27 | 555.25 | −38 | 1 | 1 | Hyp | 1.36 | 0.35 | 0.96 | 0.24 | 3 | |
| Proenkephalin | Met-Enkephalin | .GGEILAKR | YGGFM | KKDADEGD. | 573.23 | 573.23 | 7 | 1 | 1 | Amy | 1.86 | 0.14 | 1.71 | 0.27 | nsa, **b | 3 |
| Proenkephalin | Met-Enkephalin | .GGEILAKR | YGGFM | KKDADEGD. | 573.23 | 573.20 | −49 | 1 | 1 | Hyp | 1.37 | 0.41 | 0.98 | 0.36 | 3 | |
| Proenkephalin | Met-Enkephalin | .GGEILAKR | YGGFM | KKDADEGD. | 573.23 | 573.23 | −1 | 1 | 1 | Stri | 0.99 | 0.32 | 0.90 | 0.33 | 3 | |
| Proenkephalin | Octapeptide | .NDEDMSKR | YGGFMRSL | KRSPQLED. | 929.45 | 929.47 | 22 | 2 | 1 | Hyp | 1.39 | 0.24 | 1.84 | 0.36 | 3 | |
| Proenkephalin | Octapeptide | .NDEDMSKR | YGGFMRSL | KRSPQLED. | 929.45 | 929.42 | −29 | 2 | 1 | Stri | 1.02 | 0.27 | 1.15 | 0.23 | 3 | |
| Proenkephalin | 197–208 | .GFMRSLKR | SPQLEDEAKELQ | KRYGGFMR. | 1385.67 | 1385.67 | −1 | 3 | 2 | Amy | 1.97 | 0.19 | 2.32 | 0.39 | nsa, **b | 3 |
| Proenkephalin | 197–208 | .GFMRSLKR | SPQLEDEAKELQ | KRYGGFMR. | 1385.67 | 1385.68 | 9 | 2 | 2 | Hyp | 1.56 | 0.77 | 0.78 | 0.22 | nsa | 2 |
| Proenkephalin | 197–208 | .GFMRSLKR | SPQLEDEAKELQ | KRYGGFMR. | 1385.67 | 1385.66 | −2 | 3 | 2 | Stri | 1.07 | 0.18 | 1.08 | 0.27 | 3 | |
| Prohormone convertase 1 |
619–628 | . NTVQNDRR | GVEKMVNVVE | KRPTQKSL. | 1102.57 | 1102.53 | −33 | 2 | 2 | Hyp | 0.53 | 0.19 | <0.10 | **a | 3 | |
| Promelanin concentrating hormone |
Neuropeptide EI | .AESTQEKR | EIGDEENSAKFPI-amide | GRRDFDML. | 1446.68 | 1446.71 | 15 | 3 | 2 | Amy | 1.20 | 0.07 | 1.58 | 0.28 | nsa, **b | 3 |
| Promelanin concentrating hormone |
Neuropeptide EI | .AESTQEKR | EIGDEENSAKFPI-amide | GRRDFDML. | 1446.68 | 1446.68 | −5 | 2 | 2 | Hyp | 1.31 | 0.04 | 1.54 | 0.41 | nsa,b | 3 |
| Promelanin concentrating hormone |
Neuropeptide EI | .AESTQEKR | EIGDEENSAKFPI-amide | GRRDFDML. | 1446.68 | 1446.72 | 23 | 2 | 2 | Stri | 1.08 | 0.22 | 0.69 | 0.04 | nsa, **b | 3 |
| Proneurotensin | Neuromedin N | .EKEEVIKR | KIPYIL | KRQLYENK. | 745.47 | 745.44 | −39 | 2 | 2 | Hyp | 1.15 | 0.19 | 0.93 | 0.17 | 3 | |
| Proneurotensin | Neurotensin | .KIPYILKR | pELYENKPRRPYIL | KRGSYYY* | 1671.91 | 1671.93 | 12 | 3 | 1 | Hyp | 0.98 | 0.19 | 0.72 | 0.13 | 2 | |
| ProPACAP | 111–128 | .YLQSVVAR | GAGENLGGSAVDDPAPLT | KRHSDGIF. | 1639.77 | 1639.83 | 35 | 2 | 1 | Hyp | 1.06 | 0.15 | 0.87 | 0.16 | 3 | |
| ProSAAS | KEP 1–7 | signal peptide | (ARPVKEP) | RSLSAASA. | 795.46 | 795.44 | 25 | 3 | 2 | Hyp | 1.36 | 0.05 | 0.73 | 0.07 | **a | 3 |
| ProSAAS | KEP | signal peptide | (ARPVKEPR) | SLSAASAP. | 951.56 | 951.55 | −7 | 3 | 2 | Hyp | 1.44 | 0.18 | 0.80 | 0.07 | **a | 3 |
| ProSAAS | Little LEN | .GALLRVKR | LENPSPQAPA | RRLLPP* | 1022.50 | 1022.51 | 12 | 2 | 1 | Amy | 1.15 | 0.01 | 0.45 | 0.02 | **a | 3 |
| ProSAAS | Little LEN | .GALLRVKR | LENPSPQAPA | RRLLPP* | 1022.50 | 1022.52 | 20 | 2 | 1 | Hyp | 1.29 | 0.16 | 0.31 | 0.08 | *a | 2 |
| ProSAAS | Little LEN | .GALLRVKR | LENPSPQAPA | RRLLPP* | 1022.50 | 1022.51 | 7 | 2 | 1 | Stri | 0.90 | 0.19 | 0.34 | 0.11 | *a | 3 |
| ProSAAS | Little SAAS 5–16 | .KEPRSLSA | ASAPLVETSTPL | RLRRAVPR. | 1184.63 | 1184.64 | 6 | 2 | 1 | Hyp | 1.37 | 0.10 | 0.90 | 0.17 | 2 | |
| ProSAAS | Little SAAS 1–16 | ARPVKEPR | SLSAASAPLVETSTPL | RLRRAVPR. | 1542.81 | 1542.82 | 6 | 2 | 1 | Amy | 1.25 | 0.11 | 1.08 | 0.13 | 3 | |
| ProSAAS | Little SAAS 1–16 | ARPVKEPR | SLSAASAPLVETSTPL | RLRRAVPR. | 1542.81 | 1542.85 | 24 | 2 | 1 | Hyp | 1.53 | 0.34 | 0.82 | 0.19 | 3 | |
| ProSAAS | Little SAAS 1–16 | ARPVKEPR | SLSAASAPLVETSTPL | RLRRAVPR. | 1542.81 | 1542.81 | −3 | 2 | 1 | Stri | 1.10 | 0.26 | 0.59 | 0.09 | nsa,b | 3 |
| ProSAAS | Big LEN | .GALLRVKR | LENPSPQAPARRLLPP | * | 1754.98 | 1754.99 | 4 | 3 | 1 | Amy | 1.50 | 0.13 | 2.44 | 0.10 | **a | 3 |
| ProSAAS | Big LEN | .GALLRVKR | LENPSPQAPARRLLPP | * | 1754.98 | 1755.02 | 25 | 3 | 1 | Hyp | 1.76 | 0.30 | 2.70 | 0.45 | nsa, **b | 3 |
| ProSAAS | Big LEN | .GALLRVKR | LENPSPQAPARRLLPP | * | 1754.98 | 1754.99 | 6 | 3 | 1 | Stri | 1.12 | 0.17 | 1.06 | 0.08 | 3 | |
| ProSAAS | Little SAAS | ARPVKEPR | SLSAASAPLVETSTPLRL | RRAVPRGE. | 1812.01 | 1812.04 | 21 | 3 | 1 | Hyp | 1.32 | 0.15 | 0.70 | 0.10 | *a | 3 |
| ProSAAS | Big SAAS 1–24 | signal peptide | ARPVKEPRSLSAASAPLV ETSTPL | RLRRAVPR. | 2476.37 | 2476.41 | 18 | 4 | 2 | Hyp | 1.95 | 0.55 | 5.00 | 1.00 | nsa, **b | 2 |
| Prosomatostatin | Somatostatin 28–14 | .EMRLELQR | SANSNPAMAPRE | RKAGCKNF. | 1243.56 | 1243.57 | 9 | 2 | 1 | Amy | 1.01 | 0.04 | 0.32 | 0.02 | **a | 3 |
| Prosomatostatin | Somatostatin 28–14 w/ MetOx | .EMRLELQR | (SANSNPAMoxAPRE) | RKAGCKNF. | 1259.56 | 1259.58 | 12 | 2 | 1 | Amy | 1.12 | 0.13 | 0.24 | 0.02 | **a | 3 |
| Prosomatostatin | Somatostatin 28–14 w/ MetOx | .EMRLELQR | (SANSNPAMoxAPRE) | RKAGCKNF. | 1259.56 | 1259.55 | −9 | 2 | 1 | Stri | 0.80 | 0.14 | 0.21 | 0.01 | *a | 2 |
| ProVasoactive Intestinal Peptide |
111–122 | .LESLIGKR | ISSSISEDPVPI | KRHSDAVF. | 1242.63 | 1242.63 | 1 | 2 | 1 | Stri | 0.67 | 0.67 | 1 | |||
| Provasopressin | 154-end | .LLLRLVQL | AGTRESVDSAKPRVY | * | 1634.84 | 1634.85 | 10 | 4 | 2 | Hyp | 1.22 | 2.33 | 1 | |||
| Provasopressin | 151-end | .ARALLLRL | VQLAGTRESVDSAKPRVY | * | 1975.05 | 1975.10 | 28 | 4 | 2 | Hyp | 0.70 | 2.10 | 1 | |||
| Secretogranin III | 23–36 | signal peptide | FPKPEGSQDKSLHN | RELSAERP. | 1582.77 | 1582.78 | 6 | 4 | 3 | Amy | 1.14 | 0.12 | 0.57 | 0.05 | **a | 3 |
| Secretogranin III | 23–36 | signal peptide | FPKPEGSQDKSLHN | RELSAERP. | 1582.77 | 1582.76 | −7 | 4 | 3 | Hyp | 1.38 | 0.25 | <0.20 | **a | 3 | |
| VGF | 489–507 | .EEKRKRKK | NAPPEPVPPPRAAPAPTHV | RSPQPPPP. | 1914.01 | 1914.01 | 1 | 3 | 1 | Amy | 0.94 | 0.12 | 0.88 | 0.08 | 3 | |
| VGF | 489–507 | .EEKRKRKK | NAPPEPVPPPRAAPAPTHV | RSPQPPPP. | 1914.01 | 1913.99 | −10 | 3 | 1 | Stri | 0.94 | 0.11 | 0.55 | 0.06 | *a | 3 |
Precursor and peptide names are indicated in columns 1 and 2. Amino acid sequences around the cleavage the cleavage sites are indicated in column 3 (upstream) and column 5 (downstream). Most peptide sequences listed in column 4 were derived from MS/MS sequence information, in addition to several other criteria (mass within 0.005% of a known peptide, and the correct number of isotopic tags incorporated and charge state based on amino acid composition; see Materials and Methods) . However, for some peptides there was insufficient MS/MS information and these peptides are therefore only tentatively identified by the matches to known peptides; these tentatively identified peptides are indicated by parentheses surrounding the sequence. Abbreviations: Theor. Mass, theoretical monoisotopic mass in Da; Obs Mass, the observed monoisotopic molecular mass (in Da) of the unprotonated peptide after subtracting mass of tags; ppm, the difference in parts per million between the observed and theoretical masses; z, charge state;
T, number of TMAB labels observed on the peptides; Reg., brain region where the peptides was found: Amy, Amygdala; Hyp, Hypothalamus; and Stri, Striatum. PC1 Het/WT ratio, the ratio of peptide levels in extracts of PC1/3 heterozygous brain regions relative to the levels in extracts of wild-type mice. PC1 KO/WT ratio, the ratio of peptide levels in extracts of PC1/3 null brain regions relative to the levels in extracts of wild-type mice. Avg.and SEM indicate the average and standard error of the mean of ratios among the runs in which peptides were found. Sig., level of significance of PC1 KO/WT compared to PC1 Het/WT or wild-type/wild-type ratios: ns, not significant;
p ≤0.05;
p ≤0.01;
statistical significance versus PC1 Het/WT ratios;
statistical significance versus wild-type/wild-type ratios previously published (Zhang et al. 2008). n, number of runs in which the peptide was found. Note: SAAS, and VGF are not abbreviations; these are peptide names. CART, cocaine- and amphetamine-regulated transcript; PACAP, pituitary adenylate cyclase-activating polypeptide. Mox, or MetOx, oxidized methionine. pS, phosophoserine.
Many of the peptides showed generally similar results for the three replicates (Supplemental Figure S2). However, some showed large variation between the replicates, as reflected in the large standard error in Table 1. Some peptides were detected in multiple brain regions, and for most of these the changes were generally similar among the different regions. For example, the peptide little LEN decreased 55%, 69%, and 66% in the amygdala, hypothalamus, and striatum, respectively (Table 1 and Figure S3C, D). A small number of peptides showed variability among brain regions. For example, big LEN showed an increase of 144% in amygdala of PC1/3 KO mice (KO/WT ratio 2.44), an increase of 170% in hypothalamus (ratio 2.70), but no major change in striatum (ratio 1.06). The proenkephalin peptides Met-enkephalin and SPQLEDEAKELQ are increased 71% and 132% (ratios 1.71 and 2.32, respectively) in amygdala of the PC1/3 KO mice but not changed in the other two brain regions examined (Table 1). Because it is unlikely that these proenkephalin peptides are substrates of PC1/3, a simpler explanation of these data is that proenkephalin production is elevated in the amygdala of the KO mice. Because big LEN is a potential substrate of PC1/3, and because little LEN is decreased in the PC1/3 KO mice, it is likely that big LEN is a substrate of this enzyme. The proSAAS derived peptide big SAAS 1–24 is also a potential substrate of PC1/3, based on the large increase observed for this peptide in the PC1/3 KO mouse hypothalamus (Table 1).
PC1/3 and PC2 have been proposed to play complementary and overlapping roles in the processing of many neuropeptides. To explore the relationship, the peptidomics data from the present study was compared to recent peptidomics data on PC2 KO mice (Figure 1). Only those peptides found in the same brain region in both of these studies were considered; the brain area was taken into account because there may be regional differences in expression that could affect the relative contribution of each processing enzyme among brain regions. One major difference between the peptidomics results of the two different KO mice is that PC2 KO mice show many peptides decreased by >50%, whereas the PC1/3 KO show only a handful of peptides decreased by>50% (Figure 1 and supplemental Table S1). This suggests that PC2 has a unique role in the processing of a number of substrates, and that without this enzyme the products are not detected. In contrast, this would suggest that PC1/3 has fewer unique substrates and that other enzymes (such as PC2) can also cleave many of the PC1/3 substrates. Another observation from the comparison between the PC1/3 and PC2 KO peptidomics studies is that only a single peptide is substantially decreased in both mutant animals (Figure 1 and supplemental Table S1). Not surprisingly, this peptide is a fragment of PC1/3 that was previously proposed to require PC2 for the processing. Several peptides showed intermediate values in both KOs, and would therefore appear to require both enzymes for efficient processing. For example, the chromogranin B peptide LLDEGHYPV shows a KO/WT ratio of 0.23 for the PC1/3 hypothalamus and 0.71 for the PC2 hypothalamus; in the striatum, these ratios are 0.37 and 0.43, respectively. Thus, it would appear that the combination of the two PCs is required for the optimal production of this peptide. A large number of peptides show partial decreases in each of the studies, but unlike the chromogranin B substrate the sum of the ratios is >1.0, suggesting that the two enzymes are partially redundant.
Figure 1.
Comparison of results from analyses of PC1/3 null mice versus PC2 null mice. Neuropeptides detected in both the present analysis of PC1/3 null mice and previously published analysis of PC2 null mice are shown. The relative levels of peptides in PC1/3 and PC2 null mice, compared to their respective WT counterparts in each experiment, are indicated. The individual peptides used for this analysis are listed in supplementary Table S1, supplement.
DISCUSSION
The present study is the first to describe the peptidomic analysis of PC1/3 null mouse brain. Due to the reduced perinatal survival of the PC1/3 null mice, relatively few animals were available for analysis. Therefore multiple PC1/3 null mice were not pooled as typically done for peptidomics analysis, but instead individual mice were analyzed in separate LC/MS runs which resulted in weaker signals, fewer peptides detected, and more variability among replicates. Despite this, a number of peptides were detected in this analysis and the majority of the replicates were consistent within and between different brain regions. Some peptides did vary between regions such as Big LEN, a proSAAS derived peptide which increased over twofold versus WT mice in the amygdala and hypothalamus of the PC1/3 null mice while showing no increase in the KO mouse striatum. This is in contrast to Little LEN, a Big LEN derivative that decreased in the PC1/3 null mice in all brain regions, suggesting that Little LEN is a PC1/3 product in all regions examined (Figure S3). This apparent paradox can be explained by either a varying level of Big LEN expression or processing among different brain regions. Big LEN may be so abundant in the striatum that any differential processing of the peptide to Little LEN is not reflected in a change in the overall amount of Big LEN in the striatum, hence the relative decrease in the PC1/3 cleavage product Little LEN with no apparent increase in Big LEN in this brain region. Alternatively, it is possible that the changes in peptide levels are the indirect result of the disruption of the PC1/3 gene. PC 1/3 null mice have previously been shown to exhibit decreases in the activity and viability of certain cell types; anterior pituitary somatotrophs were found to be shrunken and inactive, presumably due to lack of stimulation (Zhu et al. 2002b). It is also possible that changes in peptide levels in PC1/3 null mice are due in part to changes in PC2 activity, although previous studies have found that the overall levels of PC2 are not significantly altered in the brain of the PC1/3 null mouse (Pan et al. 2005).
ProSAAS was originally identified as a PC1/3 inhibitor (Fricker et al. 2000), and the inhibitory region of proSAAS was found to be the 6–8 amino acid region surrounding the junction of PEN and LEN (Cameron et al. 2000;Basak et al. 2001;Qian et al. 2000). PC1/3 cleaves the Lys-Arg within the inhibitory region of proSAAS, although cleavage is very slow and the peptide binds tightly to PC1/3 (Cameron et al. 2000). Thus, a key question has been whether PC1/3 is responsible for the processing of proSAAS peptides, or whether another PC (or another endopeptidase) performs these cleavages. In the present study, it is clear that processing of proSAAS peptides is affected by the loss of PC1/3 in the regions studied. Big LEN, Little LEN, Big SAAS and Little SAAS levels are all affected by the loss of PC1/3, indicating that PC1/3 is instrumental in processing of these peptides in the brain regions studied. In contrast, most of these peptides are not affected by the absence of PC2 activity, except for a small decrease in Big LEN seen in PC2 null striatum (Zhang et al. 2010). There are similarities between the inhibition of PC1/3 activity by proSAAS and the inhibition of PC2 by the neuroendocrine protein 7B2, which contains a 31 residue peptide that inhibits PC2 in a tight-binding, highly specific manner (Muller and Lindberg 1999). Inactivation of the 7B2 C-terminal peptide is thought to be accomplished by PC2-mediated hydrolysis at the inhibitory dibasic pair, followed by carboxypeptidase action, much like the inactivation of proSAAS by the slow action of PC1/3 (Zhu et al. 1996).
A previous study reported a slight decrease in the level of Met-enkephalin and an increase in the level of Met-enkephalin precursors in PC1/3 null mice (Pan et al. 2005). These studies were carried out using radioimmunoassays to examine levels in extracts from whole brains. The brains of these animals were not microwave irradiated (which greatly reduces postmortem peptide breakdown) and the extraction was performed in boiling acetic acid, which has been found to lead to protein/peptide degradation (Che et al. 2005b;Che et al. 2007). The current study used striatum, amygdala and hypothalamus, prevented peptide breakdown by microwave irradiation, and analyzed the results using a mass spectrometry-based quantitative peptidomics technique. Because of these major differences between the previous and current experimental protocols it is difficult to compare the results. The previous study by Pan et al used a similar quantitative peptidomics technique to study peptide processing in PC1/3 null mouse pituitary. Three peptides were found in both the Pan et al study and the present peptidomic study. One peptide detected in both studies, which corresponds to the C-terminal region of provasopressin (VQLAGTRESVDSAKPRVY), does not require PCs and so the changes in peptide levels observed in the present study likely reflect secondary changes. The two other peptides both require PC cleavages. One of these, the chromogranin B-derived peptide (LLDEGHYPV) was found to decrease substantially in the PC1/3 null mouse hypothalamus, striatum and pituitary. The other, derived from chromogranin A (AYGFRDPGPQL) increased in PC1/3 null hypothalamus but decreased in pituitary. Thus, there are both similarities and differences in the effect of the absence of PC1/3 activity in the levels of peptides in pituitary versus brain.
Although there is evidence that cathepsin L could be responsible for endoproteolytic cleavage of neuropeptide precursors in secretory vesicles (Funkelstein et al. 2008;Hook 2006b;Beinfeld et al. 2009;Minokadeh et al. 2009), PC1/3 and 2 certainly are major enzymes for processing many peptides. The results of the present study are in agreement with many previous studies showing that PC1/3 plays a critical role in the production of many neuropeptides (Benjannet et al. 1991;Zhu et al. 2002b;Furuta et al. 1997;Furuta et al. 2001;Furuta et al. 1998;Johanning et al. 1998;Berman et al. 2000;Allen et al. 2001;Dey et al. 2003). However, levels of some peptides are not changed by the absence of either PC1/3 or PC2 alone. One explanation is that these two enzymes are redundant and can both cleave a wide range of substrates, which is supported by in vitro studies with purified enzyme and peptides (Benjannet et al. 1991;Zhou and Lindberg 1993;Dupuy et al. 1994;Johanning et al. 1998;Day et al. 1998). However, it cannot be ruled out that cathepsin L also contributes in part to the processing of a subset of these peptides, especially those that are not substantially decreased in the absence of either PC1/3 or PC2. However, studies on the Cpefat/fat mice, which found nearly all peptides to be greatly decreased in mice that lack CPE activity, argue that the major processing route is via a pathway that involves PCs and CPs rather than cathepsin and aminopeptidases (Lim et al. 2006;Zhang et al. 2008)
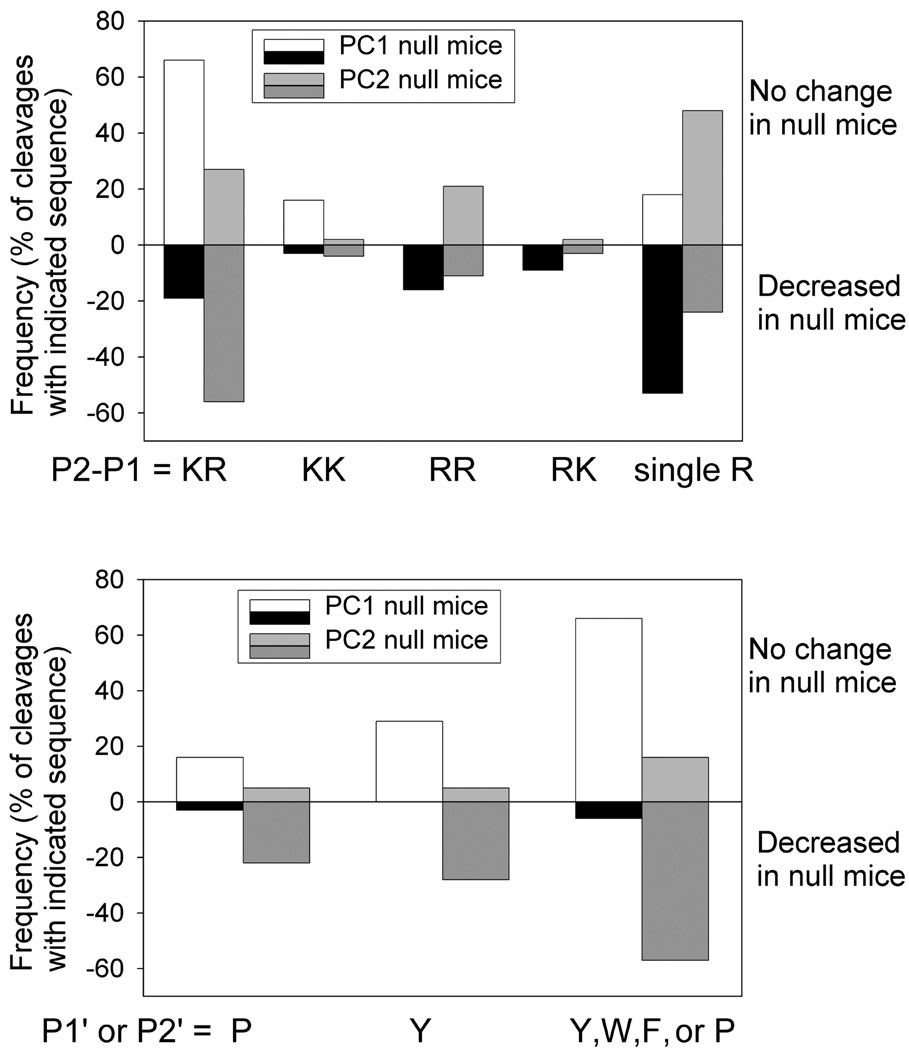
Although PC1/3 and PC2 are known to have overlapping specificities, they are not entirely redundant and each has a preference for particular cleavage sequences. Studies testing the enzyme’s activities in vitro have revealed preferences for PC1/3 or PC2 (Cameron et al. 2001;Remacle et al. 2008). A recent peptidomics study on PC2 KO mice found some of these preferences in an analysis of approximately 150 peptides in multiple brain regions (Pan et al. 2006;Zhang et al. 2010). Some of these preferences for PC2 selectivity, such as the presence of a Pro, Tyr, Trp, or Phe in the P1’or P2’ positions, were found in the present study to show a strong correlation in whether a peptide was cleaved in the PC1/3 KO mice (Pan et al. 2006;Zhang et al. 2010). Cleavage sites of peptides found to change in the current study on PC1/3 null mice were compared to previous results from peptidomics studies of peptides in PC2 KO mouse brain (Pan et al. 2006;Zhang et al. 2010); for this analysis, the previous PC2 KO mouse brain data was re-examined using the same search criteria. Lys-Arg is the most frequent combination of residues in the P2-P1 positions of the cleavage site, being present in ~80% of the cleavage sites (Figure 2, top, and Table S2). The majority of the peptides that contain Lys-Arg in the cleavage site are not affected by the absence of PC1/3 activity, while the majority of peptides with Lys-Arg are decreased in the PC2 null mice (Figure 2, top). Although there are only a small number of cleavage sites with Lys-Lys, the trend for preference by PC1/3 and PC2 is similar as with the Lys-Arg sites (Figure 2, top). In contrast, all of the peptides that contain cleavage sites with an Arg in the P2 position, either Arg-Arg or Arg-Lys, are decreased in the PC1/3 null mice (Figure 2, top). This result implies that Lys-Arg and Lys-Lys are more likely to be cleaved in vivo by PC2 than by PC1, while Arg-Arg and Arg-Lys are more likely to be cleaved by PC1/3. Sites containing a single Arg in the P1 position, without a Lys or Arg in the P2 position, are more commonly found to be decreased in the PC1/3 null mice and unchanged in the PC2 null mice, although there was overlap between these groups (Figure 2, top).
Figure 2.
Cleavage site analysis for PC1/3 null and PC2 null mice. Graphs show frequency at which peptides are cleaved at specific residues in peptides that either decrease or show no change in PC1/3 null or PC2 null mice relative to wild-type mice. Graph A indicates these frequencies in P2 and P1 positions. “Single R” refers to cleavages with a non-basic residue in the P2 position, but does not take into account if there is a basic residue in the P4 or P6 positions. Graph B indicates frequencies of selected residues in the P1’ and P2’ positions. The individual peptides used for this analysis are listed in Table 1, and the numbers of peptides in each category are indicated in Table S2, supplement.
Analysis of the residues to the C-terminal side of the cleavage site also suggests that residues in these positions contribute to the preference of PC1/3 versus PC2 (Figure 2, bottom). As described above, previous studies on the PC2 KO mice as well as analysis of in vitro enzyme activities suggested a Pro or aromatic residue in either the P1’ or P2’ position gave a preference for cleavage by PC2 and reduced the cleavage by PC1/3. Consistent with these previous observations, the majority of cleavage sites with a Pro in the P1’ or P2’ positions were not affected by the absence of PC1/3 but were decreased in the PC2 null mice (Figure 2, bottom). Similarly, Tyr in the P1’ or P2’ positions was also predictive of cleavages that were not affected in the PC1/3 null mice but which were affected in the PC2 null mice (Figure 2, bottom). The inclusion of other bulky aromatic groups, Trp and Phe, in this analysis showed the strongest correlation with cleavage in the two null PC lines, with the vast majority of the peptides unaffected in the PC1/3 null mice containing a Tyr, Trp, Phe, or Pro in the P1’ or P2’ position, while in the PC2 null mice these cleavages were decreased (Figure 2, bottom). These data support previous findings that PC1/3 and PC2 have overlapping but distinct substrate specificities (Benjannet et al. 1991;Seidah and Chretien 2004b;Seidah and Chretien 2004a;Cameron et al. 2001;Dey et al. 2003).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported primarily by National Institutes of Health grant DA-04494 (L.D.F.), and also by DK-13914 (D.F.S.), by the Howard Hughes Medical Institute (D.F.S.) and by the Canadian Institutes of Health Research (R.D.). R.D. is a member of the FRSQ funded Centre de recherche clinique Étienne-Le Bel. Mass spectrometry was performed through the Rede de Proteoma do Estado de São Paulo in the Laboratório Nacional de Luz Sincrotron, Campinas, SP, Brazil, and was funded in part by Fundação de Amparo a Pesquisa do Estado de São Paulo 04/04933-2, 04/14846-0 and Financiadora de Estudos e Projetos A-03/134 (to Emer S. Ferro), and a fellowship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (L.M.C.). We are thankful to Prof. Fabio Gozzo, Universidade de Campinas, for his immeasurable support with mass spectrometry.
REFERENCES
- Allen RG, Peng B, Pellegrino MJ, Miller ED, Grandy DK, Lundblad JR, Washburn CL, Pintar JE. Altered processing of pro-orphanin FQ/nociceptin and pro-opiomelanocortin-derived peptides in the brains of mice expressing defective prohormone convertase 2. J Neurosci. 2001;21:5864–5870. doi: 10.1523/JNEUROSCI.21-16-05864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggerman G, Verleyen P, Clynen E, Huybrechts J, De Loof A, Schoofs L. Peptidomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;803:3–16. doi: 10.1016/j.jchromb.2003.07.019. [DOI] [PubMed] [Google Scholar]
- Basak A, Koch P, Dupelle M, Fricker LD, Devi LA, Chretien M, Seidah NG. Inhibitory specificity and potency of proSAAS-derived peptides toward proprotein convertase 1. J Biol Chem. 2001;276:32720–32728. doi: 10.1074/jbc.M104064200. [DOI] [PubMed] [Google Scholar]
- Beinfeld MC, Funkelstein L, Foulon T, Cadel S, Kitagawa K, Toneff T, Reinheckel T, Peters C, Hook V. Cathepsin L plays a major role in cholecystokinin production in mouse brain cortex and in pituitary AtT-20 cells: protease gene knockout and inhibitor studies. Peptides. 2009;30:1882–1891. doi: 10.1016/j.peptides.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjannet S, Rondeau N, Day R, Chretien M, Seidah NG. PC1 and PC2 are proprotein convertases capable of cleaving proopiomelanocortin at distinct pairs of basic residues. Proc Natl Acad Sci USA. 1991;88:3564–3568. doi: 10.1073/pnas.88.9.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman Y, Mzhavia N, Polonskaia A, Furuta M, Steiner DF, Pintar JE, Devi LA. Defective prodynorphin processing in mice lacking prohormone convertase PC2. J Neurochem. 2000;75:1763–1770. doi: 10.1046/j.1471-4159.2000.0751763.x. [DOI] [PubMed] [Google Scholar]
- Berti DA, Morano C, Russo LC, Castro LM, Cunha FM, Zhang X, Sironi J, Klitzke CF, Ferro ES, Fricker LD. Analysis of intracellular substrates and products of thimet oligopeptidase (EC 3.4.24.15) in human embryonic kidney 293 cells. J Biol Chem. 2009;284:14105–14116. doi: 10.1074/jbc.M807916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslin MB, Lindberg I, Benjannet S, Mathis JP, Lazure C, Seidah NG. Differential processing of proenkephalin by prohormone convertases 1(3) and 2 and furin. J Biol Chem. 1993;268:27084–27093. [PubMed] [Google Scholar]
- Cameron A, Apletalina EV, Lindberg I. The Enzymology of PC1 and PC2. Enzymes. 2001;22:291–332. [Google Scholar]
- Cameron A, Fortenberry Y, Lindberg I. The SAAS granin exhibits structural and functional homology to 7B2 and contains a highly potent hexapeptide inhibitor of PC1. FEBS Lett. 2000;473:135–138. doi: 10.1016/s0014-5793(00)01511-8. [DOI] [PubMed] [Google Scholar]
- Che F-Y, Biswas R, Fricker LD. Relative quantitation of peptides in wild type and Cpefat/fat mouse pituitary using stable isotopic tags and mass spectrometry. J Mass Spectrom. 2005a;40:227–237. doi: 10.1002/jms.742. [DOI] [PubMed] [Google Scholar]
- Che F-Y, Fricker LD. Quantitative peptidomics of mouse pituitary: Comparison of different stable isotopic tags. J Mass Spectrom. 2005;40:238–249. doi: 10.1002/jms.743. [DOI] [PubMed] [Google Scholar]
- Che F-Y, Lim J, Biswas R, Pan H, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics. 2005b;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- Che FY, Zhang X, Berezniuk I, Callaway M, Lim J, Fricker LD. Optimization of neuropeptide extraction from the mouse hypothalamus. J Proteome Res. 2007;6:4667–4676. doi: 10.1021/pr060690r. [DOI] [PubMed] [Google Scholar]
- Day R, Lazure C, Basak A, Boudreault A, Limperis P, Dong W, Lindberg I. Prodynorphin processing by proprotein convertase 2: Cleavage at single basic residues and enhanced processing in the presence of carboxypeptidase activity. J Biol Chem. 1998;273:829–836. doi: 10.1074/jbc.273.2.829. [DOI] [PubMed] [Google Scholar]
- Dey A, Xhu X, Carroll R, Turck CW, Stein J, Steiner DF. Biological processing of the cocaine and amphetamine-regulated transcript precursors by prohormone convertases, PC2 and PC1/3. J Biol Chem. 2003;278:15007–15014. doi: 10.1074/jbc.M212128200. [DOI] [PubMed] [Google Scholar]
- Dupuy A, Lindberg I, Zhou Y, Akil H, Lazure C, Chretien M, Seidah NG, Day R. Processing of prodynorphin by the prohormone convertase PC1 results in high molecular weight intermediate forms. Cleavage at a single arginine residue. FEBS Lett. 1994;337:60–65. doi: 10.1016/0014-5793(94)80630-6. [DOI] [PubMed] [Google Scholar]
- Fricker LD, Lim J, Pan H, Che F-Y. Peptidomics: Identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom Rev. 2006;25:327–344. doi: 10.1002/mas.20079. [DOI] [PubMed] [Google Scholar]
- Fricker LD, McKinzie AA, Sun J, Curran E, Qian Y, Yan L, Patterson SD, Courchesne PL, Richards B, Levin N, Mzhavia N, Devi LA, Douglass J. Identification and characterization of proSAAS, a granin-like neuroendocrine peptide precursor that inhibits prohormone processing. J Neurosci. 2000;20:639–648. doi: 10.1523/JNEUROSCI.20-02-00639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funkelstein L, Toneff T, Mosier C, Hwang SR, Beuschlein F, Lichtenauer UD, Reinheckel T, Peters C, Hook V. Major role of cathepsin L for producing the peptide hormones ACTH, beta-endorphin, and alpha-MSH, illustrated by protease gene knockout and expression. J Biol Chem. 2008;283:35652–35659. doi: 10.1074/jbc.M709010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta M, Carroll R, Martin S, Swift HH, Ravazzola M, Orci L, Steiner DF. Incomplete processing of proinsulin to insulin accompanied by elevation of Des-31,32 proinsulin intermediates in islets of mice lacking active PC2. J Biol Chem. 1998;273:3431–3437. doi: 10.1074/jbc.273.6.3431. [DOI] [PubMed] [Google Scholar]
- Furuta M, Yano H, Zhou A, Rouille Y, Holst JJ, Carroll R, Ravazzola M, Orci L, Furata H, Steiner DF. Defective prohormone processing and altered pancreatic islet morphology in mice lacking active SPC2. Proc Natl Acad Sci USA. 1997;94:6646–6651. doi: 10.1073/pnas.94.13.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta M, Zhou A, Webb G, Carroll R, Ravazzola M, Orci L, Steiner DF. Severe defect in proglucagon processing in islet A-cells of prohormone convertase 2 null mice. J Biol Chem. 2001;276:27197–27202. doi: 10.1074/jbc.M103362200. [DOI] [PubMed] [Google Scholar]
- Hatcher NG, Atkins N, Jr, Annangudi SP, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV. Mass spectrometry-based discovery of circadian peptides. Proc Natl Acad Sci U S A. 2008;105:12527–12532. doi: 10.1073/pnas.0804340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook VY. Protease pathways in peptide neurotransmission and neurodegenerative diseases. Cell Mol Neurobiol. 2006a;26:449–469. doi: 10.1007/s10571-006-9047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook VY. Unique neuronal functions of cathepsin L and cathepsin B in secretory vesicles: biosynthesis of peptides in neurotransmission and neurodegenerative disease. Biol Chem. 2006b;387:1429–1439. doi: 10.1515/BC.2006.179. [DOI] [PubMed] [Google Scholar]
- Johanning K, Juliano MA, Juliano L, Lazure C, Lamango NS, Steiner DF, Lindberg I. Specificity of prohormone convertase 2 on proenkephalin and proenkephalin-related substrates. J Biol Chem. 1998;273:22672–22680. doi: 10.1074/jbc.273.35.22672. [DOI] [PubMed] [Google Scholar]
- Lim J, Berezniuk I, Che F-Y, Parikh R, Biswas R, Pan H, Fricker LD. Altered neuropeptide processing in prefrontal cortex of Cpefat/fat mice: Implications for neuropeptide discovery. J Neurochem. 2006;96:1169–1181. doi: 10.1111/j.1471-4159.2005.03614.x. [DOI] [PubMed] [Google Scholar]
- Mathis JP, Lindberg I. Posttranslational processing of proenkephalin in AtT-20 cells: Evidence for cleavage at Lys-Lys site. Endocrinol. 1992;131:2287–2296. doi: 10.1210/endo.131.5.1425427. [DOI] [PubMed] [Google Scholar]
- Minokadeh A, Funkelstein L, Toneff T, Hwang SR, Beinfeld M, Reinheckel T, Peters C, Zadina J, Hook V. Cathepsin L participates in dynorphin production in brain cortex, illustrated by protease gene knockout and expression. Mol Cell Neurosci. 2009 doi: 10.1016/j.mcn.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Morano C, Zhang X, Fricker LD. Multiple Isotopic Labels for Quantitative Mass Spectrometry. Anal Chem. 2008;80:9298–9309. doi: 10.1021/ac801654h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller L, Lindberg I. The cell biology of the prohormone convertases PC1 and PC2. Prog Nucleic Acid Res Mol Biol. 1999;63:69–108. doi: 10.1016/s0079-6603(08)60720-5. [DOI] [PubMed] [Google Scholar]
- Pan H, Che FY, Peng B, Steiner DF, Pintar JE, Fricker LD. The role of prohormone convertase-2 in hypothalamic neuropeptide processing: a quantitative neuropeptidomic study. J Neurochem. 2006;98:1763–1777. doi: 10.1111/j.1471-4159.2006.04067.x. [DOI] [PubMed] [Google Scholar]
- Pan H, Nanno D, Che FY, Zhu X, Salton SR, Steiner DF, Fricker LD, Devi LA. Neuropeptide Processing Profile in Mice Lacking Prohormone Convertase-1. Biochemistry. 2005;44:4939–4948. doi: 10.1021/bi047852m. [DOI] [PubMed] [Google Scholar]
- Paquet L, Zhou A, Chang EY, Mains RE. Peptide biosynthetic processing: distinguishing prohormone convertases PC1 and PC2. Mol Cell Endocrinol. 1996;120:161–168. doi: 10.1016/0303-7207(96)03834-8. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. San Diego: Academic Press; 2001. [Google Scholar]
- Prigge ST, Mains RE, Eipper BA, Amzel LM. New insights into copper monooxygenases and peptide amidation: structure, mechanism and function. Cell Mol Life Sci. 2000;57:1236–1259. doi: 10.1007/PL00000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Devi LA, Mzhavia N, Munzer S, Seidah NG, Fricker LD. The C-terminal region of proSAAS is a potent inhibitor of prohormone convertase 1. J Biol Chem. 2000;275:23596–23601. doi: 10.1074/jbc.M001583200. [DOI] [PubMed] [Google Scholar]
- Remacle AG, Shiryaev SA, Oh ES, Cieplak P, Srinivasan A, Wei G, Liddington RC, Ratnikov BI, Parent A, Desjardins R, Day R, Smith JW, Lebl M, Strongin AY. Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J Biol Chem. 2008;283:20897–20906. doi: 10.1074/jbc.M803762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik SE, Fricker LD. Carboxypeptidases from A to Z: implications in embryonic development and Wnt binding. Cell Mol Life Sci. 2001;58:1790–1804. doi: 10.1007/PL00000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah NG, Chretien M. Eukaryotic protein processing: endoproteolysis of precursor proteins. Curr Opin Biotechnol. 1997;8:602–607. doi: 10.1016/s0958-1669(97)80036-5. [DOI] [PubMed] [Google Scholar]
- Seidah NG, Chretien M. Proprotein convertase 2. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. San Diego: Academic Press; 2004a. pp. 1865–1868. [Google Scholar]
- Seidah NG, Chretien M. Proprotein convertase I. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. San Diego: Academic Press; 2004b. pp. 1861–1864. [Google Scholar]
- Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- Svensson M, Skold K, Nilsson A, Falth M, Nydahl K, Svenningsson P, Andren PE. Neuropeptidomics: MS applied to the discovery of novel peptides from the brain. Anal Chem. 2007;79:15–21. doi: 10.1021/ac071856q. [DOI] [PubMed] [Google Scholar]
- Zhang X, Che FY, Berezniuk I, Sonmez K, Toll L, Fricker LD. Peptidomics of Cpe(fat/fat) mouse brain regions: implications for neuropeptide processing. J Neurochem. 2008;107:1596–1613. doi: 10.1111/j.1471-4159.2008.05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Pan H, Peng B, Steiner DF, Pintar JE, Fricker LD. Neuropeptidomic analysis establishes a major role for prohormone convertase-2 in neuropeptide biosynthesis. J Neurochem. 2010;112:1168–1179. doi: 10.1111/j.1471-4159.2009.06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Mains RE. Endoproteolytic processing of proopiomelanocortin and prohormone convertases 1 and 2 in neuroendocrine cells overexpressing prohormone convertases 1 or 2. J Biol Chem. 1994;269:17440–17447. [PubMed] [Google Scholar]
- Zhou A, Webb G, Zhu X, Steiner DF. Proteolytic processing in the secretory pathway. J Biol Chem. 1999;274:20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lindberg I. Purification and characterization of the prohormone convertase PC1 (PC3) J Biol Chem. 1993;268:5615–5623. [PubMed] [Google Scholar]
- Zhu X, Orci L, Carroll R, Norrbom C, Ravazzola M, Steiner DF. Severe block in processing of proinsulin to insulin accompanied by elevation of des-64,65 proinsulin intermediates in islets of mice lacking prohormone convertase 1/3. Proc Natl Acad Sci USA. 2002a;99:10299–10304. doi: 10.1073/pnas.162352799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Rouille Y, Lamango NS, Steiner DF, Lindberg I. Internal cleavage of the inhibitory 7B2 CT peptide by PC2: A potential mechanism for its inactivation. Proc Natl Acad Sci USA. 1996;93:4919–4924. doi: 10.1073/pnas.93.10.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhou A, Dey A, Norrbom C, Carroll R, Zhang C, Laurent V, Lindberg I, Ugleholdt R, Holst JJ, Steiner DF. Disruption of PC1/3 expression in mice causes dwarfism and multiple neuroendocrine peptide processing defects. Proc Natl Acad Sci USA. 2002b;99:10293–10298. doi: 10.1073/pnas.162352599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.