Abstract
Intranasal MHV-1 infection of susceptible mouse strains mimics some important pathologic features observed in the lungs of SARS-CoV infected humans. The pathogenesis of Severe Acute Respiratory Syndrome (SARS) remains poorly understood, although increasing evidence suggests that immunopathology could play an important role. We previously reported that the adaptive immune response plays an important protective role in MHV-1 infected resistant B6 mice and that both CD4 and CD8 T-cells play a significant role in the development of morbidity and lung pathology following intranasal MHV-infection of susceptible C3H/HeJ and A/J mice. In this study we identified novel CD4 and CD8 epitopes in MHV-1 infected susceptible and resistant strains of mice. Susceptible C3H/HeJ mice mount robust and broad MHV-1 specific CD4 T-cell responses while in resistant B6 mice, antigen-specific CD8 T-cell responses dominate. We also show that previously immunized susceptible C3H/HeJ mice do not develop any morbidity and are completely protected following a lethal dose MHV-1 challenge despite mounting only a modest secondary T-cell response. Finally we demonstrate that the resistance displayed by B6 mice is not solely accounted for by the elaboration of a broad and vigorous MHV-1-specific CD8 T-cell response as MHV-1 infection of C3.SW-H2b/SnJ mice, which mount an equally robust CD8 T-cell response of the same specificity, is still associated with significant morbidity. Thus, identification of novel CD4 and CD8 T-cell epitopes for MHV-1 permitted high resolution analyses of pulmonary T-cell responses in a mouse model of SARS.
Introduction
Severe acute respiratory syndrome (SARS) is a clinical manifestation of infection by a human respiratory coronavirus SARS-CoV (1). Although the virus is capable of establishing a systemic infection, the most striking pathology is observed in the lungs (1). There appears to be an emerging consensus that immunopathology may play an important role in mediating the morbidity and mortality associated with SARS-CoV infection (2-4). The early phase of SARS-CoV infection is associated with elevated levels of pro-inflammatory cytokines (2, 3) however, the exact role of specific components of the immune response to SARS-CoV infection that could be involved in the development of pathology is still unclear and remains an area of active investigation. While some reports have suggested a link between the severity of disease and an inability of the host to mount an optimal adaptive immune response following infection, others have reported a correlation between increased disease severity and robust antigen-specific CD4 and CD8 T cell responses (2, 3). In addition, one of the major limitations of human studies is their restriction to analyzing responses in peripheral blood mononuclear cells (PBMC) and the inability to sample immune responses occurring in the tissue of interest.
MHV-1 infection of susceptible strains of mice has been shown to be a clinically relevant model to study SARS (5-7). Using this model we have recently shown that both CD4 and CD8 T cells contribute to the development of morbidity and lung pathology in susceptible strains of mice following primary sublethal MHV-1 infection (6). Additionally, we see enhancement of disease in naïve recipients that are adoptively transferred with purified memory CD4 and CD8 T cells obtained from donors previously immunized with MHV-1 (6). In concordance with other published reports (8-11), we have also shown that primary MHV-1 infection of susceptible strains of mice induces robust neutralizing antibody responses and transfer of immune serum into naïve recipients that are subsequently challenged with MHV-1 reduces systemic viral burden and morbidity (6). On the other hand, B6 mice that are resistant to MHV-1-induced disease experience increased morbidity in the absence of T and B cells (6). Collectively, our data from MHV-1 infection of susceptible and resistant strains of mice suggest that the adaptive immune response can play an important role in infection control but can also mediate immunopathology in hosts with specific genotypes.
In this present study we have delineated the antigen-specific CD4 and CD8 T cell responses to MHV-1 structural proteins in susceptible C3H/HeJ and resistant B6 mice in order to define the role of the T cell response in mediating pathology and protection following MHV-1 infection. In addition, we have also examined the importance of non-MHC-linked genes in mediating susceptibility to MHV-1-induced respiratory disease.
Materials and Methods
Peptide Library
420 peptides encompassing the spike (S), nucleocapsid (N), membrane (M) and envelope (E) were synthesized by Mimotopes (Victoria, Australia) based on the MHV-1 structural protein sequence obtained from the NCBI database (accession number EF682498). Each peptide was a 15mer with an overlap of 10 amino acids (aa) and an offset of 5aa. The crude peptides (>65% pure by HPLC) were reconstituted using 100% sterile DMSO (Sigma-Aldrich, St. Louis, MO) to a concentration of 2mM. A sub-stock was prepared that was diluted to a concentration of 25μM using sterile 1X-Dulbecco's PBS (GIBCO, Grand Island, NY). Peptides were screened for potential epitopes by performing standard intracellular cytokine staining for IFNγ production from splenocytes obtained from mice 7 days after intraperitoneal infection with MHV-1 (described below). Following identification of the relevant epitopes, independent synthesis of candidate peptides was undertaken (Biosynthesis Inc, Lewisville, TX).
Mice
5-7 week old female A/J (H-2a), C3H/HeJ (H-2k), BALB/c (H-2d), C57BL/6J (H-2b) were purchased from the National Cancer Institute (NCI, Frederick, MD). C3.SW-H2b/SnJ mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Kb-/- mOva mice were obtained from Dr. Stephen P. Schoenberger (La Jolla Institute of Allergy and Immunology, San Diego, CA). All mice were housed under SPF conditions at the University of Iowa (Iowa City, IA) animal care unit until the time of infection at which point the mice were transferred to housing at the appropriate biosafety level. All animals were maintained in accredited facilities at the University of Iowa (Iowa City, IA) and used in accordance with the guidelines established by the University of Iowa animal care and use committee. Mice were infected at 8-9 weeks of age.
Virus
Parent stock of MHV-1 was obtained from the American Type Culture Collection, (ATCC, Manassas, VA). The virus was propagated and titered as previously described (6).
Virus infection of mice
For intranasal infections, mice were anesthetized with avertin (2,2,2-tribromoethanol; Aldrich, Milwaukee, WI) intraperitoneally (i.p.) and administered a sublethal dose of MHV-1 intranasally in a volume of 50μl. The sublethal dose used for infecting C3H/HeJ mice was 5 × 103 PFU/mouse and 105 PFU/mouse for B6 mice. This dosing was based on morbidity and mortality patterns described for susceptible and resistant strains previously (6). For screening the peptide library, splenocytes were obtained from mice that had been previously infected with 2 × 105 PFU of MHV-1 i.p.
Analysis of FoxP3+CD4+ T cells
Single cell suspensions obtained from spleens, lungs and draining lymph nodes of naïve B6 and C3H/HeJ control mice and B6 and C3H/HeJ mice infected intranasally with MHV-1 were stained for Foxp3 using the mouse regulatory T cell staining buffer kit (eBioscience, San Diego, CA) according to the manufacturer's instructions. Briefly, following cell surface staining and fixation, cells were stained with optimal concentrations of mAb specific to Foxp3 (clone FJK-16s; eBioscience, San Diego, CA). Cells were then washed twice with 1X permeabilization buffer and resuspended in staining buffer.
Intracellular Cytokine Staining
Intracellular cytokine staining was performed as previously described (12, 13). For the peptide library screen and analysis of Kb-/-mOva mice, single-cell suspensions from the spleens of MHV-1 infected mice were either left untreated or stimulated with MHV-1 derived peptide epitopes at a concentration of 1μM for 5 hours at 37°C. The intracellular accumulation of IFNγ was facilitated by the addition of brefeldin A (Golgiplug; BD Pharmingen). Infected mice were perfused with 10 ml PBS and explanted lungs were treated with Collagenase (100U/ml) (GIBCO)/DNAse I (1mg/ml) (Sigma, St. Louis) for 45 minutes. Single-cell suspensions were prepared using wire mesh screens and erythrocytes were removed by treatment with ACK lysis buffer and counted using 0.1% trypan blue. Antigen presenting cells (APCs) were left either untreated or incubated with appropriate MHV-1-derived peptides at a concentration of 1μM for CD8 peptide epitopes and 10μg/ml for CD4 peptide epitopes at 37°C for 1 hour, washed and added to each tube along with the lung cell suspensions followed by incubation for 5 hours at 37°C. For B6 mice, the mouse B cell line CHB3 was utilized as APCs. In the case of C3H/HeJ mice, APCs were derived from splenocytes of naïve mice that were left either untreated or incubated with appropriate MHV-1 peptide epitopes at 37°C for 1 hour, washed and then labeled with 2μM CFSE for 10 min at 37°C in PBS to facilitate discrimination from lung cells during flow cytometric analysis. For assays describing the identification of MHC restriction element for CD8 peptide epitopes in B6 mice, APCs used were either P815 cells (H-2d MHC) or P815 cells transfected with cDNA encoding either the Db or Kb molecules. Total numbers of antigen-specific CD4 and CD8 T cells in the spleens and lungs were determined by multiplying the frequency of IFNγ+ cells with total numbers of CD4 or CD8 T cells present in that tissue. Surface and intracellular staining was performed using the following mAbs, anti-CD8 (clone 53-6.7), anti-CD4 (clone RM4-5), anti-CD90.2 (clone 30-H12), anti-IFNγ (clone XMG1.2), anti-TNFα (clone MP6-XT22) and anti-IL2 (clone JES6-5H4). Samples were acquired using a FACSCalibur and FACS Canto (BD Biosciences, San Jose, CA) and analyzed using FlowJo software (Tree Star Inc., OR).
Evaluation of morbidity and mortality
For the evaluation of weight loss after infection, the weight of each mouse was normalized to 100% at the time they were infected (D0) and subsequent weight measurements were recorded at defined time points after infection. Weight data are presented as the mean percentage of the starting weight±SEM. Survival data for mice are represented by Kaplan-Meier curves indicating the percentage of mice that survived the viral challenge.
Lung Function
Baseline airway resistance as determined by the measurement of the parameter “enhanced pause” (Penh) was evaluated using a whole body plethysmograph from Buxco Electronics, Inc (Troy, NY).
Statistical Analyses
All statistical analyses were performed using GraphPad software (San Diego, CA). For weight loss and cytokine analysis data, statistical significance was determined using an unpaired Student's t test. A p value <0.05 was considered statistically significant. For survival data, statistical significance was determined using a two-tailed Fisher's exact test. Once again, a p value <0.05 was considered statistically significant.
Results
Identification of MHV-1-specific CD4 and CD8 T cell epitopes in susceptible and resistant mice
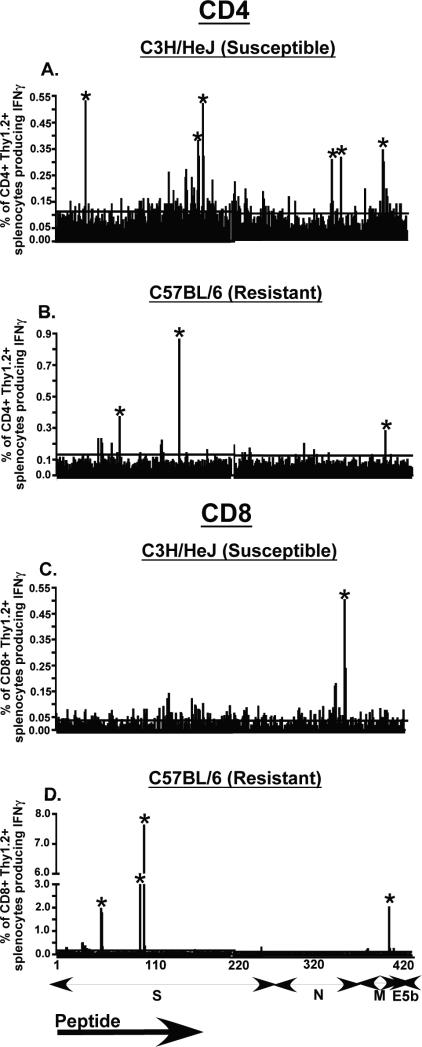
Previously published data from our lab showed that T cells contribute to morbidity and lung pathology observed in MHV-1 susceptible strains of mice (6). We have also demonstrated that resistance to MHV-1-induced disease is compromised in B6-Rag1-KO mice indicating the importance of the adaptive immune response in infection control (6). Based on these data we wanted to examine antigen-specific T cell responses to MHV-1 in susceptible and resistant strains of mice. T cell epitopes in other coronaviruses have been primarily localized to structural proteins (3, 14-20), thus we obtained protein sequence information for the spike (S), nucleoprotein (N), membrane (M) and envelope (E) structural proteins of MHV-1 from the NCBI database (accession number EF682498) to screen a peptide library consisting of 420 overlapping peptides encompassing the four proteins mentioned above. Each peptide was a 15mer with an overlap of 10 amino acids (aa) and an offset of 5 aa. To identify MHV-1-specific T cell epitopes, each peptide was individually screened in an intracellular cytokine staining assay to determine IFNγ production by peptide-pulsed splenocytes harvested and pooled from mice 7 days after intraperitoneal MHV-1 infection. The large number of peptides to be screened necessitated the use of splenocytes instead of lung cells in our initial analysis. Responses to peptide stimulation that resulted in IFNγ production at least 2.5-fold over that observed in no-peptide controls were considered positive. We were able to identify several novel CD4 and CD8 T cell epitopes in both susceptible (C3H/HeJ) and resistant (B6) mice (Fig.1A-D and Table 1). In the susceptible C3H/HeJ strain of mice we identified six novel CD4 epitopes, three of which mapped to the spike protein, two to the nucleocapsid protein and one to the membrane protein. The screen revealed a single CD8 epitope in these susceptible mice and it was localized in the nucleocapsid protein. Based on the analysis of the peptide library screen for the resistant B6 mice we identified four CD8 epitopes and three CD4 epitopes. Once again the majority of CD4 and CD8 epitopes were found in the spike protein. None of the epitopes identified in the resistant B6 mice mapped to the nucleocapsid protein. In addition we also identified several novel MHV-1-specific CD4 and CD8 T cell epitopes in susceptible A/J mice and BALB/c mice that display an intermediate susceptibility phenotype (data not shown). These analyses revealed that MHV-1 infection induced a more robust and broader virus-specific CD4 T cell response in the susceptible C3H/HeJ mice whereas the resistant B6 mice mounted a broad and vigorous virus-specific CD8 T cell response with the S587-594 specific response clearly being the most dominant.
Figure 1. Screening of the peptide library.
C3H/HeJ and B6 mice were infected with MHV-1 i.p. (2 × 105 PFU/mouse) and 7 days after infection spleens were harvested and processed to generate single cell suspensions and pooled to be used as effectors in an intracellular cytokine assay. To induce IFNγ production, effector splenocytes were stimulated with each of the 420 individual overlapping peptides separately. These 420 peptides encompassed the four main structural proteins, the spike (S; peptides 1-271), nucleocapsid (N; peptides 272-360), membrane (M; peptides 361-404) and envelope (E; peptides 405-420). Data are shown as % of CD4+Thy1.2+ splenocytes producing IFNγ for C3H/HeJ mice (A) and B6 mice (B) and % of CD8+Thy1.2+ splenocytes producing IFNγ for C3H/HeJ mice (C) and B6 mice (D). Responses that were at least 2.5 fold above those of no-peptide controls were scored as positive (indicated by an asterisk). The line parallel to the x-axis indicates the magnitude of the background response observed in no-peptide controls.
Table 1.
Identification of MHV-1 specific T cell epitopes in susceptible C3H/HeJ and resistant B6 mice
| C3H/HeJ | |||
|---|---|---|---|
| 15 mer | CD4 or CD8 Epitope | Predicted 8 mer | Predicted MHC Restriction |
| M196-210 | CD4 | I-Ek | |
| S171-185 | CD4 | I-Ek | |
| S881-895 | CD4 | I-Ek | |
| S921-935 | CD4 | I-Ek | |
| N346-360 | CD4 | I-Ek | |
| N376-390 | CD4 | I-Ek | |
| N421-435 | CD8 | N421-428 | Dk |
| C57BL/6 | ||||
|---|---|---|---|---|
| 15 mer | CD4 or CD8 Epitope | Predicted 8 mer | Predicted MHC Restriction | Confirmed MHC Restriction |
| M131-145 | CD4 | I-Ab | ||
| S361-375 | CD4 | I-Ab | ||
| S766-780 | CD4 | I-Ab | ||
| M184-198 | CD8 | M184-191 | Kb | Kb |
| S317-331 | CD8 | S317-324 | Kb | Kb |
| S532-546 | CD8 | S532-539 | Kb | Kb |
| S587-601 | CD8 | S587-595 | Kb | Kb |
Prediction of MHC-restriction elements and minimal epitopes of newly identified MHV-1-specific epitopes
The peptides identified as those containing CD4 T cell epitopes were further analyzed using the SYFPEITHI epitope-prediction software to determine putative MHC-restriction elements (21). Based on this analysis all of the CD4 epitopes identified in susceptible C3H/HeJ mice were predicted to bind to I-Ek and since the resistant B6 mice lack the I-Eb allele we expected all 3 CD4 epitopes identified in these mice to be I-Ab-restricted and this was also confirmed using the software program. Given the variability in the lengths of CD4 epitopes in general, further definition of the CD4 epitopes were not attempted.
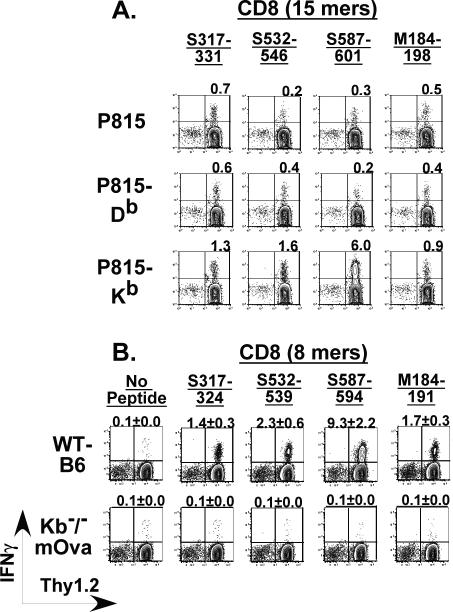
The CD8 peptides identified as those containing potential epitopes by the peptide library screen were similarly analyzed by the SYFPEITHI epitope-prediction software program (21). The lone CD8 epitope in the C3H/HeJ was determined to be an octamer expressed in the context of Dk based on the presence of the arginine residue at position 2 and leucine at position 8 (22). The four CD8 peptides that came up positive for potential epitopes in B6 mice were similarly predicted to be octamers restricted by Kb. These predictions regarding MHC restriction for CD8 epitopes identified in B6 mice were formally confirmed using P815 cells transfected with either the Db or Kb molecules in intracellular cytokine staining assays where P815-Db or P815-Kb cells coated with 15mers were used to stimulate splenocytes obtained from MHV-1-infected B6 mice (Figure 2A). These results were also separately confirmed by intracellular cytokine staining for IFNγ production using splenocytes from MHV-1-infected wild-type B6 and Kb-/-mOva B6 mice (Figure 2B) that do not express the Kb MHC molecule on the surface of their cells but are fully capable of presenting epitopes that bind to the Db molecule (unpublished observations). Based on these data we obtained the minimal predicted CD8 epitopes for both C3H/HeJ and B6 mice.
Figure 2. Identification of MHC-restriction element for CD8 epitopes identified in MHV-1 infected B6 mice.
(A) 15 mers identified by the peptide library screen to contain potential epitopes recognized by CD8 T cells from MHV-1 infected mice were further analyzed to identify the restricting MHC molecule. Single cell suspensions derived from the spleens of B6 mice infected with MHV-1 (2 × 105 PFU/mouse; i.p.) 7 days previously were utilized as effectors in an intracellular cytokine assay. To induce IFNγ production by the splenocytes we utilized peptide-pulsed P815 cells (H-2d) that expressed either Db or Kb as antigen-presenting cells. Non-transfected P815 cells (H-2d) were included in the assay as controls. IFNγ production against non-peptide pulsed P815, P815-Db and P815-Kb cells served as additional background controls (not shown). Representative contour plots gated on CD8+ T cells are shown and the numbers over each plot depict the magnitude of the IFNγ+ response to the indicated peptide. (B) Octamers identified as minimal epitopes based on the SYFPEITHI epitope-prediction software program, were additionally tested to conclusively identify the MHC-restriction element. Wild-type B6 mice and Kb-/-mOva mice that lack the Kb molecule were infected intranasally with MHV-1 (1 × 105 PFU/mouse) and one week after infection, the mice were sacrificed and spleens were harvested and processed to generate single cell suspensions that were assessed for IFNγ production in an intracellular cytokine assay. Representative contour plots gated on CD8+ T cells are shown and the numbers over each plot depict the magnitude of the IFNγ+ response to the indicated octamer.
Kinetics of MHV-1-specific CD4 and CD8 T cell responses in the lungs of susceptible C3H/HeJ and resistant B6 mice
As mentioned above our analysis of the peptide library was performed using splenocytes from mice infected intraperitoneally with MHV-1. Intraperitoneal MHV-1 infection of mice is asymptomatic while intranasal infection is associated with pulmonary pathology and morbidity that is most severe in susceptible C3H/HeJ mice and minimal in the resistant B6 mice (6, 7). Given these differences in morbidity after intranasal MHV-1 infection we were interested in determining antigen-specific T cell responses in the lungs of both susceptible and resistant strains of mice after sublethal infections, as previously defined for each mouse strain (6). Virus-specific CD4 and CD8 T cell responses in the lungs of C3H/HeJ mice (Figure 3) and B6 mice (Figure 4) were determined by intracellular cytokine staining following peptide stimulation at the indicated time points after MHV-1 infection and total numbers of antigen-specific T cells in the lungs were also enumerated. MHV-1 infection induced a broad virus-specific CD4 T cell response in the lungs of C3H/HeJ mice.
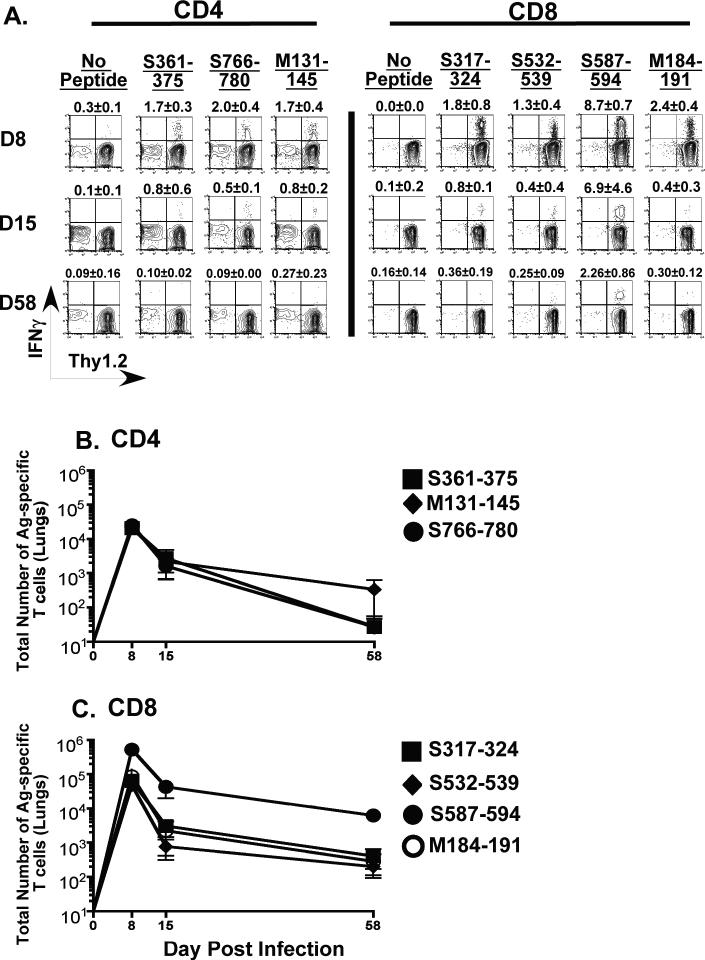
Figure 3. Kinetics of MHV-1 specific CD4 and CD8 T cell responses in the lungs of susceptible C3H/HeJ mice.
Naïve C3H/HeJ mice were intranasally infected with MHV-1 (5 × 103 PFU/mouse) and at the indicated time points following infection mice, lung cells were obtained as described in the Materials and Methods. MHV-1-specific CD4 and CD8 T cells in the bulk lung population were identified by peptide-stimulated intracellular cytokine staining. (A) Representative contour plots are gated on CD4 or CD8 T cells. Background staining is represented by no-peptide controls. The numerical values above each contour plot depict the frequency of the epitope-specific T cell response. The line graphs enumerate the total numbers of antigen-specific CD4 (B) and CD8 (C) T cells identified in the lungs of MHV-1 infected C3H/HeJ mice at the indicated time points.
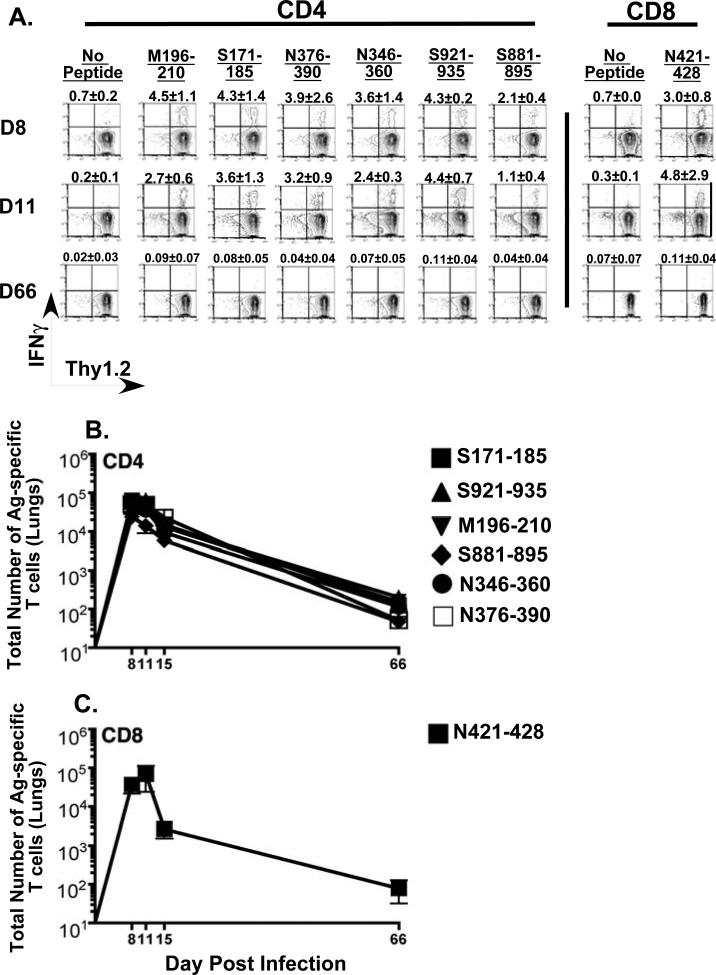
Figure 4. Kinetics of MHV-1 specific CD4 and CD8 T cell responses in the lungs of resistant B6 mice.
Naïve B6 mice were intranasally infected with MHV-1 (1 × 105 PFU/mouse) and at the indicated time points following infection mice lung cells were obtained as described in the Materials and Methods. MHV-1- specific CD4 and CD8 T cells in the bulk lung population were identified by peptide-stimulated intracellular cytokine staining. (A) Representative contour plots are gated on CD4 or CD8 T cells. Background staining is represented by no-peptide controls. The numerical values above each contour plot depict the frequency of the epitope-specific T cell response. The line graphs enumerate the total numbers of antigen-specific CD4 (B) and CD8 (C) T cells identified in the lungs of MHV-1 infected B6 mice at the indicated time points.
Between days 8-11 post-infection roughly 16-19% of the total CD4 T cells in the lungs of these mice were specific for MHV-1. The response to all six CD4 epitopes was similar in magnitude and we did not observe any significant difference in the numbers of antigen-specific CD4 T cells between days 8 and 11 post-infection. A slight reduction in numbers was discernible at day 15 post-infection and these numbers continued to decline up to day 66 post-infection, the last time point measured for this analysis. The CD8 T cell response in the lungs of the C3H/HeJ mice was more narrowly focused targeting the N421 epitope. This response peaked at day 11 post-infection after which there was clearly defined contraction phase and thereafter, similar to the CD4 T cells, the numbers of N421-specific CD8 T cells displayed a downward trend.
Clearly measurable responses to all three CD4 epitopes identified in the peptide library scan were also detected in the lungs of the resistant B6 mice with ~5% of the total CD4 T cells in the lungs of these mice being specific for MHV-1 at day 8 post-infection (Figure 4). Calculation of total numbers revealed that B6 mice had ~4.3-fold lower numbers of antigen-specific CD4 T cells (~7 × 104 cells) in the lungs compared to C3H/HeJ mice (~3 × 105 cells) at the peak of the response. The magnitude of the responses of all three CD4 epitopes in B6 mice were similar to each other and their numbers peaked at day 8 post-infection after which they declined and by day 58 post-infection were at levels similar to those observed in the lungs of C3H/HeJ mice. B6 mice also mounted broad and strong MHV-1-specific CD8 T cell responses with the S587-specific response being clearly immunodominant accounting for 72% of the total antigen-specific CD8 T cell response in the lungs of these mice. The responses directed against the other three CD8 epitopes were similar to each other in magnitude and almost 8-fold lower than that measured for the S587 epitope. Overall the MHV-1 specific CD8 T cell response in the B6 mice (~7.4 × 105 cells) was ~11-fold greater than the corresponding CD8 T cell response measured in the lungs of the susceptible C3H/HeJ mice (~7 × 104 cells). The CD8 T cell response also peaked at day 8 post-infection after which it contracted and gradually declined.
However, the S587-specific CD8 T cells were maintained at higher levels in the lungs of infected B6 mice than the other specificities.
Collectively, these data suggest that either qualitative (ratios of CD4:CD8 T cells) or quantitative (absolute numbers of CD4 or CD8 T cells) differences in the MHV-1 specific T cell response could dictate the development of lung disease and morbidity following intranasal MHV-1 infection of mice.
Evaluation of cytokine production by antigen-specific T cells and determining FoxP3+CD4+ T cell responses in MHV-1 infected B6 and C3H/HeJ mice
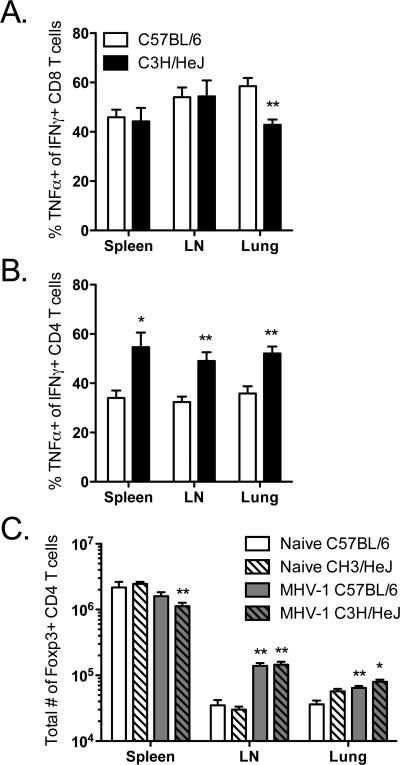
To expand on the results of the kinetics of the MHV-1 specific T cell responses described above, we wanted to determine if differences in cytokine production by MHV-1 specific T cells might also account for some of the differences in morbidity between MHV-1- infected B6 and C3H/HeJ mice. To address this possibility, we used intracellular cytokine staining to analyze the ability of MHV-1-specific CD8 and CD4 T cells to co-produce IFNγ and TNFα following MHV-1 infection of B6 and C3H/HeJ mice (Figure 5A and 5B). We observed a significant reduction in the frequency of N421-specific CD8 T cells (C3H/HeJ) co-producing IFNγ and TNFα in comparison to S587-specific CD8 T cells (B6) in the lungs. No differences were observed in the spleens and draining lymph nodes of the infected mice. CD4 T cell responses were analyzed using pooled antigen-presenting cells separately pulsed with M131, S361 and S766 peptides (B6) or M196, S171 and S921 peptides (C3H/HeJ). In contrast to the CD8 T cell data, we observed a significantly greater fraction of IFNγ-TNFα co-producing CD4 T cells in all three tissues analyzed in the susceptible C3H/HeJ mice versus the resistant B6 mice.
Figure 5. Comparison of IFNγ-TNFα co-production by antigen-specific CD8 and CD4 T cells and analysis of FoxP3+CD4+ T cells in MHV-1 infected B6 and C3H/HeJ mice.
Naïve B6 and C3H/HeJ mice were intranasally infected as described above. At day 8 post infection single cell suspensions were prepared from the spleens, mediastinal lymph nodes and lungs. (A) S587-specific {B6} and N421-specific {C3H/HeJ} CD8 T cells were evaluated for IFNγ-TNFα co-production by intracellular cytokine staining. (B) For examining IFNγ-TNFα co-production by MHV-1 specific CD4 T cells, single cell suspensions were stimulated using the following peptide epitope pools: M131-145 + S361-375 + S766-780 {B6} and M196-210 + S171-185 + S921-935 {C3H/HeJ}. (C) FoxP3+CD4+ cells were enumerated in the spleens, mediastinal lymph nodes and lungs following the staining protocol outlined in the Materials and Methods. These results are representative of two independent experiments and a minimum of 3 mice per group were analyzed in each experiment. Statistically significant differences between the two groups were determined using an unpaired Student's t test and are indicated by an asterisk (*p<0.05; **p<0.01).
In addition to looking at TNFα production, a preliminary examination of IL2 production by MHV-1-specific CD4 T cells showed that while the C3H/HeJ mice had a higher fraction of IFNγ-IL2 co-producers in the spleens and draining lymph nodes versus the B6 mice, this trend was reversed in the lungs (data not shown).
In an effort to further dissect the qualitative properties of the immune response to MHV-1 in the resistant B6 and susceptible C3H/HeJ mice we also analyzed FoxP3+CD4+ regulatory T cells. Published evidence from animal models and human studies of viral infections suggest a crucial role for FoxP3+CD4+ T cells in the development of symptomatic disease (23, 24). Therefore we wanted to determine if differences in FoxP3+CD4+ T cell responses might account for the difference in morbidity between the resistant B6 and susceptible C3H/HeJ mice following intranasal MHV-1 infection. We examined the spleens, draining lymph nodes and lungs of infected B6 and C3H/HeJ mice for the presence of FoxP3+CD4+ T cells at day 8 post-infection, a time-point at which morbidity differences between the two strains are clearly evident (Figure 5C). Our data revealed no significant differences in the total numbers of FoxP3+CD4+ T cells detected in all three of the tissues examined between the two strains of mice.
Overall these results demonstrate the presence of subtle yet significant differences in the cytokine profiles of MHV-1-specific CD8 and CD4 T cells that might potentially influence development of symptomatic disease in the susceptible C3H/HeJ mice following intranasal MHV-1 infection. Furthermore, MHV-1-induced lung disease in susceptible C3H/HeJ mice is not due to numerical deficiencies in FoxP3+ regulatory CD4 T cells.
Previously immunized C3H/HeJ mice display no morbidity and are completely protected following a lethal MHV-1 challenge
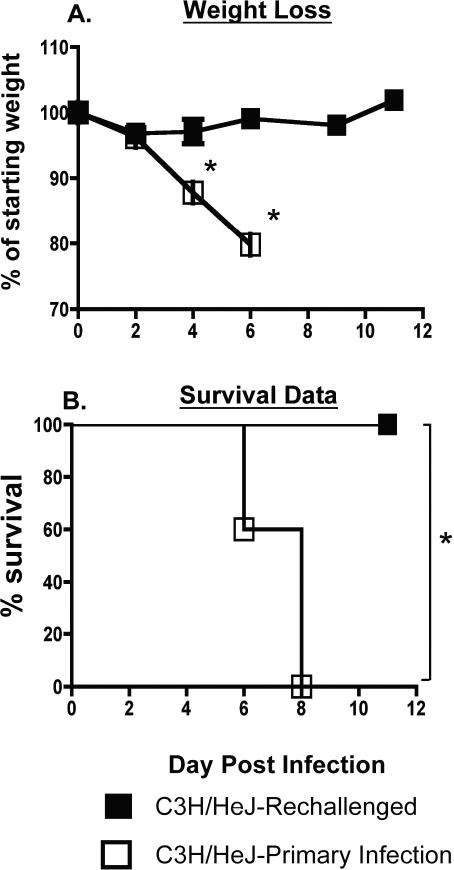
Intranasal MHV-1 infection of naïve C3H/HeJ mice induces vigorous antigen-specific T cell responses in the lungs as well as significant morbidity (6). Additionally, we have previously shown that naïve C3H/HeJ mice adoptively transferred with memory CD4 or CD8 T cells obtained from previously MHV-1 immunized syngeneic donors display enhanced morbidity and mortality following intranasal MHV-1 infection (6). Given these data, we were also interested in determining the outcome of an MHV-1 challenge in previously immunized C3H/HeJ mice that have an established pool of memory T cells. Significant lethality is observed in naïve C3H/HeJ mice at doses greater than 5 × 103 PFU/mouse of MHV-1 administered intranasally (6, 7). Thus, we administered a 10-fold higher dose to C3H/HeJ mice that had previously been immunized with a sublethal dose of MHV-1. As controls we also included naïve C3H/HeJ mice that also received the same high dose challenge as the immunized mice. Both sets of mice were followed for the development of morbidity following infection. As expected, naïve controls progressively lost weight following infection and all mice in this group died by day 8 post-infection. In contrast, previously immunized mice experienced no morbidity and all mice survived the lethal MHV-1 challenge (Figure 6). This result was noteworthy considering the propensity of adoptively transferred memory T cell responses to enhance morbidity and pathology in naïve mice following exposure to MHV-1 and prompted us to examine secondary T cell responses in previously immunized C3H/HeJ mice following a lethal MHV-1 challenge.
Figure 6. Assessment of morbidity and mortality in previously immunized C3H/HeJ mice rechallenged with a lethal dose of MHV-1.
C3H/HeJ mice that had been intranasally infected with MHV-1 (5 × 103 PFU/mouse) at least 3-6 months previously were rechallenged with a lethal dose (5 × 104 PFU/mouse) of MHV-1 delivered intranasally. Naïve C3H/HeJ mice infected with 5 × 104 PFU/mouse of MHV-1 were included as controls. (A) Development of morbidity was monitored by tracking changes in body weight at the indicated time points after infection. Statistically significant differences between the mean weights of rechallenged mice and those undergoing a primary infection were determined using an unpaired Student's t test and are indicated by an asterisk (*). (B) Kaplan Meier survival curves depict the percentage of surviving mice in each group at the indicated time points following infection. Statistically significant differences between the survival rates of rechallenged mice and those undergoing a primary infection were determined using a Fisher's exact test and are indicated by an asterisk (*). 5 mice/group were analyzed and data are representative of two independent experiments.
Suboptimal expansion of secondary T cell responses in lethally challenged immune C3H/HeJ mice
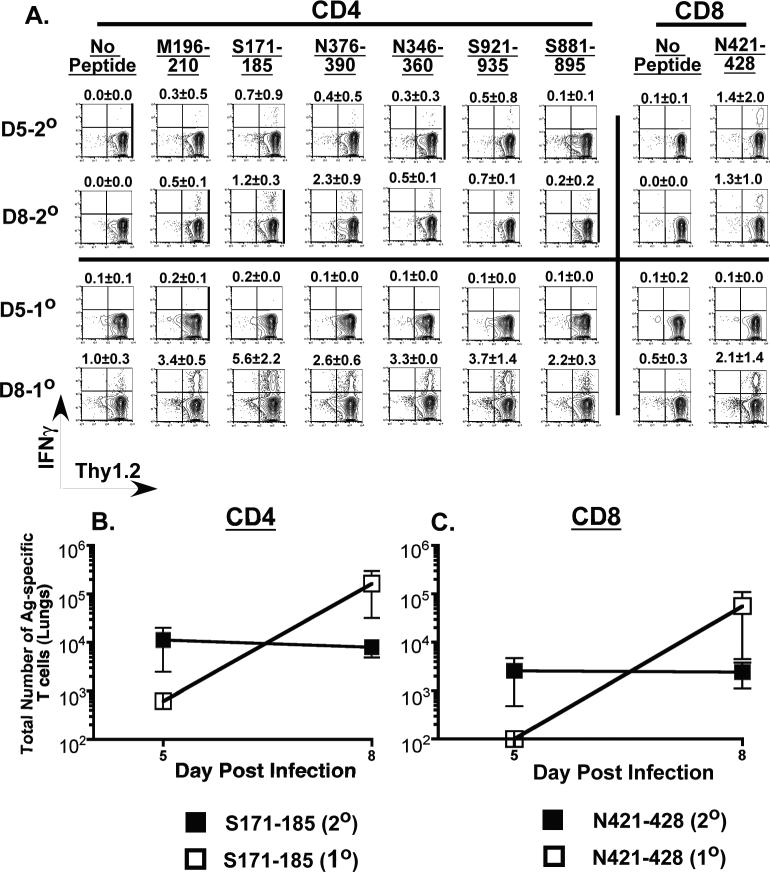
Our data from the lethal MHV-1 challenge of previously immunized C3H/HeJ mice showed that these mice experienced no morbidity and mortality leading us to speculate that the memory T cells in these mice were unable to exert their pathologic effects. In order to examine this issue we evaluated antigen-specific T cell responses in the lungs of previously immunized C3H/HeJ following rechallenge with a lethal dose (5 × 104 PFU/mouse) of MHV-1. As controls we included naïve C3H/HeJ mice that received a sub-lethal dose of MHV-1 (5 × 103 PFU/mouse). This difference in viral input doses was necessary to ensure survival of naïve mice to allow us to evaluate T cell responses in their lungs at day 8 post-infection. Analysis of the secondary S171-specific CD4 and N421-specific CD8 T cell responses revealed that their numbers remained fairly static between days 5 and 8 post rechallenge while enumeration of cells participating in a primary response in naïve control mice that received a 10-fold lower dose of MHV-1 displayed a marked expansion in numbers between days 5 and 8 after infection (Figure 7A-C). In addition, the control mice developed characteristic signs and symptoms of MHV-1 induced disease (weight loss, ruffled fur, hunched posture, reduced activity) while no morbidity was observed in the rechallenged group. The numbers for all of the remaining five specificities of MHV-1-specific CD4 T cells tested were also 5 to10-fold higher for the primary responders at day 8 in comparison to the secondary responders (data not shown). C3H/HeJ mice exposed to MHV-1 develop high titer neutralizing antibodies 15-20 days after primary MHV-1 infection (6, 8-11). The neutralizing antibody response likely accounts for the attenuated secondary T cell response in rechallenged mice and a consequence of this is the reduced morbidity observed in rechallenged mice. Control mice undergoing a primary MHV-1 infection lack pre-existing neutralizing antibodies, and mount a vigorous T cell response that help control the infection but also induce significant immunopathology (6).
Figure 7. Measurement of secondary T cell responses in the lungs of previously immunized C3H/HeJ mice rechallenged with a lethal dose of MHV-1.
C3H/HeJ mice that had been intranasally infected with MHV-1 (5 × 103 PFU/mouse) at least 6 months previously were rechallenged with a lethal dose (5 × 104 PFU/mouse) of MHV-1 delivered intranasally. Naïve C3H/HeJ mice undergoing a primary MHV-1 infection (5 × 103 PFU/mouse) were included as controls. Antigen-specific CD4 and CD8 T cell responses in the lungs of both groups of mice (2° = secondary infection; 1° = primary infection) were evaluated by peptide-stimulated intracellular cytokine staining on days 5 and 8 post infection. (A) Representative contour plots are gated on CD4 or CD8 T cells. Background staining is represented by no-peptide controls. The numerical values above each contour plot depict the frequency of the epitope-specific T cell response. The line graphs enumerate the total numbers of S171-specific CD4 T cells (B) and NP421-specific CD8 T cells (C) identified in the lungs of MHV-1 infected C3H/HeJ mice at the indicated time points.
Elaboration of a broad and vigorous antigen-specific CD8 T cell response to MHV-1 does not prevent the development of morbidity in susceptible C3.SW-H2b/SnJ mice
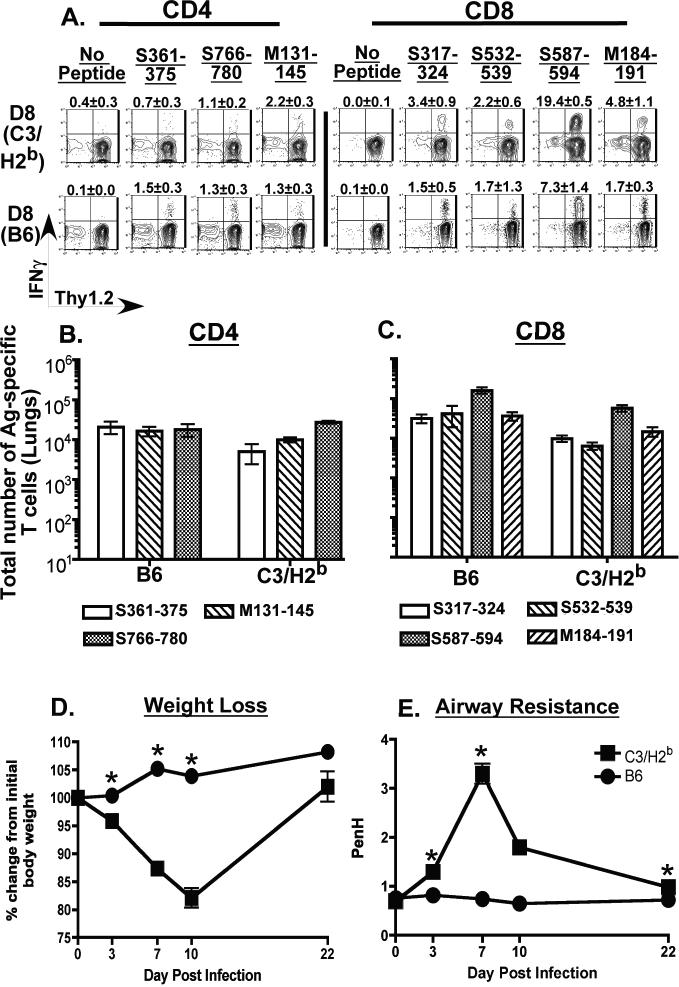
One of the key differences we observed between the susceptible C3H/HeJ mice and the resistant B6 mice was that MHV-1 infection elicited a broad and vigorous virus-specific CD8 T cell response in the lungs of the resistant B6 mice. Thus, we were interested in determining if elaboration of a similarly broad and vigorous virus-specific CD8 T cell response in susceptible background mice would minimize morbidity. Hence we examined morbidity and antigen-specific T cell responses in the lungs of congenic C3.SW-H2b/SnJ mice that express the H-2b MHC-haplotype of the B6 mice (25) following infection with a sub-lethal dose of MHV-1 (5 × 103 PFU/mouse). As controls we included wild-type B6 mice that were similarly infected. Analysis of the antigen-specific T cell responses in the lungs of the congenic C3.SW-H2b/SnJ mice at day 8 post-infection demonstrated that they elaborated strong antigen-specific CD8 and CD4 T cell responses that were not significantly different from the responses observed in the lungs of similarly infected control B6 mice (Figure 8A-C). Interestingly however, development of morbidity assessed in terms of weight loss and development of airway resistance revealed that, despite elaboration of antigen-specific T cell responses similar to those observed in the resistant B6 mice, the C3.SW-H2b/SnJ mice nonetheless lost a significant amount of weight and showed significantly elevated airway resistance compared to controls following intranasal MHV-1 infection (Figure 8D and 8E). These data demonstrate that elaboration of vigorous antigen-specific CD8 T cell responses on their own are insufficient to confer a resistant phenotype following intranasal MHV-1 infection.
Figure 8. Evaluation of morbidity and antigen-specific T cell responses in the lungs of MHV-1 infected C3.SW-H2b/SnJ mice.
Congenic C3.SW-H2b/SnJ mice that express the H-2b MHC locus were intranasally infected with 5 × 103 PFU/mouse of MHV-1. Similarly infected wild-type B6 mice were included as controls in the analysis. Antigen-specific CD4 and CD8 T cell responses in the lungs of both groups of mice were evaluated by intracellular cytokine staining on day 8 post infection. (A) Representative contour plots are gated on CD4 or CD8 T cells. Background staining is represented by no-peptide controls. The numerical values above each contour plot depict the frequency of the epitope-specific T cell response. The bar graphs enumerate the total numbers of MHV-1-specific CD4 T cells (B) and CD8 T cells (C) identified in the lungs of MHV-1 infected C3H/HeJ mice. Development of morbidity was monitored by tracking changes in body weight (D) and airway resistance (Penh) (E) at the indicated time points after infection. 10-20 mice per group were evaluated at each time point and statistically significant differences between the mean weights and Penh values of the C3.SW-H2b/SnJ mice and wild-type B6 mice were determined using an unpaired Student's t test and are indicated by an asterisk (*).
Discussion
In this report we have expanded on the findings of our previous study where we examined the protective and pathologic roles of the immune response to MHV-1 in resistant and susceptible strains of mice (6). In the previous report we had demonstrated that the adaptive immune response plays a protective role in the resistant B6 mice while conversely, both CD4 and CD8 T cells contributed significantly to the development of lung pathology following intranasal MHV-1 infection of susceptible C3H/HeJ mice (6). Given these contrasting roles of the adaptive immune response to MHV-1 infection we were interested in carrying out a more comprehensive examination of the T cell response to MHV-1. Screening of an overlapping peptide library encompassing the four main structural MHV-1 proteins led to the identification of several novel CD4 and CD8 T cell epitopes in both susceptible and resistant strains of mice. Identification of these epitopes allowed us to examine the kinetics of the antigen-specific T cell responses in the lungs of these mice following both a primary as well as a secondary infection. One of the differences between the susceptible and resistant strains was the fact that MHV-1 infection induced robust and broad antigen-specific CD4 T cell responses in the susceptible mice while the antigen-specific CD8 T cell responses dominated in terms of magnitude and breadth in the resistant B6 mice. Although primary MHV-1 infection was associated with significant morbidity and T-cell-mediated immunopathology in susceptible strains (6), re-exposure to lethal doses of MHV-1 following recovery from primary infection was marked by the complete absence of any morbidity and these mice were fully protected and an analysis of the secondary T cell responses in the rechallenged mice revealed a lack of robust expansion of these cells. Finally we also demonstrate that vigorous expansion and elaboration of a broad set of MHV-1-specific CD8 T cells, that is the hallmark of the resistant B6 mice, fails to induce a similar resistant phenotype in C3H mice engineered to express the B6 haplotype.
The limited availability of genomic and T cell epitope information for MHV-1 necessitated the generation and screening of a peptide library encompassing the four main viral structural proteins, the spike (S), membrane (M), envelope (E) and nucleocapsid (N). The reason to exclude analysis of the non-structural proteins at this juncture was based on historical data suggesting that T cell epitopes for coronaviruses reside primarily in structural proteins (3, 14-20). This also appears to be the case for SARS-CoV where a recently published report showed 70% of the responses were localized to the four main structural proteins with the spike protein being highly immunogenic accounting for 41% of the total antigen-specific T cell response (3). In contrast the non-structural replicase protein that spans two-thirds of the viral proteome accounted for only 13% of the total antigen-specific response (3). Screening of our peptide library revealed that resistant B6 mice had a broader MHV-1-specific CD8 T cell response while the susceptible C3H/HeJ mice had a broader MHV-1-specific CD4 T cell response with a majority of the of the epitopes mapping to the spike protein. Following the identification of the epitopes we ran the sequences of the specific 15 mers through the SYFPEITHI epitope-prediction software program (21) to identify the minimal CD8 epitopes as well as the MHC restriction element for the particular epitope. MHC restriction element binding was also separately confirmed as described in the results section. Overall these results seem consistent with those observed for SARS-CoV where analysis of PBMC samples from SARS-CoV infected patients revealed that patients that had more severe disease, like the susceptible C3H/HeJ mice in our analysis, tended to have stronger CD4 T cell responses that were primarily directed against epitopes identified in the spike protein (3).
As T cell-mediated immunopathology in the lungs is an important feature of intranasal MHV-1 infection of mice (6) we were interested in examining the magnitude and kinetics of the antigen-specific T cell responses in the lungs of the infected mice. Although a number of studies have examined T cell responses to SARS both in human and murine systems, they are beset with certain limitations (2, 3, 14, 26-31). One of the major limitations of human studies is that they are often restricted to analyzing immune responses from PBMC samples that may not reflect the quality and magnitude of the immune response occurring in situ at the primary site of infection or pathology. And the limitation of a majority of small animal studies examining immune responses to SARS is the fact that the human SARS-CoV infection of mice does not recapitulate all of the pathologic features of the disease observed in humans (1, 32). Hence measurement of immune responses in the lungs of mice following human SARS-CoV infection may not provide a complete picture of the immunopathology that is believed to play an important role in the pathogenesis of the disease. However, the use of MHV-1 infection of susceptible and resistant mouse strains (5-7) and the availability now of mouse-adapted SARS-CoV (33) provide a useful tool to carry-out rapid throughput analyses to examine pathology and the role of the immune response in the pathogenesis of respiratory coronaviral diseases such as SARS. Our analysis revealed that intranasal MHV-1 infection of susceptible C3H/HeJ mice resulted in the induction of a robust antigen-specific CD4 T cell response in the lungs of these mice and between days 8-11 post infection. Overall based on the available epitope information, the total numbers of antigen-specific CD4 T cells (~3 × 105) far outnumbered the antigen-specific CD8 T cells (7 × 104) in the lungs of the C3H/HeJ mice. In contrast to this picture, intranasal MHV-1 infection of the resistant B6 mice induced a very strong and multi-epitope specific CD8 T cell response in the lungs of these mice with S587-594 clearly emerging as the immunodominant epitope. The S587-specific CD8 T cells also had a significantly greater proportion of IFNγ-TNFα co-producers in the lungs versus the N421-specific CD8 T cell response elaborated in the lungs of the susceptible C3H/HeJ mice. This trend was reversed for the CD4 T cells where the susceptible C3H/HeJ mice had a significantly greater proportion of IFNγ-TNFα co-producers in the lungs, spleen and mediastinal lymph nodes in comparison to the resistant B6 mice. Our preliminary data also indicate the presence of fewer IFNγ-IL2 co-producing CD4 T cells in the lungs of the susceptible C3H/HeJ mice. We also examined the role of FoxP3+CD4+ T cells in regulating MHV-1-infection associated morbidity in the susceptible and resistant strains. We had speculated that susceptible strains would mount a suboptimal regulatory T cell response thereby allowing the development of immunopathology while the resistant strains of mice would mount an optimal regulatory T cell response that keeps T-cell mediated immunopathology in check. However our data reveal no difference in the magnitude of the FoxP3+CD4+ T cell response in the spleens, lungs or mediastinal lymph nodes measured at day 8 post infection, indicating that MHV-1 infection-associated disease in susceptible mouse strains is not due to decreased regulatory CD4 T cell responses. The data up until this point suggested that differences in magnitude and cytokine profile of the antigen-specific CD4 and CD8 T cell responses to MHV-1 might account for the differences in morbidity observed between the susceptible and resistant mouse strains.
We have previously reported that both CD4 and CD8 T cells play an important role in mediating acute disease and lung pathology observed in the susceptible C3H/HeJ mice following primary intranasal infection (6). Moreover these T cells retain their pathologic properties as they differentiate into memory T cells (6). Adoptive transfer of these memory T cells into naïve syngeneic hosts that are subsequently intranasally infected with MHV-1 results in greater morbidity and mortality than that observed during primary MHV-1 infection (6). These findings suggested that re-exposure to the virus could potentially amplify the pathologic behavior of the MHV-1 specific T cells. In order to examine this issue in a system that would be more representative of what would occur in a natural setting we rechallenged C3H/HeJ mice that had been previously immunized with MHV-1 at least 3-6 months previously with a lethal dose of MHV-1 and assessed morbidity and mortality at various time points following re-infection. In contrast to what we had observed in the adoptive transfer experiment, interestingly the rechallenged mice did not develop any morbidity and all of the mice survived the lethal dose challenge. We have previously shown that primary MHV-1 infection of susceptible C3H/HeJ mice induces robust neutralizing antibody responses by day 15-20 post-infection, and adoptive transfer of immune serum into naïve syngeneic recipients is very effective at minimizing systemic viral burden and morbidity following MHV-1 challenge (6). Neutralizing antibodies would also be present in previously immunized C3H/HeJ that were subsequently rechallenged with MHV-1. Given this information we wanted to closely examine the magnitude of the secondary T cell response in the rechallenged mice. One of the hallmarks of a secondary T cell response is to respond very vigorously and rapidly following re-exposure to the pathogen (34). Our analysis of the secondary T cell response in the lungs of the rechallenged mice at days 5 and 8 after infection revealed that the memory T cells failed to expand optimally and their numbers remained unchanged between these two time-points. Overall these findings suggested a putative link between that the absence of morbidity in the previously immunized C3H/HeJ mice that were rechallenged with a lethal dose of MHV-1 and the blunted secondary T cell response. Although it is formally possible that the secondary T cell response may have peaked prior to day 5, given the pathologic properties of these T cells and the failure to detect any morbidity during the course of this analysis, argues more in favor of a weak secondary T cell response. It is likely that neutralizing antibody responses dominate the recall response in previously immunized C3H/HeJ mice after re-exposure to the virus, effectively and rapidly controlling the rechallenge inoculum thereby reducing the numbers of memory T cells that get recruited into the secondary response. This could be a mechanism to try and curb T cell-mediated immunopathology in susceptible hosts that survive primary MHV-1 infection induced disease.
Our data show that C3H/HeJ mice that are highly susceptible to MHV-1-induced disease mount a vigorous and antigen-specific CD4 T cell response directed against at least six epitopes in the lungs, while B6 mice that are resistant to MHV-1-induced disease generate a strong and multi-epitope-specific CD8 T cell response in the lungs following intranasal MHV-1 infection. These findings led us to ask the question if the susceptible phenotype could be modulated by inducing a strong, multi-epitope-specific CD8 T cell response similar to that observed in the resistant B6 mice. We addressed this question by analyzing T cell responses in the lungs of C3.SW-H2b/SnJ mice following intranasal MHV-1 infection. This congenic strain of mice expresses the H-2b MHC haplotype similar to the B6 mice instead of the H-2k haplotype of the parent strain, C3H/HeSnJ, which is also shared by the susceptible C3H/HeJ mice (25). As we expected, the C3.SW-H2b/SnJ mounted an antigen-specific T cell response very similar in magnitude to that of wild-type B6 controls characterized by a strong and broadly defined antigen-specific CD8 T cell response that was dominated by the S587-specific CD8 T cells. However, these mice did experience morbidity as evidenced by weight loss and development of airway resistance following primary MHV-1 infection. These results suggest that the fine specificity of the antigen-specific T cell response does not control disease phenotype in the context of MHV-1 infection. In the larger context, these findings additionally suggest that host background genes play a significant role in determining host resistance/susceptibility to disease. It is formally possible that qualitative differences in innate immune responses and functional properties of FoxP3+CD4+ T cells influenced by host background genes could affect development of disease following MHV-1 infection. The crosstalk between background genes and the immune response genes may be crucial in programming the host immune response into one that induces pathology and morbidity (as observed in the C3H/HeJ mice and C3.SW-H2b/SnJ mice) versus one that does not (as observed in the B6 mice).
In conclusion, in this report we have identified novel CD4 and CD8 T cell epitopes specific to MHV-1 in both susceptible and resistant strains of mice. Our data demonstrate the divergence in the quality and quantity of the antigen-specific T cell response between susceptible and resistant hosts following primary MHV-1 infection. This report also shows that re-exposure to MHV-1 in susceptible hosts will most likely be associated with minimal morbidity and a favorable outcome for the host. Reduced morbidity in this setting will most likely be due to a dominant neutralizing antibody response that reduces viral burden and curtails an overwhelming and potentially lethal secondary T cell response. Finally our data also highlight the potentially important contribution that host background genes make in shaping the host response to infection that alters the balance between disease and lack thereof. Overall these findings have important implications for trying to understand the complex, multi-factorial nature of coronaviral disease pathogenesis and suggest the importance of focusing on an Ab-based versus a T-cell-based vaccine approach to combat respiratory coronaviral infections.
Acknowledgements
We would like to thank Stanley Perlman for critical review of the manuscript.
This work was supported by the National Institutes of Health Program Project Grant PO1 AI-060699 (to JTH and SMV).
Footnotes
Disclosures
The authors have no financial conflict of interest.
References
- 1.Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10:S88–97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cameron MJ, Ran L, Xu L, Danesh A, Bermejo-Martin JF, Cameron CM, Muller MP, Gold WL, Richardson SE, Poutanen SM, Willey BM, DeVries ME, Fang Y, Seneviratne C, Bosinger SE, Persad D, Wilkinson P, Greller LD, Somogyi R, Humar A, Keshavjee S, Louie M, Loeb MB, Brunton J, McGeer AJ, Kelvin DJ. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li CK, Wu H, Yan H, Ma S, Wang L, Zhang M, Tang X, Temperton NJ, Weiss RA, Brenchley JM, Douek DC, Mongkolsapaya J, Tran BH, Lin CL, Screaton GR, Hou JL, McMichael AJ, Xu XN. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perlman S, Dandekar AA. Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Albuquerque N, Baig E, Ma X, Zhang J, He W, Rowe A, Habal M, Liu M, Shalev I, Downey GP, Gorczynski R, Butany J, Leibowitz J, Weiss SR, McGilvray ID, Phillips MJ, Fish EN, Levy GA. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J mice. J Virol. 2006;80:10382–10394. doi: 10.1128/JVI.00747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khanolkar A, Hartwig SM, Haag BA, Meyerholz DK, Epping LL, Haring JS, Varga SM, Harty JT. Protective and Pathologic roles of the Immune Response to Mouse Hepatitis Virus-1 (MHV-1): Implications for Severe Acute Respiratory Syndrome. J Virol. 2009;83:9258–9272. doi: 10.1128/JVI.00355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khanolkar A, Hartwig SM, Haag BA, Meyerholz DK, Harty JT, Varga SM. Toll-Like Receptor 4 Deficiency Increases Disease and Mortality after Mouse Hepatitis Virus Type 1 Infection of Susceptible C3H Mice. J Virol. 2009;83:8946–8956. doi: 10.1128/JVI.00355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bisht H, Roberts A, Vogel L, Bukreyev A, Collins PL, Murphy BR, Subbarao K, Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci U S A. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchholz UJ, Bukreyev A, Yang L, Lamirande EW, Murphy BR, Subbarao K, Collins PL. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci U S A. 2004;101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Zhang L, Qin C, Ba L, Yi CE, Zhang F, Wei Q, He T, Yu W, Yu J, Gao H, Tu X, Gettie A, Farzan M, Yuen KY, Ho DD. Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J Virol. 2005;79:2678–2688. doi: 10.1128/JVI.79.5.2678-2688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du L, He Y, Wang Y, Zhang H, Ma S, Wong CK, Wu SH, Ng F, Huang JD, Yuen KY, Jiang S, Zhou Y, Zheng BJ. Recombinant adeno-associated virus expressing the receptor-binding domain of severe acute respiratory syndrome coronavirus S protein elicits neutralizing antibodies: Implication for developing SARS vaccines. Virology. 2006;353:6–16. doi: 10.1016/j.virol.2006.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khanolkar A, Fuller MJ, Zajac AJ. CD4 T cell-dependent CD8 T cell maturation. J Immunol. 2004;172:2834–2844. doi: 10.4049/jimmunol.172.5.2834. [DOI] [PubMed] [Google Scholar]
- 13.Fulton RB, Olson MR, Varga SM. Regulation of cytokine production by virus-specific CD8 T cells in the lungs. J Virol. 2008;82:7799–7811. doi: 10.1128/JVI.00840-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhi Y, Kobinger GP, Jordan H, Suchma K, Weiss SR, Shen H, Schumer G, Gao G, Boyer JL, Crystal RG, Wilson JM. Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein. Virology. 2005;335:34–45. doi: 10.1016/j.virol.2005.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergmann CC, Marten NW, Hinton DR, Parra B, Stohlman SA. CD8 T cell mediated immunity to neurotropic MHV infection. Adv Exp Med Biol. 2001;494:299–308. doi: 10.1007/978-1-4615-1325-4_46. [DOI] [PubMed] [Google Scholar]
- 16.Bergmann CC, Yao Q, Lin M, Stohlman SA. The JHM strain of mouse hepatitis virus induces a spike protein-specific Db-restricted cytotoxic T cell response. J Gen Virol. 1996;77(Pt 2):315–325. doi: 10.1099/0022-1317-77-2-315. [DOI] [PubMed] [Google Scholar]
- 17.Kyuwa S, Stohlman SA. Establishment of MHV-A59 S protein specific cytotoxic T lymphocyte clones. Adv Exp Med Biol. 1993;342:471–472. doi: 10.1007/978-1-4615-2996-5_75. [DOI] [PubMed] [Google Scholar]
- 18.Stohlman SA, Kyuwa S, Cohen M, Bergmann C, Polo JM, Yeh J, Anthony R, Keck JG. Mouse hepatitis virus nucleocapsid protein-specific cytotoxic T lymphocytes are Ld restricted and specific for the carboxy terminus. Virology. 1992;189:217–224. doi: 10.1016/0042-6822(92)90697-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleming JO, Wang FI, Trousdale MD, Hinton DR, Stohlman SA. Immunopathogenesis of demyelination induced by MHV-4. Adv Exp Med Biol. 1990;276:565–572. doi: 10.1007/978-1-4684-5823-7_78. [DOI] [PubMed] [Google Scholar]
- 20.Woodward JG, Matsushima G, Frelinger JA, Stohlman SA. Production and characterization of T cell clones specific for mouse hepatitis virus, strain JHM: in vivo and in vitro analysis. J Immunol. 1984;133:1016–1021. [PubMed] [Google Scholar]
- 21.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 22.Wilson CS, Moser JM, Altman JD, Jensen PE, Lukacher AE. Cross-recognition of two middle T protein epitopes by immunodominant polyoma virus-specific CTL. J Immunol. 1999;162:3933–3941. [PubMed] [Google Scholar]
- 23.Lund JM, Hsing L, Pham TT, Rudensky AY. Coordination of early protective immunity to viral infection by regulatory T cells. Science. 2008;320:1220–1224. doi: 10.1126/science.1155209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanteri MC, O'Brien KM, Purtha WE, Cameron MJ, Lund JM, Owen RE, Heitman JW, Custer B, Hirschkorn DF, Tobler LH, Kiely N, Prince HE, Ndhlovu LC, Nixon DF, Kamel HT, Kelvin DJ, Busch MP, Rudensky AY, Diamond MS, Norris PJ. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J Clin Invest. 2009;119:3266–3277. doi: 10.1172/JCI39387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer Lindahl K. On naming H2 haplotypes: functional significance of MHC class Ib alleles. Immunogenetics. 1997;46:53–62. doi: 10.1007/s002510050242. [DOI] [PubMed] [Google Scholar]
- 26.Du L, Zhao G, Lin Y, Sui H, Chan C, Ma S, He Y, Jiang S, Wu C, Yuen KY, Jin DY, Zhou Y, Zheng BJ. Intranasal vaccination of recombinant adeno-associated virus encoding receptor-binding domain of severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein induces strong mucosal immune responses and provides long-term protection against SARS-CoV infection. J Immunol. 2008;180:948–956. doi: 10.4049/jimmunol.180.2.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Cao Y, Du J, Bu X, Ma R, Wu C. Priming with SARS CoV S DNA and boosting with SARS CoV S epitopes specific for CD4+ and CD8+ T cells promote cellular immune responses. Vaccine. 2007;25:6981–6991. doi: 10.1016/j.vaccine.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Ma R, Wu CY. Immunization with SARS-CoV S DNA vaccine generates memory CD4+ and CD8+ T cell immune responses. Vaccine. 2006;24:4905–4913. doi: 10.1016/j.vaccine.2006.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng H, Yang LT, Li J, Lu ZQ, Wang LY, Koup RA, Bailer RT, Wu CY. Human memory T cell responses to SARS-CoV E protein. Microbes Infect. 2006;8:2424–2431. doi: 10.1016/j.micinf.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng H, Yang LT, Wang LY, Li J, Huang J, Lu ZQ, Koup RA, Bailer RT, Wu CY. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology. 2006;351:466–475. doi: 10.1016/j.virol.2006.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang LT, Peng H, Zhu ZL, Li G, Huang ZT, Zhao ZX, Koup RA, Bailer RT, Wu CY. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin Immunol. 2006;120:171–178. doi: 10.1016/j.clim.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts A, Lamirande EW, Vogel L, Jackson JP, Paddock CD, Guarner J, Zaki SR, Sheahan T, Baric R, Subbarao K. Animal models and vaccines for SARS-CoV infection. Virus Res. 2008;133:20–32. doi: 10.1016/j.virusres.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts A, Deming D, Paddock CD, Cheng A, Yount B, Vogel L, Herman BD, Sheahan T, Heise M, Genrich GL, Zaki SR, Baric R, Subbarao K. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Badovinac VP, Harty JT. Programming, demarcating, and manipulating CD8+ T-cell memory. Immunol Rev. 2006;211:67–80. doi: 10.1111/j.0105-2896.2006.00384.x. [DOI] [PubMed] [Google Scholar]