Abstract
Purposes
Glucuronidation via UDP-glucuronosyltransferases (or UGTs) is a major metabolic pathway. The purposes of this study are to determine the UGT-isoform specific metabolic fingerprint (or GSMF) of wogonin and oroxylin A, and to use isoform-specific metabolism rates and kinetics to determine and describe their glucuronidation behaviors in tissue microsomes.
Methods
In vitro glucuronidation rates and profiles were measured using expressed UGTs and human intestinal and liver microsomes.
Results
GSMF experiments indicated that both flavonoids were metabolized mainly by UGT1As, with major contributions from UGT1A3 and UGT1A7-1A10. Isoform-specific metabolism showed that kinetic profiles obtained using expressed UGT1A3 and UGT1A7-1A10 could fit to known kinetic models. Glucuronidation of both flavonoids in human intestinal and liver microsomes followed simple Michaelis-Menten kinetics. A comparison of the kinetic parameters and profiles suggests that UGT1A9 is likely the main isoform responsible for liver metabolism. In contrast, a combination of UGT1As with a major contribution from UGT1A10 contributed to their intestinal metabolism. Correlation studies clearly showed that UGT isoform-specific metabolism could describe their metabolism rates and profiles in human liver and intestinal microsomes.
Conclusion
GSMF and isoform-specific metabolism profiles can determine and describe glucuronidation rates and profiles in human tissue microsomes.
Keywords: UGT, wogonin, flavonoids, intestine, liver
Introduction
Glucuronidation is a major metabolic pathway for the metabolism of a large number of drugs and phytochemicals (1). Characterization of a compound’s glucuronidation behaviors typically includes the determination if the pathway is its major metabolic pathway, kinetics of glucuronidation (2), and more recently, UGT-isoform specific metabolic fingerprint (GSMF) (3). Although kinetic studies of glucuronidation using tissue-specific microsomes are widely available in published literatures, the scientific values of those kinetic parameters are not very well-defined. For example, they do not always predict the excretion rates of glucuronides very well (4). However, since rapid metabolism via glucuronidation in tissue microsomes usually means metabolism is rapid in that organ, efforts to characterize glucuronidation represent an important part of ongoing work delineating a new drug candidate’s biopharmaceutical and pharmacokinetic properties. GSMF is an important part of the glucuronidation characterization work, and here we presented new evidence to support additional utility of this characterization work to determine and describe rates and kinetics of tissue (organ) -specific glucuronidation of phytochemicals using two flavonoids, wogonin and oroxylin A, as the model compounds.
These two flavones were chosen for the present study because they, especially wogonin, could become promising anti-tumor agents. These two flavonoids possess potent activities against cancer in vitro and in vivo, and have attractive safety profiles. Recently, it was shown that wogonin could induce apoptosis in malignant T-cells in vitro and suppress growth of human T-cell leukemia xenografts in vivo, but did not affect T lymphocytes from healthy donors (5). Wogonin was also demonstrated to be active against both estrogen receptor-positive and -negative human breast cancer cell lines in vitro and in nude mice xenografts in vivo (6). Oroxylin A, with a structure similar to wogonin, was similarly active and was shown to be a potent inducer of apoptosis in human hepatoma carcinoma cells (HepG2 and SMMC-7721) (7) and human cervical cancer cells (HeLa) (8). Oroxylin A also has potentially anti-metastatic effects in vitro (9). Furthermore, subchronic toxicity in beagle dogs demonstrated that wogonin offered a wide margin of safety and had no apparent organ toxicity after chronic intravenous administration (10). Therefore, these two flavonoids especially wogonin are very good candidates to be developed into cancer chemopreventive agents.
Wogonin and oroxylin A (Fig.1) are richly concentrated in the dry root of Scutellaria baicalensis (Chinese name: Huang-Qin), a popular and multi-functional herb used in China, Japan and several Asian countries. The extract of Scutellaria radix has been recently shown to possess cytostatic effect on several cancer cells in vitro (11, 12) and also in mouse tumor models in vivo (13, 14). The basis of this anti-tumor effect was attributed to the presence of bioactive flavones in this plant (15, 16).
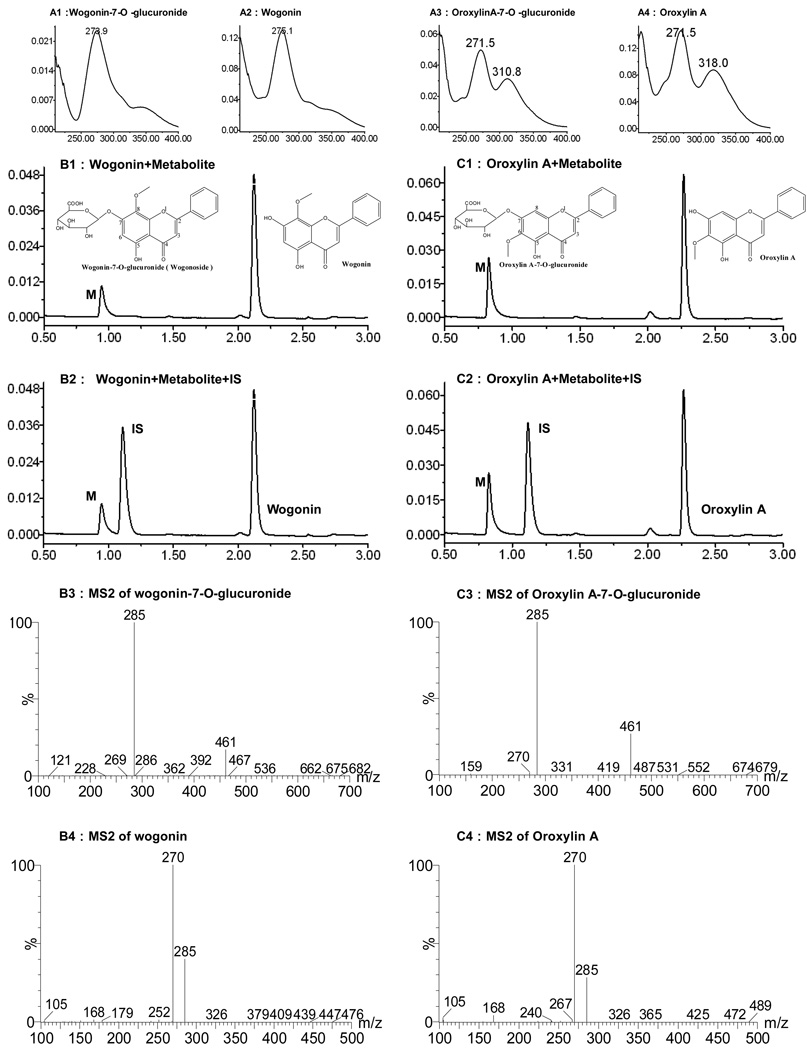
Fig.1. Structure of wogonin and oroxylin A, UPLC and LC-MS/MS analysis of two flavones and their mono-glucuronides.
Panels A1–A4 are the UV spectra of two flavones and their corresponding metabolites (one each per flavone). Panels B1, B2, C1, C2 are the UPLC chromatograms that showed the retention time, chemical structure of each flavone and its respective metabolite (M), as well as the retention time of internal standard (IS). Panels B3, C3 show the MS2 scan of LC-MS/MS for mono-glucuronide of each flavone, and panels B4, C4 show the MS2 scan for each flavone. The metabolite was wogonin-7-O-glucuronide and oroxylin A-7-O-glucuronide, since parent and metabolite displayed almost identical UV spectra. If the metabolite was 5-O-glucuronide, the spectra would have changed significantly in that maximal absorbance wavelength would have blue shifted.
Despite of many beneficial properties and demonstrated preclinical activities of these two flavones, their low bioavailabilities have impeded their development into chemopreventive agents. Many studies including our own have demonstrated that extensive first-pass metabolism by phase II enzymes including UGTs and SULTs is the most important reason for flavonoids’ poor bioavailabilities (17). A few published works have shown that after intravenous administration, wogonin pharmacokinetics showed a “biphasic phenomenon” in its plasma concentration-time profile (18). After oral administration of a purified extract of Scutellaria baicalensis, only plasma concentrations of wogonin were measurable, whereas those of oroxylin A were below limit of quantitation (19). In another study, human urinary excretion studies showed that the glucuronides and sulfates of wogonin were found after oral administration of the extract of Scutellariae Radix. Moreover, its sulfate level was comparable to the corresponding glucuronide level (20). Effects of wogonin on modulating phase II metabolic enzymes were delineated in C57BL/6J mice, and the results showed that the ingestion of wogonin could modulate drug-metabolizing enzymes in a tissue-specific manner (21).
The purpose of this study is to measure the rates and kinetics of UGT-catalyzed phase II reactions using expressed UGT isoforms, and investigate if the rates so obtained will determine and describe phase II metabolism using microsomes prepared from human intestine and liver, two major organs responsible for the first-pass metabolism (22). Twelve UGT isoforms were used in the current study because they are commercially available. Some of these isoforms are well expressed in nearly all the tissues (e.g., UGT1A1), some are present in the liver but not the intestine (e.g., UGT1A9), whereas others only expressed in the intestine but not liver (e.g., UGT1A8 and 1A10) (23).
Materials and Methods
Materials
Wogonin, wogonoside and oroxylin A (purity >98%, HPLC grade, confirmed by LC/MS/MS) were purchased from Chengdu Mansite Pharmaceutical Co. LTD (Chengdu, China). Expressed human UGT isoforms (supersomes), pooled male human liver and intestinal microsomes were purchased from BD Biosciences (Woburn, MA). Uridine diphosphoglucuronic acid (UDPGA), alamethicin, D-saccharic-1,4-lactone monohydrate, magnesium chloride, and β-glucuronidase (Type HP-2 from helix pomatia) were purchased from Sigma-Aldrich (St Louis, MO). All other materials (typically analytical grade or better) were used as received.
Enzymatic Activities of Expressed UGTs and Organ Microsomes
The incubation procedures for measuring enzyme’s activities using tissue microsomes or expressed UGTs (or Supersomes™) were essentially the same as the previous publications from University of Houston (24, 25). Briefly, the procedures were as follows: (1) mix microsomes/supersomes (final concentration ≈ in range of 0.0053~0.053 mg protein per ml as optimum for the reaction), magnesium chloride (0.88 mM), saccharolactone (4.4 mM), alamethicin (0.022 mg/ml), different concentrations of substrates in a 50 mM potassium phosphate buffer (pH 7.4), and UDPGA (3.5 mM, add last) to a final volume of 680 µl; (2) separate the 680 µl mixture to three equal portions with the volume of 200 µl, and incubate all three portions at 37°C simultaneously for a predetermined period of time (10 to 60 min); and (3) stop the reaction by the addition of 100 µl of 94% acetonitrile/6% glacial acetic acid containing 90 µM acetophenone as the internal standard. The reaction mixture was centrifuged at 13,000 rpm for 15min and the supernatant was directly subjected to UPLC for analysis.
To profile UGT’s activities, three substrate concentrations, 2.5, 10 and 35 µM were used. To profile kinetics of glucuronidation of wogonin and oroxylin A by UGT 1A1, 1A3 and 1A7-1A10, 9–11 substrate concentrations in the range of 1.25–35 µM (0.5–35 µM) were used. To investigate the differences in the flavone glucuronidation rates between single UGT and combined UGTs, two substrate concentrations of 2.5 and 10 µM were used, and reactions were catalyzed using an expressed UGT 1A3, 1A8, 1A9 or a mixture of the three previous UGTs, each at 1/3 of original amounts, which meant that the total protein concentration of microsomes stayed the same.
UPLC Analysis of Two Flavones and Their Glucuronides
Wogonin, oroxylin A as well as their corresponding glucuronides were analyzed by a common chromatographic method: system, Waters Acquity UPLC with photodiode array detector and Empower software; column, BEH C18, 1.7 µm, 2.1 × 50 mm; mobile phase B, 100% acetonitrile, mobile phase A, 100% aqueous buffer (0.1%, v/v formic acid, pH 2.5); flow rate 0.4 ml/min; gradient, 0 to 1.5min, 30–40% B, 1.5 to 2.5 min, 40–70% B, 2.50 to 3.0 min, 70-30% B, wavelength, 280 nm for flavones and their respective glucuronides and acetophenone; and injection volume, 10 µl. The test linear response range was 0.313–50 µM (total 9 concentrations) for wogonin and oroxylin A. Analytical methods for each compound were validated for inter-day and intra-day variation using 6 samples at three concentrations (40, 10 and 1.25 µM). Precision and accuracy for both compounds were in the acceptable range of 0.11%–3.97%, and at 88.19% −104.62%, respectively.
Determination of Conversion Factor for Oroxylin A Glucuronide
Although wogonoside (or wogonin-7-glucuronic acid) could be obtained commercially, pure glucuronide of oroxylin A was not. To quantify glucuronide more precisely, previously published method was adapted (3, 26). In this method, the peak area increase in aglycone was compared with the peak area decrease in glucuronide after hydrolysis by β-glucuronidase. Since 1 mol metabolite generates 1 mol aglycone after hydrolysis, the change in concentrations as the consequence of hydrolysis can be expressed as:
| (1) |
where Δ PDHFG is the change in peak areas of di-hydroxyl flavone glucuronide (or DHFG), Δ PDHF is the change in the peak area of its corresponding flavone aglycone (or DHF) obtained from the extracted samples before and after hydrolysis, and αDHF and αDHFG are the slopes of aglycone and glucuronide calibration curve going through the origin.
Then, equation (1) can be rearranged to equation (2), since glucuronides of most flavones were not commercially available:
| (2) |
where K represents the conversion factor of molar extinction coefficients of glucuronides to their corresponding aglycones. Now, substitute various K values into the following equation, the true concentrations of metabolite (CDHFG) can be quantified by:
| (3) |
where PDHFG is the metabolite peak area in the chromatogram. The same experiments were performed simultaneously at three different substrate concentrations (2.5, 10 and 35 µM) to obtain an average K value. Then, the concentrations of glucuronides (e.g., oroxylin A-7-O-glucuronide) can be calculated by using the standard curve of aglycones (e.g., oroxylin A).
Here, the experiment procedures for obtaining the conversion factor (K) were as follows: (1) react oroxylin A with the most active UGT isoform using the incubation procedures described previously, (2) extract the aqueous samples containing the glucuronide with dichloromethane (sample/dichloromethane = 2:5, v/v) twice to remove the aglycone, (3) divide one extracted aqueous sample into two equal portions, where one portion was subjected to UPLC for analysis directly, and the other was analyzed after hydrolysis by β--glucuronidase (800 units/ml) at 37°C for 1 h (10 h for oroxylin A). The conversion factor of oroxylin A obtained was 1.29 ± 0.05 (wavelength 280 nm). The conversion factor of wogonin was derived using the standard curve of both wogonin and wogonoside, and it was 1.09 ± 0.04, also at 280 nm.
Confirmation of Flavone Glucuronide Structure by LC-MS/MS
The separation, detection and analysis of flavones and their glucuronides were achieved by Waters Micromass Quattro Premier XE, operated in the positive ion mode. The main mass working parameters for the mass spectrometers were set as follows: capillary voltage, 3KV; cone voltage, 35V; ion source temperature, 100°C; desolvation temperature, 350°C; cone gas flow, 50 l/hr; desolvation temperature gas flow, 600 l/hr. Data acquisition and analysis were performed using a MassLynx V4.1 software (Waters Corp, Milford, MA, USA).
Kinetic of Glucuronidation
Rates of flavone metabolism by expressed human UGT isoforms, human liver and intestine microsomes were expressed as amounts of metabolites formed per min per mg protein (nmol/min/mg). Kinetic parameters were then obtained according to the profile of Eadie-Hofstee plots (4, 25). If the Eadie-Hofstee plot was linear, formation rates (V) of flavone glucuronides at respective substrate concentrations (C) were fit to the standard Michaelis-Menten equation:
| (4) |
where Km is the Michaelis-Menten constant and Vmax is the maximum rate of forming glucuronides.
When Eadie-Hofstee plots showed characteristic profiles of atypical kinetics (autoactivation and biphasic kinetics) (27, 28), the data from these atypical profiles were fit to equations (5) and (6), using the ADAPT II program (29). To determine the best-fit model, the model candidates were discriminated using the Akaike's information criterion (AIC) (30), and the rule of parsimony was applied. Therefore, using this minimum AIC estimation (MAICE), a negative AIC value (e.g., −37.54) would be considered a better representation of the data versus a set of data having a positive AIC value (e.g., 3.17) (31).
| (5) |
where Vmax-0 is maximal intrinsic enzyme activity, Vmax-d is maximal inducible enzyme activity, R is rate of enzyme activity induction, C is concentration of substrate, and Km is concentration of substrate to achieve 50% of (Vmax-0 + Vmax-d).
| (6) |
where Vmax1 is maximum enzyme activity, C is concentration of substrate, Km1 is concentration of substrate to achieve 50% of Vmax, and Ksi is substrate inhibition constant.
Use of Expressed UGTs to Determine and Describe Flavone Glucuronidation in Human Intestinal and Liver Microsomes
Because each tissue expressed different UGTs and/or different quantities of the same UGTs (23), we first determined the main isoforms responsible for the metabolism of each flavone and then use a combination of several major active isoforms to determine substrate metabolism profiles in human intestinal and liver microsomes. The combination was achieved based on weighted mean average expression levels (23). Taking wogonin as an example, UGT1A3 contributes greatly to wogonin glucuronidation both in liver and intestine, and liver is UGT 1A1- and 1A9-rich whereas intestine is UGT1A8- and 1A10-rich. Therefore, the major isoforms chosen for the determination of liver metabolism of wogonin were UGT1A1, 1A3 and 1A9 and the weighted expression ratio was UGT1A1 (51.60): UGT1A3(4.73): UGT1A9(43.67). Similarly, the major isoforms chosen to determine its intestinal metabolism were UGT 1A3(2.30): 1A8(6.59): 1A10(91.11). Similarly, ratios of UGT 1A1(51.51): 1A3(4.72): 1A7(0.18): 1A9(43.59) and UGT 1A3(2.24): 1A7(2.42): 1A8(6.43): 1A10(88.91) were for glucuronidation of oroxylin A in human liver and intestinal microsomes, respectively. Taking the ratios as weighing coefficients, the combined glucuronidation rates were calculated using rates obtained experimentally from each UGT isoform. Subsequently, the glucuronidation profiles obtained using the combined glucuronidation rates versus flavone concentrations were used to obtain the apparent kinetic parameters after corresponding Eadie-Hofstee plots were generated. Moreover, linear regression was applied to derive apparent correlations between rates of reaction obtained using a combination of main UGT isoforms and those obtained using human intestinal and liver microsomes.
Statistical Analysis
One-way ANOVA with or without Tukey-Kramer multiple comparison (posthoc) tests were used to evaluate statistical differences. Differences were considered significant when p values were less than 0.05.
Results
Confirmation of Flavone Glucuronide Structure by LC-MS/MS
Based on a straightforward LC-MS/MS analysis of the metabolites, all glucuronides generated in the present study were mono-glucuronides (Fig.1), and no di-glucuronide was found. Based on the use of authentic standard and UV absorbance spectrum as described (32), one mono-glucuronide each was identified as wogonin-7-O- glucuronide and oroxylin A-O-glucuronide, respectively. Moreover, the internal standard peak didn’t overlap any glucuronide that may have been present (Fig.1).
Main Isoforms Responsible for the Metabolism of Wogonin and Oroxylin A
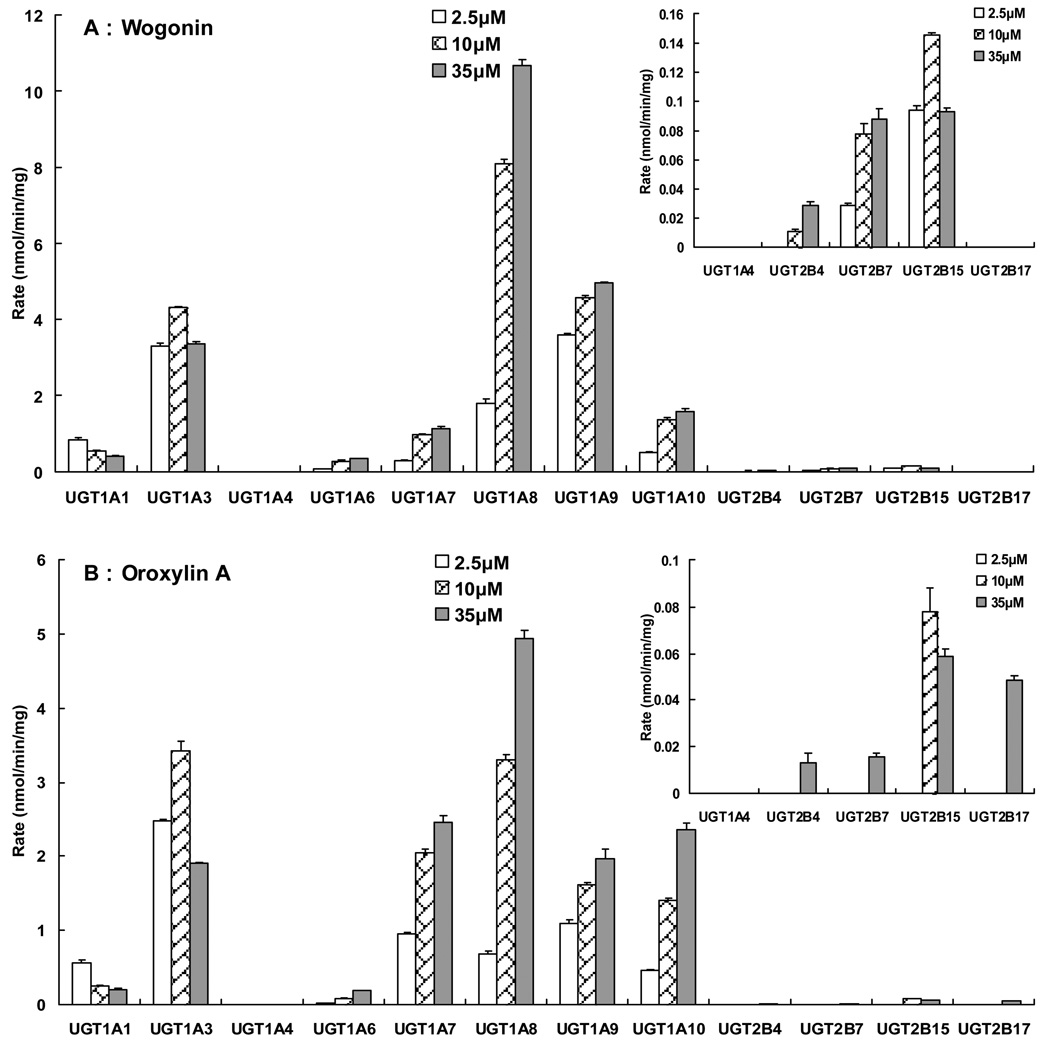
In order to determine the main UGT isoform(s) responsible for metabolizing wogonin and oroxylin A, incubation experiments at three concentrations (2.5, 10 and 35 µM) were conducted at 37 °C for up to 1 hr, after considering the thermostability studies of specific UGT isoforms (24). Glucuronidation rates by various UGT isoforms were, as expected, significantly different for each flavone (p<0.05, one way ANOVA), and the top isoform changed with flavone and flavone concentration. However, the isoforms playing key roles (i.e., caused rapid glucuronidation) were always the same: UGT1A3, 1A8, 1A9 and 1A10. Moreover, UGT1A7 was found to rapidly glucuronidate oroxylin A. As for other UGT isoforms, 1A1, 1A6 slowly metabolize these two flavones, whereas 1A4 did not glucuronidate any of these two flavones. For UGT2Bs, tested isoforms (i.e., 2B4, 2B7, 2B15 and 2B17) showed slow and often negligible rates of glucuronidation (Fig.2).
Fig.2. Glucuronidation of wogonin, oroxylin A by expressed human UGTs.
Three concentrations (2.5 µM, 10 µM and 35 µM) substrates were incubated with 12 commercially available expressed human UGTs at 37 °C for up to 1 hr. Glucuronides formed in the incubation sample were quantified by UPLC and rates of glucuronidation were calculated as nmol/min/mg of protein. Each bar is the average of three determinations, and the error bars are the standard deviations of the mean (n=3).
It was reported previously that the human UGT1A locus could be considered in terms of sequence similarity to four gene clusters: UGT1A1, UGT1A6, UGT1A2P–1A5, and UGT1A7–1A13P. Isoforms in the same cluster have greater sequence similarities (33). Therefore, most of the highly active UGT isoforms belong to the fourth cluster (UGT1A7–1A13P), and only one (UGT1A3) belongs to a different cluster. Taken together, UGT1As are the main subfamily responsible for metabolism of wogonin and oroxylin A. Among this subfamily, UGT1A3 and 1A9 were the two most active isoforms at a low concentration of 2.5 µM, whereas at a middle concentration of 10 µM, the most active isoforms were UGT1A3, 1A9 and 1A8 (1A7 for oroxylin A), followed by UGT 1A10. At a high concentration of 35 µM, the most active isoform was UGT 1A8 for both flavones, and the relative contribution of UGT1A3 declined (Fig.2).
Kinetics of Wogonin and Oroxylin A Glucuronidation by Five Major UGT Isoforms and UGT 1A1
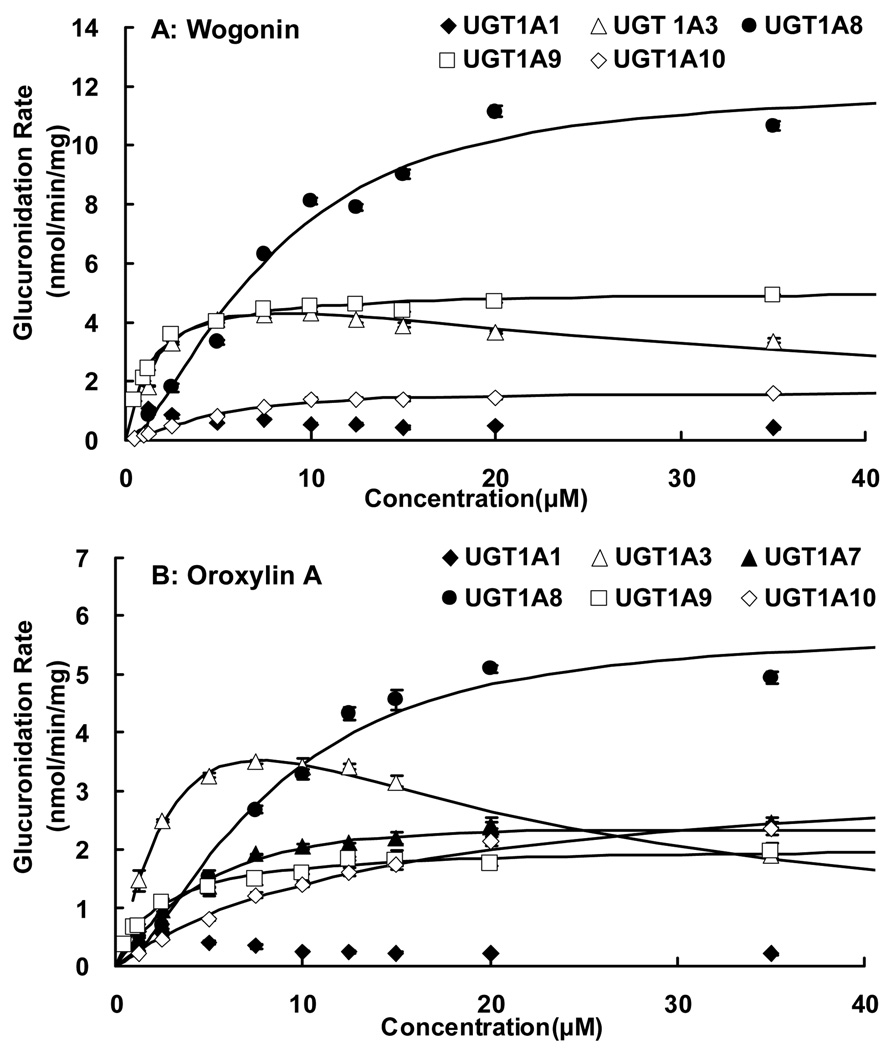
Delineation of the metabolic kinetics of wogonin and oroxylin A by the most active UGT1As holds the premise of determining and describing their metabolism rates and kinetic profiles by human intestinal and liver microsomes. Therefore, the kinetic parameters of glucuronidation by top 5 or 6 most active human UGT isoforms along with UGT1A1 were determined (Fig.3 and Fig.4). UGT1A1 was studied because it is highly expressed in both liver and intestine (23).
Fig.3. Kinetics of wogonin (A) and oroxylin A (B) glucuronidation by UGT1A1, UGT1A3, UGT1A7 (B), UGT1A8, UGT1A9 and UGT1A10 (n = 3).
The glucuronidation rates of two flavones were measured at substrate concentrations ranging from 1.25 to 35µM. The reaction time was controlled so that substrate concentration did not decrease substantially (usually less than 30%) at the end of experiments, which lasted for of up to 1 hr. For UGT 1A9, substrate concentrations were in the range of 0.5 −35 µM, and for wogonin glucuronidation by UGT1A10, its concentrations were in the range of 0.5 −35 µM. Each data point is the average of three determinations and the error bar represents the standard deviation of the mean. The data points in figures are observed glucuronidation rates, and the curves are plotted using the fitted kinetic parameters (Table II and Table III). The UGT1A1-mediated glucuronidation displayed unusual metabolic patterns that did not fit any known model, and hence kinetic parameters were not obtained.
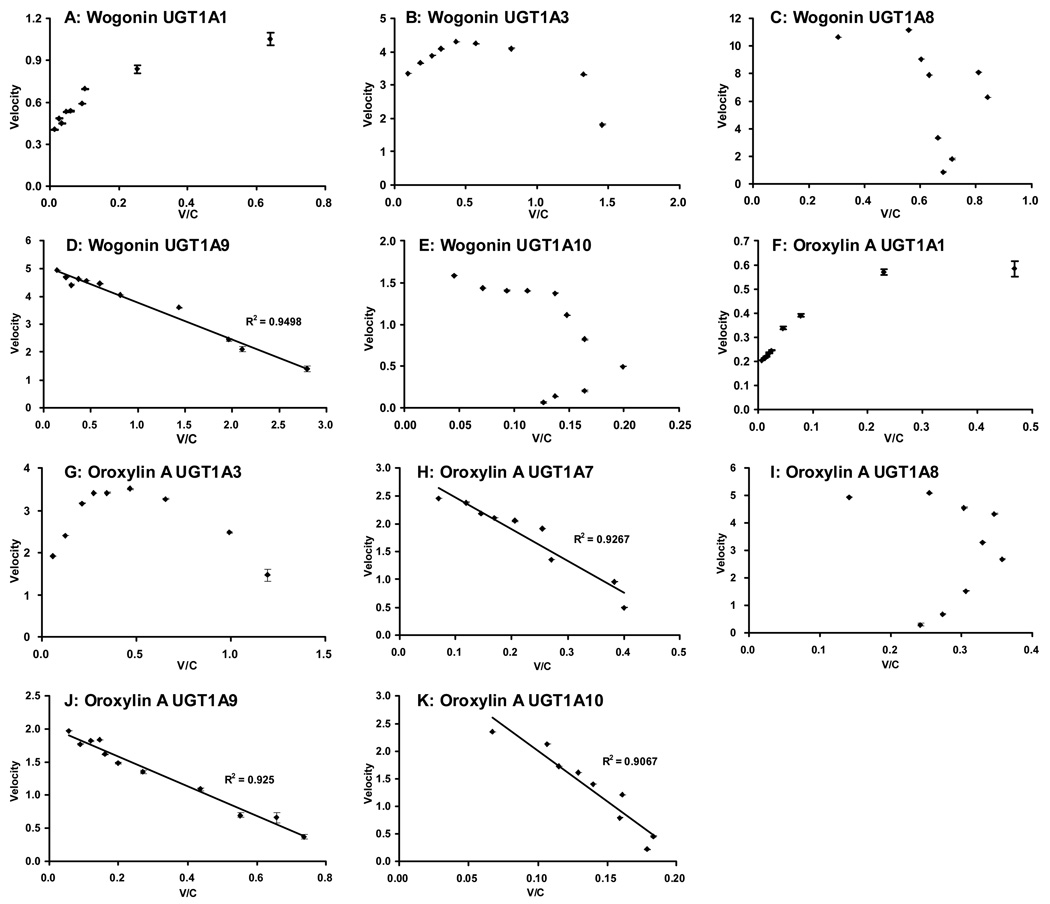
Fig.4. Eadie-Hofstee plots of glucuronidation profiles shown in Fig.3.
To determine the best-fit equation, Eadie-Hofstee plots were generated and formation rates of glucuronides at 9 (11) substrate concentrations were calculated as described in the Method section. For wogonin, UGT1A9 showed classic MM kinetics, UGT1A8, 1A10 showed autoactivation kinetics, and UGT1A3 showed substrate inhibition kinetics. For oroxylin A, UGT1A7, UGT1A9 and UGT1A10 showed classic MM kinetics, UGT1A8 showed autoactivation kinetics, and UGT1A3 also showed substrate inhibition kinetics. Eadie-Hofstee plots related to the glucuronidation of these two flavones by UGT1A1 did not conform to any known model.
For wogonin, the UGT isoforms used were UGT1A1, 1A3, 1A8, 1A9, and 1A10. Among these five isoforms, UGT1A9-mediated glucuronidation followed the classic Michaelis-Menten kinetics, UGT1A3-mediated glucuronidation of wogonin followed substrate inhibition kinetics, and UGT1A8- and 1A10-mediated glucuronidation followed autoactivation kinetics (Fig. 3A, Fig. 4B–E). In contrast, UGT1A1-mediated glucuronidation of wogonin displayed kinetic profile that was unusual and did not conform to any known kinetic model (Fig. 3A, Fig. 4A). The kinetic parameters of various UGT isoform catalyzed glucuronidation of wogonin were shown in (Table I, Table II). Among the four UGT isoforms whose enzyme kinetics can be described by known kinetic equation, the results indicated that their Km values were bunched together, in the range of 1.3–3.4 µM (<3-fold), indicating that these isoforms have similar affinity to the flavone. The Vmax values on the other hand, were more scattered, and ranged from 1.6 to 12.4 nmol/min/mg protein (nearly 8-fold).
Table I. Apparent kinetic parameters of glucuronidation of two flavones by UGT1A3.
| Wogonin | Oroxylin A | |
|---|---|---|
| Km(µM) | 2.30 | 7.41 |
| Ksi (µM) | 32.00 | 7.86 |
| Vmax (nmol/min/mg) | 6.60 | 10.38 |
| R2 | 0.95 | 0.98 |
| AIC | −6.68 | −13.18 |
Table II. Apparent kinetic parameters of wogonin glucuronidation into wogonoside or wogonin-7-O-glucuronide by UGT1A8, 1A9, 1A10, human liver microsomes (HLM), human intestinal microsomes (HIM), and combined UGTs.
The combined glucuronidation rates were obtained via calculating weighted mean of determinations at various substrate concentrations as described in “Materials and Methods” section. Moreover, the weighted expression ratios of UGT 1A1(54.17): 1A9(45.83) and UGT 1A8(6.74): 1A10(93.26) were for glucuronidation of wogonin in human liver and intestinal microsome respectively, when only the two isoforms were considered. All parameters were calculated based on curve fitting using Michaelis-Menten and autoactivation enzyme kinetics models as described in “Materials and Methods” section. Only kinetic parameters derived from the best-fit model as determined using the smallest AIC value were presented here.
| Kinetic Parameters | HLM | HIM | UGT1A8 | UGT1A9 | UGT1A10 | UGT1A1+1A9 | UGT1A1+1A3+1A9 | UGT1A8+1A10 | UGT1A3+1A8+1A10 |
|---|---|---|---|---|---|---|---|---|---|
| Km (µM) | 1.67 | 0.70 | 3.41 | 1.27 | 1.34 | 0.55 | 0.58 | 1.35 | 0.90 |
| Vmax ( nmol/min/mg ) | 7.88 | 6.53 | 12.38 | 5.07 | 1.61 | 2.49 | 2.57 | 2.29 | 2.28 |
| Vmax / Km( ml/min/mg ) | 4.72 | 9.33 | 3.60 | 3.99 | 1.20 | 4.53 | 4.43 | 1.70 | 2.53 |
| R2 | 0.98 | 0.99 | 0.98 | 0.99 | 0.99 | 0.92 | 0.92 | 0.99 | 0.99 |
| AIC | 3.17 | −21.33 | 15.47 | −15.78 | −37.54 | −25.28 | −24.19 | −25.46 | −25.67 |
| R (autoactivation only) | - | - | 0.16 | - | 0.26 | - | - | 0.21 | 0.20 |
| Vmax-0 (autoactivation only) (nmol/min/mg) |
- | - | 1.07*10−6 | - | 0.09 | - | - | 0.13 | 0.16 |
| Vmax-d (autoactivation only) (nmol/min/mg) |
- | - | 12.38 | - | 1.56 | - | - | 2.21 | 2.18 |
Eadie-Hofstee plot of wogonin by UGT1A1 dose not conform to any known model. Therefore, apparent kinetic parameters were not calculated.
For oroxylin A, the isoforms used were UGT1A1, 1A3, 1A7, 1A8, 1A9 and 1A10. Among these six isoforms, UGT1A7-, UGT 1A9- and 1A10-mediated glucuronidation followed classic Michaelis-Menten kinetics, whereas UGT 1A8-mediated glucuronidation followed autoactivation kinetics (Fig. 3B, Fig. 4H–K). UGT1A3-mediated glucuronidation of oroxylin A decreased at high substrate concentration, and their Eadie-Hofstee plot followed substrate inhibition kinetics (Fig. 3B, 4G). UGT1A1-mediated glucuronidation of oroxylin A decreased as concentration increased, and their Eadie-Hofstee plot did not match with any known kinetic profile (Fig. 3B, 4F). The kinetic parameters were determined for Michaelis-Menten, autoactivation and substrate inhibition kinetics using Adapt II kinetic modeling software and the results were shown in Table I and III. The Km values for oroxylin A were more variable than wogonin, with a range of 2.4 to 14.7 µM (6-fold), whereas its Vmax values fell within the tighter range of 2.1 to 6.0 (3-fold).
Table III. Apparent kinetic parameters of oroxylin A glucuronidation into oroxylin A-7-O-glucuronide by UGT1A7, 1A8, 1A9, 1A10, human liver microsomes (HLM), human intestinal microsomes(HIM), and combined UGTs.
The combined glucuronidation rates were calculated by the same method as described in Table. 1. The weighted expression ratios was as described in “Materials and Methods” section, or the same as wogonin when only UGT 1A1, 1A9 and UGT 1A8, 1A10were considered for the glucuronidation in human liver and intestinal microsome respectively. All parameters were calculated based on curve fitting using Michaelis-Menten and autoactivation enzyme kinetics models as described in “Materials and Methods” section. Only kinetic parameters derived from the best-fit model as determined using the smallest AIC value were presented here.
| Kinetic Parameters | HLM | HIM | UGT1A7 | UGT1A8 | UGT1A9 | UGT1A10 | UGT1A1+1A9 | UGT1A1+1A3+1A7+1A9 | UGT1A8+1A10 | UGT1A3+1A7+1A8+1A10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Km (µM) | 2.70 | 1.50 | 4.93 | 4.17 | 2.38 | 14.73 | 0.69 | 0.70 | 14.52 | 12.56 |
| Vmax (nmol/min/mg) | 4.33 | 7.38 | 2.93 | 6.03 | 2.06 | 3.46 | 0.98 | 1.09 | 3.75 | 3.57 |
| Vmax / Km ( ml/min/mg ) | 1.60 | 4.92 | 0.59 | 1.45 | 0.87 | 0.23 | 1.42 | 1.56 | 0.26 | 0.28 |
| R2 | 0.99 | 0.99 | 0.98 | 0.98 | 0.99 | 0.99 | 0.90 | 0.93 | 0.99 | 0.99 |
| AIC | −20.31 | −10.66 | −20.47 | 1.48 | −33.77 | −24.95 | −37.17 | −38.78 | −20.00 | −21.77 |
| R (autoactivation only) | - | - | - | 0.17 | - | - | - | - | - | - |
|
Vmax-0 (autoactivation only) (nmol/min/mg) |
- | - | - | 1.75*10−6 | - | - | - | - | - | - |
|
Vmax-d (autoactivation only) (nmol/min/mg) |
- | - | - | 6.03 | - | - | - | - | - | - |
Eadie-Hofstee plot of oroxylin A by UGT1A1 did not conform to any known model. Therefore, apparent kinetic parameters were not calculated.
Kinetics of Wogonin and Oroxylin A Glucuronidation by Human Liver and Intestinal Microsomes
The rates of glucuronidation of wogonin and oroxylin A by human liver and intestinal microsomes were determined at 11 concentrations in the range from 0.5 to 35 µM (Fig.5). Based on the established LC-MS/MS method of detecting the metabolites, only one mono-glucuronide was formed for each flavone after incubation with the microsomes.
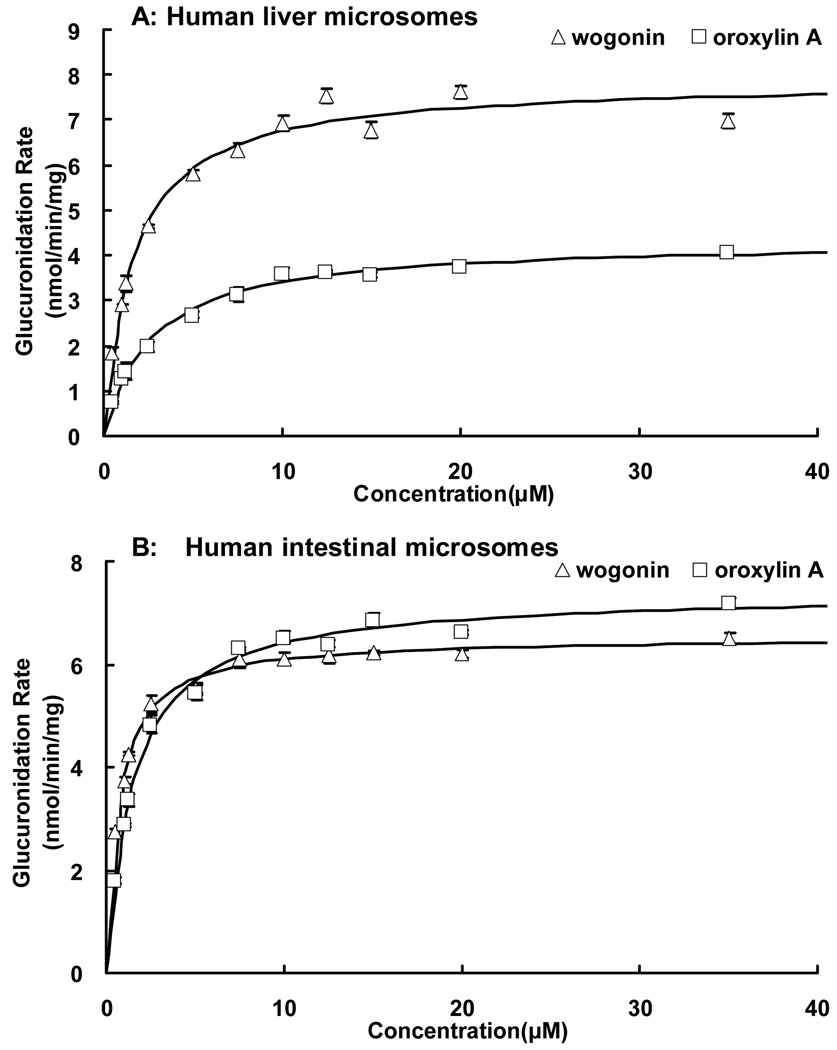
Fig.5. Kinetics of wogonin and oroxylin A glucuronidation by pooled human liver (A) and intestinal microsomes (B) (n = 3).
The glucuronidation rates of two flavones were determined at a concentration range of 0.5 to 35 µM. Each data point is the average of three determinations and the error bar represents the standard deviation of the mean. The points in panels are the flavone glucuronidation rates, and the curves are plotted using calculated rates derived from fitted parameters shown in Table II and Table III. The software applied was as described in the Method section.
For both flavones, human liver microsomes-mediated glucuronidation followed classic Michaelis-Menten kinetics (Fig. 5A, Fig. 6A, C). The Km value was 1.67 for wogonin, and 2.70 µM for oroxylin A. In contrast, the UGT1A9 mediated glucuronidation had a Km value of 1.27 µM for wogonin and 2.38 µM for oroxylin A (Table II, Table III).
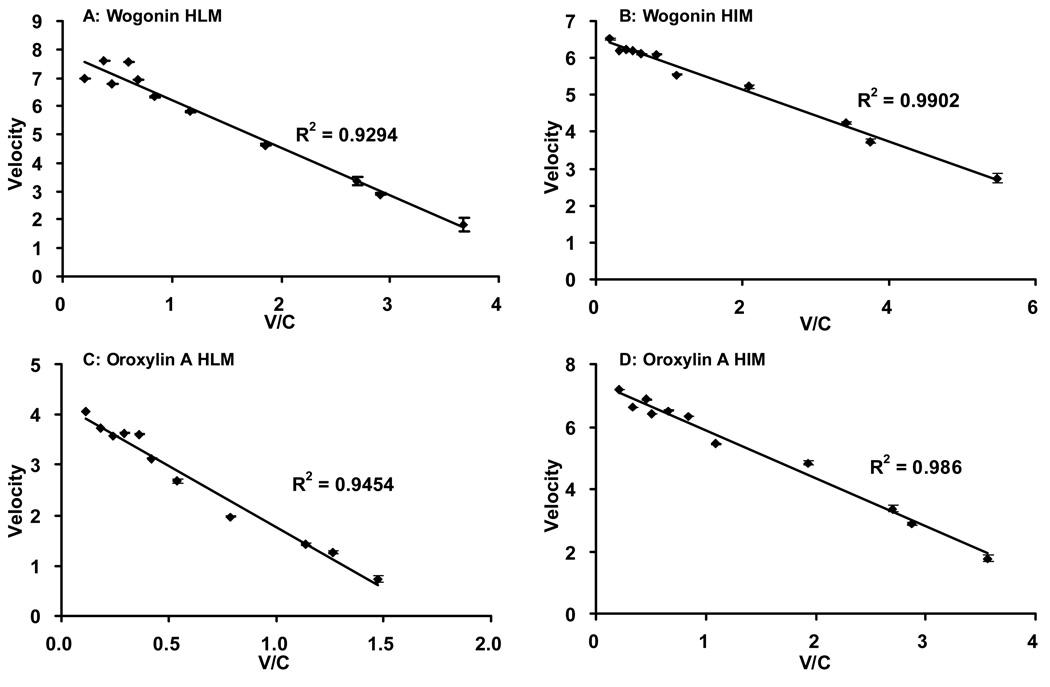
Fig.6. Eadie-Hofstee plots of glucuronidation profiles shown in Fig.5.
To determine the best-fit equation, Eadie-Hofstee plots were generated and formation rates of glucuronides at 11 substrate concentrations were calculated by software described in the Method section. For wogonin and oroxylin A, both human liver microsomes (HLM) and human intestinal microsomes (HIM) showed classic Michaelis-Menten kinetics.
UGT-mediated glucuronidation in human intestinal microsomes also matched classic Michaelis-Menten kinetics (Fig. 5B, Fig. 6B, D). The Km value was 0.7 µM for wogonin, and 1.50 µM for oroxylin A, different from that of UGT1A8 and 1A10, two isoforms selectively and highly expressed in intestine (Table II, Table III).
Kinetics of Wogonin and Oroxylin A Glucuronidation by Combined UGT 1As
A combination of UGT isoform glucuronidation rates was used to resolve if the profile derived from the combination rates could determine and explain or describe the metabolic profile found in a particular type of human liver or intestinal microsomes. This was done based on UGT expression in human liver and intestine (23) and contribution of every UGT isoform to the metabolism of these two flavones in previous experiments (Fig. 2). Hence, UGT1A9 and 1A1 were chosen as the most important isoforms for liver metabolism. Similarly, UGT1A8 was chosen as the most important for intestine, followed by UGT1A10. UGT1A3 and 1A7 (UGT1A7 was only for oroxylin A) were also chosen because they also made major contributions to glucuronidation and UGT1A3 was expressed extensively in both liver and intestine (23).
For wogonin, a combination of the glucuronidation rates by UGT1A1 and UGT1A9 (ratio≈54:46 slightly favoring UGT1A1) or UGT1A1+UGT1A3+UGT1A9 (~52:5:43) generated a “combinational” profile that was demonstrated to conform to classic Michaelis-Menten kinetics, similar to the profile generated by human liver microsomes (Table II, Fig.7A, Fig.8A, Fig.8B), On the other hand, a combination of UGT1A8 and UGT1A10 (~7:93) or UGT1A3+UGT1A8+UGT1A10 (2:7:91) followed autoactivation kinetics, different from those of intestinal microsomes (Table II, Fig.7A, Fig.8C, Fig.8D). The Km value (0.9 µM) of combined UGTs (1A8+1A10+1A3) was almost the same as that of human intestine microsomes (0.7 µM), whereas the Km value of UGT1A9 alone was closer to that of human liver microsomes than the combined UGT1As (Table II).
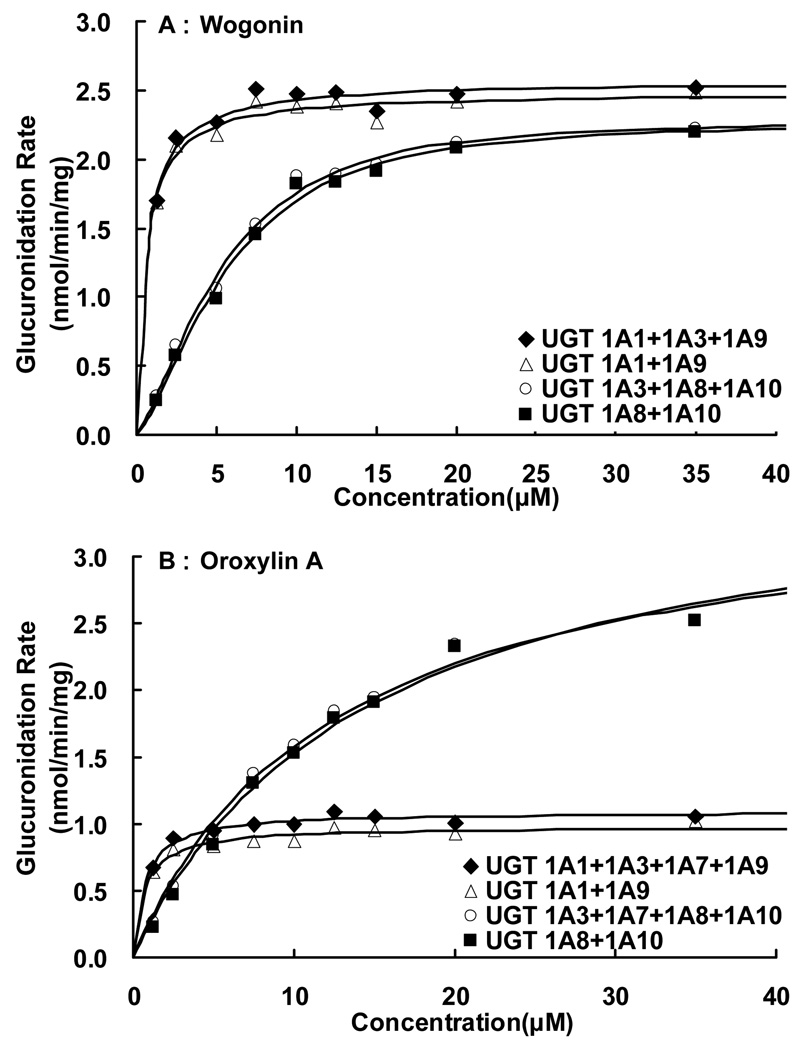
Fig. 7. Apparent kinetic profiles of wogonin and oroxylin A glucuronidation by combined UGT1As.
The combined glucuronidation rates were calculated based on weighted expression level calculation as described in the Methods. Kinetic profiles of wogonin glucuronidation by combined UGT1As were shown in panel A, and kinetic profiles of oroxylin A glucuronidation by combined UGT1As were shown in panel B. The points in panels are the flavone glucuronidation rates calculated based on both reaction rates and expression level described in the Method, and the curves are plotted using fitted parameters shown in Table II and III.
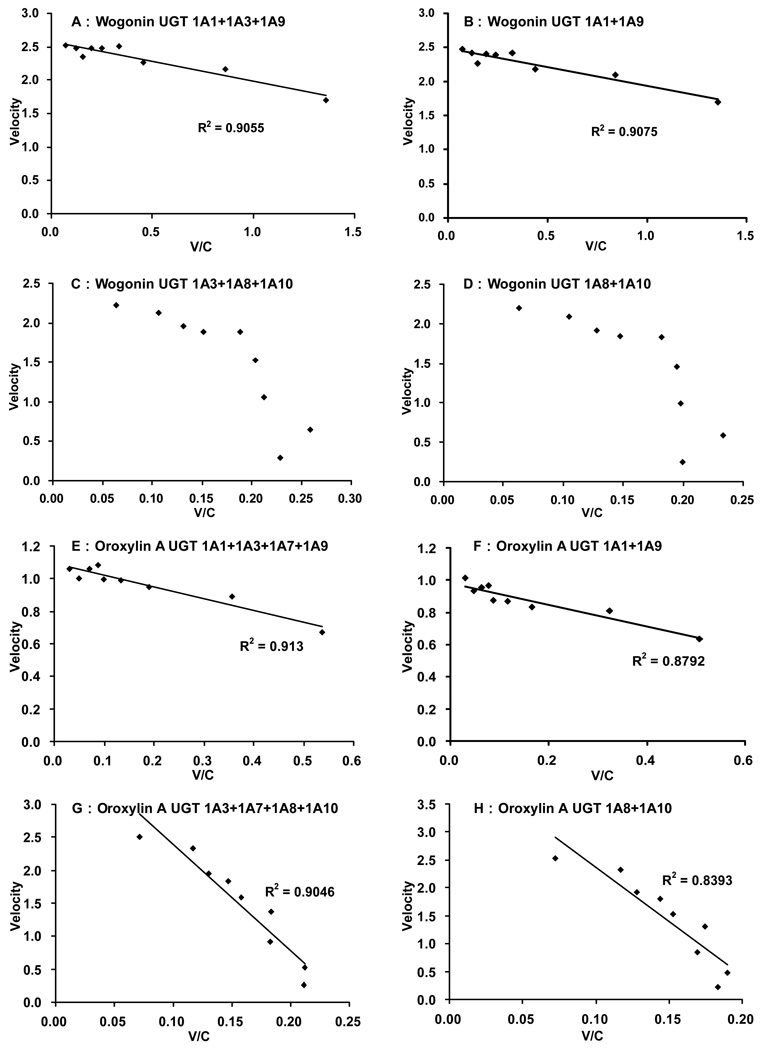
Fig.8. Eadie-Hofstee plots of glucuronidation kinetics shown in Fig.7.
To determine the best-fit equation, Eadie-Hofstee plots were generated and formation rates of glucuronides at 9 substrate concentrations were calculated by software described in the Method section. For wogonin, combined glucuronidation rates using UGT1A1+1A9±1A3 showed classic Michaelis-Menten kinetics, while combined glucuronidation rates using UGT1A8+1A10±1A3 showed autoactivation kinetics. For oroxylin A, combined glucuronidation rates using UGT1A1+1A9±(1A3 and/or 1A7), or using UGT1A8+1A10±(1A3 and/or 1A7) displayed classic Michaelis-Menten kinetics.
For oroxylin A, a combined rates from UGT1A1+UGT1A9 or UGT1A1+UGT1A9+ UGT1A3+UGT1A7 generated a kinetic profile conforming to classic Michaelis-Menten kinetics, as did combined rates from UGT1A8+UGT1A10 or UGT1A8+UGT1A10+ UGT1A3+UGT1A7 (Fig.7B, Fig.8E–H). These kinetic profiles were the same as the kinetic profiles generated from intestinal and liver microsomes. Here, the Km value derived from combined UGT1A8+UGT1A10 (14.5 µM) or UGT1A8+UGT1A10+UGT1A3+UGT1A7 (12.6 µM) differed significantly from human intestine microsomes (1.50 µM). Once again, the Km value of UGT 1A9 was closer to that of human liver microsomes than the combined UGT1As (Table III).
Correlation of Isoform-Determined Rates with Rates Derived from Human Liver and Intestinal Microsomes
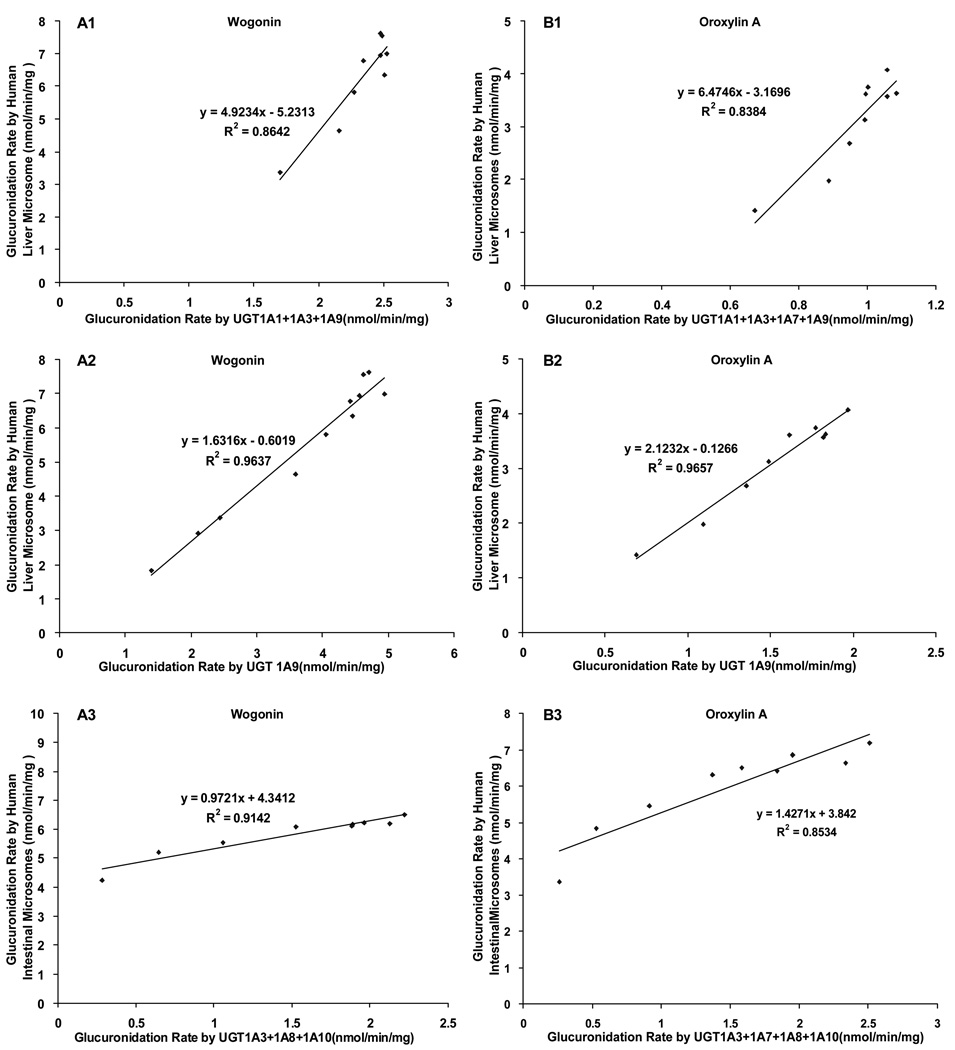
Linear regression was applied to derive apparent correlations between the glucuronidation rates of a single UGT1As or several UGT1As combined and human intestinal and liver microsomes. For wogonin, the glucuronidation rates of human liver microsomes have an apparent correlation with those of a single UGT1A or UGT1A9 or a combination of UGT1As (UGT1A1+ 1A3+1A9). Similarly, the glucuronidation rates of wogonin in human intestinal microsomes also have an apparent correlation with those derived from a combination of UGT1As (UGT1A3+1A8+1A10) (Fig.9 A1–A3).
Fig. 9. Correlation of glucuronidation rates of wogonin (A) and oroxylin A (B) obtained from major UGT1A isoform(s) with those from human liver microsomes and human intestinal microsomes.
Linear regression was used to derive apparent correlations. The combined glucuronidation rates for major UGT 1As were calculated the same as described in Fig. 7. For wogonin, the correlation between combined UGT1As and human liver microsomes (R2=0.8642) is shown in panel A1, the correlation between UGT1A9 and human liver microsomes (R2=0.9637) in panel A2, and the correlations between combined main UGT1As and human intestinal microsomes (R2=0.9142) in panel A3. For oroxylin A, correlations between human liver microsomes and combined major UGT1As (R2=0.8384) or UGT1A9 (R2=0.9657) are shown in panels B1 and B2, respectively; and the correlation between combined main UGT1As and human intestinal microsomes (R2=0.8534) in panel B3.
For oroxylin A, obvious correlations were obtained between its glucuronidation rates in human liver microsomes and those of UGT1A9 alone or combined UGT1As. Obvious correlations were again obtained between its glucuronidation rates in human intestine microsomes and those derived from a combination of UGT1As (Fig.9 B1–B3).
Comparison of Glucuronidation Rates of Wogonin and Oroxylin A by a Single UGT1A or a Mixture of UGT1As
The influence of mixing UGT1As on the flavone glucuronidation rates were investigated in order to determine if rates of individual UGT isoforms are essentially the same as those generated from the most active UGT1As mixed together. The results indicated the Km values of combined UGTs or human organ microsomes were mostly higher than those of individual UGT isoforms. One possible reason is that a combinational approach can usually generate a Km value that is higher than the UGT isoform with the smallest Km values. Another possible reason was that glucuronidation rates decreased after enzymes were mixed, especially at low substrate concentrations. To determine if the second reason was the case, experiments were performed to determine the total microsomal protein concentrations.
Lastly, for wogonin, at a low substrate concentration of 2.5 µM, the glucuronidation rates in a three UGT mixture were 2.46 ± 0.02 nmol/min/mg, which is similar to the average glucuronidation rate of the three individual UGTs (2.44 nmol/min/mg). Similarly, the glucuronidation rates at 10 µM were 4.91±0.13 nmol/min/mg, which was slightly above the average glucuronidation rate of the three individual UGTs (4.72 nmol/min/mg).
For oroxylin A at a low substrate concentration of 2.5 µM, the glucuronidation rates in a mixture of three UGT1As were 1.61±0.02 nmol/min/mg, which was somewhat faster (p>0.05) than the average glucuronidation rate of the three individual UGTs (1.38 nmol/min/mg). At 10 µM concentration, the glucuronidation rates of the three UGT mixture were 2.14±0.03 nmol/min/mg, which was significantly less than (p<0.05) the average glucuronidation rate in the three individual UGT1As (2.61 nmol/min/mg), although the extent of the difference was not very large (Table IV).
Table IV. Comparison of glucuronidation rates of wogonin and oroxylin A by individual main UGT isoforms and a mixture of three main UGT isoforms.
The glucuronidation rates of two flavones were measured at substrate concentrations of 2.5 and 10 µM after reaction of up to 1 hr. The reactions were catalyzed by UGT1A3, 1A8, 1A9 or a mixture of the three previous UGTs present at equal proportion (each at 1/3 of the original protein concentration), respectively. The glucuronidation rates are the average of three determinations, and data represent mean±SEM.
| Analyte | Concentration (µM) |
Glucuronidation Rate (nmol/min/mg) |
|||
|---|---|---|---|---|---|
| UGT1A3 | UGT1A8 | UGT1A9 | UGT mixture | ||
| Wogonin | 2.5 | 2.15±0.04 | 1.90±0.13 | 3.26±0.06 | 2.46±0.02 |
| 10 | 3.95±0.05 | 6.03±0.16 | 4.17±0.13 | 4.91±0.13 | |
| Oroxylin A | 2.5 | 2.27±0.15 | 0.89±0.01 | 0.99±0.05 | 1.61±0.02 |
| 10 | 3.12±0.16 | 3.45±0.11 | 1.26±0.02 | 2.14±0.03 | |
Discussion
A systematic metabolic profiling study of two flavonoids was conducted here to demonstrate for the first time that rates and profiles derived from multiple UGT isoforms may be used to determine and describe the rates and profile their glucuronidation in human intestinal and liver microsomes. The results of this study indicate that UGT1A9 was likely the main isoform responsible for their liver metabolism, whereas a combination of UGT1As worked together (with the largest contribution from UGT1A10) to enable the intestine glucuronidation of wogonin. The studies also showed that even minor structural differences between wogonin and oroxylin A could significantly impact flavone glucuronidation.
Importance or perhaps dominance of UGT1A9 in the liver metabolism of these two flavones was substantiated by multiple lines of evidence. Among these, the most important is that UGT1A9 was expressed at a high level in liver (23). Although UGT1A1 is also expressed at a very high level (23), its ability to glucuronidate these two flavones was very weak compared to UGT1A9 (Fig.2). Secondly, kinetics study of two compounds showed that glucuronidation in pooled human liver microsomes and UGT1A9 all followed classic Michaelis-Menten kinetics with very similar Eadie-Hofstee plots (Fig.3, Fig.4 D, J, Fig.5A, Fig.6A,C), and the Km values were also similar (Table II, Table III). Thirdly, although UGT1A9 has a maximum weight of 46% in the multiple isoform studies, the profile derived from the combined UGT1As followed that of UGT1A9 (Table II and Table III). Lastly, correlation coefficients between rates of glucuronidation by UGT1A9 and those by human liver microsomes were quite satisfactory, better than those obtained from combined UGTs (Fig.9). These suggested that rates and profiles derived from UGT 1A9 could be used to determine metabolism of these two compounds in human liver microsomes.
Importance of UGT1A10 in the human intestinal glucuronidation of these two flavones was almost equal to that of UGT1A9 in human liver microsomes. Because of the high expression level of UGT1A10 (>90% relative abundance), it also dominates the rates and profiles of mixed UGT reactions (Fig.7, Fig.8, Table II and Table III). However, only glucuronidation of oroxylin A followed the classical Michaelis-Menten kinetics (Fig.7B, Fig.8G, H), whereas that of wogonin did not (Fig.7A, Fig.8C, D). Surprisingly, for wogonin, the Km value derived from a combination of UGT1A8, UGT1A10, and UGT 1A3 was almost identical with that of human intestinal microsomes (Table I), but for oroxylin A, the Km values were very different. On the other hand, correlation between glucuronidation rates derived from combined UGTs and those from human intestinal microsomes were above 0.9 for wogonin, and above 0.8 for oroxylin A (Fig.9, A3, B3). Therefore, combined UGT1As appeared to be able to determine rates but did not fully describe the profile of wogonin glucuronidation in human intestinal microsomes (Table II). In contrast, the same combination only adequately described the profile but did not precisely determine the rates of oroxylin A glucuronidation in the human intestinal microsomes.
The above analysis clearly shows for the first time that rates and profiles of glucuronidation derived from UGT isoforms could be used to reasonably describe the profiles of glucuronidation of two flavones wogonin and oroxylin A in human liver and intestinal microsomes. Coupled together with the results that glucuronidation rates derived from a single dominant UGT isoform or a mixture of UGT isoforms correlate well with the rates of metabolism in human liver and intestinal microsomes (Fig.9), the current study represents an important advance in determining organ-/tissue- specific glucuronidation using expressed UGT isoforms. Whereas this type of studies was performed routinely for CYP catalyzed metabolism (34), our present study is the first one of its kind in the study of UGT metabolism. Previous glucuronidation studies have focused on determining rates of metabolism (3, 24), but this study aims to determine rates and to describe profiles of glucuronidation. The results showed that our approach could provide accurate determination with the exception of oroxylin A metabolism in human intestine, which showed about 8-fold difference in Km values (Table III). We believe that the main reason is the dominant expression levels of UGT1A8 and UGT1A10 in our combination calculation, which makes Km values for the combination of isoform approaching the large Km value of UGT1A10 (14.52 µM). Another important factor is the presence of an alternative UGT isoform that was not used in the present study. We have observed this phenomenon previously, and attributed it to the absence of UGT3A1 from our panel of UGT isoforms, which also metabolizes phenolics (35). Other contributing factors include a moderate decrease in glucuronidation rates of oroxylin A when multiple UGT isoforms were present (Table IV). Further studies would be needed to further clarify the reason for this moderate discrepancy, and additional substrates would also be helpful.
The present approach to determine and describe rates and profiles of glucuronidation may have several important applications. First of all, this approach can be used to determine the main organs responsible for the metabolism of these two flavones. Using the correlation data, isoform fingerprint and levels of expression in a particular tissue, it would be relatively easy to resolve which isoform(s) to use to determine and describe the rates and profile of metabolism in a particular organ. In this case, the rates and profiles for liver metabolism of both flavones and for intestinal metabolism of wogonin were well described and predetermined using the UGT isoform kinetic data.
Second, the approach may be used to resolve which UGT genotype or phenotype may be potential predictors of response and toxicity in patients. In other words, if we knew which isoform is mainly responsible for the metabolism of a particular compound in a particular organ, then we could project the consequence of a genotype or phenotype change. For example, UGT1A9 genotypes was predictor for metastatic colorectal cancer patients treated with capecitabine plus irinotecan (which is also glucuronidated), and patients with genotypes conferring low UGT1A9 (dT)9/9 genotype were particularly likely to exhibit greater antitumor response with little toxicity (36).
In conclusion, our study demonstrated for the first time that the novel approach of UGT isoform specific metabolic fingerprinting (GSMF) combined with kinetic profiling of individual UGT isoforms may be used to determine and describe the rates and profiles of flavone glucuronidation in human tissues such as intestine and liver. It is likely that a detailed characterization study like the one conducted here can also be potentially used to predict a variety of other pharmacologically important responses including drug response and toxicity as well as drug interactions. Further studies are needed to demonstrate that this approach is broadly applicable to other compounds undergoing glucuronidation.
Acknowledgements
The authorz would like to thank Dr. Vincent Tam of University of Houston College of Pharmacy for his help in rendering the program to analyze the kinetics of glucuronidation by UGT 1A3. The authors would like to acknowledge Ms. Lin Ye for her technical assistance in the measurement of metabolites using LC-MS/MS. Additional technical assistance by Ms. Jie Zhao in the determination of conversion factor is also acknowledged here.
This work was mainly supported by the Ministry of Science and Technology of the People’s Republic of China Grants 2006BAT11B08-4, and the Grant of Science and Technology of Guangzhou 2006Z1-E6021, both to ZL. MH was also supported by NIH GM070737.
ABBREVIATIONS
- UGTs
UDP-glucuronosyltransferases
- UDPGA
Uridine diphosphate glucuronic acid
- GSMF
UGT-isoform specific metabolic fingerprint
- DHFG
di-hydroxyl flavone glucuronide
- DHF
di-hydroxyl flavone
- AIC
Akaike’s information criterion
- MAICE
minimum AIC estimation
- HLM
human liver microsomes
- HIM
human intestinal microsomes.
References
- 1.Kiang TK, Ensom MH, Chang TK. UDP-glucuronosyltransferases and clinical drug-drug interactions. Pharmacol Ther. 2005;106:97–132. doi: 10.1016/j.pharmthera.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Andoand Y, Hasegawa Y. Clinical pharmacogenetics of irinotecan (CPT-11) Drug Metab Rev. 2005;37:565–574. doi: 10.1080/03602530500316254. [DOI] [PubMed] [Google Scholar]
- 3.Tang L, Singh R, Liu Z, Hu M. Structure and concentration changes affect characterization of UGT isoform-specific metabolism of isoflavones. Mol Pharm. 2009;6:1466–1482. doi: 10.1021/mp8002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang SW, Chen J, Jia X, Tam VH, Hu M. Disposition of flavonoids via enteric recycling: structural effects and lack of correlations between in vitro and in situ metabolic properties. Drug Metab Dispos. 2006;34:1837–1848. doi: 10.1124/dmd.106.009910. [DOI] [PubMed] [Google Scholar]
- 5.Baumann S, Fas SC, Giaisi M, Muller WW, Merling A, Gulow K, Edler L, Krammer PH, Li-Weber M. Wogonin preferentially kills malignant lymphocytes and suppresses T-cell tumor growth by inducing PLCgamma1- and Ca2+-dependent apoptosis. Blood. 2008;111:2354–2363. doi: 10.1182/blood-2007-06-096198. [DOI] [PubMed] [Google Scholar]
- 6.Chung H, Jung YM, Shin DH, Lee JY, Oh MY, Kim HJ, Jang KS, Jeon SJ, Son KH, Kong G. Anticancer effects of wogonin in both estrogen receptor-positive and -negative human breast cancer cell lines in vitro and in nude mice xenografts. Int J Cancer. 2008;122:816–822. doi: 10.1002/ijc.23182. [DOI] [PubMed] [Google Scholar]
- 7.Hu Y, Yang Y, You QD, Liu W, Gu HY, Zhao L, Zhang K, Wang W, Wang XT, Guo QL. Oroxylin A induced apoptosis of human hepatocellular carcinoma cell line HepG2 was involved in its antitumor activity. Biochem Biophys Res Commun. 2006;351:521–527. doi: 10.1016/j.bbrc.2006.10.064. [DOI] [PubMed] [Google Scholar]
- 8.Li HN, Nie FF, Liu W, Dai QS, Lu N, Qi Q, Li ZY, You QD, Guo QL. Apoptosis induction of oroxylin A in human cervical cancer HeLa cell line in vitro and in vivo. Toxicology. 2009;257:80–85. doi: 10.1016/j.tox.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y, Lu N, Ling Y, Gao Y, Chen Y, Wang L, Hu R, Qi Q, Liu W, Yang Y, You Q, Guo Q. Oroxylin A suppresses invasion through down-regulating the expression of matrix metalloproteinase-2/9 in MDA-MB-435 human breast cancer cells. Eur J Pharmacol. 2009;603:22–28. doi: 10.1016/j.ejphar.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Peng J, Qi Q, You Q, Hu R, Liu W, Feng F, Wang G, Guo Q. Subchronic toxicity and plasma pharmacokinetic studies on wogonin, a natural flavonoid, in Beagle dogs. J Ethnopharmacol. 2009;124:257–262. doi: 10.1016/j.jep.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 11.Yano H, Mizoguchi A, Fukuda K, Haramaki M, Ogasawara S, Momosaki S, Kojiro M. The herbal medicine sho-saiko-to inhibits proliferation of cancer cell lines by inducing apoptosis and arrest at the G0/G1 phase. Cancer Res. 1994;54:448–454. [PubMed] [Google Scholar]
- 12.Bonham M, Posakony J, Coleman I, Montgomery B, Simon J, Nelson PS. Characterization of chemical constituents in Scutellaria baicalensis with antiandrogenic and growth-inhibitory activities toward prostate carcinoma. Clin Cancer Res. 2005;11:3905–3914. doi: 10.1158/1078-0432.CCR-04-1974. [DOI] [PubMed] [Google Scholar]
- 13.Zhang DY, Wu J, Ye F, Xue L, Jiang S, Yi J, Zhang W, Wei H, Sung M, Wang W, Li X. Inhibition of cancer cell proliferation and prostaglandin E2 synthesis by Scutellaria baicalensis. Cancer Res. 2003;63:4037–4043. [PubMed] [Google Scholar]
- 14.Yu J, Liu H, Lei J, Tan W, Hu X, Zou G. Antitumor activity of chloroform fraction of Scutellaria barbata and its active constituents. Phytother Res. 2007;21:817–822. doi: 10.1002/ptr.2062. [DOI] [PubMed] [Google Scholar]
- 15.Li-Weber M. New therapeutic aspects of flavones: the anticancer properties of Scutellaria and its main active constituents Wogonin, Baicalein and Baicalin. Cancer Treat Rev. 2009;35:57–68. doi: 10.1016/j.ctrv.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Mu R, Qi Q, Gu H, Wang J, Yang Y, Rong J, Liu W, Lu N, You Q, Guo Q. Involvement of p53 in oroxylin A-induced apoptosis in cancer cells. Mol Carcinog. 2009 doi: 10.1002/mc.20570. [DOI] [PubMed] [Google Scholar]
- 17.Hu M. Commentary: bioavailability of flavonoids and polyphenols: call to arms. Mol Pharm. 2007;4:803–806. doi: 10.1021/mp7001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai TH, Chou CJ, Tsai TR, Chen CF. Determination of wogonin in rat plasma by liquid chromatography and its pharmacokinetic application. Planta Med. 1996;62:263–266. doi: 10.1055/s-2006-957874. [DOI] [PubMed] [Google Scholar]
- 19.Kim YH, Jeong DW, Kim YC, Sohn DH, Park ES, Lee HS. Pharmacokinetics of baicalein, baicalin and wogonin after oral administration of a standardized extract of Scutellaria baicalensis, PF-2405 in rats. Arch Pharm Res. 2007;30:260–265. doi: 10.1007/BF02977703. [DOI] [PubMed] [Google Scholar]
- 20.Lai MY, Hsiu SL, Chen CC, Hou YC, Chao PD. Urinary pharmacokinetics of baicalein, wogonin and their glycosides after oral administration of Scutellariae Radix in humans. Biol Pharm Bull. 2003;26:79–83. doi: 10.1248/bpb.26.79. [DOI] [PubMed] [Google Scholar]
- 21.Ueng YF, Shyu CC, Lin YL, Park SS, Liao JF, Chen CF. Effects of baicalein and wogonin on drug-metabolizing enzymes in C57BL/6J mice. Life Sci. 2000;67:2189–2200. doi: 10.1016/s0024-3205(00)00809-2. [DOI] [PubMed] [Google Scholar]
- 22.Jeong EJ, Liu X, Jia X, Chen J, Hu M. Coupling of conjugating enzymes and efflux transporters: impact on bioavailability and drug interactions. Curr Drug Metab. 2005;6:455–468. doi: 10.2174/138920005774330657. [DOI] [PubMed] [Google Scholar]
- 23.Ohno S, Nakajin S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos. 2009;37:32–40. doi: 10.1124/dmd.108.023598. [DOI] [PubMed] [Google Scholar]
- 24.Joseph TB, Wang SW, Liu X, Kulkarni KH, Wang J, Xu H, Hu M. Disposition of flavonoids via enteric recycling: enzyme stability affects characterization of prunetin glucuronidation across species, organs, and UGT isoforms. Mol Pharm. 2007;4:883–894. doi: 10.1021/mp700135a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Tam VH, Hu M. Disposition of flavonoids via enteric recycling: determination of the UDP-glucuronosyltransferase isoforms responsible for the metabolism of flavonoids in intact Caco-2 TC7 cells using siRNA. Mol Pharm. 2007;4:873–882. doi: 10.1021/mp0601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Lin G, Zuo Z. Position preference on glucuronidation of mono-hydroxylflavones in human intestine. Life Sci. 2006;78:2772–2780. doi: 10.1016/j.lfs.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 27.Houstonand KE, Kenworthy JB. In vitro-in vivo scaling of CYP kinetic data not consistent with the classical Michaelis-Menten model. Drug Metab Dispos. 2000;28:246–254. [PubMed] [Google Scholar]
- 28.Hutzlerand TS, Tracy JM. Atypical kinetic profiles in drug metabolism reactions. Drug Metab Dispos. 2002;30:355–362. doi: 10.1124/dmd.30.4.355. [DOI] [PubMed] [Google Scholar]
- 29.Akaike H. Information theory and an extension of the maximum likelihood principle. In: PBaC F, editor. 2nd International Symposium on Information Theory; Akademia Kiado, Budapest. 1973. pp. 267–281. [Google Scholar]
- 30.Yamaoka K, Nakagawa T, Uno T. Application of Akaike's information criterion (AIC) in the evaluation of linear pharmacokinetic equations. J Pharmacokinet Biopharm. 1978;6:165–175. doi: 10.1007/BF01117450. [DOI] [PubMed] [Google Scholar]
- 31.D'Argenio DZ, Schumitzky A. ADAPT II User's Guide: Pharmacokinetic/ Pharmacodynamic Systems Analysis Software. Los Angeles: University of Southern California; 1997. [Google Scholar]
- 32.Chen X, Wang H, Du Y, Zhong D. Quantitation of the flavonoid wogonin and its major metabolite wogonin-7 beta-D-glucuronide in rat plasma by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;775:169–178. doi: 10.1016/s1570-0232(02)00288-x. [DOI] [PubMed] [Google Scholar]
- 33.Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- 34.Gibson CR, Bergman A, Lu P, Kesisoglou F, Denney WS, Mulrooney E. Prediction of Phase I single-dose pharmacokinetics using recombinant cytochromes P450 and physiologically based modelling. Xenobiotica. 2009;39:637–648. doi: 10.1080/00498250902954296. [DOI] [PubMed] [Google Scholar]
- 35.Mackenzie PI, Rogers A, Treloar J, Jorgensen BR, Miners JO, Meech R. Identification of UDP glycosyltransferase 3A1 as a UDP N-acetylglucosaminyltransferase. J Biol Chem. 2008 doi: 10.1074/jbc.M807961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlini LE, Meropol NJ, Bever J, Andria ML, Hill T, Gold P, Rogatko A, Wang H, Blanchard RL. UGT1A7 and UGT1A9 polymorphisms predict response and toxicity in colorectal cancer patients treated with capecitabine/irinotecan. Clin Cancer Res. 2005;11:1226–1236. [PubMed] [Google Scholar]