Short abstract
Future sequencing of the human microbiota will require greater breadth rather than depth.
Abstract
Culture-independent studies of human microbiota by direct genomic sequencing reveal quite distinct differences among communities, indicating that improved sequencing capacity can be most wisely utilized to study more samples, rather than more sequences per sample.
Review
In the past few years, the availability of improved sequencing methods, including pyrosequencing [1], has revolutionized what we know about the microbes that inhabit our bodies. Although it has been known for decades that our microbial symbionts outnumber our own cellsby about a factor of 10 [2], the differences in the repertoires ofsymbiontsharbored by different healthy individuals, different siteswithin the individual, and by individuals over time are only now coming to light. Initially, it was assumed that a 'core microbiome' existed; that is, that a substantial number of microbial species was shared in each body habitat in all or most humans, and that the genomes of these core species could be used as scaffolds to assemble fragmentary data from short-read shotgun sequencing of microbial community DNA [3].
The first three individuals whose gut microbiomes were surveyed using substantial numbers of 16S rRNA genesequences shared few of their species, however [4]. Similarly, observations that a person's left and right hands have only 17% of bacterial species in common, and that two different people's hands share only 13% [5], cast doubt on the concept of a substantial core set of microbial species shared by all or most people. This doubt has been reinforced by recent work that redefines core lineages or genes as 'core' even if shared by relatively few people [6,7]. In fact, on the basis of 16S rRNA geneanalyses we can rule out the possibility that, even within relatively homogeneous small populations of fewer than 100 individuals, everyone's skin-surface communities or gut communities share more than a tiny fraction of species [6-8]. This unanticipated variability in shared community membership, and also in other important aspects of the human microbiome, poses substantial conceptual and computational challenges.
Of particular importance for microbiome studies is the following question: what is the effect size? That is, using standard terminology from statistics, how distinguishable are two communities or groups of communities? Obtaining an answer is essential for addressing many practical concerns with experimental design. For example, the effect size determineshow many individuals need to be recruited for a given study, and how many sequences need to be collected per sample to observe differences if they exist. These considerations are particularly importantfor the study ofsystemic disorders such as diabetes or some autoimmune disorders, which are expected to influence the microbiomein multiple body habitats. We need a sense of how much variation exists among different body habitats, how much variation is observed among healthy individuals for the same body habitat, and how much of a shift occurs due to a pathophysiologic state. It is also importantto define the most appropriate method for determining the magnitude of similarity or difference between communities, as the choice of method has a large influence on the results of community comparisons [9-12]. A general discussion of the pros and cons of different metrics of community overlap is beyond the scope of this paper (see [9-12] for reviews). Here, we summarize the types and sizes of effects found in studies that used various methods of comparing groups of samples, and look for large-scale patterns that can give information on the number of individuals and sequences that are needed to observe different types of effects (Figure 1).
Figure 1.
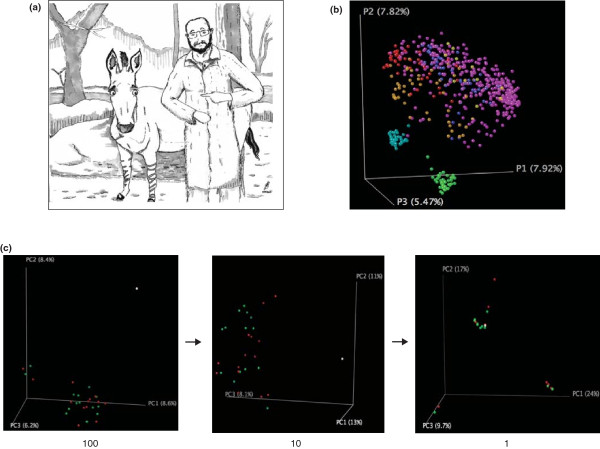
The problem of distinguishing between sequences. (a) An investigator contemplating the problem of distinguishing between sequences from the gut of Equus asinus and the volar forearm of humans. (b) Our solution; guess the effect size based on the effect sizes reported in published studies; perform simulations based on these effect sizes as shown in Figure 2, and then acquire sufficient sequences to resolve microbial community differences of the expected magnitude. (c) When comparing the Equus asinus gut (white point) to human forearms (red and green points represent left and right arms, respectively), 100 or even 10 sequences per sample provide sufficient resolution, but one sequence per sample does not.
Figure 2.
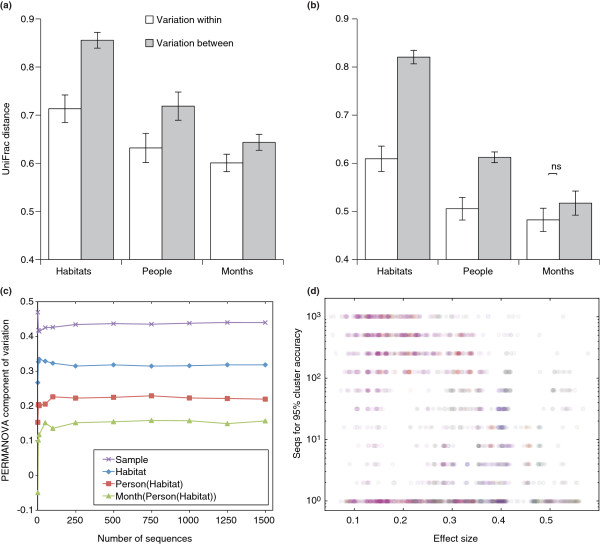
Variation in human body habitats within and between people. (a) The full dataset (approximately 1,500 sequences per sample); (b) the dataset sampled at only 10 sequences per sample, showing the same pattern; (c) the relationship between sequencing depth and the PERMANOVA component of variation. The amount of variation explained by the factors plateaus at relatively shallow sequencing depths. Note that the proportion of variation captured by differences between the samples (that is, residual variation) is still highest despite the explanatory values of the three factors examined. (d) Effect size determines the number of sequences required for sample identification. Each point in the figure represents a specific sample selected from a pair of body sites, and the number of sequences required to correctly distinguish which site the sample originated from. The point is colored according to the two body sites under consideration, the center's color represents the broad category the selected sample originated from, the border color represents the other broad category under consideration. Many body sites share the same broad category, and thus some points have the same border and center coloring. Red, external ear canal; yellow, hair; green, oral cavity; blue, gut; magenta, skin; gray, nostril. ns, not significant.
A variety of interrelated features differentiate microbial communities. These features include the the relative abundance of specific taxa (the proportion of the bacteria in the sample that are Firmicutes, for example), the level of species richness or diversity observed within a community (alpha diversity), and the degree to which different communities share membership or structure (beta diversity). A major challenge in comparing studies is that there is no consistent way in which the size of community differences is reported, as the type of difference that is relevant depends on the study. For example, lean and obese mice and humans differ in their ratios of prominent bacterial phyla (Bacteroidetes (which include the common gut commensal Bacteroides), Firmicutes (Gram-positive bacteria, including Lactobacillus and Clostridium), and Actinobacteria (which include Corynebacteria and Mycobacteria) [13-15]); men's and women's hands differ in the number of species-level phylotypes (defined as organisms with 16S sequence identity >97%) observed on average [5]; and samples from the same or similar sites on the bodies of different individuals cluster together using UniFrac-based principal coordinates analysis [4,16,17]. UniFrac is a metric for comparing microbial communities using phylogenetic information, which has been implemented in several tools.
Because of the diverse ways in which microbial communities respond to various environmental factors, it is difficult to compare effect sizes across different studies or systems, as an analysis that highlights differences in one system may obscure them in another. Thus, in what follows, we review effect types and sizes as reported by the authors of individual studies. We focus on variation in human-associated microbial community diversity as assessed by 16S rRNA gene sequence surveys of abundant lineages, using various measures of both within- and between-sample diversity (alpha and beta diversity, respectively). We review comparisons of microbial communities in relationship toboth sampling depth (that is, number of sequences per sample) and breadth (that is, number of samples or individuals). We then perform simulations using an atlas of microbes associated with different sites in the human body to ask how many sequences per sample are needed in order to detect differences across individuals, time, and locations within the body.
Reported effect sizes between and within different body habitats
Table 1a provides an illustrative (though not exhaustive) overview of the literature regarding differences observed in different body habitats and locations in healthy individuals, and the number of subjects and sequences that were used to identify these differences. Although metagenomic studies that examine all the genes in the genome are also of immense interest, shotgun metagenomic data are so far available only from the gut and for a relatively few samples, and so the range of questions that can be addressed at present is substantially more limited than for 16S rRNA-based surveys, the type of survey we consider here. One robust finding that exemplifies relative effect sizes is that there appears to be a greater degree of variation in microbial community composition between individuals than within the same individual over time (Table 1a). This has been found to be true in multiple studies and over a wide range of body habitats. For example, gut community composition is relatively stable in the same individual across a period of months when diet is consistent [6,16], and even to a certain degree when diet is altered. (Changes in the Firmicutes:Bacteroidetes ratio have been reported in individuals who lost weight, whether they were consuming low-calorie fat- or carbohydrate-restricted diets, but despite these shifts in relative abundance, interpersonal variation was the largest effect observed using phylogenetic comparisons of the communities [14].) Likewise, skin community composition is more similar within a subject than between subjects over a period of months [16,18], as are oral, nasal and external auditory canal communities [16]. These results indicate that you are likely to be more similar to yourself in 3 months time than to your friend today in terms of the bacteria you harbor.
Table 1.
Variations observed among different types of microbial communities, and the extent of sequencing and sampling used
| Topic | Number of subjects | Number of samples sequenced | Total number of 16S sequences in final analysis | Average number of sequences per sample | Study conclusions | Reference |
|---|---|---|---|---|---|---|
| (a) Microbial communities associated with healthy humans | ||||||
| Oral (saliva) |
120 | 120 | 14,115 | 118 | Collected saliva from 10 individuals at each of 12 globally widespread locations. They attributed approximately 13.5% of the total variation in the distribution of genera to differences between individuals and found little evidence for geographic structure: 11.7% of the variation was among individuals from the same location while just 1.8% was among individuals from different locations | [38] |
| Oral (tooth, tongue, buccal mucosa, palate) |
3 | 29 | 298,261 | 10,285 | Collected samples from various oral niches of three individuals; 26% of the unique sequences and 47% of species-level phylotypes found in the study were found in all three subjects. Bacterial community composition was shaped primarily by oral niche: principal components analysis differentiated communities from shedding (tongue, cheek, palate) versus tooth surfaces | [39] |
| Skin (right and left volar forearm) | 6 | 20 | 2,038 | 102 | Sampled the superficial left and right volar forearms of six healthy subjects (four of whom were sampled again 8 to 10 months later). Samples from the same subject at the same time point (left versus right) were not significantly different, whereas samples from the same subject at different time points could be significantly different | [40] |
| Skin (right and left palms) |
51 | 102 | 351,630 | 3,251 | Collected skin swabs from the left and right palms of 51 volunteers. On average, individuals shared only 17% of species-level phylotypes between their right and left palms, while only 13% of species-level phylotypes were shared between different individuals. (UniFrac similarity between hands from different individuals = 0.30, and the same individual = 0.36 to 0.38.) Palm surface bacterial community structure was determined by handedness, time since washing, and the individual's sex | [5] |
| Skin (20 skin sites, including moist, dry, and sebaceous sites) |
10 | 300 | 112,283 | 374 | Obtained samples from 20 skin sites on each of 10 individuals (half of whom were sampled twice). They found that interpersonal variation in community membership and structure depended on skin site, and that subjects were more similar to themselves (site-to-site) than to others. Four of the five re-sampled subjects were also more similar to themselves over time than they were to other volunteers. Bacterial community composition was shaped by microhabitat: sebaceous, moist, or dry | [18] |
| Gut | 3 | 18 | 11,831 | 657 | Interpersonal and site-to-site variation in three subjects at six sites. Between subject dissimilarity was greater than within subject dissimilarity | [4] |
| Gut | 154 | 281 | 1,947,381 | 6,930 | Interpersonal variation was found to be largest between unrelated individuals, smaller between children and their mothers, still smaller between twins, and dramatically smaller in the same individual over time. (Average UniFrac distance over time within-individual = 0.69 and between unrelated individuals = 0.80) | [6] |
| (b) Microbial communities and human disease | ||||||
| Obesity | 12 subjects 2 controls |
50 | 18,348 | 367 | Obese people have fewer Bacteroidetes (5%; P < 0.001) and more Firmicutes (85%; P = 0.002) than lean controls (25% Bacteroidetes and 75% Firmicutes). During the diet, the relative abundance of Bacteroidetes increased from 5 to 20% (P < 0.001) and the abundance of Firmicutes decreased from 85 to 75% (P = 0.002). Increased abundance of Bacteroidetes correlated with percentage loss of body weight (R2 = 0.8 for the CARB-R diet and 0.5 for the FAT-R diet, P < 0.05), and not with changes in dietary calorie content over time (R2 = 0.06 for the CARB-R diet and 0.09 for the FAT-R diet) | [14] |
| Diabetes | 10 Diabetic patients 10 healthy subjects* |
20 | 382,229 357,782 |
37,001 | The proportion of Firmicutes was significantly higher (P = 0.03) in the controls (mean 56.4%) compared to the diabetic group (mean 36.8%). Accordingly, phyla Bacteroidetes and Proteobacteria were somewhat but not significantly enriched in the diabetic group (50.4 and 4.1% in the diabetic group compared with 35.1 and 2.7% in the healthy group, respectively) | [41] |
| Crohn's disease (CD) and ulcerative colitis (UC) | 6 CD patients 5 UC patients 5 healthy subjects |
16 | 1,590 678 1,037 |
207 | Proteobacteria were significantly (P = 0.0007) increased in CD patients (13%) versus UC patients (9.4%) or healthy subjects (8.5%). Bacteroidetes were far less diverse than Firmicutes, containing only 32 phylotypes, versus 87 species-level phylotypes in the latter phylum, but were nevertheless the most abundant, representing over 70% of total clones. Bacteroidetes were significantly increased (75%) in CD patients versus UC patients (64.3%) or healthy subjects (67.4%) The increase in Bacteroidetes and Proteobacteria was accompanied by a significant (P = 0.0001) decrease in Firmicutes (CD,10%; UC, 25.8%; healthy subjects, 24%), all belonging to the class Clostridia in the CD group | [42] |
| CD and UC | 20 CD patients 15 UC patients 14 healthy subjects |
49 | 809 691 235 |
35 | The results obtained from CD and healthy subject samples did not differ (P > 0.05). Bacterial numbers associated with non-inflamed and inflamed mucosa within CD and UC groups did not differ (P > 0.05). The ratio of Actinobacteria:Bacteroidetes:Firmicutes: Proteobacteria differed between healthy (approximately 1:27:53:6%), UC (approximately 0.3:34:48:7%) and CD subjects (approximately 0.5:34:40.5:6%) | [43] |
| CD and UC | 190 CD, UC or healthy patients (around equal numbers) | 190 | 15,172 | 80 | Bacteroidetes (10%, P = 0.001) and Firmicutes (20%, P = 0.001) were greatly depleted while Actinobacteria (10%, P = 0.001) and Proteobacteria (50%, P = 0.001) were substantially more abundant in the inflammatory bowel disease (IBD) subset samples, relative to control subset samples (approximately 20% Bacteroidetes, approximately 50% Firmicutes, approximately 5% Actinobacteria, approximately 10% Proteobacteria) | [44] |
| Necrotizing enterocolitis (NEC) | 10 infants with NEC and 10 healthy infants | 21 | 5,354 | 255 | For the control infants four phyla were present: Proteobacteria, (34.97% relative abundance), Firmicutes (57.79%), Bacteroidetes (2.45%) and Fusobacteria (0.54%) with 4.25% unclassified bacteria. However, NEC patients had only two phyla, Proteobacteria (90.72%) and Firmicutes (9.12%) with 0.16% unclassified bacteria. The average proportion of Proteobacteria was significantly increased and the average proportion of Firmicutes was significantly decreased compared to controls (P = 0.001) | [45] |
| Clostridium difficile-associated diarrhea (CDAD) | 4 ICD patients 3 RCD patients 3 healthy subjects |
10 | 581 447 399 |
143 | Using rarefaction curves, species richness in the patients with ICD (initial episode of antibiotic-associated diarrhea due to C. difficile) was similar to that in the control subjects, with the shape of the curve revealing that the total richness of the microbial community had not been completely sampled (minimum of 20 phylotypes). However, the species richness in the patients with RCD (recurrent antibiotic associated diarrhea due to C. difficile) was consistently lower (around ten phylotypes) than both that in the patients with ICD and that in the control subjects | [46] |
| Gastric cancer | 10 non-cardia gastric cancer patients 5 control patients |
15 | 140 | 9 | No significant differences in microbial compositions were found between cancer patients and controls | [47] |
| Helicobacter pylori colonization | 19 H. pylori (+) subjects 4 H. pylori (-) subjects |
23 | 1,833 | 80 | Subjects negative for H. pylori had twice as many Fusobacteria as H. pylori-positive subjects (10% compared to 5%, respectively). Twenty percent of the clone libraries derived from H. pylori-positive patients were non-H. pylori Proteobacteria compared with 10% in the control subjects; this was also the case for Bacteroidetes (20% compared with 10% in the control) | [48] |
| (c) Experimentally manipulated microbial communities | ||||||
| Restoration of wetland soils | 3 agriculture wetlands, 3 restored wetlands and 3 reference wetlands | 13 | 1,235 | 95 | A significant difference in the Proteobacteria:Acidobacteria ratio from around 0.6 to around 0.4 was observed between agricultural and reference wetlands, respectively (P < 0.001). A difference was also found in the relative abundance of β-Proteobacteria from 14 to 3% in the same soils (P < 0.001) | [22] |
| Soil moisture | 4 wet and 4 dry soils | 8 | 665 | 83 | The relative abundance of Proteobacteria decreased from 48 to 36% in wet versus dry plots (P < 0.05). Acidobacteria increased in relative abundance from 7 to 23% in the same soils (P < 0.01) | [21] |
| Antibiotic effects on piglet gut microbiota | 6 control pigs and 6 pigs treated with chlor-tetracycline | 12 | 1,900 | 171 | An effect of antibiotics was seen on the overall community composition (P < 0.03) | [23] |
| Effects of a 24-hour fast on mouse gut microbiota | 4 to 5 fasted and control mice | 38 | 145,428 | 3,827 | The fast resulted in a significant increase in the proportion of Bacteroidetes (approximately 21 to approximately 42%, P = 0.01) and a significant decrease in the fraction of Firmicutes (approximately 77 to around 53%, P = 0.007) within the gut microbial community | [49] |
| Effects of diet and genotype on murine gut microbiota | 5 individuals from 2 genotypes fed standard or low-fat chow | 20 | 25,790 | 1,290 | The relative abundance of Bacteroidetes decreased (around 90% versus around 40%) in animals fed the high-fat diet regardless of genotype (P < 0.001). Likewise, mice fed the standard chow diet showed a lower relative abundance of Firmicutes (around 7 versus around 42) independent of genotype (P < 0.001) | [50] |
| Antibiotic effects on canine gut microbiota | 5 dogs sampled three times | 15 | 44,096 | 2,940 | Enterococcus-like organisms, Pasteurella species, and Dietzia species all increased significantly (P < 0.05) following tylosin treatment | [51] |
*The entire study consisted of 36 subjects of which only 20 were selected for pyrosequencing.
Microbial community changes in human disease and environmental samples
Although a wide range of studies in healthy subjects have identified substantial interpersonal variation in overall microbial community composition, how do these effect sizes compare with differences correlated with disease, or in response to treatments ofvarious environmental samples? To address this question, we reviewed culture-independent, 16S rRNAgene-based surveys associated with different physiological conditions (Table 1b) and associated with experimental manipulations in non-human environments (which were surprisingly scarce; Table 1c).
One of the best-characterized effects of health status on the gut microbiome is the association between obesity and the proportional representation of Bacteroidetes, Firmicutes and Actinobacteria [6,13-15]. Studies in mice indicate that the microbiota contributes to the obese state by providing the host with a greater amount of energy from the diet compared with the microbiota of a lean host [15], as well as by manipulating host genes that regulate the deposition of energy in adipocytes [19]. The obesity-associated microbiomes of humans (and mice) are enriched in functional genes for certain types of carbohydrate metabolism, and this is directly attributable to the reduction in the numbers of genomes of members of the Bacteroidetes [6,15].
However, even the size of the differences in gut bacterial community composition of obese versus lean hosts is debated, as different studies using different methodologies have returned varied results [20]. The impact of methodology is particularly evident in a study of twins concordant for obesity or leanness, in which the observed relative abundances of Bacteroidetes, Actinobacteria and Firmicutes, as judged by sequencing of different regions of 16S rRNA clones, depended on the sequencing approach - pyrosequencing of PCR products, Sanger sequencing of 16S rRNA clones, or shotgun sequencing and phylogenetic classification of reads [6]. However, the direction of the effect was consistent across methodologies, and detectable with as few as a couple of hundred sequences per sample.
Observable phenotypes such as obesity may be caused by a variety of underlying factors, and which of those factors is responsible for shifts in the host's microbiota is difficult to address in such correlative studies. Experimental manipulations of microbial communities, however, allow determination of the relative effects of specific variables on overall community composition or the abundance of particular taxa, and as such, allow researchers to draw conclusions regarding cause and effect. Examples of experimental manipulations of non-human environments that used 16S rRNA gene sequencing approaches (either clone libraries or pyrosequencing) and that were well enough replicated to allow statistical analysis are shown in Table 1c. For soil samples, three to four replicates with 70 to 100 sequences were sufficient to observe differences in microbial communities due to land use and moisture regimes [21,22]. For piglet gut microbiota, the effects of antibiotics on overall community composition were evident with as few as 96 sequences per sample [23]. It would be fascinating to test whether similar antibiotic-induced effects in outbred populations of humans with diverse diets [24] can be found with relatively few sequences. Similarly, it would be important to consider sampling depth under human physiological conditions in cases where the effect size is known to be large, for example, in the development of the infant gut microbiota [25].
Has the depth of sequencing used up to now really been necessary?
The literature reviewed in Table 1 reports how many sequences were used to reveal a variety of different effects. Could the same results have been achieved with less sequencing? To begin to address this question, we carried out a limited reanalysis of a study of multiple body habitats by Costello et al. [16], which encompasses variability explained by nested factors with different effect sizes (Box 1).
Box 1.
How many sequences does it take...? .
In conclusion, the results described here, and previously reported [8,37], show that arbitrarily choosing to generate large numbers of sequences may not be the most cost-effective way to identify changes in microbial communities associated with different physiological or pathophysiological states. Instead, we call for a few standardized methods to assess differences among microbial communities, which will allow for effect size and power calculations, and therefore a considered assessment of the number of individuals and sequences required to differentiate among given communities. The following four methods have been successful in a range of studies: differences in alpha diversity (number of phylotypes observed or extrapolated); differences in abundance of specific lineages; differences in location on a principal coordinates plot obtained from UniFrac distances or other metrics; and the FST measure described in the previous section.
The rapid increase in sequencing capacity provides a spectacular opportunity to advance the field in ways that were unimaginable even 3 years ago. How can individual investigators, or groups of investigators, use these resources most wisely at this unique moment of democratization of the ability to perform sequence-based studies? The data summarized here suggest that study designs consisting of tens of thousands of samples sequenced at shallow coverage will be highly informative (depending on the effect size), and such studies are possible with the instruments available today. Given recent observations that inter-habitat and inter-personal variations are large effects, we believe that individual researchers can and should sieze the opportunity provided by these findings to analyze vast numbers of samples at low-coverage (for example, 100 to 1,000 sequences). At this number of samples, detailed exploration of spatial and temporal dynamics of microbial communities will be possible, as will comparisons of large patient populations. In addition, replicate samples can be acquired and analyzed without too strongly impairing the breadth of an investigation, allowing more robust experimental designs to be implemented. One can envisage that perhaps within the next few years, a group of motivated high-school students might, for a science-fair project, be able to track movements in microbes between humans and their pets and livestock across the planet. These studies, especially when combined with hypothesis-driven approches to understanding the effects of factors such as diet and antibiotic exposure, could go far beyond even the largest purely observational studies being contemplated today.
Such studies will yield an overall map of variation within the human microbial ecosystem, and relate differences to specific physiological states within and between individuals in a manner that is replicated across individuals. These studies will serve as a framework to identify and compare the shifts that take place in the microbial community that are related to specific disorders.
Acknowledgements
We thank the Crohn's and Colitis Foundation of America, the Bill and Melinda Gates Foundation, the HHMI and the NIH for support of work by the authors cited in this review.
References
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houte J, Gibbons RJ. Studies of the cultivable flora of normal human feces. Antonie Van Leeuwenhoek. 1966;32:212–222. doi: 10.1007/BF02097463. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, A human gut microbial gene catalogue established by metagenomic sequencing. Nature. pp. 59–65. [DOI] [PMC free article] [PubMed]
- Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Gallagher ED. Ecologically meaningful transformations for ordinations of species data. Oecologia. 2001;129:271–280. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008;32:557–578. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magurran AE. Measuring Biological Diversity. Oxford: Blackwell; 2004. [Google Scholar]
- Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial Community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R. Forensic identification using skin bacterial communities. Proc Natl Acad Sci USA. 2010;107:6477–6481. doi: 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC. NISC Comparative Sequencing Program. Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA.. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. pp. 5–11. [DOI] [PubMed]
- Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW. Soil microbial community responses to multiple experimental climate change drivers. Appl Environ Microbiol. 2010;76:999–1007. doi: 10.1128/AEM.02874-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman WH, Richardson CJ, Vilgalys R, Bruland GL. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc Natl Acad Sci USA. 2008;105:17842–17847. doi: 10.1073/pnas.0808254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettedal E, Vilain S, Lindblom S, Lehnert K, Scofield C, George S, Clay S, Kaushik RS, Rosa AJ, Francis D, Brözel VS. Alteration of the ileal microbiota of weanling piglets by the growth-promoting antibiotic chlortetracycline. Appl Environ Microbiol. 2009;75:5489–5495. doi: 10.1128/AEM.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR, Gorley RN. Primer v6. http://www.primer-e.com/
- Anderson MJ. Distance-based tests for homogeneity of multivariate dispersions. Biometrics. 2006;62:245–253. doi: 10.1111/j.1541-0420.2005.00440.x. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamames J, Abellan JJ, Pignatelli M, Camacho A, Moya A. Environmental distribution of prokaryotic taxa. BMC Microbiol. 2010;10:85. doi: 10.1186/1471-2180-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auguet JC, Barberan A, Casamayor EO. Global ecological patterns in uncultured Archaea. ISME J. 2010;4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nat Rev Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. Inbreeding coefficients and coalescence times. Genet Res. 1991;58:167–175. doi: 10.1017/S0016672300029827. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored 'rare biosphere'. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy 'core microbiome' of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N, Vogensen FK, Berg FW van den, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. p. e9085. [DOI] [PMC free article] [PubMed]
- Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibiloni R, Mangold M, Madsen KL, Fedorak RN, Tannock GW. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn's disease and ulcerative colitis patients. J Med Microbiol. 2006;55:1141–1149. doi: 10.1099/jmm.0.46498-0. [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, Antonopoulos DA, Chang EB, Claud EC. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol. 2009;58:509–516. doi: 10.1099/jmm.0.007302-0. [DOI] [PubMed] [Google Scholar]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford PA, Crowley JR, Sambandam N, Muegge BD, Costello EK, Hamady M, Knight R, Gordon JI. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci USA. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski JS, Dowd SE, Westermarck E, Steiner JM, Wolcott RD, Spillmann T, Harmoinen JA. The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rRNA gene sequencing. BMC Microbiol. 2009;9:210. doi: 10.1186/1471-2180-9-210. [DOI] [PMC free article] [PubMed] [Google Scholar]