Short abstract
Whole-genome analysis of human tumors has identified some unsuspected tumor-associated genes
Abstract
Unbiased sequencing and analysis of human tumors is revealing unsuspected somatic changes that, upon further study, are elucidating aspects of tumor biology and identifying new biomarkers.
Whole-genome sequencing and analysis of tumor and matched normal genomes with next-generation sequencing platforms has begun to illuminate commonly mutated genes and transcript-level events that contribute to the underlying tumor biology. To elucidate the role of frequent somatic mutations, the mutant proteins have been biochemically characterized and the results interpreted in terms of the selective advantages these variants may confer on the tumor. Certain somatic alterations have demonstrable prognostic value for specific tumor types in which they commonly occur, although their downstream metabolic signatures may obviate genotyping to identify their mutational status. The metabolic signature is a direct result of the mutation's impact on a given protein/enzyme; therefore, rather than performing sequencing to detect whether a mutation is present, metabolic profiling may be more straightforward, cheaper, and have a lower error rate, for example. New insights into the relationship between a primary tumor and its fatal metastatic disease are also beginning to emerge from genomic comparisons, with the fine detail afforded by next-generation sequencing enabling these comparisons.
The transcriptomes of cancer cells also have their own unique somatic complexities, which often result from structural perturbations to the genome, but can be due to transcription-level events such as alternative splicing, RNA editing or transcript fusion. These types of alterations may explain certain aspects of tumor biology and may also be corroborated by cytogenetic phenomena. In this review, I will describe some tumor-specific alterations that were discovered as a result of analyses of unbiased genome or transcriptome sequencing data (unbiased sequencing does not select for portions of the genome or transcriptome in advance, and the entire genome or transcriptome is therefore surveyed) and then illustrate how these discoveries were pursued further to reveal insights into tumor biology that have enhanced our clinical diagnosis of cancer and our concepts of how best to treat it.
Genome-based discovery in cancer
In a 1956 paper [1], Otto Warburg observed that the predominant mode of energy production in cancer cells was by aerobic glycolysis rather than by oxidation of pyruvate in mitochondria, as in normal cells. This observation led Warburg to postulate that this change in metabolism was a fundamental cause of cancer. Years later, in 1986, Renato Dulbecco opined that studying the cellular genome should be pursued to learn more about cancer, either by taking a 'piecemeal' approach of looking gene by gene, or by sequencing the whole genome [2].
Somatic mutations in the genes IDH1 and IDH2
In the current era of cancer genomics, one of the most interesting and unexpected discoveries to result from unbiased sequencing of matched tumor and normal samples is the somatic point mutations found in the genes for two isocitrate dehydrogenase isoenzymes, IDH1 and IDH2. First discovered by sequencing the exome (the exons collectively) of the glial brain tumor glioblastoma multiforme (GBM) [3], mutated IDH1 was found in 12% of tumors analyzed. The general approach to exome capture and analysis is shown in Figure 1. Subsequent focused re-sequencing refined the occurrence of mutations at arginine 132 (R132) of IDH1, which are found in more than 80% of secondary GBMs (GBMs that initially present as low-grade astrocytomas (tumors of astrocytes)), and less than 10% of primary GBMs [4-6]. Subsequent work, explained later in this review, identified the biological impact of these mutations on enzyme function. On examining gliomas, including GBMs, negative for IDH1 mutations, recurrent somatic mutations of IDH2 at the analogous R172 residue were identified [4,7]. Not only are the IDH1 and IDH2 mutations frequent, but studies by several laboratories have established that the mutation in IDH1 occurs early in glioma progression [8]. Notably, the mutations affect only one allele at the given locus (of the two alleles of either IDH1 or IDH2, but not both in the same tumor), which is puzzling considering the evidence that they are selected for early in tumorigenesis. Analysis of the correlation between mutation in IDH1 or IDH2 and various clinical features has revealed interesting associations between the presence of mutation and an early age of disease onset and overall longer survival time in GBMs and in anaplastic astrocytomas (another type of glial tumor, distinct from GBMs) [4].
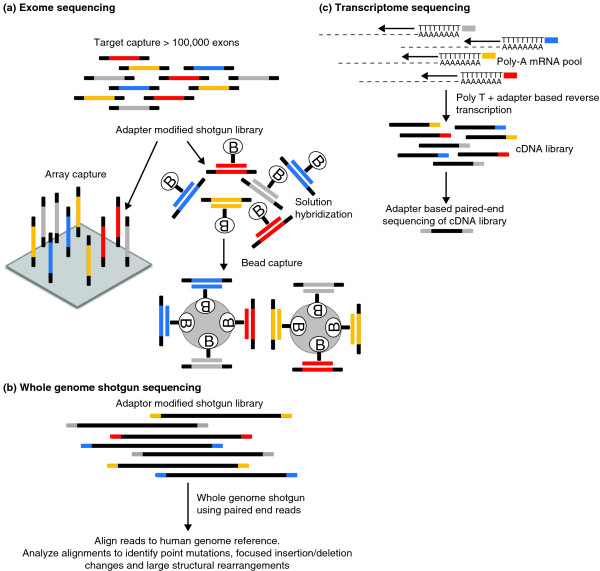
Figure 1.
General schema for targeted exome capture, whole genome sequencing, and transcriptome sequencing. (a) In exome capture, a random library of genomic fragments, each containing platform-specific adapters on each end, is combined with a set of probes that define the human exome. Following hybridization, the probe:genomic library fragment hybrids are captured using magnetic beads and isolated from solution by the application of a magnet, or by solid phase capture. Denaturing conditions are used to elute the captured genomic library fragment population from the hybrids, and prepared for sequencing. (b) In whole genome sequencing, the same random fragment library is constructed as in (a), but the resulting fragments are sequenced directly without a capture step. (c) In transcriptome sequencing, the RNA is converted to cDNA, the resulting cDNAs are fragmented, and the library adapters are ligated to the resulting fragments, followed by sequencing. Panel (a) reproduced with permission from [27].
The IDH enzymes play a key role in cellular metabolism, catalyzing the conversion of isocitrate to α-ketoglutarate (α-KG) and generating NADPH from NADP+ in the process (Figure 2). The crystal structure of IDH1 [9] predicts that the amino acid substitutions found at the R132 position will impair the interaction of the enzyme with its isocitrate substrate, and functional and biochemical studies of the mutant proteins by several groups have provided critical insights into this [4,10,11]. Zhao and colleagues [10] evaluated the in vitro enzymatic activities of three tumor-derived IDH1 mutants - R132H, R132C and R132S - by expressing mutant constructs in transformed human embryonic kidney 293T cells. They observed all three mutants to have a more than 80% reduction in the ability to convert isocitrate to α-KG compared with the wild-type enzyme, and further kinetic analyses revealed a dramatically reduced affinity for isocitrate in all three mutants. As IDH1 functions as a homodimeric complex, Zhao et al. [10] isolated IDH1 dimers expressed from the R132H mutant and wild-type genes introduced into Escherichia coli. Three dimer combinations were identified, the wild-type:R132H heterodimer exhibited only 4% of the wild-type dimer enzyme activity, while R132H:R132H homodimers were almost completely inactive.
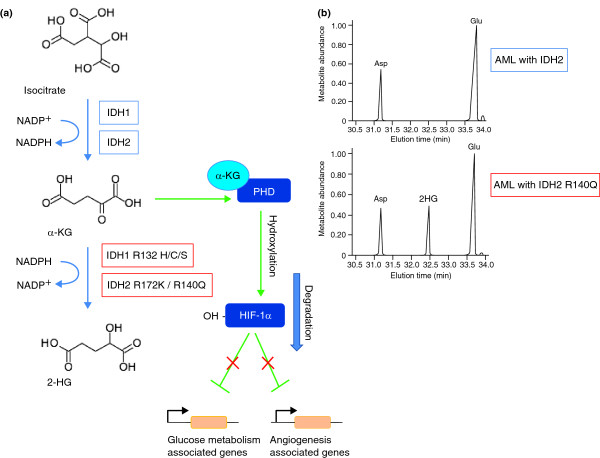
Figure 2.
Impact of IDH1/2 mutations on tumor cell biology. (a) In normal cells, the role of IDH1 and IDH2 enzymes is to convert isocitrate to α-ketoglutarate (α-KG), converting NADP+ to NADPH. The presence of α-KG regulates prolylhydroxylases (PHD) that, in turn, promote the degradation of hypoxia-inducible factor 1α (HIF-1α). HIF-1α is a transcription factor that regulates the expression of genes related to glucose metabolism, angiogenesis, and other signaling pathways by sensing low cellular oxygen levels. The mutant IDH enzymes convert α-KG to 2-hydroxyglutarate (2-HG), leading to the build up of this oncometabolite. (b) Comparison of metabolomic profiling of IDH wild-type (upper panel) and mutant (lower panel) cells, indicating the increased levels of 2-HG associated with the mutation. 2-HG is absent in IDH wild-type cells. Panel (b) reproduced with permission from [15].
What are the metabolic consequences of IDH1 mutations? Using the U-87 MG human glioblastoma cell line, Zhao et al. [10] demonstrated a concomitant reduction in cellular α-KG levels after knocking down endogenous IDH1, and because α-KG is required by prolylhydroxylases, enzymes that hydroxylate and promote the degradation of hypoxia-inducible factor 1α (HIF-1α), the intracellular levels of HIF-1α were also characterized. Zhao et al. showed that when wild-type IDH1 is knocked down by RNA interference using short hairpin RNA, HIF-1α is elevated, and when IDH1 is overexpressed, HIF-1α levels are reduced. HIF-1α is a component of HIF-1, a transcription factor that regulates the expression of genes related to glucose metabolism, angiogenesis, and other signaling pathways by sensing low cellular oxygen levels. By performing quantitative PCR to measure the transcripts of three known HIF-1 target genes - glucose transporter 1 (Glut1), vascular endothelial growth factor (VEGF), and phosphoglycerate kinase (PGK1) - Zhao et al. demonstrated induced expression of these genes as a consequence of either the knockdown of wild-type IDH1 or the expression of the IDH1 R132H mutant. On staining glioma samples for HIF-1α, those tumors with previously identified R132H mutations showed a statistically stronger staining signal than those without mutations. Thus, this body of evidence has demonstrated that when IDH1 is mutated, its function is reduced and the downstream impact of that reduced function (the consequential upregulation of HIF-1α) contributes to the cell's progression to cancer, thereby indicating that a likely function of IDH1 is that of a tumor suppressor gene. Further experimentation is needed to support the claim that IDH2 may be a tumor suppressor gene also.
Building on the initial characterizations of IDH1 mutations in gliomas, Dang et al. [11] took a metabolomics-based approach to identify further changes in associated metabolite levels when an IDH1 mutation is present [11]. They found 2-hydroxyglutarate (2-HG) to be the only metabolite with significantly increased abundance in cells expressing R132H mutant IDH1. In a clever series of experiments, the increase in 2-HG was shown to result from the NADPH-dependent reduction of α-KG by mutant IDH1, a new function that is enabled by the mutation at R132. The authors demonstrated a similar gain of function for the R132C, R132L and R132S mutations. Their X-ray crystallographic studies showed that the R132H mutation in IDH1 results in the formation of an active site distinct from that of the wild-type enzyme. With the aim of improving diagnostic efficacy, Dang et al. examined 12 GBM tumors with various R132 mutations in IDH1, and found 2-HG levels 100-fold greater or more than in tumors with wild-type IDH1; the measured decrease in α-KG was, however, not statistically different in mutant versus wild-type IDH1 tumors. This finding indicates that in the clinic, detecting patients with increased 2-HG levels would identify GBMs with IDH1 mutations, predicting an overall longer survival time. Indeed, since secondary GBMs develop from lower-grade gliomas, therapeutic inhibition of 2-HG production might slow the transition time to GBM development, offering an improved survival benefit as a result.
In our laboratory we have been using a whole-genome shotgun approach to sequence tumor genomes. In the second case of acute myeloid leukemia (AML) we sequenced, we discovered an IDH1 R132 mutation that was subsequently found in about 8% of our 187 banked AML patient samples, showing that this mutation is not restricted to gliomas [12]. In these tumor genomes, we detected R132C, R132H and R132S substitutions, with R132C being most common (8 of 16). PCR assays designed to detect IDH2 R172 mutations in the 188 AML samples did not reveal any. Correlation analyses of clinical data and mutational status for AML patients with IDH1 mutations indicated that the presence of IDH1 mutations, in cases with normal cytogenetics and with wild-type nucleophosmin-1 (NPM1), a commonly mutated gene in AML that has been known for some time, predicted a worse prognosis, although we did not reach statistical significance with our cohort. Soon after, Schnittger et al. [13] reported an analysis of a cohort of 999 AML patients, finding that IDH1 mutations were frequent (in 9.3% of samples), that the R132C mutant was the most common, and that in the presence of wild-type NPM1 and intermediate cytogenetics (a cytogenetic evaluation of the leukemia cells revealing no clues as to the patient's prognosis), patients with IDH1 mutations had a significantly unfavorable prognosis (P = 0.038).
A subsequent study by Gross et al. [14] examined an additional 145 AML biopsies, identifying 11 IDH1 R132 mutant samples [14]. Four IDH1-mutant primary samples had relapse samples that also carried the IDH1 mutation. AML cells carrying the R132 mutant of IDH1 were found by gas chromatography-mass spectrometry to have 2-HG levels around 50-fold greater than in samples with wild-type IDH1. Similarly, higher 2-HG levels were detected in sera from patients positive for the IDH1 R132 mutation. Two wild-type IDH1 samples had elevated 2-HG levels and were found to be carrying IDH2 R172 mutations, the first report of these in AML. Importantly, this paper reinforced the fact that metabolite screening rather than mutational screening can be an important diagnostic approach for the detection of elevated 2-HG levels and, by inference, IDH1/2 mutations. Because of the apparent predominance in AML of the IDH1 R132C mutation over R132H (which is more predominant in gliomas), Gross et al. [14] looked at the kinetics of the R132C mutant enzyme. The R132C enzyme showed a dramatic loss of affinity for isocitrate (resulting in a reduction in KM) and a drop of more than six orders of magnitude in net efficiency (Kcat/KM) of isocitrate metabolism.
In another recent study, our understanding of IDH mutations and their detection has been extended. Ward et al. [15] have determined that the gain of function seen in the IDH1 R132 mutants (that is, the ability to reduce α-KG) is also found in the IDH2 R172K mutant. Metabolic profiling of cells expressing IDH2 R172K revealed an approximately 100-fold increase in intracellular 2-HG compared with cells overexpressing wild-type IDH2, and this finding was extended to leukemia cells carrying the IDH2 R172K mutation. Ward et al. [15] also screened AML samples with normal cytogenetics but unknown IDH mutational status for increased levels of 2-HG, and then evaluated the mutational status based on the result of the screening assay. In this test, 2-HG measurement was found to predict mutational status with high accuracy. In addition, a new IDH2 mutation, R140Q, was identified in five samples. In a second evaluation of 78 AML samples, IDH2 R140Q mutations were found to be more frequent than either IDH1 R132 mutations or IDH2 R172K [15].
Despite some differences among sample sets, this body of work, aiming to characterize the impact of IDH mutations on tumor cell biology has led to the conclusion that all mutations discovered so far enable a gain of function in α-KG reduction with a concomitant increase in the tumor-specific metabolite, or oncometabolite, 2-HG. Although the contribution of 2-HG to tumor cell biology remains speculative, Ward et al. noted that all IDH mutation-containing tumor types identified so far (leukemias and gliomas) are distinguished by proliferation of a relatively undifferentiated cell population, and in this context the effect of 2-HG in the tumor and its microenvironment is to block cellular differentiation [15].
Whole-genome comparisons of matched primary and metastatic cancers
One intriguing aspect of cancer genomics, for which published examples are few, involves comparing genome-wide alterations between the matched primary and metastatic cancer genomes from the same patient as a way of elucidating both their inter-relationships and the metastatic process. The first such study, by Shah et al. [16], focused whole-genome and whole-transcriptome sequencing on a metastatic tumor genome from an estrogen-receptor-positive, invasive lobular breast cancer that occurred 9 years after the patient's initial diagnosis and treatment. After a combined analysis to identify somatic mutations in both the genomic and transcriptomic data, the primary tumor taken 9 years earlier and the matched blood normal genomes were queried in the light of these findings. The aim was to search the primary tumor genome for the 30 mutations that had been found and validated as tumor-associated in the metastatic genome, and to establish the somatic or germline nature of the variants by comparing their occurrence in blood cells. Variants that were found in the primary tumor data were deeply sampled by sequencing to provide an estimate of the allele frequency for the somatic mutation in the primary tumor. Because of the 9-year interval between diagnosis of the primary tumor and metastasis, significant differences in mutational load were present; only 3 of the 28 tested mutations were prevalent in the primary cancer cells, 6 had an allele frequency of 1 to 13%, and 19 were not detected and were therefore metastasis-specific. Genes found in this analysis to have somatic point mutations were tested for frequency of mutation in 192 breast cancers, revealing that both the gene for the receptor kinase HER2 (3 of 192) and for the HAUS augmin-like complex, subunit 3 (HAUS3); 2 of 192) were mutated [16]. A second interesting finding from this study was the detection of two nonsynonymous variants that were introduced by RNA editing, perhaps the first description of this phenomenon from next-generation sequencing data and a strong testament to the importance of transcriptomic data in broadening the range of variant discovery from cancer genomics.
A recent study by our laboratory used next-generation whole-genome resequencing, analysis and comparison of the genomes of a matched primary breast tumor, metastatic brain tumor, and blood normal from an African-American patient with basal subtype breast cancer [17] This estrogen-receptor-negative tumor represents one the most aggressive types of breast cancer, and in this patient only 8 months elapsed between diagnosis of the primary tumor and a diagnosis of metastatic disease. We also sequenced the genome of a human-in-mouse xenograft [18] passage of the patient's primary tumor, taken by core biopsy procedure before adjuvant chemotherapy and placed into the fat pad of a NOD/SCID female mouse. Briefly, we found 48 mutations shared across all 3 tumors, and only 2 metastasis-specific point mutations. Two mutations, identified in this analysis, in the kinase JAK2 (affecting the JAK-STAT signaling pathway) and in the gene CSMD1 (CUB and Sushi multiple domains 1; loss of CSMD1 expression is associated with poor survival in invasive ductal breast carcinoma [19]) were also found mutated in other types of breast cancer samples. Interestingly, we established that the frequency of 16 mutations in the primary tumor cell population was lower than 10% but rose to very high frequency in both the metastatic and xenograft samples.
In addition to point mutations, 7 inter-chromosomal translocations, 6 inversions and 28 large deletion events were detected and validated as tumor-associated. One of the large deletions was particularly interesting, and nicely illustrates the exquisite resolution afforded by next-generation sequencing as well as emphasizing the important role of large structural rearrangements in tumor biology. A large (more than 500 kb) biallelic deletion, shown in Figure 3, was detected by the BreakDancer algorithm [20], which identifies read pair sequences that map to the reference genome at an unexpected distance or orientation relative to one another, and hence identifies a putative site of structural variation. The assembly of paired reads defining the deleted region resulted in two contigs with distinctly different breakpoints, both of which were confirmed by PCR and sequencing. Annotation of the region indicated that the gene CTNNA1 was completely deleted on both alleles. CTNNA1 encodes an α-catenin, loss of which has been shown to lead to global loss of cell adhesion in human breast cancer cells [21]. Figure 3 shows that there are an increasing number of cells containing the bi-allelic deletion in the transition from primary to metastatic disease, and that the xenograft tumor cells carry only the bi-allelic deletion in their genomes - trends reminiscent of the somatic point mutations mentioned in the previous paragraph.
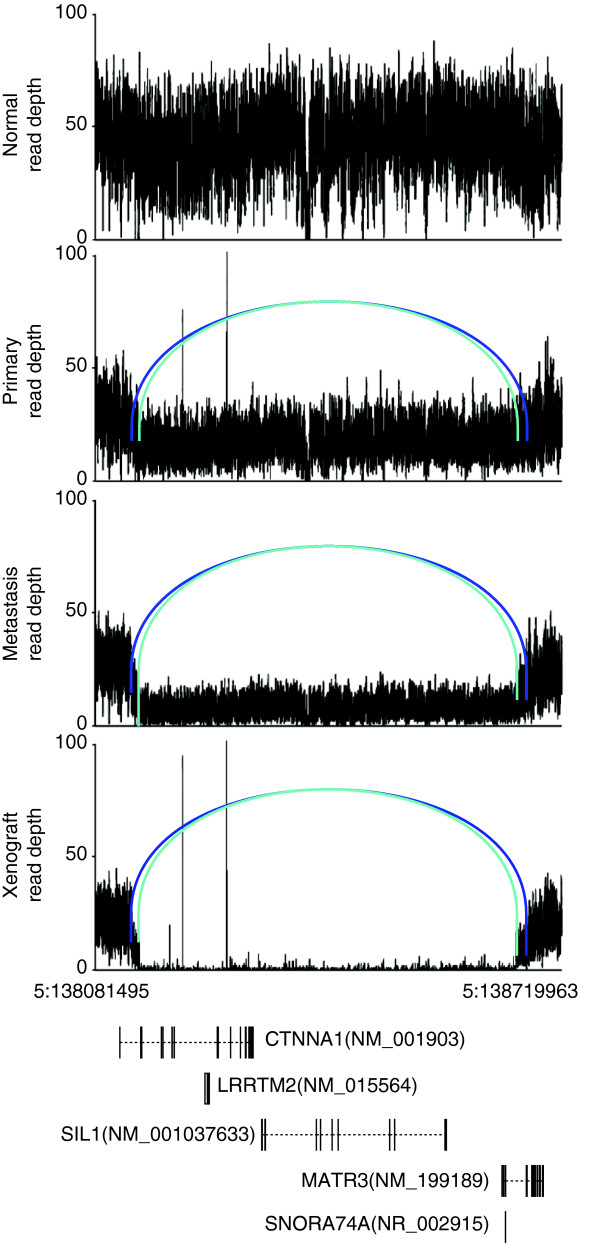
Figure 3.
Two overlapping CTNNA1 deletions on chromosome 5 in three tumors. A graph of sequence depths, read pairs and genes in a 638,468-bp region containing two overlapping deletions. The top four panels display the read depths at each base, and the reads within the region whose mates mapped at an abnormal distance are displayed as bars, with matched pairs connected by arcs. Two different shades of blue indicate the two separate allelic deletion events (538,467 bp and 515,465 bp in length). The bottom panel displays genes annotated in this genomic region. Reproduced with permission from [17].
Monitoring tumor-specific DNA biomarkers
At the interface between variant discovery and the transition from primary to metastatic disease lies the possibility of identifying personalized biomarkers that might enable an oncologist to monitor the progression or remission of a cancer. This approach has been elegantly developed by Leary et al. [22], who utilized long-distance mate-pair sequencing (a variation of paired end sequencing involving circularization of long DNA fragments (>1 kb) and ligation to an internal adapter DNA of known sequence) of an approximately 1.4 kbp region on the SOLiD platform to detect and characterize tumor-specific structural variations in two colon and two breast tumors [22]. Each sample was found to carry at least four validated somatic rearrangements that were then used to design PCR assays by which they could be detected. Patients' sera were assayed by PCR and this revealed that the amount of DNA circulating in plasma was sufficient to detect the tumor DNA rearrangements. In one case, serum samples were taken before and after tumor resection, and the levels of tumor-specific biomarker DNA in the plasma mirrored these procedures. This remarkable demonstration may transform our clinical approach to monitoring the course of cancer with minimally invasive methods.
Transcriptome-based discovery in cancer
Novel fusion transcripts in prostate tumors
Several groups now have pioneered efforts at unbiased transcriptome discovery using next-generation sequencing of mRNA (RNA-seq) from the tumor cell population and a variety of approaches to analyze the data. Although algorithmically complex to detect, there are a number of unique transcription-level processes that modify the encoded genome, including alternative splicing, RNA editing, and the formation of chimeric transcripts. Maher et al. have published two studies [23,24] that illustrate the development and implementation of RNA-seq analytical approaches to discover novel fusion transcripts in prostate tumors, which are often regulated by androgen levels. Initially, a dual-platform strategy combined longer read-length RNA-seq data from the Roche/454 platform with shorter RNA-seq fragment reads from the Illumina platform, and resulted in the discovery of a novel chimeric transcript, SLC45A3-ELK4, in prostate tumor samples [23]. The second approach [24] took advantage of Illumina paired-end RNA-seq data and a different algorithmic filtering of mapped paired ends to identify putative chimeric transcripts. When combining these read pairs with unmapped reads that span the fusion boundary, fusion transcripts previously identified in prostate cancer cells, such as TMPRSS2-ERG, were detected, as were novel fusion transcripts such as HERPUD1-ERG. These discoveries not only enhance our understanding of fusion transcripts in cancer, but have led to experiments to interrogate the role of hormonal signaling by androgens in inducing chromosomal movements that bring two genes that participate in a detected fusion event into close proximity [25].
FOXL2 mutations in granulosa-cell ovarian tumors
In a recent study, Shah et al. [26] evaluated the transcriptomes of four adult granulosa-cell tumors (GCTs) of the ovary, identifying putative variants involved in tumorigenesis by shared analysis of all four tumors. A single missense point mutation in FOXL2 was identified (C134W), and was subsequently found in an additional 86 out of 89 cases of adult GCTs. The gene was not mutated in other ovarian tumors of different types, nor in breast cancers that were tested. FOXL2 is a transcription factor in the forkhead-winged-helix family and is required for the normal development of granulosa cells. Although loss-of-function mutations in FOXL2 have been described for germline genomes, this was the first description of FOXL2 somatic mutations in ovarian tumors. We await the results of downstream functional studies, which will be required to reveal the impact of the cysteine-to-tryptophan amino acid change on the activity of this transcription factor, as well as altered transcription of genes bound by FOXL2.
In conclusion, cancer genomics, largely due to the unbiased and comprehensive nature of data that can be produced by next-generation sequencing platforms, is being applied to unravel the DNA- and RNA-level somatic alterations that determine tumor development and progression. It has been remarkable to see the pursuit of enzymatic, biochemical, functional and diagnostic implications of the earliest discoveries afforded by these methods. Hopefully, these important efforts will scale to accommodate the wave of next-generation-based discovery that is imminent, and the ultimate benefactors of our enhanced knowledge will be the patients and families whose lives are touched by this disease.
References
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Dulbecco R. A turning point in cancer research: sequencing the human genome. Science. 1986;231:1055–1056. doi: 10.1126/science.3945817. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G. et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, Frattini M, Molinari F, Knowles M, Cerrato A, Rodolfo M, Scarpa A, Felicioni L, Buttitta F, Malatesta S, Marchetti A, Bardelli A. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, Weller M, Herold-Mende C, Unterberg A, Jeuken JW, Wesseling P, Reifenberger G, von Deimling A. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, Ding J. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279:33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, Ding J, Lei Q, Guan KL, Xiong Y. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Heiden MG Vander, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF. et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IDH1 mutations are detected in 9.3% of all AML and are strongly associated with intermediate risk karyotype and unfavourable prognosis: a study of 999 patients. American Society of Hematology. http://ash.confex.com/ash/2009/webprogram/Paper25668.html
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, Dang L, Fantin VR, Mak TW. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, Steidl C, Holt RA, Jones S, Sun M, Leung G, Moore R, Severson T, Taylor GA, Teschendorff AE, Tse K, Turashvili G, Varhol R, Warren RL, Watson P, Zhao Y, Caldas C, Huntsman D, Hirst M, Marra MA, Aparicio S. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Ling L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, DeSchryver K, Davies S, Guintoli T, Lin L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, Richardson A, Weinberg RA. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci USA. 2004;101:4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal M, Shaaban AM, Zhang L, Walker C, Gray S, Thakker N, Toomes C, Speirs V, Bell SM. Loss of CSMD1 expression is associated with high tumour grade and poor survival in invasive ductal breast carcinoma. Breast Cancer Res Treat. 2009. doi: 10.1007/s10549-009-0500-4. [DOI] [PubMed]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, Shi X, Fulton RS, Ley TJ, Wilson RK, Ding L, Mardis ER. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajpai S, Feng Y, Krishnamurthy R, Longmore GD, Wirtz D. Loss of alpha-catenin decreases the strength of single E-cadherin bonds between human cancer cells. J Biol Chem. 2009;284:18252–18259. doi: 10.1074/jbc.M109.000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary RJ, I Kinde, F Diehl, K Schmidt, C Clouser, C Duncan, A Antipova, C Lee, K McKernan, FM De La Vega, KW Kinzler, B Vogelstein, LA Diaz Jr, VE Velculescu. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, Khrebtukova I, Barrette TR, Grasso C, Yu J, Lonigro RJ, Schroth G, Kumar-Sinha C, Chinnaiyan AM. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci USA. 2009;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Köbel M, Senz J, Morin RD, Clarke BA, Wiegand KC, Leung G, Zayed A, Mehl E, Kalloger SE, Sun M, Giuliany R, Yorida E, Jones S, Varhol R, Swenerton KD, Miller D, Clement PB, Crane C, Madore J, Provencher D, Leung P, DeFazio A, Khattra J, Turashvili G, Zhao Y, Zeng T, Glover JN, Vanderhyden B, Zhao C. et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med. 2009;360:2719–2729. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]