Abstract
Viruses recruit host proteins to secure viral genome maintenance and replication. However, whether they modify host histones directly to interfere with chromatin-based transcription is unknown. Here we report that Paramecium bursaria chlorella virus 1 (PBCV-1) encodes a functional SET domain histone Lys methyltransferase (HKMTase) termed vSET, which is linked to rapid inhibition of host transcription after viral infection. We show that vSET is packaged in the PBCV-1 virion, and that it contains a nuclear localization signal and probably represses host transcription by methylating histone H3 at Lys 27 (H3K27), a modification known to trigger gene silencing in eukaryotes. We also show that vSET induces cell accumulation at the G2/M phase by recruiting the Polycomb repressive complex CBX8 to the methylated H3K27 site in a heterologous system. vSET-like proteins that have H3K27 methylation activity are conserved in chlorella viruses. Our findings suggest a viral mechanism to repress gene transcription by direct modification of chromatin by PBCV-1 vSET.
Introduction
Dynamic control of H3K27 methylation 1, 2, 3 is central to gene silencing in development,4, 5, 6, 7 X-inactivation,8 stem-cell pluripotency,9, 10 cancer,11, 12 and inflammation.13 So far, only the mammalian EZH2 of the Polycomb repressive complex 2 (PRC2) is known to catalyze this important epigenetic histone modification.14, 15 H3K27 methylation as a viral strategy to regulate host transcription has not been reported. Viruses recruit chromatin-associated transcription proteins for maintenance of their genome and replication. For example, the papillomavirus E2 protein binds Brd4, tethering the viral genome to mitotic chromosomes to ensure persistence of viral episomes in infected cells.16 The adenovirus E1A protein interacts with the retinoblastoma protein p130 to silence E2F-responsive genes in quiescent cells.17 The HIV trans-activator Tat recruits histone acetyltransferases for chromatin acetylation and remodeling for transcriptional activation of the provirus.18, 19 Despite the ability of viruses to recruit cellular histone-modifying enzymes to facilitate viral transcription, no viral enzymes have been identified that directly modify host histones and modulate transcription. Of the large family of SET domain HKMTases,20, 21 a small subset of SET proteins is encoded by viruses and bacteria.22, 23 The cellular functions of these proteins are unknown but their presence in viruses raises questions about whether they are able to modify histones and, if so, how this would affect the host cells. To address these questions, we have characterized SET domain proteins from chlorella viruses.
We first focused on the function of vSET, a SET domain protein encoded by PBCV-1, which is a large double-stranded DNA virus that replicates in the unicellular, eukaryotic green algae Chlorella NC64A.24 The 330-kb PBCV-1 genome has 366 non-overlapping protein-encoding genes and 11 tRNA genes.24 Despite its large genome, PBCV-1 lacks an RNA polymerase gene, suggesting that it is dependent on the host transcription machinery. Notably, PBCV-1 infection causes a rapid inhibition of total RNA synthesis in Chlorella cells, achieving 60–80% inhibition by 20–40 min after infection (Figure 1a). PBCV-1 transcripts can be detected 5–10 min after infection, indicating that transition from host to virus transcription occurs rapidly.24 These results led us to postulate that a factor(s) packaged in the virion may be responsible for this rapid inhibition of host RNA synthesis. vSET is a possible candidate as it methylates H3K27, which has been linked to PRC-mediated gene silencing in eukaryotes.1, 2, 3
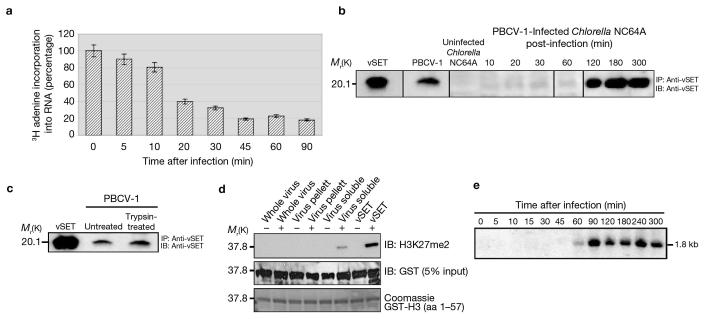
Figure 1. Presence of vSET in PBCV-1 virions.
(a) Inhibition of host transcription by virus infection. Incorporation of 3H-adenine into RNA in PBCV-1 infected Chlorella NC64A cells, compared with mock-infected cells. The data are mean ± s.d. of triplicate assays. (b) Detection of vSET in PBCV-1 virions and virus-infected Chlorella cells by immunoprecipitation (IP) and then immunoblotting (IB) with anti-vSET antibodies. (c) The presence of vSET in PBCV-1 virions is not due to external contamination, as demonstrated by trypsin pre-treatment of purified PBCV-1 virus particles. (d) H3K27 methylation activity of vSET in the lysate from disrupted PBCV-1 virions as demonstrated by western blotting with anti-H3K27me2 antibodies. (e) Northern blot analysis establishes that vset is expressed as a transcript of about 1.8 kb in PBCV-1 infected Chlorella NC64A cells 60 min after infection. For full scans of western blots in b and d, see Supplementary Information, Figure S5.
To investigate vSET function, we generated an anti-vSET antibody to immunoprecipitate vSET from PBCV-1 infected Chlorella cell extracts. vSET is present in mature virions and a small amount was detected as early as 10 min after infection of Chlorella cells, presumably, released from PBCV-1 (Figure 1b). Expression of vSET was markedly enhanced 120 min after infection. Pretreatment of isolated virus particles with trypsin, which removes any exogenous vSET contamination on the virus surface, had no effect on viral infectivity or the amount of vSET detected in PBCV-1 particles (Figure 1c). The presence of vSET in PBCV-1 virions was confirmed by Q-TOF mass spectrometry (data not shown). By comparing western blots from a known number of virus particles with blots containing different concentrations of vSET, we estimated that four vSET molecules are packaged per virion (data not shown). Furthermore, vSET in the disrupted virions possess H3K27 methylation activity (Figure 1d), which indicates that PBCV-1 virions contain active vSET.
To detect vset transcripts during PBCV-1 replication, RNA from infected Chlorella cells was hybridized with the vset gene. The probe hybridized to an RNA of about 1.8 kb, beginning approximately 60 min after infection (Figure 1e). This transcript is larger than expected for a 119-residue protein and may be a bicistronic transcript encoding the co-linear genes a609l (approximately 1.2 kb) and a612l (approximately 0.4 kb). The latter gene encodes vSET. This result is consistent with vSET expression occurring about 120 min after infection (Figure 1b).24
vSET adopts a core β-barrel structure,25, 26 a fold that is conserved in eukaryotic SET domain HKMTases.21 To test whether vSET functions as a bona fide HKMTase in Chlorella cells, we cloned histone H3 from Chlorella NC64A and found that it has high sequence identity with human H3 (Figure 2a). We measured vSET methylation activity using both the nucleosome and individual core histones and confirmed its activity on full-length free H3 and H3 within the nucleosome, but not on other core histones (Figure 2b). To identify vSET methylation site(s) in H3, we expressed a series of GST–H3 peptides (residues 1–57) with Arg substitutions at Lys methylation sites: Lys 4, Lys 9, Lys 27, Lys 36 and Lys 37 (Supplementary Information, Table S2). The purified GST–H3 peptides produced two bands when subjected to SDS–PAGE, which corresponded to an intact H3 peptide of residues 1–57 (plus SGRIVTD from the expression vector) and a truncated H3 of residues 1–55, as confirmed by MALDI-TOF mass spectrometry (Figure 2c). The methylation assay showed that vSET only methylated H3 with an intact Lys 27 site (Figure 2c). Using site- and state-specific antibodies against Lys-methylated histone H3 (Supplementary Information, Figure S1a, b), we confirmed that vSET catalyses H3K27 dimethylation (H3K27me2) and, to a much less extent, mono- and trimethylation. No methylation occurred at Lys 4, Lys 9 or Lys 36 sites in H3 (Figure 2d). These results are consistent with the kinetic analyses of vSET methylation of H3K27 peptide, showing that monomethylation and mono- to dimethylation are very rapid, whereas di- to trimethylation is about 10 times slower than mono- to dimethylation (Supplementary Information, Figure S1c, d). Finally, we confirmed that histone H3 is the major substrate of vSET in an in vitro methylation assay using heat-inactivated extracts from HeLa cells depleted of EZH2 by RNA interference (RNAi) and uninfected Chlorella cells (Supplementary Information, Figure S2a–d). Collectively, these results establish that vSET is a H3K27-specific methyltransferase.
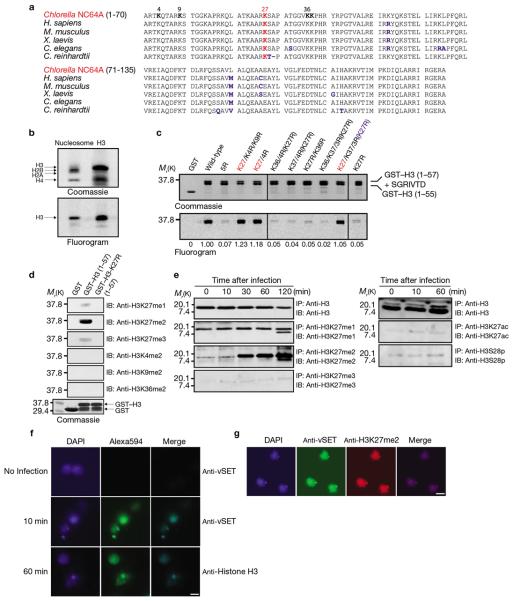
Figure 2. Methylation of Chlorella NC64A histone H3K27 by vSET.
(a) Amino acid sequence alignment of H3 from Chlorella NC64A, Homo sapiens, Mus musculus, Xenopus laevis, Caenorhabditis elegans, and Chlamydomonas reinhardtii. Residues that differ in H3 are shown in blue. (b) vSET methylates H3 in nucleosomes. Native histone H3 was used as a control (right lane). (c) HKMTase activity of vSET targets H3K27, as illustrated by methyl transfer from 14C-methyl-SAM to GST-fusion H3 peptides (residues 1–57). Fluorescent values relative to wild-type enzyme activity are indicated under the lower panel. Relative amounts of GST–H3 in the assays are shown in SDS–PAGE (upper panel). (d) Western blot analysis of vSET-treated GST, GST–H3 wild-type and K27R mutant peptides using anti-H3K27me1, 2, 3, anti-H3K4me2, anti-H3K9me2, and anti-H3K36me2 antibodies. (e) Western blot analyses of Chlorella NC64A H3 before and after PBCV-1 infection at specified times using antibodies against histone H3 with different modifications. (f) vSET is localized in the nucleus of PBCV-1 infected Chlorella cells collected 60 min after infection, as illustrated by immunofluorescence microscopy. Chlorella cells (uninfected or 10 and 60 min after infection) were probed with anti-vSET (uninfected and 10 min) or anti-H3 antibodies (60 min). (g) vSET and host Chlorella H3K27me2 colocalize in the nucleus 60 min after infection, as shown by immunofluorescence microscopy. Chlorella cells were probed with anti-vSET and anti-H3K27me2 antibodies and visualized by immunofluorescence microscopy using Alexa488 and Alexa594 dyes, respectively. Scale bars are 10 μm. For full scans of western blots in d and e see Supplementary Information, Figure S5. H3 peptide sequences of c are shown in Supplementary Information, Table S2.
We next assessed the effects of PBCV-1 infection on H3K27 methylation in Chlorella cells. Western blot analysis showed that H3K27me1 was unchanged, whereas H3K27me2 increased markedly as early as 30 min after infection, and H3K27me3 was slightly enhanced (Figure 2e). This observation correlates with in vitro vSET state-specific methylation activity at H3K27 (Figure 2d). Immunofluoresence showed that vSET was present in the nucleus of Chlorella cells after PBCV-1 infection (Figure 2f), and vSET colocalized with the H3K27me2 (Figure 2g). These results confirm that vSET is a bona fide HKMTase and causes increased H3K27me2 in Chlorella cells during virus replication.
Technical difficulties in manipulating the PBCV-1 genome and genetic transformation of the host Chlorella cells prevented us from conducting a cellular study of vSET in its native host. But given the highly conserved H3 sequence and H3K27 methylation in eukaryotes, we reasoned that mammalian cells could serve as a suitable model system to study the biological function of vSET, which has enzymatic activity similar to that of EZH2 (ref. 3).
If vSET participates in early suppression of host transcription, it must move to the host nucleus on viral infection. To assess whether a solvent-exposed KRMR motif in vSET (residues 85–88, see Figure 5b) functions as a nuclear localization signal (NLS), NIH-3T3 cells were transfected with a GFP–vSET or a triple mutant in which KR(M)R was changed to AA(M)A. Fluorescence microscopy showed that wild-type GFP–vSET was localized to the nucleus, whereas the triple mutant remained in the cytoplasm (Figure 3a), confirming that the KR(M)R motif is an NLS in vSET. vSET H3K27 methylation activity was not compromised in this mutant (Supplementary Information, Figure S3a–c). A classical NLS sequence is absent in most eukaryotic SET domains, except for yeast SET1, which has an RRIV motif located at a site analogous to the KRMR motif in vSET.
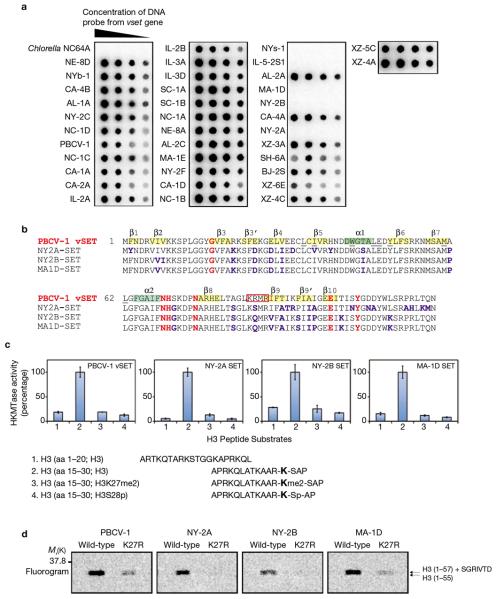
Figure 5. HKMT activity of SET proteins from chlorella viruses.
(a) Southern dot blot analysis of 37 chlorella viruses confirmed the presence of a vset-like gene in 32 viruses. Vset-like genes from three of the five viruses that did not exhibit positive hybridization signals: NY-2A, NY-2B and MA-1D were detected using low stringency Southern hybridization (data not shown). (b) Sequence alignment of vSET and SET proteins from chlorella viruses NY-2A, NY-2B and MA-1D. Positions of ? helices (grey) and β strands (yellow) are highlighted in the vSET sequence. Residues conserved among all known SET proteins are in red; residues in the three viral SET proteins that differ from vSET are in blue. Residues of vSET at the dimer interface are underlined. The NLS sequence in PBCV-1 is boxed. (c) Methylation activity of the SET proteins from viruses NY-2A, NY-2B and MA-1D measured with H3 peptides using 14C-methyl-SAM. Data are mean ± s.d. of triplicate assays. (d) HKMTase activity of the viral SET proteins using wild-type H3 (residues 1–57) or the K27R mutant. The fluorogram shows that NY-2A, NY-2B and MA-1D SET proteins methylate wild-type H3 but not the K27R mutant. H3 modified by vSET was used as a control.
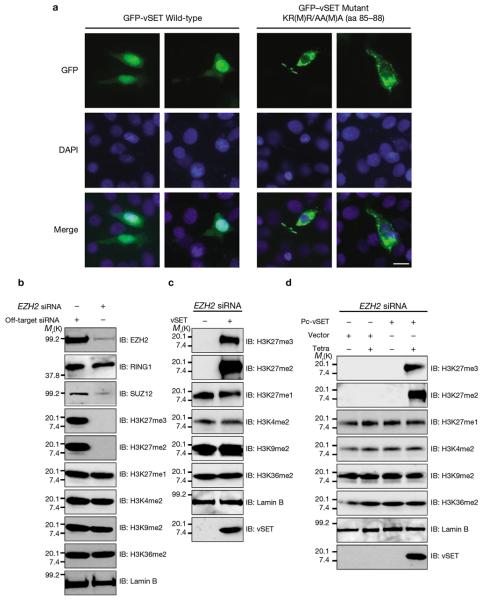
Figure 3. Nuclear localization and H3K27 methylation activity of vSET.
(a) Nuclear localization of GFP–vSET in transiently transfected NIH-3T3 cells. Fluorescence microscopy images show that wild-type vSET is located in the nucleus, whereas a triple mutant, KR(M)R/AA(M)A (amino acids 85–88) is in the cytoplasm. Cells were counterstained with DAPI to highlight nuclei. Scale bar is 10 μm. (b) Western blot analysis of HeLa cells showing effects of EZH2-specific- and mock-siRNAs treatment on protein expression of EZH2, RING1, SUZ12, and histone H3 with different modifications. Lamin B was used as a control. (c) Western blot analysis of the nuclear extract from HeLa cells treated with EZH2 siRNA and subjected to in vitro methylation by vSET. (d) Western blot analysis showing di-and trimethylation of H3K27 after tetracycline-induced expression of stably transfected vSET in HeLa cells 72 h after EZH2 RNAi knockdown. For full scans of western blots in d see Supplementary Information, Figure S5. EZH2 siRNA sequences are shown in Supplementary Information, Table S3.
H3K27 methylation in eukaryotes is catalyzed by the EZH2 of PRC2 and recruits the PRC1 complex, leading to gene silencing.1, 14 To investigate whether vSET can methylate H3K27 in mammalian cells, we knocked down EZH2 using the RNAi method in HeLa cells. When treated with EZH2-specific siRNA, EZH2 expression was reduced by more than 90%, compared with control cells (Figure 3b). EZH2 knockdown caused a reduction of SUZ12, a component of PRC2, as well as near complete loss of H3K27me2 and H3K27me3, and a slight reduction of H3K27me1. No significant changes were observed with RING1 of PRC1, H3K4, H3K9, and H3K36 dimethylation. These results confirm that EZH2 is responsible for H3K27 di- and trimethylation in HeLa cells. vSET treatment of the nuclear extract from EZH2 knockdown HeLa cells restored H3K27me2 and H3K27me3, and resulted in a slight reduction of H3K27me1, possibly because of its conversion to H3K27me2 and H3K27me3 by vSET (Figure 3c). Similar results were obtained when tetracycline was used to induce vSET expression in the EZH2 knockdown HeLa cells (Figure 3d). Thus, these results demonstrate that vSET can mimic EZH2 to methylate H3K27 in mammalian cells.
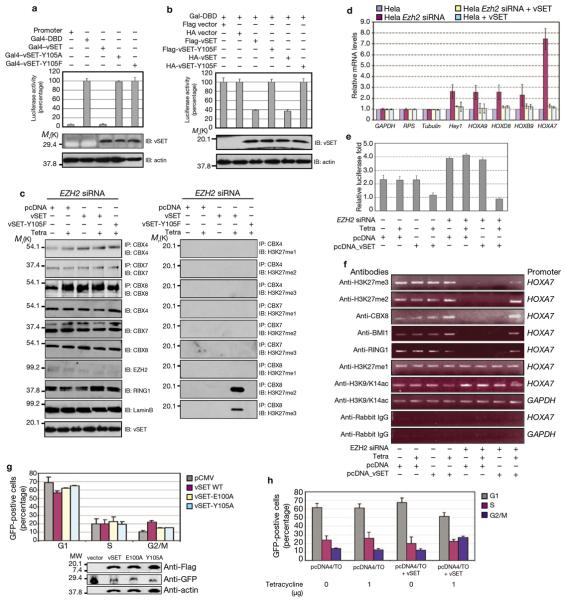
We next examined the effect of vSET methylation activity on gene transcription in a luciferase-based reporter assay. In this assay, we first transfected the 293T cells with vSET fused to a Gal4 DNA-binding domain (DBD) and a luciferase reporter gene encoding HSV-tk promoter (plus Gal4 DNA-binding sites). Wild-type vSET repressed luciferase gene expression by about 95%, whereas the inactive mutants Y105A or Y105F had no vSET gene-silencing ability (Figure 4a). Notably, expression of a Flag- or HA-tagged vSET in the 293T cells caused repression of approximately 60%, compared with the vectors containing only Flag- or HA-tag, or inactive mutant Y105F (Figure 4b). The lesser repressive activity of Flag- or HA-tagged vSET on host transcription, compared with that of vSET–Gal4-DBD may be due, at least in part, to the presence of H3K27ac and/or H3S28p in host cells (Figure 2e), which would prevent H3K27 methylation by vSET 26 (Figure 5c). In a luciferase-based assay, we also observed that Flag–vSET co-transfected with HIV-Tat in 293T cells (Supplementary Information, Figure S4a) or HeLa cells (Supplementary Information, Figure S4b) repressed transcription of the HIV LTR promoter,18, 19 confirming repression of chromatin-mediated transcription by vSET.
Figure 4. Repression of gene transcription by vSET H3K27 methylation activity.
(a) Transcriptional repression of a reporter luciferase gene by vSET and its inactive mutants Y105A or Y105F in 293T cells; vSET was fused to the Gal4–DNA binding domain. (b) Transcriptional repression of the reporter luciferase gene by Flag- or HA-tagged vSET and its inactive mutant Y105F transiently transfected in 293T cells. Data are mean ± s.d. of triplicate assays (a, b). (c) Western blot analysis of PRC1 and PRC2 protein association with methylated H3K27 after induction of stably transfected vSET in the EZH2 knockdown HeLa cells. The inactive vSET-Y105F mutant was used as a control. (d) Quantitative RT–PCR measurements showing relative mRNA levels of five Polycomb target genes of Hey1, HOXA9, HOXD8, HOXB9, and HOXA7, and three house keeping genes (GAPDH, RPS, tubulin) in normal HeLa cells (grey) and EZH2 knockdown HeLa cells with (yellow) or without (red) induction of the transiently transfected vSET, as well as HeLa cells transfected with vSET (blue). (e) Luciferase-based reporter assay of HOX7A promoter in the presence of tetracycline-induced vSET in normal and EZH2 depleted HeLa cells. Data are mean ± s.d. of triplicate assays (d, e). (f) ChIP analysis at the HOXA7 promoter on induction of vSET in the HeLa cells with and without EZH2 siRNA treatment. (g) vSET induces G2/M phase cell accumulation in NIH-3T3 cells co-transfected with vSET and Us9–GFP expression plasmids. Western blots showing relative equal expression of vSET and its mutants. (h) G2/M accumulation of HeLa cells stabilized on vSET induction with tetracycline. The DNA content of the gated GFP-positive cells was determined by propidium iodide staining and FACS analysis. DNA content of cell-cycle phases is shown (g, h, mean ± s.d. of at least three independent transfection experiments). For full scans of western blots in a–c see Supplementary Information, Figure S5. RT–PCR and ChIP primer sequences of d and f are available in Supplementary Information, Table S3.
To examine the biological consequence of H3K27 methylation by vSET, we assessed Polycomb group protein occupation at the H3K27me site on tetracycline-induction of vSET in the EZH2 knockdown HeLa cells. Although their protein levels were unaltered, vSET induction enhanced occupation of the PRC1 complex protein CBX8, but not CBX4 and CBX7 at the H3K27me2 and H3K27me3 sites (Figure 4c). This CBX8/H3K27me2/3 association is probably facilitated by the chromodomain of CBX8, which binds methyl-Lysine.27, 28 Interestingly, a search of the SMART database reveals that a virus PBCV-1 encoded protein (accession number NP_049022) seems to contain the chromodomain as in CBX8. Therefore, we conclude that vSET induces transcription repression by H3K27me, and facilitates accumulation of the PRC1 complex at the H3K27me site.
Using a quantitative RT–PCR assay, we examined whether vSET methylation activity can modulate Polycomb target genes.29 The analysis was performed in HeLa cells on five representative Polycomb target genes HOXA7, HOXA9, HOXB9, HOXD8 and Hey1, and three general housekeeping genes GAPDH, RPS and tubulin. The effect on the three housekeeping genes was negligible; however EZH2 knockdown by siRNA caused a 2.5-fold increase in transcription of HOXA9, HOXB9, HOXD8 and Hey1, and a 7.5-fold increase in HOXA7 expression (Figure 4d). The increase in the Polycomb target gene expression was reversed by tetracycline-induced expression of vSET to a level similar to or slightly higher than that of the control cells. Moreover, vSET induction in the normal HeLa cells did not alter the expression level of the five HOX genes when compared to the controls.
We next characterized vSET-dependent transcriptional repression of the HOXA7 promoter in luciferase gene expression and chromatin immunoprecipitation (ChIP) assays. Induction of vSET effectively repressed luciferase expression at the HOXA7 promoter in both EZH2 siRNA-treated and untreated HeLa cells (Figure 4e; Supplementary Information, Figure S4c). The ChIP analysis confirmed that vSET specifically di- and tri-methylates H3K27 at the HOXA7 promoter, which recruits CBX8 of the PRC1 and causes gene repression (Figure 4f). We obtained similar results for the characterization of vSET-mediated transcription repression of HIV-Tat-dependent LTR promoter at the nucleosome Nuc2 (Supplementary Information, Figure S4d, e). These results establish that vSET can modulate transcription of Polycomb target genes through its H3K27 methylation activity.
We then examined the effect of vSET expression on the cell cycle. Flow cytometric analysis indicated that vSET caused cell accumulation at the G2/M phase in transiently transfected NIH-3T3 cells, whereas active-site mutants E100A and Y105A were less effective (Figure 4g). The vSET effect on the G2/M phase in the cell-cycle was confirmed in the HeLa cells that were stably co-transfected with vSET in a tetracycline-controlled expression vector (Figure 4h). Collectively, our data indicate that vSET H3K27 methylation activity results in the recruitment of the Polycomb repressive complexes and modulation of their target genes, leading to transcription repression and accumulation of cells at the G2/M phase.
To determine whether vSET is unique to viral PBCV-1 or is present in other chlorella viruses, we hybridized genomic DNA from 36 other chlorella viruses with a vset gene probe. Thirty-one of these viruses hybridized to the probe (Figure 5a). The probe did not hybridize to DNA from 5 viruses or the host DNA. However, a vset gene was identified in 3 of these 5 viruses using low stringency hybridization. The vSET proteins from these three viruses, NY-2A, NY-2B and MA-1D, have 85% amino acid similarity to PBCV-1 vSET, including the conserved active-site residues in the SET domain family (Figure 5b). We suspect the remaining two viruses, NYs-1 and IL-5-2S1, also have a vSET gene but we did not examine them further.
Using histones H2A, H2B, H3 and H4 as substrates, we established that the SET proteins from these three viruses selectively methylate H3 (data not shown). As with vSET, these viral SET proteins only methylated a H3 peptide containing residues 15–30 but not one containing H3 residues 1–20 (Figure 5c). They showed little methyltransferase activity towards H3 peptide that was previously dimethylated at Lys 27 or phosphorylated at Ser 28, suggesting a preference for the H3K27 site. The selective methylation at H3K27 was confirmed using GST–H3 (residues 1–57), in which these viral SET proteins had robust methylation activity on wild-type H3 but diminished activity on the K27R mutant (Figure 5d). These results establish that SET proteins from the chlorella viruses possess selective H3K27 methylation activity, and suggest that vSET function in host chromatin modification and transcription repression is conserved among the chlorella viruses.
Histone modifications provide epigenetic control of gene transcription. Viruses recruit host proteins to either integrate their genomes into host chromatin 16, 17 or to facilitate viral transcription and replication.18, 19 Here, we show for the first time that a virus regulates the host transcriptional machinery through direct modifications of chromatin. Our study reveals that vSET is packaged in the PBCV-1 virion and can directly methylate host H3K27, an important epigenetic modification in eukaryotes that is functionally linked to Hox gene silencing,4, 5, 30, 31 X-inactivation,8 and stem-cell pluripotency.9, 10 Our results establish that vSET H3K27 methylation promotes the recruitment of the PRC1 complex through a molecular interaction between CBX8 and di- and/or trimethylated H3K27. This interaction results in modulation of Polycomb target genes, transcription repression and cell accumulation at the G2/M phase of the cell cycle in continuously dividing cells. Our study supports the view that vSET proteins, which are encoded by all chlorella viruses tested, function to repress transcription in infected chlorella cells. Collectively, our findings suggest a unique and powerful mechanism by which some viruses commandeer the host transcription machinery through direct modifications of histones and in this way govern host chromatin-mediated cellular processes.
Methods
Growth of cells and viruses, and preparation of Chlorella cell extracts
Growth of Chlorella NC64A on MBBM medium, plaque assay, production of viruses and isolation of viral DNA were carried out as described previously.32 Actively growing Chlorella NC64A cells, concentrated to 2 × 108 cells ml−1, were mock-infected or infected with PBCV-1 at a multiplicity of infection (MOI) of 5 for the indicated times. Samples were pelleted, disrupted in lysis buffer by vortexing with glass beads and clarified by centrifugation. Purified PBCV-1 virus particles (200 μl of 6 mg ml−1 total virus) were treated with 200 U ml−1 sequencing-grade modified trypsin (Promega) before further purification.
RNA analysis of infected Chlorella
Actively growing Chlorella NC64A cells (1–2 × 107 ml−1) were incubated with 3H-adenine (5 μl ml−1, 37 MBq ml−1) for 10 min and centrifuged. Cells were then resuspended in 50 mM Tris-HCl, pH 7.8, (5 × 107 cells ml−1) containing PBCV-1(MOI, 5). Samples of 1 ml were collected at the appropriate times and mixed with equal volumes of ice-cold 10% trichloroacetic acid (TCA). Cells were collected on glass filters, washed three times with 5% TCA, once with 80% ethanol and dried. RNA was hydrolyzed by placing the filters in 5 ml of NaOH (0.5 M) overnight at 37 °C, collected by filtering through a fresh filter, neutralized with TCA, and analyzed by scintillation counting. Total RNA was isolated from PBCV-1 infected Chlorella cells at various times after viral infection using the Trizol reagent (Invitrogen). Total RNA was electrophoresed and hybridized with a 32P-labelled gene probe using a random-primer DNA labeling kit (Invitrogen). Viral DNAs used for dot blots were denatured, applied to nylon membranes, fixed by UV cross-linking and hybridized with the gene probe used for the northern analysis.
DNA cloning, protein purification and HKMTase assays
Full-length histone H3 from Chlorella NC64A was cloned with degenerate primers based on H3 from Chlamydomonas reinhardtii. The SET gene was isolated from chlorella viruses MA-1D, NY-2A and NY-2B using an established protocol.33 The DNA bands that hybridized weakly with the PBCV-1 vSET probe were excised from the gel, recovered with a QIAEX II gel extraction kit (Qiagen) and cloned into pGEM-7Zf+. Primers were designed according to the vSET homologues identified from these hybridized DNA fragments. The vSET homologues from these three viruses were amplified by PCR and subcloned into pET-15b (Novagen). All DNA constructs and vSET mutants prepared using the QuikChange mutagenesis kit (Stratagene) were confirmed by DNA sequencing. Recombinant proteins were expressed in Escherichia coli strain BL21. The in vitro HKMTase reaction was performed by following methyl transfer from S-adenosyl-14C-methyl-L-methionine (Amersham) to histone peptide or nucleosome substrates, or by mass spectroscopy analysis of the histone peptide substrates.26 Upstream and downstream primer sequences used for histone H3 are 5′-ATGGCCCGCACCAAG and 5′-TTAGGCGCGCTCGCC, respectively.
Western blot and histone immunoprecipitation analyses
PBCV-1 infected Chlorella cells were collected at various times after infection. Western blot analysis of the immunoprecipitated protein was performed using anti-vSET antibodies (Supplementary Information, Table S1), generated in rabbits against recombinant vSET (Covance). Histone immunoprecipitation was performed on Chlorella cells collected at 0–120 min after PBCV-1 infection. The assay was carried out using antibodies against histone H3, H3K27me1, H3K27me2, H3K27me3, H3K27ac, and H3S28p (Millipore) with cells cross-linked with paraformaldehyde and disrupted by sonication. To assess the PRC proteins at H3K27, HeLa cells stably transfected with vSET were induced with tetracycline, subjected to cross-linking and immunoprecipitated using antibodies against EZH2 and Polycomb complex proteins, RING1, SUZ12, CBX4, CBX7, and CBX8, followed by western blot analyses using anti-H3 antibodies (see Information, Supplementary Table S1).
Nuclear localization and co-localization
vSET nuclear localization was analyzed in NIH-3T3 cells transiently transfected with a pEGFP-N1 vector (Clontech) that encodes a C-terminal GFP fusion to vSET. The analysis in chlorella cells include fixation, permeablization, and immunofluorescence microscopy. Typically, the exponentially growing chlorella cells were collected and suspended in 2% paraformaldehyde in PBS buffer of pH 7.4 at 4 °C for 6 h. The fixed cells were pelleted by centrifugation, resuspended in 5 ml chilled methanol, washed with PBS, spotted onto a slide and air-dried. Cells that adhered to the slides were incubated for 15 min at 4 °C in DMSO (0.5% vol/vol in PBS). For fluorescence microscopy analysis, cells were blocked with 0.5% bovine serum album in PBS and incubated with anti-vSET and anti-histone antibodies (Abcam) for 1 h at 25 °C. Cells were washed with PBS and incubated with secondary fluorescein-labeled antibody (Alexa 488), mounted on a coverslip and analyzed using a Zeiss Axioplan2 microscope. For the colocalization assay, cells were stained for rabbit polyclonal anti-vSET and anti-rat monoclonal H3K27me2 antibodies. The respective immunocomplexes were detected using Alexa488 and 594 (Molecular Probes).
Transcription reporter assay and cell-cycle analysis
A luciferase reporter gene transcription assay was performed as described previously34 (see Supplementary Information, Methods). Briefly, 293T cells were co-transfected with 2 μg of pcDNA3–Gal4-DBD–vSET (or the mutants) and 1 μg of HSV–tk–promoter plus Gal4 binding site and luciferase gene constructs. After 48 h, cells were lysed and assayed for luciferase activity using the Stop–Glo luciferase assay system (Promega). A luciferase assay on HOX7A promoter was also performed after co-transfection with Renilla luciferase in HeLa cells with or without EZH2 knockdown by siRNA, as described below. Each assay was performed in duplicate and repeated five times. The vSET effect on the cell cycle was analyzed in NIH-3T3 cells transiently transfected with vSET in a pCMV-tag2B vector (Stratagene) and a Us9–GFP encoding plasmid, or HeLa cells stably co-transfected with vSET in a tetracycline-controlled vector pcDNA4/TO (Invitrogen) and the Us9–GFP plasmid. Post-transfection cells were treated with tetracycline (1 μg ml−1), collected after 24 h by trypsinization, washed with PBS and fixed in chilled 70% ethanol in PBS. One hour before acquiring the data, cells were washed again with PBS and stained with propidium iodide. Cell-cycle analysis was performed with a Calibur flow cytometer (Becton Dickinson) after GFP and propidium iodide staining.
EZH2 RNAi knockdown, quantitative RT–PCR and ChIP analyses
EZH2 knockdown by RNAi was performed in HeLa cells that were transfected with target-specific and smart-pool siRNAs (Dharmacon) according to manufacturer's instructions (Supplementary Information, Table S3). EZH2 RNAi knockdown was evaluated 72 h after transfection by western blot analysis using an anti-EZH2 antibody (Cell Signaling). Methylation states of Lys 4, Lys 9, Lys 27, and Lys 36 in histone H3 were evaluated using various antibodies obtained from Millpore and Abcam. For RT–PCR analysis, RNA was extracted using RNAeasy kit (Qiagen) from HeLa cells that were transfected with EZH2 siRNA and/or vSET. The primers for the select Polycomb target genes and house-keeping genes for analysis were designed based on the published sequences.35 cDNA was generated by RT–PCR using the affinity script from Stratagene. Reactions were determined using the SYBR Green I detection chemistry system (Applied Biosystems) with an ABI Prism 7300 Sequence Detection System. Chromatin immunoprecipitation (ChIP) analysis was performed for the HOXA7 gene using the EZ-ChIP kit (Millipore) following the manufacturer's instruction.13
Supplementary Material
Acknowledgments
We thank L. W. Enquist for the Us9–GFP construct, M. Ptashne for the Gal4-DBD construct, P. Traber for the firefly luciferase construct, R. Slany for the HOX7A luciferase construct, I. V. Agarkova for the results used in Figure 1a, and M. Walsh for helpful discussions. Fluorescence microscopy imaging was performed at the Mount Sinai Microscopy Shared Resource Facility supported in part with funding from NIH-NCI shared resources grant. This work was supported by grants from the National Institutes of Health (M.-M. Z. and J. L. V. E.).
References
- 1.Schuettengruber B, et al. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 3.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 5.Kuzmichev A, et al. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 7.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 8.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein B, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 10.Boyer L, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 11.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature Rev. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 12.Kleer C, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of Polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 14.Whitcomb S, Basu A, Allis C, Bernstein E. Polycomb group proteins: an evolutionary perspective. Trends Genet. 2007;23:494–502. doi: 10.1016/j.tig.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Gil J, Bernard D, Peters G. Role of polycomb group proteins in stem cell self-renewal and cancer. DNA Cell Biol. 2005;24:117–125. doi: 10.1089/dna.2005.24.117. [DOI] [PubMed] [Google Scholar]
- 16.You J, et al. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh M, Harter M. A viral mechanism for remodeling chromatin structure in G0 cells. Mol. Cell. 2003;12:255–260. doi: 10.1016/s1097-2765(03)00225-9. [DOI] [PubMed] [Google Scholar]
- 18.Mujtaba S, et al. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol. Cell. 2002;9:575–586. doi: 10.1016/s1097-2765(02)00483-5. [DOI] [PubMed] [Google Scholar]
- 19.Dorr A, et al. Transcriptional synergy between Tat and PCAF is dependent on the binding of acetylated Tat to the PCAF bromodomain. EMBO J. 2002;21:2715–2723. doi: 10.1093/emboj/21.11.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lachner M, O'Sullivan R, Jenuwein T. An epigenetic road map for histone lysine methylation. J. Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 21.Qian C, Zhou M. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol. Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manzur K, Zhou M. An archaeal SET domain protein exhibits distinct lysine methyltransferase activity towards DNA-associated protein MC1-α. FEBS Lett. 2005;579:3859–3865. doi: 10.1016/j.febslet.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez-Venegas R, Sadder M, Tikhonov A, Avramova Z. Origin of the bacterial SET domain genes: vertical or horizontal? Mol. Biol. Evol. 2007;24:482–497. doi: 10.1093/molbev/msl184. [DOI] [PubMed] [Google Scholar]
- 24.Van Etten J. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 2003;37:153–195. doi: 10.1146/annurev.genet.37.110801.143915. [DOI] [PubMed] [Google Scholar]
- 25.Manzur K, et al. A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3. Nature Struct. Biol. 2003;10:187–196. doi: 10.1038/nsb898. [DOI] [PubMed] [Google Scholar]
- 26.Qian C, et al. Structural insights of the specificity and catalysis of a viral histone H3 lysine 27 methyltransferase. J. Mol. Biol. 2006;359:86–96. doi: 10.1016/j.jmb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 27.Min J, Zhang Y, Xu R. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yap K, Zhou M. Structure and function of protein modules in chromatin biology. Results Probl. Cell Differ. 2006;41:1–23. [PubMed] [Google Scholar]
- 29.Bracken AP, et al. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 31.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 32.Schuster A, et al. Characterization of viruses infecting a eukaryotic chlorella-like green alga. Virology. 1986;150:170–177. doi: 10.1016/0042-6822(86)90276-x. [DOI] [PubMed] [Google Scholar]
- 33.Kang M, et al. Genetic diversity in chlorella viruses flanking kcv, a gene that encodes a potassium ion channel protein. Virology. 2004;326:150–159. doi: 10.1016/j.virol.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 34.Nishio H, Walsh M. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc. Natl. Acad. Sci. USA. 2004;101:11257–11262. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svingen T, Tonissen KF. Altered HOX gene expression in human skin and breast cancer cells. Cancer Biol. Ther. 2003;2:518–523. doi: 10.4161/cbt.2.5.441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.