Abstract
The process of assembly and egress of human cytomegalovirus (HCMV) virions requires significant morphological alterations of the nuclear and cytoplasmic architecture. In the studies presented we show that the nuclear periphery is dramatically altered, especially near the cytoplasmic assembly compartment, where the nuclear lamina is specifically rearranged, the outer nuclear membrane is altered, and the nucleus becomes permeable to large molecules. In addition, the tethering of the inner and outer nuclear membranes is lost during infection due to a decrease in levels of the SUN domain proteins. We previously demonstrated that the endoplasmic reticulum protein BiP functions as a component of the assembly compartment and disruption of BiP causes the loss of assembly compartment integrity. In this study we show that the depletion of BiP, and the loss of assembly compartment integrity, results in the loss of virally induced lamina rearrangement and morphology of the nucleus that is characteristic of HCMV infection. BiP functions in lamina rearrangement through its ability to affect lamin phosphorylation. Depletion of BiP and disruption of the assembly compartment result in the loss of lamin phosphorylation. The dependency of lamin phosphorylation on BiP correlates with an interaction between BiP and UL50. Finally, we confirm previous data (S. V. Indran, M. E. Ballestas, and W. J. Britt, J. Virol. 84:3162-3177, 2010) suggesting an involvement of dynein in assembly compartment formation and extend this observation by showing that when dynein is inhibited, the nuclear morphology characteristic of an HCMV infection is lost. Our data suggest a highly integrated assembly-egress continuum.
The process of assembly and egress of human cytomegalovirus (HCMV) virions requires significant morphological alterations of the nuclear and cytoplasmic architecture. A striking change is the enlargement of the nucleus, which is often seen to take on a characteristic concave, kidney-like shape. Nestled against the concave surface of the nucleus, a perinuclear structure referred to as the assembly compartment (AC) forms (12, 38, 40). The diagram in Fig. 1 A illustrates the nuclear periphery, including the inner and outer nuclear membranes (INM and ONM, respectively) and the nuclear lamina which lies inside the INM. Figure 1B illustrates the placement of the assembly compartment immediately adjacent to the concave side of the nucleus. There appears to be an intimate relationship between the concave nucleus and the assembly compartment, since perturbing the perinuclear position of the assembly compartment results in the nucleus regaining its normal shape and size (1, 5, 18, 34). Interestingly, these same conditions often abolish cytoplasmic viral activity, suggesting that the remodeled nucleus, the assembly compartment, and cytoplasmic assembly-egress processes make up a highly interconnected assembly-egress continuum.
FIG. 1.
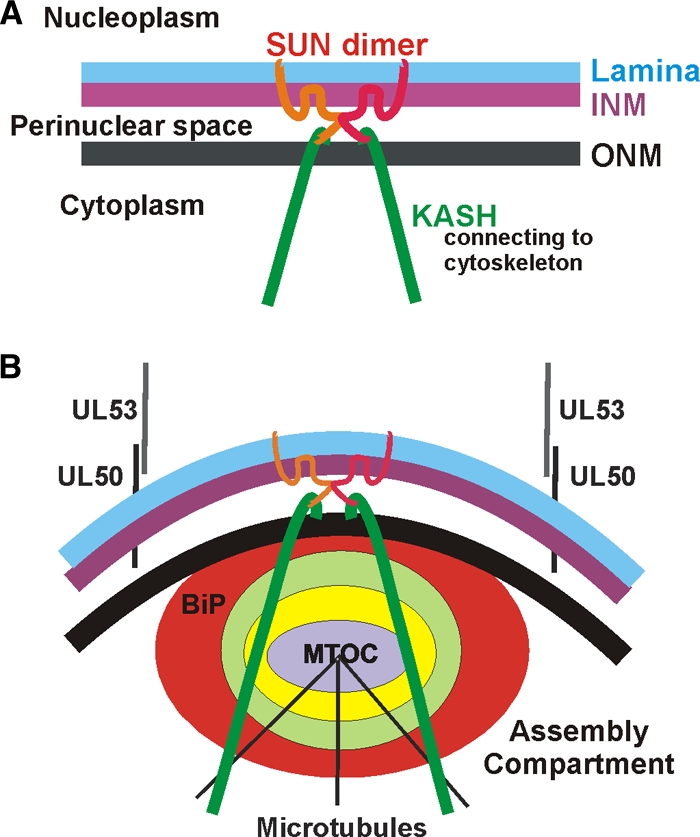
Models of the nuclear periphery in normal human fibroblasts (A) and HCMV-infected human fibroblasts (B). See the introduction for details.
We have shown previously that during HCMV infection some of the endoplasmic reticulum (ER) chaperone BiP (immunoglobulin binding protein; also known as glucose-regulated protein 78) is relocated from the lumen of the ER to a novel position within the assembly compartment (Fig. 1B) (5). Our studies have shown that depletion of BiP from infected cells (i) disrupts the assembly compartment and the formation of the concave nucleus, (ii) clears all viral cytopathic effects normally seen in the cytoplasm, and (iii) prevents the formation of infectious virions (5, 6). Thus, BiP plays an important role in maintaining the assembly compartment and in the assembly-egress continuum.
In addition to BiP, evidence suggests that the assembly compartment is derived from the trans-Golgi network (TGN) and early endosomes. However, defining the exact origin of this compartment has been complicated. While specific organellar markers can be detected in and around the assembly compartment, other markers of the same organelles are not (11, 12, 20). This suggests that the virus recruits and relocates specific cellular factors to form the assembly compartment. Numerous viral proteins have been identified as part of this structure, for example, tegument proteins (pp28 and pp65) (38) and viral glycoproteins (gB, gH, gL, gO, and gp65) (11, 38, 44). A rigorous study of assembly compartment structure (12) resulted in a three-dimensional model proposing that the assembly compartment is cylindrical and composed of organelle-specific vesicles (Golgi complex, TGN, and early endosomes) which form nested cylinders, making ordered layers of the assembly compartment (represented by the concentric colored rings in Fig. 1B). Each layer is proposed to contain a specific set of tegument proteins which are transferred to nucleocapsids as they move toward the center of the assembly compartment (12). In agreement with this model, ultrastructural studies suggest that full tegumentation requires the nucleocapsids to migrate through circularly arrayed vesicles (33, 41, 42).
These observations of nuclear structure and assembly compartment formation/function must be considered in the light of the current model for HCMV assembly and egress. HCMV nucleocapsids are formed in the nucleus. While the precise mechanism of egress to the cytoplasm remains speculative, it is proposed that nucleocapsids acquire an envelope by budding through the inner nuclear membrane (INM). It has been suggested that this occurs at infoldings of the nuclear membrane (7, 16, 29, 41, 42). The nucleocapsids enveloped in the INM then move into the lumen of the nuclear envelope (the perinuclear space) where the envelope fuses with the outer nuclear membrane (ONM), releasing naked nucleocapsids into the cytoplasm (16, 33, 41, 42). Nucleocapsids then become mature virions by gaining their full complement of tegument and envelope in the cytoplasm. It is believed that nucleocapsids are actively transported through these processes, particularly through the complexity of the assembly compartment (12). Such movement may be mediated via attachment to microtubule-associated motors. In this regard, the assembly compartment forms at a microtubule organizing center (MTOC; Fig. 1B) (38). Additionally, a role for the molecular motor dynein in locating HCMV protein pp150 to the assembly compartment has recently been described (21).
The above observations and models indicate extensive interplay between the nucleus, the nuclear envelope, the assembly compartment, molecular motors, and the cytoskeleton. The steps in the egress process, from the nucleus to the assembly compartment and on to the cytoplasm for envelopment and cellular egress (reviewed in reference 15), suggest a highly orchestrated, interdependent assembly-egress continuum. Our previous studies suggest that BiP, in its novel assembly compartment location, plays a central role in directing this coordinated effort (5, 6).
Each step in the assembly-egress continuum is accompanied by significant alterations in nuclear and cytoplasmic architecture. For example, a significant barrier to nucleocapsid access to the nuclear envelope is the nuclear lamina, a network of predominantly insoluble cellular proteins located on the nucleoplasmic side of the inner nuclear membrane (Fig. 1A). The lamina provides (i) structural support for the nuclear membrane, (ii) attachment sites for chromatin, (iii) mediation of nuclear assembly following mitosis, and (iv) facilitation of DNA replication and transcription (17, 43). During mitosis the nuclear lamina disassembles and reassembles following metaphase. The precise mechanism of nuclear lamina rearrangement has not been fully delineated, but it has been suggested that the mitosis-promoting kinase p34cdc2 and protein kinase C (PKC) are involved in phosphorylating lamins, resulting in their disassembly (13, 14). It has been proposed that a similar mechanism is used in infected cells to rearrange the lamina to allow nucleocapsid nuclear egress (18, 24, 25, 39). A virus-specific nuclear egress complex has been proposed which includes the HCMV viral proteins UL50 and UL53, the viral kinase UL97, and the cellular proteins p32 and protein kinase C (26). A similar egress function has been described for the homologs of UL50 and UL53 in herpes simplex virus type 1 (HSV-1) (30, 36) and in mouse CMV (28). UL50 is anchored in the INM (Fig. 1B) and believed to associate with the nuclear lamina (25). UL53 binds UL50, and it is proposed that these proteins attract the protein kinases which phosphorylate the lamins; this modification appears to be necessary for lamina rearrangement that occurs during infection (8, 24, 25). Data presented here suggest that lamina rearrangement is facilitated by BiP localization in the assembly compartment (Fig. 1B).
Additional nuclear membrane-localized proteins that may affect nucleocapsid egress and virion formation include the Sad1/UNC-84 homology (SUN) and Klarsicht, Anc-1, Syne homology (KASH) domain-containing proteins (Fig. 1A). SUN domain proteins form homo- and heterodimers; the N termini cross the inner nuclear membrane and anchor in the nuclear lamina. The C-terminal domains of the SUN proteins are located in the perinuclear space where they interact with the KASH domains of nesprin family proteins (reviewed in references 46 and 47), which are anchored in the outer nuclear membrane (Fig. 1A). The association of SUN and KASH domain proteins in the perinuclear space is believed to keep the inner and outer nuclear membranes tethered and close together, thus limiting the size of the perinuclear space. When SUN domain proteins are depleted, the space between the INM and ONM increases (9).
The very large nesprin (KASH domain) proteins span the outer nuclear membrane and extend well into the cytoplasm, where their N termini interact with the cytoskeleton via direct interactions with microtubules and actin, or through adaptor proteins, such as plectin (Fig. 1A and B) (reviewed in reference 47). In the context of HCMV infection, it is interesting that the nesprins bind to both the microtubule organizing center (MTOC) and the Golgi complex (3, 47). Thus, it is not difficult to imagine a connection with the assembly compartment and a role for these proteins in altering nuclear morphology, positioning the assembly compartment, and providing connection between the nucleus, cytoskeleton, and the plasma membrane for the facilitation of virion egress.
In the studies presented we show that during infection the nuclear periphery is dramatically altered, especially near the assembly compartment, where the nuclear lamina is rearranged, the outer nuclear membrane is disrupted, and the nucleus becomes permeable to large molecules. In addition, the tethering of the inner and outer nuclear membranes is lost during infection due to the decrease in SUN domain protein levels. As mentioned above, we previously demonstrated that BiP functions as a component of the assembly compartment and that disruption of BiP causes the loss of assembly compartment integrity (5). In this study, we show that the depletion of BiP and the loss of assembly compartment integrity result in the loss of virus-induced lamina rearrangement and the virus-specific nuclear morphology. BiP's role in lamina rearrangement is, at least in part, due to BiP's effects on phosphorylation of the lamins. Depletion of BiP after it has localized to the assembly compartment causes the loss of lamin phosphorylation. The dependency of lamin phosphorylation on BiP correlates with an interaction between BiP and UL50. Finally, we confirm previous data (21) suggesting an involvement of dynein in assembly compartment formation and extend this observation, showing that the inhibition of dynein's function results in the loss of the nuclear morphology characteristic of HCMV infection. Models are discussed integrating these observations in the assembly-egress continuum.
MATERIALS AND METHODS
Tissue culture, reagents, plasmids, and primary antibodies.
Low-passage-number life-extended human foreskin fibroblasts (HFFs) (4) were cultured at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (DMEM10), 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM GlutaMax. SubAB toxin and its nontoxic derivative SubAA272B (31) were generously provided by Adrienne and James Paton (University of Adelaide) and added at 100 ng/ml to cultured cells. The CC1-mCherry expression plasmid, which expresses the coiled-coil domain 1 (CC1, amino acids [aa] 217 to 548) of p150Glued, was a gift from Erika Holzbaur (University of Pennsylvania).
Primary antibodies against lamin B (C-20), pp28 (5C3), lamin A/C (N-18), and UL44 (CH13) were purchased from Santa Cruz. The antibody that recognizes the common exons 2 and 3 of the HCMV major immediate-early proteins (MIEPs) has been previously described (19). The antibody that recognizes phosphorylated lamin A/C on serine 22 was purchased from Cell Signaling. Antibodies against SUN1 and SUN2 were from Sigma-Aldrich. The BiP antibody used in Western analysis was from BD Biosciences. For immunoprecipitation analysis, the BiP and KDEL antibodies were purchased from Abcam and MBL, respectively. Anti-pp65 and gB antibodies, used for both Western and immunoprecipitation analyses, were purchased from US Biological and Abcam, respectively. The antibody that recognizes UL50 was produced by Open Biosystems (Huntsville, AL) in rabbits using a peptide containing UL50 amino acids 152 to 170 (GPENEGEYENLLRELYAKK); the antibody was affinity purified using the same peptide.
Virus preparation, titration, and infections.
Virus stocks (Towne strain) were prepared as previously described (23), and titers were determined using the 50% tissue culture infective dose method. All experiments were performed using a multiplicity of infection of 3.
Indirect immunofluorescence (IF).
Coverslips containing either mock- or HCMV-infected human foreskin fibroblasts were washed in phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde at room temperature. Cells were permeabilized in PBS containing 0.5% Triton X-100 and blocked in PBS containing 5% human serum and 0.5% Tween 20. Primary and secondary antibodies conjugated with Alexa Fluor 594 and 647 (Invitrogen) were diluted in blocking buffer. Coverslips were washed in PBS, rinsed in H2O, and mounted on slides using Vectashield (Vector Laboratories) mounting medium containing 4′,6′-diamidino-2-phenylindole (DAPI). Slides were examined by wide-field fluorescence using a Nikon Eclipse E600 microscope or by confocal microscopy using a Leica DM 6000 confocal microscope. Micrographs were acquired, processed by deconvolution, and analyzed using Image-Pro 6.3 and Autoquant X2 software (MediaCybernetics) or Leica confocal software (Leica Microsystems).
Western analysis.
Western blot samples were added to 3× loading solution (187.5 mM Tris-HCl [pH 6.8], 6% sodium dodecyl sulfate, 30% glycerol, 0.3% bromophenol blue, 467 mM β-mercaptoethanol) and boiled for 5 min. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (8 or 10% gels). Following electrophoresis, the gels were transferred to nitrocellulose in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) under a constant current. Membranes were blocked in Tris-buffered saline containing 1% Tween 20 (TBST) plus 5% nonfat dry milk. Primary antibodies were diluted in TBST containing 2% bovine serum albumin (BSA); horseradish peroxidase-conjugated secondary antibodies (Thermo) were diluted in TBST containing 5% milk. Membranes were washed in TBST and developed with Lumi-Light Western blotting substrate (Roche).
EM.
Following infection and treatment of cells, HFFs were washed with phosphate-buffered saline and fixed with electron microscopy (EM) fixative (2.5% glutaraldehyde, 2% paraformaldehyde in sodium cacodylate). Cells were prepared for EM by the Penn Biomedical Imaging Facility. Briefly, cells were pelleted in Eppendorf tubes and postfixed with 1% aqueous OsO4 for 1 h. The cell pellets were dehydrated with ethanol and propylene oxide, embedded in epoxy resin, and polymerized at 65°C for 28 h. Ultrathin (∼80-nm-thick) sections were cut with a diamond knife, mounted on single-slot grids, stained with uranyl acetate and bismuth, and examined with an FEI Tecnai T12 transmission electron microscope.
Immunoprecipitation.
Mock- and HCMV-infected cell lysates were prepared at 96 hours postinfection (hpi) by being harvested in 1% Triton buffer (20 mM HEPES [pH 7.5], 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM EDTA, 10 mM tetrasodium pyrophosphate, 100 mM sodium fluoride, 17.5 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 1.25 μg/ml pepstatin, 8.5 μg/ml aprotinin). Immunoprecipitation reactions were performed using Dynabeads Protein G (Invitrogen) according to the manufacturer's instructions. One hundred micrograms of either mock- or HCMV-infected lysates and 20 μl of Dynabeads were used for each reaction. Antibody amounts varied according to the manufacturer's recommendations. After immunoprecipitation, protein was removed from the beads using 20 μl of 1× loading dye containing 2× β-mercaptoethanol. Samples were then prepared for Western analysis as described above.
Dextran staining.
Cells which had been mock or HCMV infected for 48 h were loaded with tetramethyl rhodamine isocyanate (TRITC)-labeled dextran (average molecular weight [MW], 155,000; Sigma-Aldrich) using a modification of a reported protocol (45). Briefly, cells were removed from the plate by trypsinization and pelleted at 1,200 rpm (250 × g) for 5 min at 4°C. The cell pellet was washed once in PBS and once in hypertonic buffer (0.5 M sucrose, 10% polyethylene glycol 1000, 20 mM HEPES, pH 7.2). The cells were then incubated for 10 min at 37°C in hypertonic buffer containing 10 mg/ml TRITC-dextran. After addition of 80 volumes of DMEM10, the cells were incubated for an additional 10 min at 37°C. The cells were twice pelleted and washed in DMEM10 and then plated on coverslips. At 72 and 96 hpi, live cells were imaged for TRITC immunofluorescence.
RESULTS
The nuclear periphery is altered in HCMV-infected cells.
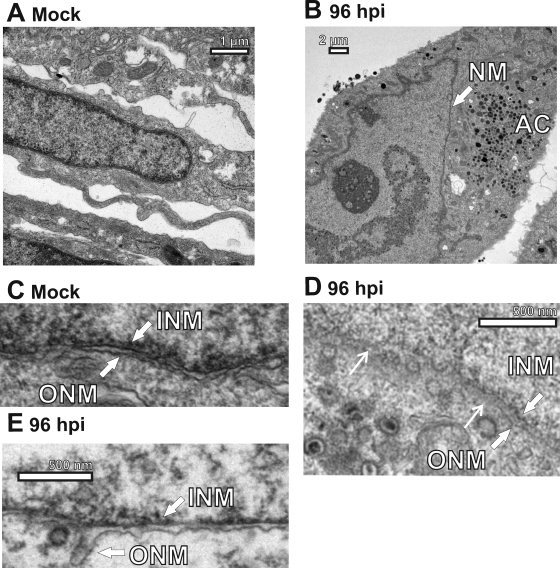
As described in the introduction, there are many alterations to the nuclear periphery during HCMV infection. Figure 2 A and B show electron micrographs of mock- and HCMV-infected (96 hpi) cells, respectively. The nucleus of the mock-infected cell shows a relatively uniform nuclear periphery with the nuclear membrane backed by a darker region that is likely to represent heterochromatin. The periphery of the infected cell nucleus is more varied (Fig. 2B). In the region adjacent to the assembly compartment, it is much thinner (arrow), appearing to be only nuclear membrane. Other areas of the infected cell nuclear periphery, not adjacent to the assembly compartment, appear thicker. These data suggest that the nuclear periphery adjacent to the assembly compartment is significantly altered; it is likely that this is due to the removal of heterochromatin and rearrangement of the lamina (7, 18, 24, 25, 39). Such thinning of the periphery in this region would aid in nucleocapsid egress to the assembly compartment.
FIG. 2.
Electron micrographs of mock- or HCMV-infected human fibroblasts. (A) A mock-infected cell highlighting the nuclear periphery. (B) An infected cell (96 hpi) highlighting the thin nuclear membrane (NM; arrow) adjacent to the assembly compartment (AC). (C) A mock-infected cell highlighting the inner and outer nuclear membranes (INM and ONM, respectively). (D) An infected cell (96 hpi) highlighting the INM and ONM, showing disruptions in the ONM, e.g., between the two thin arrows. (E) An infected cell (96 hpi) showing the irregularity of the separation between the inner and outer nuclear membranes, indicative of the loss of SUN domain proteins.
Additional examination of the nuclear envelope of mock- and HCMV-infected cells shows a number of other morphologies that are specific to infected cell nuclei. Figure 2C shows the nuclear envelope of a mock-infected cell: the inner nuclear membrane and the outer nuclear membrane are distinct (INM and ONM arrows, respectively), and the perinuclear space between them is relatively uniform. Figure 2D shows the nuclear envelope of an infected cell: inner and outer nuclear membranes can be seen on the right side of the micrograph (thick white arrows), but toward the left the ONM becomes indistinct and disrupted (between the two thin white arrows) with the appearance of vesicles as well as nucleocapsids that had recently exited the nucleus.
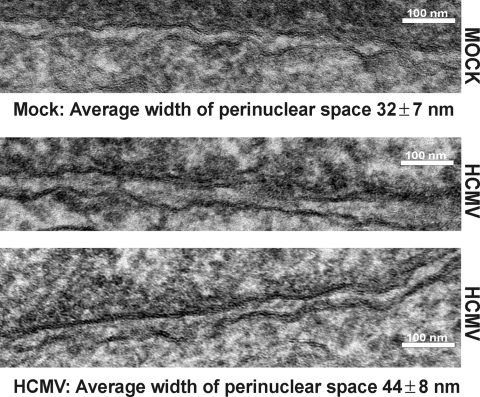
Figure 2E shows another morphology specific to infected cell nuclei: the perinuclear space is quite variable in size, i.e., the distance between the INM and the ONM is not uniform and there are occasional large bulges of the ONM (arrow). To document the increase in the perinuclear space, we measured the distance between the inner and outer nuclear membranes in mock- and HCMV-infected samples. We chose regions of the nuclei where the INM and ONM could be readily detected for measurement. Figure 3 shows the electron micrographs of the nuclear membranes measured using ImagePro 6.3 (MediaCybernetics) software. We made 30 measurements between the membranes in each micrograph shown and found that indeed the space increased from 32 ± 7 nm in mock-infected cells to 44 ± 8 nm in infected cells. These measurements are representative of measurements taken from multiple fields of mock- and HCVM-infected cells. However, our measurements of infected cells are likely to represent a minimal level of INM/ONM separation since they were made in regions where distinct inner and outer nuclear membranes could be detected, i.e., in regions where there was minimal viral egress activity.
FIG. 3.
The distance between the inner and outer nuclear membranes is increased in infected cells. Mock- or HCMV-infected HFFs were prepared for EM analysis at 96 hpi. The two HCMV samples are from similar regions of two different infected nuclei. Thirty measurements of the perinuclear space were digitally taken on the micrographs shown using ImagePro 6.3 software.
SUN domain proteins decrease during HCMV infection, accounting for separation of the inner and outer nuclear membranes.
The increase in space between the INM and ONM and the detection of bulges in the outer nuclear membrane have been reported in normal cells when the SUN domain proteins were depleted (9). In Fig. 4 A and B we show that both SUN1 and SUN2 levels are significantly decreased beginning at 60 hpi for SUN1 and at 36 hpi for SUN2. This loss of SUN proteins can account for the separation of the outer and inner nuclear membranes and the increase in size of the perinuclear space. Such separation of the two nuclear membranes would allow nucleocapsids the ability to push the two membranes apart as they move into the perinuclear space during nuclear egress (Fig. 4C).
FIG. 4.
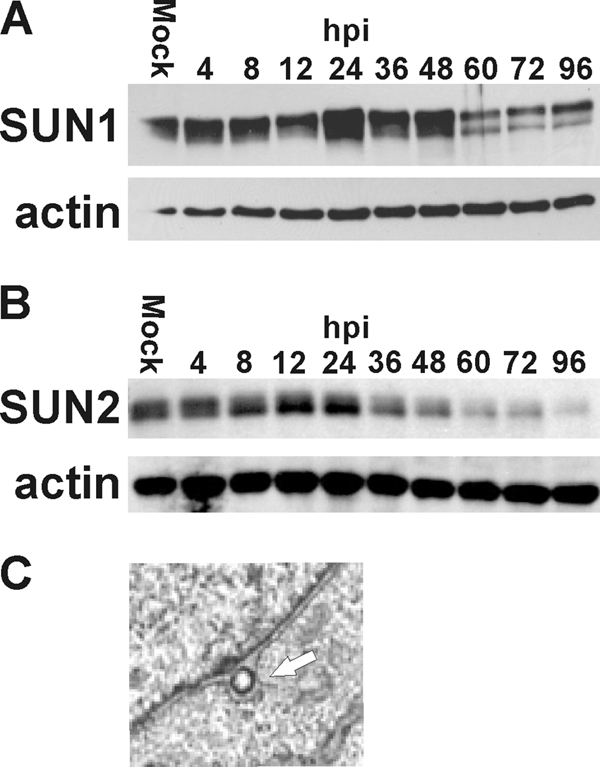
The levels of SUN proteins are reduced during HCMV infection. (A and B) Proteins were harvested from mock- and HCMV-infected life-extended (LE) HFFs at the indicated hours postinfection (hpi) and assessed by Western analysis using antibodies that detect SUN1 (A) and SUN2 (B). Actin was used as a loading control. (C) A nucleocapsid in the perinuclear space causing distension of the outer nuclear membrane (white arrow).
The data in Fig. 2, 3, and 4 suggest that the nuclear periphery and nuclear envelope are significantly altered during infection and that these alterations vary from position to position around the nucleus. The most significant alterations appear at regions adjacent to the assembly compartment, where active viral egress is occurring. One aspect of nuclear membrane alterations that was not detected here was large nuclear membrane infoldings that have been reported to penetrate deep into the infected cell nuclei, putatively providing nucleocapsids access to the membrane for egress (7, 16, 29, 41, 42).
Nuclear lamina rearrangement is dependent on the presence of BiP in the assembly compartment.
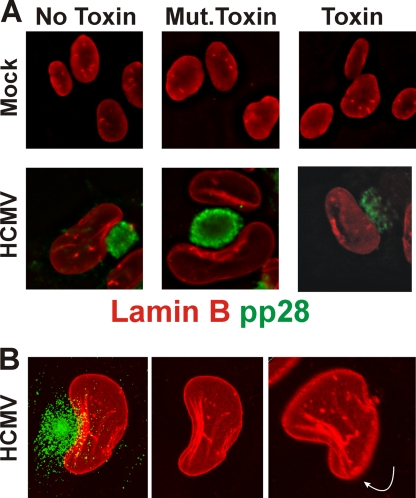
In addition to alterations in the nuclear periphery described above, previous studies have described nuclear lamina alteration and reorganization during HCMV infection of human fibroblasts (7, 8, 18, 24-26, 39). In our previous studies of BiP and the assembly compartment, we have used the SubAB cytotoxin, which specifically cleaves and inactivates BiP within 1 h of addition (31, 32). Using the toxin, we have shown that BiP depletion caused assembly compartment disruption and the cessation of cytoplasmic viral activity in the infected cells (5, 6). In Fig. 5 A (top panels), we show the lamina staining (lamin B, red) in mock-infected cells that were either untreated (No Toxin), treated for 12 h with a mutant form of the toxin that cannot cleave BiP (Mut.Toxin), or treated for 12 h with the toxin that cleaves BiP. In all cases the lamina staining remains the same. In the bottom panels of Fig. 5A we show the same experiment done in infected cells at 96 hpi. In these experiments we visualized the assembly compartment using anti-pp28 (green). In both the untreated cells and the mutant toxin-treated cells, we see that the lamina is reorganized, especially in the region of the nucleus next to the assembly compartment; a similar type of rearrangement has been previously observed (8). Treatment of the cells with toxin for 12 h (84 to 96 hpi) caused not only the previously characterized disruption of the assembly compartment but also the disruption of the virally induced lamina rearrangement. These data suggest that the presence of BiP and the integrity of the assembly compartment are needed to maintain the virally induced lamina rearrangement.
FIG. 5.
Nuclear lamina rearrangement is disrupted when BiP is depleted. (A) Mock- or HCMV-infected human fibroblasts were treated with SubAB (Toxin) or SubAA272B (Mut.Toxin) or left untreated (No Toxin) at 84 hpi and prepared for immunofluorescence analysis at 96 hpi by staining with antibodies that detect pp28 (green) and lamin B (red). (B) HCMV-infected human fibroblasts were stained as in panel A at 96 hpi and examined by confocal microscopy. Maximum projections of a z-axis series are shown; the far right panel was enlarged slightly and rotated as indicated by the white arrow.
In Fig. 5B we further examined the rearrangement of the lamina adjacent to the assembly compartment. The confocal micrograph is a maximum projection z-axis stack showing the assembly compartment (green) and the nucleus as visualized by anti-lamin B (red). It is again seen that the most significant reorganization of the lamina occurs adjacent to the assembly compartment, where layers of lamina appear which are set back from the nuclear periphery. This is clearly seen in the middle panel, where the green channel was removed. The panel on the right shows the maximum projection slightly enlarged and rotated forward and to the left, displaying the lower side of the nucleus and a second view of the lamina layers, emphasizing their position set back from the nuclear periphery. The data in Fig. 5 were generated using anti-lamin B because it provided very good staining, better than that by anti-lamin A and C; however, similar rearrangements are seen using anti-lamin A and C (not shown).
HCMV-induced lamin phosphorylation is lost upon depletion of BiP and disruption of the assembly compartment.
The above data suggest that BiP depletion and the disruption of the assembly compartment resulted in the loss of HCMV-induced lamina rearrangements. As described in the introduction, UL50 is thought to be integral to lamin phosphorylation, which is necessary for the rearrangement needed for nucleocapsid egress (8, 18, 26, 39). Thus, we examined whether the depletion of BiP and the disruption of the assembly compartment affected lamin phosphorylation. Figure 6 A shows lamin A and C phosphorylation on serine 22 (S22) during an infection time course; phosphorylation increases very early in infection, 4 hpi; reaches a peak at 12 to 48 hpi; and then decreases modestly after 48 hpi but remains markedly higher than that in mock-infected cells at 96 hpi. Total lamin A and C levels are not altered by infection. In Fig. 6B we show the phosphorylation of S22 of lamins A and C in mock-infected cells and in HCMV-infected cells at 24 and 96 hpi. In each case cultures are left untreated or treated with (i) NGIC-1, an inhibitor of the HCMV UL97 kinase, and protein kinase C (PKC), which have both been reported to be involved in lamin phosphorylation, or (ii) with the SubAB toxin, which cleaves BiP. In untreated samples (lanes 1, 4, and 7) we see that lamin A and C phosphorylation is increased in infected cells by 24 hpi and remains elevated at 96 hpi. Treatment with NGIC-1 from 2 hpi to 24 hpi (lane 5) or from 72 to 96 hpi (lane 8) inhibited detectable S22 phosphorylation. Treatment with the toxin from 2 to 24 hpi (lane 6) depleted BiP to below detectable levels but did not affect S22 phosphorylation. However, toxin treatment between 72 and 96 hpi (lane 9) did inhibit HCMV-induced S22 phosphorylation. In all cases total lamin A and C levels were not significantly altered. In addition, HCMV UL50 levels were not altered, nor were the levels of the major immediate-early proteins (MIEPs) and the early protein p52. Interestingly, the levels of the late protein, pp28, did decrease with BiP depletion. We have previously shown that BiP and pp28 interact in the assembly compartment (5).
FIG. 6.
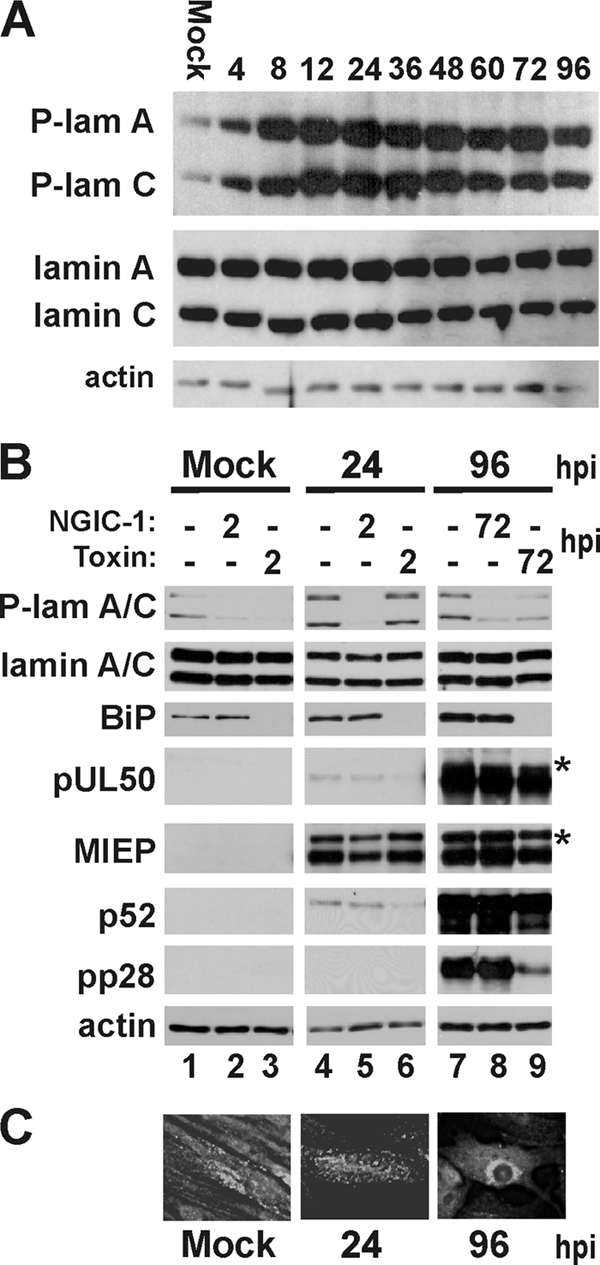
Depletion of BiP alters nuclear lamin phosphorylation. (A) Proteins were harvested from mock- and HCMV-infected life-extended (LE) HFFs at the indicated times (hours) postinfection and analyzed by Western blot analysis using antibodies that detect lamins A and C phosphorylated at serine 22 (P-lam A and P-lam C) and total lamins A and C. Actin was used as a loading control. (B) Proteins were harvested from mock- and HCMV-infected samples treated with SubAB (Toxin) or NGIC-1 and analyzed by Western analysis using antibodies that detect lamins A and C phosphorylated at serine 22 (P-lam A/C), total lamins A and C, BiP, and pUL50. Antibodies directed against the major immediate-early proteins (MIEP); an early protein, p52; and a late protein, pp28, were used to monitor viral protein expression. Actin was used as a loading control. The asterisks indicate that, for the 96-hpi pUL50 and MIEP samples, exposures are shown that are less than that for the corresponding 24-hpi samples; this is because equal exposure time resulted in uninterpretable overexposure for these samples. (C) Immunofluorescence analysis of BiP after mock infection and at 24 and 96 hpi.
These data suggest that the mechanism of virus-induced phosphorylation of lamins A and C is different at early and late times after infection. Lamin A/C phosphorylation is resistant to BiP depletion at early times but is sensitive to BiP depletion at late times. This difference correlates with BiP localization to the assembly compartment. As shown in Fig. 6C, at 24 hpi immunofluorescent detection shows that BiP has not yet relocalized significantly compared to that in mock-infected cells. However, by 96 hpi BiP has localized to the assembly compartment. Thus, lamina phosphorylation at late times correlates with BiP localization to the assembly compartment. Lamin B phosphorylation on S16 could not be measured due to the lack of an adequate antibody.
BiP interacts with HCMV UL50.
We previously showed that during infection there is a striking increase in BiP levels and it is relocalized to two distinct cytoplasmic localizations. One pool is located in condensed ER structures in the periphery of the cytoplasm which contain other ER markers. However, a second pool is relocalized from the lumen of the ER to the assembly compartment, where it is present in the absence of other ER markers (5, 6). This pattern of BiP localization can be detected because antibodies to different BiP epitopes differentially detect BiP in infected cells (5). Antibodies to the C terminus of BiP cannot detect or immunoprecipitate BiP localized in the assembly compartment. This is especially true for antibodies to the KDEL ER localization signal at the very C terminus of the protein. These data suggest that C-terminal epitopes are blocked when BiP is in the assembly compartment. Such blockage of the KDEL ER localization signal by a viral protein would explain how BiP can be diverted from its normal ER localization to the assembly compartment. However, assembly compartment-localized BiP can be specifically detected and precipitated using antibodies to the N terminus of BiP, which rather specifically detect BiP in the assembly compartment in infected cells (for example, Fig. 6C at 96 hpi) (5).
We used one such N-terminal antibody (Santa Cruz; GRP78 N-20) to immunoprecipitate BiP and analyze the HCMV proteins that coprecipitated, since they should represent viral proteins that are primarily associated with BiP in the assembly compartment. Two separate immunoprecipitations were performed; in each case we had mass spectrometric analysis performed on Coomassie blue-stained bands that appeared in both BiP immunoprecipitates from infected cell extracts and not in immunoprecipitates from mock-infected cell extracts.
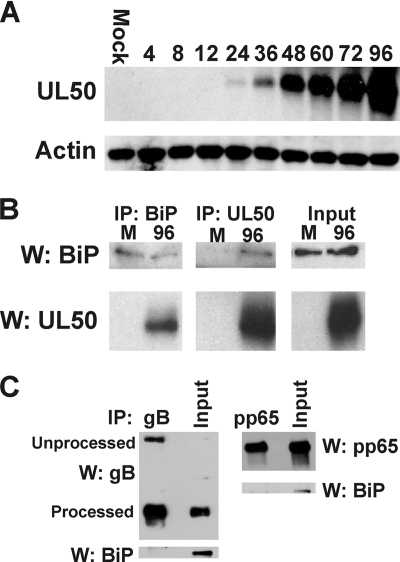
Two of the proteins identified were TRS1 and UL99 (pp28), which we previously identified as BiP-interacting proteins (5). An additional HCMV protein identified was UL50, which, as described above, is directly involved with lamina phosphorylation and reorganization. In order to study UL50 directly, we had a peptide antibody prepared commercially as described in Materials and Methods. Figure 7 A shows that by Western analysis the antibody specifically detects a protein of the predicted size of UL50 (∼45 kDa) only in infected cells. During a time course the UL50 band appears weakly at 24 hpi and significantly by 48 hpi, suggesting that UL50 is expressed with early to delayed-early kinetics. The antibody was not suitable for immunofluorescence.
FIG. 7.
BiP interacts with nuclear egress factor pUL50. (A) Proteins were harvested from mock- and HCMV-infected human fibroblasts at the indicated times (hours) postinfection and analyzed by Western analysis using the anti-UL50 antibody described in the text. Actin was used as a loading control. (B) Mock (M)- and HCMV-infected lysates, harvested at 96 hpi, were subjected to immunoprecipitation (IP) using antibodies that detect BiP or UL50. The precipitates were assessed by Western (W) analysis using anti-BiP and anti-UL50. Input represents 20% of protein lysates used for IP-Western analyses. (C) Western analysis of BiP in immunoprecipitates of gB and pp65.
Reciprocal immunoprecipitation and Western analyses (Fig. 7B) showed that the anti-N-terminal BiP antibody (specific for BiP in the assembly compartment) coprecipitates UL50 and that the anti-UL50 antibody coprecipitates BiP. These data suggest a close association between BiP in the assembly compartment and UL50. Interpretation of immunoprecipitation data involving BiP is complicated by the fact that BiP is a chaperone that, by definition, interacts weakly with many proteins. To address the specificity of the UL50 interaction, we performed immunoprecipitations using antibodies against a very abundant viral tegument protein, pp65, and glycoprotein B (gB), which traverses the ER to be posttranslationally modified. BiP did not coprecipitate with either of these viral proteins (Fig. 7C). This is consistent with our previously published data that BiP and gB do not colocalize to the same area of the assembly compartment (5). These observations support the conclusion that the BiP-UL50 association is specific.
The nuclear membrane adjacent to the assembly compartment is modified such that the dextran can penetrate into the nucleus.
The above data suggest that the nuclear periphery and the nuclear lamina are significantly remodeled during HCMV infection. The lamina reorganization and phosphorylation are dependent on BiP in the assembly compartment. The interaction between BiP and UL50 suggests a mechanism for BiP's effect on lamin phosphorylation and reorganization. However, the nature of this interaction is not clear since BiP, in the assembly compartment, and UL50, in the inner nuclear membrane, are separated by the outer nuclear membranes. However, Fig. 2D suggests that specific regions of the ONM may be disrupted in infected cells. This would allow BiP in the assembly compartment to access UL50, which is anchored in the inner nuclear membrane.
The changes in the nuclear periphery discussed above, especially the possibility of local disruption of the ONM, suggested that the permeability of the nucleus to large molecules may be altered during infection. To study nuclear permeability, dextran labeled with TRITC was introduced into mock- or HCMV-infected cells and analyzed by immunofluorescence microscopy. Dextran is a complex, branched polysaccharide, which can be used for a variety of biomedical applications, including monitoring the permeability of cellular structures. Permeation by dextran into cellular organelles is dependent on size. We used TRITC-dextran of 155 kDa, which is largely excluded from the nucleus of normal cells; this is shown in two examples of uninfected human fibroblasts in Fig. 8 A. Figure 8B shows four examples of infected cell nuclei: the two on the right are at 72 hpi and the two on the left are at 96 hpi. In all cases dextran is clearly seen in the nuclear region. The greatest accumulation is in the region of the nucleus next to the assembly compartment. The bottom left panel of Fig. 8B shows a very clear assembly compartment (AC) and a kidney-shaped nucleus to illustrate this.
FIG. 8.
High-molecular-weight dextran permeates infected cell nuclei. (A and B) Mock- (A) and HCMV-infected (B) human fibroblasts were loaded with dextran-TRITC (green) at 48 hpi as described in Materials and Methods. Cells were examined for the presence of nuclear dextran at 72 (B, right two panels) and 96 hpi (B, left two panels). The position of the assembly compartment (AC) is indicated in the lower left panel of B. (C) Using the cell in the lower left panel of B, a z-axis series was generated, and slices through the cell from top to bottom are shown. The DAPI staining of the nucleus is included (blue).
It is not clear from these two-dimensional pictures whether the dextran is within the nucleus or concentrated above or below it. To clarify this, we took a z-axis series through the cell discussed above which showed the very clear assembly compartment. Figure 8C shows four slices moving through the cell from top to bottom (left to right). In all cases the dextran is in the nucleus, not just at the top or the bottom. These data suggest that in infected cells the nuclear membrane adjacent to assembly compartment is modified such that the dextran can penetrate deep into the nucleus.
Dynein is necessary for assembly compartment formation and the alterations in nuclear shape during infection.
Recent evidence has shown that the HCMV tegument protein pp150 (ppUL32) plays a role in the cytoplasmic virion assembly and interacts with Bicaudal D1 (BicD1), a protein which is involved in trafficking within the secretory pathway and interacts with the dynein motor complex (21). These studies showed that depletion of BicD1 by short hairpin RNA (shRNA) caused decreased virus yield and defective localization of pp150 to the assembly compartment. These data also showed that a truncated BicD1 acts as a dominant negative and leads to disruption of the assembly compartment, a phenotype similar to that seen during overexpression of dynamitin. These data suggested that morphogenesis of the assembly compartment is dynein dependent (21).
Dynein is the predominant minus-end-directed microtubule motor in eukaryotic cells; it functions with a second complex, dynactin, which acts as an adaptor that allows dynein to bind its cargo. A projecting arm of dynactin, made up of a dimer of the p150Glued subunit, binds directly to dynein (22). Overexpression of p150Glued results in a wide range of motility defects. However, constructs have been made that express only the coiled-coil domain 1 (CC1, aa 217 to 548) of p150Glued. Overexpression of CC1 has far fewer adverse effects than does overexpression of p150Glued (35). Evidence suggests that CC1 binds dynein directly, acting as a competitive inhibitor for the interaction between dynein and intact dynactin; thus, dynein cannot load its cargo (35).
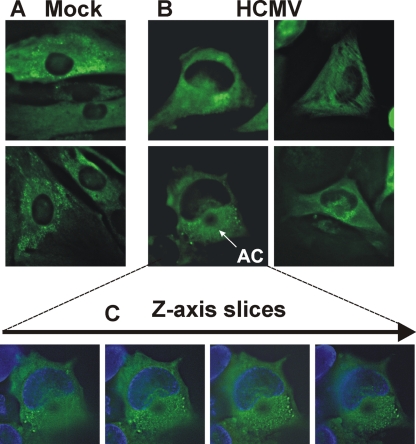
In order to further assess dynein's involvement in assembly compartment formation and nuclear structure, we infected human fibroblasts for 24 h and then electroporated a plasmid that expresses CC1 fused to mCherry. At 72 hpi (48 h postelectroporation) the cells were examined by immunofluorescence (IF) for CC1 expression (mCherry, red), pp28 (green), and nuclei (blue). Figure 9 shows a field with cells under three different conditions: (i) an uninfected cell (no green or yellow pp28) expressing CC1 (indicated by mCherry, red) that has a normal small round nucleus (blue), (ii) an HCMV-infected cell which has not been transfected (i.e., no mCherry and therefore not expressing CC1) and which shows pp28 in the assembly compartment (green) sharing two enlarged and somewhat kidney-shaped nuclei, and (iii) an infected cell with two nuclei expressing CC1 (red) and pp28 (yellow due to the overlap with CC1); the pp28 is in dispersed speckles through the cytoplasm, and there is no discernible assembly compartment. The disruption of viral assembly compartment was observed in all HCMV-infected cells expressing the CC1 inhibitor; however, the degree of disruption depended on the level of CC1 expressed. Additionally, in the infected cells expressing CC1 the nuclei are not enlarged or kidney shaped. These data agree with the previous data suggesting the importance of dynein in assembly compartment formation (21) and extend these observations to show that virus-induced alterations in nuclear size and shape correlate with dynein function and assembly compartment formation.
FIG. 9.
Inhibition of dynein causes disruption of assembly compartment formation and loss of HCMV-mediated nuclear shape alterations. Human fibroblasts were infected for 24 h and then electroporated with a plasmid that expresses CC1-mCherry. The cells were plated on coverslips and incubated for an additional 48 h (72 hpi) and then prepared for immunofluorescence. CC1-expressing cells are indicated by the mCherry (red). Anti-pp28 was used to stain infected cells and indicate the status of the assembly compartments (green or yellow where it overlaps CC1). The shape and size of the nuclei were visualized with DAPI. The arrows show an uninfected cell expressing CC1 (CC1 Uninfected), an infected cell expressing CC1 (CC1 Infected), and an infected cell not expressing CC1 (Infected).
DISCUSSION
The data presented suggest that BiP, SUN domain proteins, and dynein are necessary for the coordinated processes of remodeling the nucleus in conjunction with assembly compartment formation and placement. The manifestations of these processes include enlargement of the nucleus, formation of the kidney-shaped nuclei, alteration of the nuclear membrane, rearrangement of the nuclear lamina, and formation of the cytoplasmic assembly compartment. All of these processes appear to be highly interconnected as part of an assembly-egress continuum that provides the means for nucleocapsids to egress the nucleus, become fully tegumented, and move on into the cytoplasm for envelopment and cellular egress. The close association of the processes of the continuum is indicated by existing data and data presented here showing that disruption of any one aspect of the continuum disrupts the entire process. For example, BiP is integral to the formation and integrity of the assembly compartment and its depletion not only causes assembly compartment disruption but also disrupts nuclear remodeling, as well as the cytoplasmic phenotype of an HCMV infection (5, 6).
In the present study we show that depletion of BiP, and the accompanying disruption of the assembly compartment, also reverses the characteristic rearrangement of the nuclear lamina in infected cells. Under BiP-depleted conditions, not only is the nuclear lamina rearrangement lost but the nuclei regain a more normal shape and size. HCMV-mediated lamina rearrangement requires phosphorylation of the lamins in a manner that may mimic nuclear lamina rearrangement following metaphase. However, HCMV appears to modify this process through the utilization of a viral protein kinase, UL97, in a virus-specific nuclear egress complex which includes HCMV proteins UL50 and UL53 plus cellular proteins p32 and PKC (26). UL50 is anchored in the inner nuclear membrane and believed to associate with the nuclear lamina (25). UL53 binds UL50, and it is proposed that these proteins attract the kinases which phosphorylate the lamins, resulting in lamina rearrangement (8, 24, 25).
The very early (4-hpi) phosphorylation of serine 22 on lamins A and C was somewhat surprising considering that the expression of UL50 occurs during the early phase of infection (Fig. 7A). Interestingly, UL50, UL53, and UL97 have all been detected in virions (46a) and may function upon viral entry to phosphorylate lamins A and C. This very early phosphorylation is not dependent on BiP and is not sufficient to rearrange the lamins in the manner seen later in infection when BiP is required and the assembly compartment is forming. These data suggest that BiP and the integrity of the assembly compartment positively affect lamin phosphorylation and rearrangement. These events correlate with an interaction between BiP and UL50; thus, the effects of BiP on lamin phosphorylation and lamina rearrangement may be due to a direct effect on the function of the UL50-containing nuclear egress complex.
An interaction between BiP in the assembly compartment and UL50 anchored in the inner nuclear membrane is complicated by the outer nuclear membrane lying between the two. However, our data suggest several alterations of the nuclear membranes that may facilitate such an interaction. First, our EM data indicate that the nuclear periphery near the assembly compartment is very thin compared to other regions of infected nuclei and compared to mock-infected nuclei. In addition, the data suggest that some regions of the outer nuclear membrane are disrupted during infection. Such changes would favor increased access of BiP in the assembly compartment to UL50 in the inner nuclear membrane.
The findings that the nuclear membrane in the region of the assembly compartment is significantly altered, and potentially disrupted, are supported by our observation that 150,000-molecular-weight (MW) dextran, loaded into mock-infected and infected cells, appears inside the nuclei of infected cells, whereas it does not enter nuclei of uninfected cells. The dextran detected in infected cell nuclei was especially concentrated at sites adjacent to the assembly compartment. The data suggest that the nuclear membrane adjacent to the assembly compartment may be more penetrable to large molecules. Such penetrability also suggests a means for the BiP-UL50 interaction. The increase in dextran penetration occurs in areas of high viral activity, areas in which we have shown the nuclear periphery to be most altered. An increase in nuclear penetrability may result from the loss of nuclear membrane integrity as indicated by the disruption of the ONM. This may result from nucleocapsid budding or from alterations of nuclear pore complexes. We must also consider that some of the dextran that appears inside the nucleus may be located in deep invaginations that emanate from the region of the assembly compartment. Such invaginations have been suggested to serve as a means for nucleocapsid egress (7, 10, 16, 29, 41, 42). Overall, the data suggest that the nuclear membrane, especially in the region of the assembly compartment, is significantly altered and more permeable to large molecules, features that would support interactions between assembly compartment proteins and nuclear proteins (e.g., UL50).
In addition to the above alteration of the nuclear membrane, we have also determined that the distance between the inner and outer nuclear membranes increases in infected cells. This correlates with a decrease in the levels of SUN domain proteins during the course of an HCMV infection. The loss of SUN domain proteins has been shown to result in the separation of the inner and outer nuclear membranes (9). Specifically, dimers of SUN domain proteins, anchored in the inner nuclear membrane, interact in the perinuclear space with KASH domain proteins which are anchored in the outer nuclear membrane. This interaction tethers the inner and outer nuclear membranes and maintains a relatively uniform distance between the two (9). The loss of SUN domain proteins relieves the tethering and allows the inner and outer nuclear membranes to disassociate from one another (9) and function separately. One result of this is that large nucleocapsids in the process of nuclear egress can push the two membranes apart and fit between them, in the perinuclear space.
One of the most intriguing questions about the assembly-egress continuum is how the virus mediates the enlargement of the nucleus and its change to a kidney shape wrapped around the assembly compartment. Studies of nuclear envelope breakdown suggest that the cellular motor dynein attaches to the nuclear membrane and moves toward the centromere, generating tension on the nucleus and creating folds in the nuclear envelope (2, 37). Recent data have suggested that dynein is integral in assembly compartment formation (21); data presented here confirm these findings, showing that inhibition of dynein function causes the components of the assembly compartment to disperse throughout the cytoplasm. In these cells, the nuclear shape and size are normal, suggesting that dynein function is necessary for the virus-specific nuclear shape and enlargement. Thus, we propose that the virus uses a modified version of nuclear envelope breakdown (NEBD), in which dynein binds assembly compartment components and the nuclear membrane and moves toward the microtubule organizing center that will form the center of the assembly compartment. This pulling of the nuclear membrane toward the MTOC can account for the concave nuclear shape commonly seen forming around the assembly compartment and may promote tight association between the assembly compartment and the nucleus. During normal NEBD the stress of dynein's pulling results in tearing of the nuclear membrane and nuclear breakdown. In infected cells nuclear breakdown does not occur, but the nuclei do enlarge. A potential explanation for this is that nuclear enlargement is facilitated by the addition of new nuclear membrane to relieve the stress. This explanation is feasible given the recent observation of the dramatic increase in fatty acid synthesis for membrane formation that occurs in HCMV-infected cells (27). In addition, it is very likely that the dynein-mediated reformation of the nucleus and the rearrangement of the nuclear lamina are interrelated. In herpes simplex virus type 1 (HSV-1) an interaction between dynein and UL34, the HSV-1 homolog of UL50, has been reported (48). While an interaction of HCMV UL50 and dynein has yet to be reported, these observations suggest the intriguing possibility of an integrated mechanism involving BiP, UL50, and dynein in assembly compartment formation and nuclear restructuring.
Figure 10 presents a model of the nucleus and assembly compartment based on the data presented and the discussion above; it builds on the diagrams in Fig. 1. Based on the model of Das et al. (12), the assembly compartment is shown as concentric rings made up of specific organellar components and virion structural components, for example, tegument proteins. Our previous data suggest that BiP is in one of the outer rings, adjacent to the nucleus (5). The nuclear membrane near the assembly compartment is modified, with disruption of the outer nuclear membranes, increased permeability of the nuclear membrane, and separation of the inner and outer nuclear membranes due to the loss of the SUN domain proteins. All of these changes would increase the access of the proteins in the assembly compartment to nuclear proteins. We propose that this accounts for the ability of BiP and UL50 to interact. The model also shows the rearrangement of the nuclear lamina in the region of the assembly compartment. We have shown that this rearrangement, as well as the prerequisite phosphorylation of the lamins, requires BiP and an intact assembly compartment. The data suggest that an interaction between BiP and UL50 mediates signaling between the assembly compartment and the nucleus to direct lamina rearrangement adjacent to the assembly compartment. The model also shows the dynein-mediated formation of the assembly compartment and remodeling of nuclear shape. This appears to be facilitated by microtubules radiating from the microtubule organizing center within the assembly compartment. All of this coordinately forms the assembly-egress continuum, which provides a means for nucleocapsids to navigate through the lamina and access the nuclear membranes near the assembly compartment. As suggested by the model of Das et al. (12), the nucleocapsids move through the layers of the assembly compartment and emerge tegumented and possibly enveloped. A final aspect of the model pertains to the large KASH domain proteins, the nesprins. These proteins, which are in the ONM and bind to the microtubule organizing center, reach deep into the cytoplasm, where they interact with the cytoskeleton (reviewed in reference 47). The nesprins putatively provide the last step in the assembly-egress continuum, a connection between the assembly compartment and the plasma membranes upon which virions can be moved through the cytoplasm and on to cellular egress.
FIG. 10.
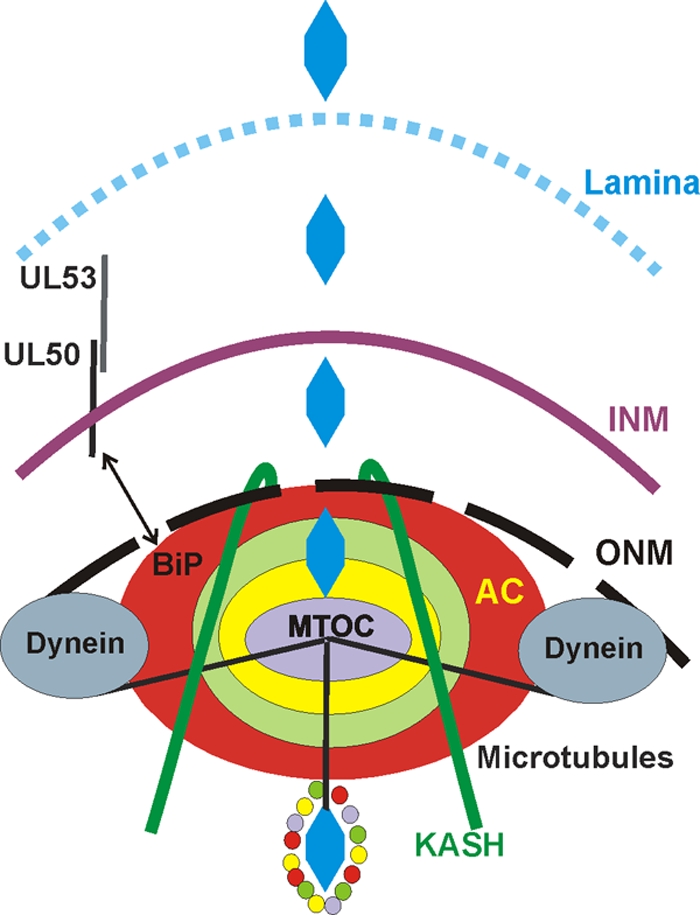
The assembly-egress continuum: a model of HCMV-mediated nuclear restructuring and assembly compartment formation/function as it relates to nucleocapsid egress and virion formation. The model is based on the data presented, previous models (12), and the Discussion. Refer also to Fig. 1, and see the text for details.
Acknowledgments
We thank the members of the Alwine lab, Sherri Adams, Alan Diehl, Brian Keith, and Erika Holzbaur for helpful discussions and advice during the course of these experiments. We thank Adrienne and James Paton for the SubAB toxin and its mutant form and Erika Holzbaur for the CC1-mCherry expression plasmid.
N.J.B. was supported by training grant T32 CA115299 awarded to Erle S. Robertson. This work was funded by NIH grant R01 CA028379-29 awarded to J.C.A.
Footnotes
Published ahead of print on 19 May 2010.
REFERENCES
- 1.Azzeh, M., A. Honigman, A. Taraboulos, A. Rouvinski, and D. G. Wolf. 2006. Structural changes in human cytomegalovirus cytoplasmic assembly sites in the absence of UL97 kinase activity. Virology 354:69-79. [DOI] [PubMed] [Google Scholar]
- 2.Beaudouin, J., D. Gerlich, N. Daigle, R. Eils, and J. Ellenberg. 2002. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108:83-96. [DOI] [PubMed] [Google Scholar]
- 3.Beck, K. A. 2005. Spectrins and the Golgi. Biochim. Biophys. Acta 1744:374-382. [DOI] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., G. E. Hultman, and T. Shenk. 2000. Replication of wild type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J. Virol. 74:10816-10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchkovich, N. J., T. G. Maguire, A. W. Paton, J. C. Paton, and J. C. Alwine. 2009. The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the HCMV assembly compartment. J. Virol. 83:11421-11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchkovich, N. J., T. G. Maguire, A. W. Paton, J. C. Paton, and J. C. Alwine. 2008. Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78 which is required for virion assembly. J. Virol. 82:31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buser, C., P. Walther, T. Mertens, and D. Michel. 2007. Cytomegalovirus primary envelopment occurs at large infoldings of the inner nuclear membrane. J. Virol. 81:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camozzi, D., S. Pignatelli, C. Valvo, G. Lattanzi, C. Capanni, P. Dal Monte, and M. P. Landini. 2008. Remodelling of the nuclear lamina during human cytomegalovirus infection: role of the viral proteins pUL50 and pUL53. J. Gen. Virol. 89:731-740. [DOI] [PubMed] [Google Scholar]
- 9.Crisp, M., Q. Liu, K. Roux, J. B. Rattner, C. Shanahan, B. Burke, P. H. Stahl, and D. Hodzic. 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dal Monte, P., S. Pignatelli, N. Zini, N. M. Maraldi, E. Perret, M. C. Prevost, and M. P. Landini. 2002. Analysis of intracellular and intraviral localization of the human cytomegalovirus UL53 protein. J. Gen. Virol. 83:1005-1012. [DOI] [PubMed] [Google Scholar]
- 11.Das, S., and P. E. Pellett. 2007. Members of the HCMV US12 family of predicted heptaspanning membrane proteins have unique intracellular distributions, including association with the cytoplasmic virion assembly complex. Virology 361:263-273. [DOI] [PubMed] [Google Scholar]
- 12.Das, S., A. Vasanji, and P. E. Pellett. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 81:11861-11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields, A. P., and L. J. Thompson. 1995. The regulation of mitotic nuclear envelope breakdown: a role for multiple lamin kinases. Prog. Cell Cycle Res. 1:271-286. [DOI] [PubMed] [Google Scholar]
- 14.Gant, T. M., and K. L. Wilson. 1997. Nuclear assembly. Annu. Rev. Cell Dev. Biol. 13:669-695. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, W. 2008. Structure and formation of the cytomegalovirus virion. Curr. Top. Microbiol. Immunol. 325:187-204. [DOI] [PubMed] [Google Scholar]
- 16.Gilloteaux, J., and M. R. Nassiri. 2000. Human bone marrow fibroblasts infected by cytomegalovirus: ultrastructural observations. J. Submicrosc. Cytol. Pathol. 32:17-45. [PubMed] [Google Scholar]
- 17.Gruenbaum, Y., K. L. Wilson, A. Harel, M. Goldberg, and M. Cohen. 2000. Review: nuclear lamins—structural proteins with fundamental functions. J. Struct. Biol. 129:313-323. [DOI] [PubMed] [Google Scholar]
- 18.Hamirally, S., J. P. Kamil, Y. M. Ndassa-Colday, A. J. Lin, W. J. Jahng, M. C. Baek, S. Noton, L. A. Silva, M. Simpson-Holley, D. M. Knipe, D. E. Golan, J. A. Marto, and D. M. Coen. 2009. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 5:e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harel, N. Y., and J. C. Alwine. 1998. Phosphorylation of the human cytomegalovirus 86-kilodalton immediate early protein IE2. J. Virol. 72:5481-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homman-Loudiyi, M., K. Hultenby, W. Britt, and C. Soderberg-Naucler. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J. Virol. 77:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Indran, S. V., M. E. Ballestas, and W. J. Britt. 2010. Bicaudal D1-dependent trafficking of human cytomegalovirus tegument protein pp150 in virus-infected cells. J. Virol. 84:3162-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karki, S., and E. L. Holzbaur. 1995. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J. Biol. Chem. 270:28806-28811. [DOI] [PubMed] [Google Scholar]
- 23.Kudchodkar, S., Y. Yu, T. Maguire, and J. C. Alwine. 2004. Human cytomegalovirus infection induces rapamycin insensitive phosphorylation of downstream effectors of mTOR kinase. J. Virol. 78:11030-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marschall, M., A. Marzi, P. aus dem Siepen, R. Jochmann, M. Kalmer, S. Auerochs, P. Lischka, M. Leis, and T. Stamminger. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280:33357-33367. [DOI] [PubMed] [Google Scholar]
- 25.Milbradt, J., S. Auerochs, and M. Marschall. 2007. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 88:2642-2650. [DOI] [PubMed] [Google Scholar]
- 26.Milbradt, J., S. Auerochs, H. Sticht, and M. Marschall. 2009. Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J. Gen. Virol. 90:579-590. [DOI] [PubMed] [Google Scholar]
- 27.Munger, J., B. D. Bennett, A. Parikh, X.-J. Feng, J. McArdle, H. A. Rabitz, S. T., and J. D. Rabinowitz. 2008. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 26:1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 29.Papadimitriou, J. M., G. R. Shellam, and T. A. Robertson. 1984. An ultrastructural investigation of cytomegalovirus replication in murine hepatocytes. J. Gen. Virol. 65:1979-1990. [DOI] [PubMed] [Google Scholar]
- 30.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paton, A. W., T. Beddoe, C. M. Thorpe, J. C. Whisstock, M. C. Wilce, J. Rossjohn, U. M. Talbot, and J. C. Paton. 2006. AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature 443:548-552. [DOI] [PubMed] [Google Scholar]
- 32.Paton, A. W., P. Srimanote, U. M. Talbot, H. Wang, and J. C. Paton. 2004. A new family of potent AB5 cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pignatelli, S., P. Dal Monte, M. P. Landini, B. Severi, R. Nassiri, J. Gilloteaux, J. M. Papadimitriou, G. R. Shellam, T. Mertens, C. Buser, D. Michel, and P. Walther. 2007. Cytomegalovirus primary envelopment at large nuclear membrane infoldings: what's new? J. Virol. 81:7320-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prichard, M. N., W. J. Britt, S. L. Daily, C. B. Hartline, and E. R. Kern. 2005. Human cytomegalovirus UL97 kinase is required for the normal intranuclear distribution of pp65 and virion morphogenesis. J. Virol. 79:15494-15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quintyne, N. J., S. R. Gill, D. M. Eckley, C. L. Crego, D. A. Compton, and T. A. Schroer. 1999. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147:321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J. Virol. 78:5564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salina, D., K. Bodoor, D. M. Eckley, T. A. Schroer, J. B. Rattner, and B. Burke. 2002. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108:97-107. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez, V., and D. H. Spector. 2002. CMV makes a timely exit. Science 297:778-779. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Severi, B., M. P. Landini, and E. Govoni. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 98:51-64. [DOI] [PubMed] [Google Scholar]
- 42.Severi, B., M. P. Landini, M. Musiani, and M. Zerbini. 1979. A study of the passage of human cytomegalovirus from the nucleus to the cytoplasm. Microbiologica 2:265-273. [Google Scholar]
- 43.Stuurman, N., S. Heins, and U. Aebi. 1998. Nuclear lamins: their structure, assembly, and interactions. J. Struct. Biol. 122:42-66. [DOI] [PubMed] [Google Scholar]
- 44.Theiler, R. N., and T. Compton. 2002. Distinct glycoprotein O complexes arise in a post-Golgi compartment of cytomegalovirus-infected cells. J. Virol. 76:2890-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapani, J. A., P. Jans, M. J. Smyth, C. J. Froelich, E. A. Williams, V. R. Sutton, and D. A. Jans. 1998. Perforin-dependent nuclear entry of granzyme B precedes apoptosis, and is not a consequence of nuclear membrane dysfunction. Cell Death Differ. 5:488-496. [DOI] [PubMed] [Google Scholar]
- 46.Tzur, Y. B., K. L. Wilson, and Y. Gruenbaum. 2006. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 7:782-788. [DOI] [PubMed] [Google Scholar]
- 46a.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasatolic, D. Wang, D. G. Camp, K. Rodland, S. Willey, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilhelmsen, K., M. Ketema, H. Truong, and A. Sonnenberg. 2006. KASH-domain proteins in nuclear migration, anchorage and other processes. J. Cell Sci. 119:5021-5029. [DOI] [PubMed] [Google Scholar]
- 48.Ye, G. J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus 1 U(L)34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]