Abstract
The morphogenesis and budding of virus particles represent an important stage in the life cycle of viruses. For Ebola virus, this process is driven by its major matrix protein, VP40. Like the matrix proteins of many other nonsegmented, negative-strand RNA viruses, VP40 has been demonstrated to oligomerize and to occur in at least two distinct oligomeric states: hexamers and octamers, which are composed of antiparallel dimers. While it has been shown that VP40 oligomers are essential for the viral life cycle, their function is completely unknown. Here we have identified two amino acids essential for oligomerization of VP40, the mutation of which blocked virus-like particle production. Consistent with this observation, oligomerization-deficient VP40 also showed impaired intracellular transport to budding sites and reduced binding to cellular membranes. However, other biological functions, such as the interaction of VP40 with the nucleoprotein, NP, remained undisturbed. Furthermore, both wild-type VP40 and oligomerization-deficient VP40 were found to negatively regulate viral genome replication, a novel function of VP40, which we have recently reported. Interestingly, while wild-type VP40 was also able to negatively regulate viral genome transcription, oligomerization-deficient VP40 was no longer able to fulfill this function, indicating that regulation of viral replication and transcription by VP40 are mechanistically distinct processes. These data indicate that VP40 oligomerization not only is a prerequisite for intracellular transport of VP40 and efficient membrane binding, and as a consequence virion morphogenesis, but also plays a critical role in the regulation of viral transcription by VP40.
Morphogenesis and budding of nonsegmented negative-sense RNA viruses (NNSVs) is facilitated by their matrix proteins (23, 25, 43). This process involves interactions of the matrix proteins with cellular components of the ESCRT (endosomal sorting complex required for transport) machinery and is relatively well studied. Matrix proteins are thought to form a lattice underneath the viral envelope, and it seems reasonable that oligomerization is involved in the formation of this lattice. Indeed, oligomerization has been observed for a number of NNSV matrix proteins, particularly for the matrix proteins of measles virus (38), Nipah virus (5), and Borna disease virus (26) as well as for the major matrix protein, VP40, of Ebola virus (EBOV) (44). However, in none of these cases has the exact function of matrix protein oligomerization been experimentally shown.
EBOV is a member of the family Filoviridae and causes severe hemorrhagic fevers, with case fatality rates of up to 90% (41). Virus particles have a characteristic thread-like appearance and consist of a central nucleocapsid containing the 19-kb RNA genome (vRNA) complexed with the nucleoprotein (NP), the polymerase (L), the polymerase cofactor (VP35), and the transcriptional activator (VP30). These proteins are necessary and sufficient for replication and transcription of the genome (31). Virus particles are further enveloped by a host-cell-derived lipid bilayer. Embedded in this envelope is the glycoprotein (GP), which is responsible for attachment and entry of virions (8). The matrix proteins VP40 and VP24 can be found in the matrix space between the envelope and the nucleocapsid. The minor matrix protein VP24 has been shown to be an interferon antagonist (39), but it also seems to be involved in nucleocapsid formation (16, 20, 34). Furthermore, it was recently shown to be a negative regulator of viral genome replication and transcription (17, 46). The major matrix protein VP40 facilitates morphogenesis and budding, which can be demonstrated by the fact that its sole expression leads to the formation of virus-like particles (VLPs) with the characteristic thread-like appearance of EBOV particles (35). This function is supported by two overlapping late-domain motifs close to the N terminus of the protein (27), but recent experiments using recombinant EBOV imply that other budding mechanisms must also exist (32). Furthermore, we have recently shown that VP40 is also involved in the regulation of replication and transcription by an as yet unknown mechanism (17).
VP40 is made up of 326 amino acids and consists of two domains connected by a flexible linker. The N-terminal domain is responsible for oligomerization of VP40, while the C-terminal domain is important for membrane binding (7, 40). VP40 has been shown to oligomerize into both hexamers and octamers (44), both of which consist of antiparallel VP40 dimers (11, 40, 42). It has been suggested that the formation of these different oligomeric forms is determined by differences in the dimer-dimer (interdimeric) interface, whereas the monomer-monomer interface within one dimer (intradimeric interface) is similar in both octamers and hexamers (33, 44) (Fig. 1 A). Oligomeric VP40 has been found in VLPs and UV-inactivated virions, as well as in mammalian cells expressing VP40, where it seems to be enriched in lipid rafts (19, 37). In addition, VP40 octamers have been shown to bind RNA in a specific manner (11). Although we have shown that octamerization of VP40 is essential for the viral life cycle (19), the functions of both the hexameric and octameric forms of VP40 remain unknown.
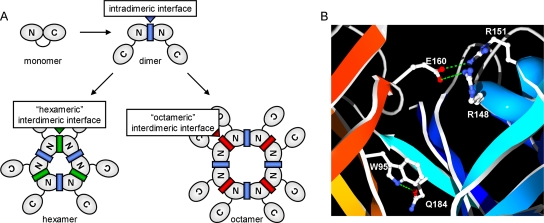
FIG. 1.
VP40 oligomeric states. (A) Schematic representation of the different oligomers formed by VP40. Shown are monomers, dimers, hexamers, and octamers, with the N- and C-terminal domains of VP40 indicated. The intradimeric protomer-protomer interface, which is predicted to be similar within hexamers and octamers, is indicated in blue. The interdimeric dimer-dimer interfaces of hexamers and octamers, which are predicted to differ substantially, are indicated in green and red, respectively. (B) Crystal structure of the VP40 intradimeric interface. Polar interactions (shown in green) at the intradimeric interface between two VP40 molecules (depicted in red and blue) in VP40 octamers. Secondary structures are shown in a ribbon representation, and the side chains of interacting amino acids are shown in an all-atom representation. The image was created using SwissPDB-Viewer 3.7 (13) and POV-Ray 3.5.
In order to better understand the contributions of matrix protein oligomerization to various virus processes, we have identified and subsequently substituted two amino acids in the intradimeric interface that are essential for dimer formation and thus also for higher-order oligomerization of EBOV VP40. Analysis of oligomerization-deficient mutants of VP40 showed that VP40 oligomerization is required for efficient membrane binding by VP40, as well as its transport to the surface and subsequent particle formation. Surprisingly, VP40 oligomerization was also involved in the regulation of viral genome transcription, but not replication.
MATERIALS AND METHODS
Cells.
293 (human embryonic kidney) cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; PAN Biotech), 2 mM l-glutamine (Q; Invitrogen), and 100 U/ml penicillin and 100 μg/ml streptomycin (P/S; Invitrogen), unless otherwise indicated, and grown at 37°C in the presence of 5% CO2. Escherichia coli XL-1 Blue strain cells were used for all routine cloning procedures.
Plasmids.
Expression plasmids for the EBOV structural proteins and the replication-competent minigenome have been described previously (14, 16), as has the replication-deficient minigenome (17). VP40 point mutants VP40-WA, VP40-EA, and VP40-WEA (carrying the W95A, E160A, and W95A E160A mutations, respectively) were created using the QuikChange site-directed mutagenesis kit (Stratagene). An N-terminal Flag tag (MDYKDDDDKK) and a C-terminal myc tag (EQKLISEEDL) were fused to VP40 and NP, respectively, using standard cloning techniques. Plasmids for the mammalian two-hybrid assay were created by amplifying VP40 or the respective mutants with the primers 5′-GCGGATCCATATGAGGCGGGTTATATTGCCTAC-3′ and 5′-CTAGCTAATTAAGAGCTCGCG-3′ and subsequent cloning into pBind or pAct (Promega) using BamHI and NotI. Plasmids for bacterial expression of VP40 were created by subcloning VP40 or the respective mutants out of the pCAGGS expression vector and into the pGEX-6P-1 vector (GE Healthcare) using EcoRI and XhoI. The sequences of all clones were confirmed prior to use.
Mammalian two-hybrid assay.
Mammalian two hybrid assays were performed using the Checkmate system (Promega), according to the manufacturer's instructions. Briefly, 293 cells at a confluence of 60% were transfected with 500 ng of each of the reporter plasmid pG5luc and plasmids expressing fusion proteins consisting of VP40 and either the GAL4 DNA-binding domain or the VP16 transactivation domain, using 3 μl TransIT LT1 (Mirus) per μg of DNA and according to the manufacturer's protocol. Forty-eight hours posttransfection (p.t.), cells were harvested and lysed in 200 μl 1× passive lysis buffer (Promega). Reporter activity was then determined using the Promega dual-luciferase reporter assay and read with a Centro LB 960 luminometer (Berthold Technologies).
Bacterial expression and cross-linking.
Bacterial expression was performed as previously described (12). Briefly, wild-type VP40 (VP40-WT) and mutants VP40-WA, VP40-EA, and VP40-WEA were expressed as fusion proteins with glutathione S-transferase (GST) in E. coli BL-21. Purification was achieved using glutathione-Sepharose 4B according to the manufacturer's directions (GE Healthcare). VP40 was eluted from the Sepharose by cleaving the linker attaching it to the GST moiety using 16 μg PreScission protease (GE Healthcare), according to the manufacturer's instructions. Eluted VP40 was loaded onto an Amicon Ultra-4 centrifugal filter unit (10,000 nominal molecular weight limit [NMWL]; Millipore), and the buffer was exchanged 3 times against phosphate-buffered saline (PBS) by centrifuging the filter unit for 30 min at 3,500 × g and 4°C and adding 3.5 ml fresh ice-cold PBS to the retained sample before each successive centrifugation step. Purified VP40-WT or VP40-WEA was then cross-linked with 0.1 mM glutaraldehyde for 5 min at room temperature, and the reaction was stopped by adding 0.2 volumes of 1 M Tris (pH 7.4).
Minigenome assays.
Minigenome assays were performed as previously described (16, 31). Briefly, 60% confluent 293 cells were transfected with the following plasmids using 3 μl TransIT LT1 (Mirus) per μg of DNA and following the manufacturer's instructions: 125 ng pCAGGS-NP, 125 ng pCAGGS-VP35, 75 ng pCAGGS-VP30, 1,000 ng pCAGGS-L, 250 ng 3E5E-minigenome, 250 ng pGL2-Control (Promega), and 250 ng pCAGGS-T7, unless otherwise indicated. In order to study the influence of VP40, 400 ng of pCAGGS-VP40 was additionally cotransfected. Differences in the absolute plasmid mass transfected were compensated for by the addition of empty pCAGGS vector. Prior to transfection, the medium on the cells was changed to 2 ml DMEM supplemented with 5% FBS and Q. Formation of the transfection complexes was carried out in 100 μl Optimem (Invitrogen). At 24 h p.t., the medium was exchanged against 4 ml of DMEM supplemented with 5% FBS, Q, and P/S. At 48 h p.t., cells were scraped into 1 ml PBS, of which 900 μl was used for minigenome RNA extraction, if necessary, while 25 μl of 5× passive lysis buffer (Promega) was added to the remaining 100 μl. This lysate was frozen once at −20°C, thawed, incubated for 15 min at room temperature, and vortexed for 5 s. Cellular debris was removed by centrifugation for 3 min at 10,000 × g. Reporter activity in the supernatant was then determined using the Promega dual-luciferase reporter assay and read on a Centro LB 960 luminometer (Berthold Technologies). Firefly luciferase, encoded by the pGL2-Control plasmid, served as an expression control.
iVLP assays.
Infectious VLP (iVLP) assays were carried out as previously described (16). Briefly, 60% confluent 293 cells (producer cells) were transfected as described above under minigenome assays but with the following plasmids: 125 ng pCAGGS-NP, 125 ng pCAGGS-VP35, 75 ng pCAGGS-VP30, 1,000 ng pCAGGS-L, 250 ng pCAGGS-GP, 250 ng pCAGGS-VP40, 60 ng pCAGGS-VP24, 250 ng 3E5E-minigenome, 250 ng pGL2-Control (Promega), and 250 ng pCAGGS-T7, unless otherwise indicated. Differences in the absolute plasmid mass transfected were compensated for by the addition of empty pCAGGS vector. At 24 h p.t., cells were washed once and the medium was exchanged against 4 ml of DMEM supplemented with 5% FBS, Q, and P/S. At 48 h p.t., 60% confluent HUH-7 cells (target cells) were transfected with the following plasmids: 250 ng pCAGGS-NP, 250 ng pCAGGS-VP35, 75 ng pCAGGS-VP30, 1,000 ng pCAGGS-L, and 100 ng pGL2-Control. At 72 h p.t. supernatant from the producer cells was cleared of cellular debris by centrifugation at 800 × g for 5 min, and cells were harvested for luciferase assay as described above under “Minigenome assays.” Next, target cells were washed once and 3 ml of producer cell supernatant, containing iVLPs, was used for infection. At 6 h postinfection (p.i.), 1 ml of DMEM supplemented with 10% FBS, Q, and P/S was added, and at 24 h p.i., medium on the target cells was exchanged against 4 ml DMEM supplemented with 5% FBS, Q, and P/S. Seventy-two hours p.i., the target cells were harvested and analyzed for luciferase assay as described above under “Minigenome assays.”
Protease K protection assay.
VLP-containing cell culture supernatant was cleared of cellular debris by centrifugation at 800 × g for 5 min. Then, 40-μl aliquots of supernatant were combined with either 12 μl of PBS, 7.2 μl PBS, and 4.8 μl protease K (150 μg/ml) or 7.2 μl PBS containing 0.1% Triton X-100 and 4.8 μl protease K. Samples were incubated for 1 h at 37°C. The protease K was then inactivated by heating at 99°C for 5 min. Subsequently, 20 μl of 4× SDS gel loading buffer with β-mercaptoethanol preheated to 99°C was added and samples were further denatured by boiling at 99°C for 5 min to allow analysis by Western blotting.
Western blotting.
SDS-PAGE and Western blotting were performed as previously described (19) using mouse monoclonal antibodies against VP40 (2C4) and NP (B1C6), both at a dilution of 1:100, and a polyclonal guinea pig serum against VP40 at a dilution of 1:2,000. As secondary antibodies, Alexa Fluor 680-coupled goat-anti-mouse (Molecular Probes) and IRDye800 goat-anti-guinea pig (Rockland) antibodies diluted 1:5,000 were used. Fluorescent signals were detected and quantified using the Odyssey infrared imaging system (Li-Cor Biosciences).
Immunofluorescense analysis.
Immunofluorescense analysis was performed as previously described (19) using a mouse monoclonal anti-VP40 antibody (2C4) at a dilution of 1:10 and a commercial rabbit anti-myc antiserum (Santa Cruz) at a dilution of 1:50. As secondary antibodies, rhodamine-coupled anti-mouse and fluorescein isothiocyanate-coupled anti-rabbit antibodies (Dianova) were used at a dilution of 1:100. Counterstaining of nuclei was performed using 4′,6-diamidino-2-phenylindole (DAPI) at a dilution of 1:10,000.
Flotation analysis.
For flotation analysis, 3 wells of a 6-well plate containing 60% confluent HUH-7 cells were transfected with 1 μg pCAGGS-VP40-WT or 1 μg pCAGGS-VP40-WEA using 3 μl TransIT LT1 (Mirus) per μg of DNA and according to the manufacturer's instructions. At 36 to 40 h p.t., cells were washed 3 times with 1 ml ice-cold lysis buffer (20 mM sucrose, 1 mM EDTA, 10 mM Tris, 1 mM phenylmethylsulfonyl fluoride [PMSF] containing one Complete miniprotease inhibitor cocktail tablet [Roche] per 10 ml of buffer) on ice. The first wash was carried out for 30 min, and subsequent washes were carried out for 10 min. Cells were then scraped into 100 μl lysis buffer/well and mechanically lysed by passing the sample repeatedly though a 26-gauge needle. Afterwards, 200 μl cell lysate was mixed with 400 μl 60% Nycodenz/TNE buffer (Progen Biotechnik) and centrifuged for 5 min at 3,500 × g to pellet any nonlysed cells. A small amount of lysate (0.5 ml) was then carefully overlaid with 3.5 ml 30% Nycodenz/TNE and 0.3 ml TNE. The resulting gradient was centrifuged for 5 h at 280,000 × g in an SW60 rotor. Five 840-μl fractions were taken from top to bottom, with membranes being visible as a white layer in the top fraction. Membrane-associated protein was also located in the top fraction, whereas soluble protein was found in the bottom fraction.
CoIP.
For coimmunoprecipitation (CoIP), 60% confluent 293 cells were transfected with 3 μl TransIT LT1 (Mirus) per μg of DNA according to the manufacturer's instructions and 1 μg of pCAGGS expression plasmids encoding NP and an N-terminal Flag-tagged VP40, as indicated. Differences in the absolute plasmid mass transfected were compensated for by the addition of empty pCAGGS vector. Forty-eight hours p.t., cells were harvested in 500 μl coimmunoprecipitation (CoIP) buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 5 mM EDTA, 1% NP-40 containing one Complete EDTA-free protease inhibitor cocktail tablet [Roche] per 25 ml buffer) with 1% Triton X-100. Cells were lysed in this buffer for 20 min, and cell debris was pelleted for 2 min at 20,000 × g. An aliquot of the supernatant was saved to control for protein expression by Western blotting, and 400 μl of the remaining supernatant was added to 35 μl of M2 anti-Flag agarose (Sigma), which had been previously washed 3 times with 1 ml CoIP buffer. Samples were then incubated for 3 h at 4°C on a rotator. Subsequently, the agarose was washed 3 times with 1 ml CoIP buffer and resuspended in 45 μl of 4× SDS sample buffer prior to boiling at 99°C for 5 min and use in SDS-PAGE and Western blotting.
vRNA quantification.
Minigenome RNA extraction and strand-specific two-step quantitative real-time PCR were performed as previously described (17). Briefly, RNA extraction was performed using the RNeasy minikit (Qiagen) and with additional DNase treatment using the RNase-free DNase set (Qiagen). RNA was then reverse transcribed using the Omniscript reverse transcription (RT) kit (Qiagen) and a vRNA-specific primer. Subsequently, quantitative real-time PCR was performed on a StepOne real-time PCR system (Applied Biosystems) using the QuantiFast SYBR green PCR kit (Qiagen). Minigenome plasmid DNA was used to standardize the genome copy numbers.
RESULTS
Construction of oligomerization-deficient mutants of VP40.
While a crystal structure is available for VP40 octamers (RCSB accession no. 1H2C and 1H2D) (11), for VP40 hexamers only a theoretical model exists (RCSB accession no. 1R32) (33), as well as single-particle reconstructions based on electron microscopy data (40, 42, 44). The antiparallel dimers in VP40 octamers, which are the building blocks for these oligomers, appear to be stabilized by polar interactions between their respective N-terminal domains, with E160 of one molecule interacting with R148 and R151 of the other molecule, as well as W95 of one molecule interacting with Q184 of the other molecule (Fig. 1B). In the theoretical hexamer model, W95 is also predicted to be involved in several interactions stabilizing the intradimeric interface. In contrast to this, the interdimeric interfaces of hexamers and octamers are predicted to show differences (33, 44), which account for the formation of both hexamers and octamers from the same dimeric subunits (Fig. 1A). Therefore, in order to impair the formation of both hexamers and octamers, we destabilized the intradimeric interface by mutating the amino acids W95 and E160 and tested the resulting mutants for their ability to homooligomerize.
As a first system to measure VP40 self-interaction, a mammalian two-hybrid assay was used. Wild-type VP40 (VP40-WT) and the VP40 W95A (VP40-WA), VP40 E160A (VP40-EA), and VP40 W95A E160A (VP40-WEA) mutants were expressed as fusion proteins consisting of VP40 and either a GAL4 DNA-binding domain or a VP16 transactivation domain in mammalian cells. Upon oligomerization of VP40, these fusion proteins form a transcription factor sufficient to promote transcription of a luciferase reporter construct and resulting in reporter activity, which reflects self-interaction of VP40. Surprisingly, when this assay was performed using VP40-WT, we were not able to detect any reporter activity compared to the positive control (pBind-Id and pAct-MyoD) (Fig. 2A), despite the fact VP40 is known to oligomerize. We, therefore, repeated this experiment using only the N-terminal oligomerization domain of VP40 (VP401-194) instead of full-length VP40. Interaction of these fusion proteins was readily detectable (Fig. 2A). Single mutation of either W95 or E160 in VP401-194 led to a 3-fold reduction in reporter activity, whereas mutation of both amino acids almost completely abolished self-interaction of VP40 (Fig. 2A), indicating that these residues are in fact critical for formation of the intradimeric interface.
FIG. 2.
W95 and E160 are essential for VP40 homooligomerization. (A) Interaction of VP40 in a mammalian two-hybrid assay. 293 cells were transfected with 500 ng each of plasmids encoding a GAL4 binding domain (pBind) and a VP16 activation domain (pAct) (negative control), pBind-Id and pAct-myo (positive control), or fusion proteins consisting of the GAL4 binding domain or the VP16 domain and VP40-WT or VP40-mutants, as indicated. Five hundred nanograms of a GAL4-driven luciferase reporter construct was also cotransfected. Cells were harvested 48 h after transfection, and luciferase reporter activity, reflecting interaction of VP40, was measured. (B) In vitro oligomerization of VP40. VP40-WT, VP40-WA, VP40-EA, and VP40-WEA were expressed as fusion proteins with glutathione S-transferase (GST) in bacteria and purified via the GST moiety. The GST was subsequently cleaved off using a specific protease, and the purified VP40 was cross-linked using glutaraldehyde. VP40 was detected after SDS-PAGE and Western blotting using VP40-specific antibodies. Monomeric and oligomeric forms of VP40 are labeled. The asterisk indicates uncleaved GST-VP40. (C) Quantitative analysis of cross-linked VP40. The average percentages of the different monomeric and oligomeric forms of VP40 after cross-linking from 3 independent experiments are shown.
To confirm these results in the context of full-length VP40 and to analyze the formation of oligomers, VP40-WT, VP40-WA, VP40-EA, and VP40-WEA were recombinantly expressed in E. coli as fusion proteins with glutathione S-transferase (GST), purified, cleaved from the GST moiety, and cross-linked. After cross-linking, we observed several oligomeric forms for VP40-WT, which accounted for more than half of the total protein (Fig. 2B and C). In SDS-PAGE, these oligomers showed apparent molecular weights consistent with dimers, tetramers, hexamers, and octamers. The single mutants VP40-WA and VP40-EA still showed formation of oligomeric forms; however, the proportion was shifted from higher-order oligomers toward dimers. The double-mutant VP40-WEA showed a strong decrease in the amount of homooligomers. While a small amount of VP40 dimer formation could still be observed, which might be due to the fact that the interdimeric interfaces of VP40-WEA were presumably still intact, no higher-order oligomers could be detected, confirming that the VP40-WEA mutant is no longer able to efficiently oligomerize.
Oligomerization-deficient VP40 is no longer able to form infectious virus-like particles.
After having experimentally shown that W95 and E160 are essential for oligomerization, the function of oligomerization-deficient VP40 in the viral life cycle was assessed. For this purpose, an infectious virus-like particle (iVLP) assay was used, which allows modeling and dissection of the EBOV life cycle under biosafety level 2 (BSL2) conditions (16, 47). This assay is an extension of a classical minigenome assay, in which a plasmid encoding for a vRNA-like minigenome, consisting of a reporter gene flanked by the EBOV noncoding terminal genome regions (leader and trailer), is expressed in mammalian cells. This vRNA minigenome is then recognized by the coexpressed viral proteins NP, VP35, VP30, and L and replicated and transcribed into mRNA. This results in reporter activity, which reflects both viral genome replication and transcription. Additionally, in an iVLP assay the other viral structural protein, VP40, VP24, and GP, are coexpressed in these cells (called producer cells). In addition to replication and transcription, this then leads to the formation of iVLPs containing minigenomes complexed with the nucleocapsid proteins. These iVLPs can then be used to infect target cells, which results in minigenome-derived reporter activity. While reporter activity in producer cells reflects viral transcription and replication of the minigenomes, activity in target cells additionally reflects particle formation in producer cells and successful infection of target cells.
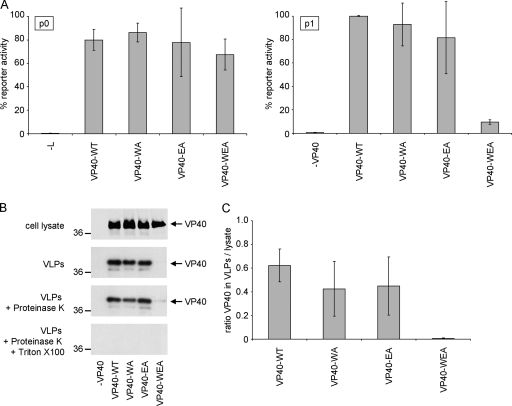
Using this assay, it was observed that reporter activity in producer cells was unchanged regardless of whether VP40-WT or any of the mutants were present (Fig. 3 A). However, in target cells, reporter activity dropped by more than 90% when oligomerization-deficient VP40-WEA was used for iVLP production instead of VP40-WT (Fig. 3A). This was despite the fact that VP40-WEA is expressed in producer cells at levels comparable to VP40-WT (Fig. 3B). In contrast, when VP40-WA or VP40-EA was transfected, which still showed some interaction in the mammalian two-hybrid assay (Fig. 2A) and some formation of higher-order oligomers (Fig. 2B and C), reporter activity in target cells did not change significantly (Fig. 3A).
FIG. 3.
Influence of VP40 oligomerization on production of infectious VLPs. (A) Viral transcription/replication in an iVLP assay. 293 producer cells (p0) were transfected with plasmids encoding all viral structural proteins as indicated, as well as expression plasmids for a minigenome, containing a Renilla luciferase reporter gene, and for T7 polymerase. Seventy-two hours after transfection, reporter activity derived from minigenome replication and transcription in these cells (p0) was determined. Supernatant of these cells was then used for infection of Vero E6 target cells previously transfected with plasmids encoding for NP, VP35, VP30, and L (p1). Seventy-two hours postinfection, reporter activity in these p1 cells was determined. The average percentages and the standard deviation of 3 independent experiments are shown. (B) Production of VLPs. 293 cells were transfected with plasmids encoding VP40 (wild-type [WT] or mutant WA, EA, or WEA, as indicated). Forty-eight hours after transfection, the cell lysate and supernatant were collected. The supernatant was cleared of cellular debris and separated into three fractions. Fraction 1 remained untreated, fraction 2 was treated with proteinase K, and fraction 3 was treated with proteinase K and Triton X-100. After 1 h at 37°C, the proteinase was heat inactivated and samples were subjected to SDS-PAGE, Western blotting, and staining using VP40-specific antibodies. (C) Quantification of released VLPs. The amount of VP40 in VLPs and in the cell lysate was quantified using the Odyssey infrared imaging system (Li-Cor) after Western blotting. Depicted is the average ratio of the signal of proteinase K-resistant VP40 in VLPs (B; VLPs + Proteinase K) divided by the VP40 signal in the cell lysate from three independent experiments. Error bars indicate standard deviation.
In order to address whether this lack of reporter activity was due to diminished infectivity of iVLPs produced in the presence of mutated VP40 or whether particle production itself was impaired, the ability of oligomerization-deficient VP40 to induce VLPs was assessed using a protease K protection assay. The amount of VLP-associated VP40 found in the supernatant of producer cells was greatly reduced in the case of VP40-WEA, whereas VP40-WA and VP40-EA were found at comparable levels to VP40-WT (Fig. 3B and C), consistent with the results of the iVLP assay. This indicates that oligomerization is essential for the production of both VLPs and iVLPs and suggests a role of VP40 oligomers either in virion formation and/or budding or at an earlier step in the viral life cycle. Since only the double mutant VP40-WEA showed a strong phenotypic difference from VP40-WT, consistent with this mutant being the most severely impaired in its oligomerization, further experiments focused on comparison of VP40-WT and VP40-WEA.
Oligomerization-deficient VP40 no longer efficiently relocalizes to the plasma membrane and is impaired in its binding to cellular membranes.
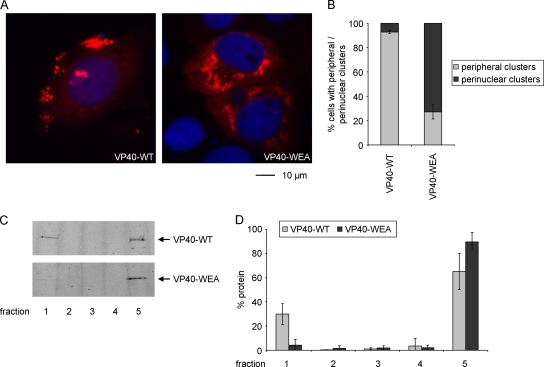
We next analyzed whether the intracellular distribution of VP40 was affected by inhibition of oligomerization. VP40-WT and oligomerization-deficient VP40-WEA were expressed in HUH-7 cells and the intracellular distribution of the proteins was analyzed by immunofluorescence microscopy. VP40-WT was found throughout the cytoplasm, but it was mostly concentrated in peripheral clusters (Fig. 4A). Quantitative analysis of the intracellular distribution of VP40 supported this observation and showed that in about 93% of cells showing VP40-WT clusters, these clusters were predominately located in the cell periphery close to the surface of the cells (Fig. 4B). In contrast, for VP40-WEA only 27% of cells showed VP40 cluster formation in the periphery, while in the other 73% of cells VP40-WEA clustered predominantly in the perinuclear region.
FIG. 4.
Intracellular localization and membrane association of VP40. (A) Intracellular localization of VP40. HUH-7 cells were transfected with expression plasmids encoding wild-type VP40 (VP40-WT) or homooligomerization-deficient VP40-WEA. Twenty-one hours posttransfection, cells were fixed in 4% paraformaldehyde, permeabilized, and stained using monoclonal mouse anti-VP40 antibodies. As secondary antibodies, rhodamine-coupled anti-mouse antibodies were used, and nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI). (B) Quantification of VP40 distribution. The phenotypes of cells in 3 independent immunofluorescence experiments as described in panel A were quantified. Shown is the percentage of cells showing predominantly peripheral clusters or predominantly perinuclear clusters. A total of 628 cells were evaluated. The average percentages and the standard deviation of 3 independent experiments are shown. (C) Flotation analysis of VP40. HUH-7 cells were transfected with expression plasmids encoding VP40 (WT or mutant WEA). Thirty-six hours after transfection, cells were lysed and the cell lysate was subjected to flotation analysis using a Nycodenz gradient. After ultracentrifugation, 5 fractions were taken off the gradient, and VP40 in these fractions was visualized by SDS-PAGE and Western blotting using VP40-specific antibodies. Membrane-associated protein is found in the top fraction (fraction 1), whereas soluble protein is located in the bottom fraction (fraction 5). (D) Quantification of flotation analysis. Experiments were performed as described in panel C. Western blot signals were quantified using an Odyssey infrared imaging system. The average and standard deviation of 3 independent experiments are shown.
In order to further analyze whether oligomerization-deficient VP40 was still able to interact with cellular membranes, we performed a flotation analysis. A significant fraction of VP40-WT coflotated with cellular membranes to the top of a density gradient, as previously reported (Fig. 4C and D) (24). However, almost none of the oligomerization-deficient VP40-WEA could be found in association with cellular membranes, although both proteins were expressed at similar levels. These results indicate that the defect observed in VP40-WEA transport to the periphery of the cells may be due to its inability to interact with cellular membranes, an observation that also explains its inability to induce the formation of iVLPs.
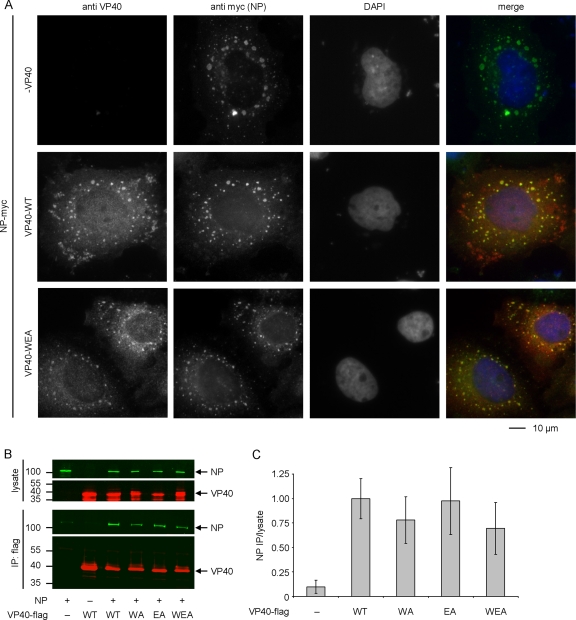
Oligomerization-deficient VP40 retains its ability to interact with NP and is recruited into NP-induced inclusions.
In addition to its role in the budding of virus particles, interaction of VP40 with the nucleoprotein NP has been described as another of its biological functions (36). In order to assess whether this interaction was also influenced by the oligomerization state of VP40, VP40-WT and VP40-WEA were coexpressed with NP and the localization of the proteins was investigated by coimmunofluorescence analysis (Fig. 5A). NP formed inclusion bodies, which are characteristic of solitary expression of this protein (12) and can also be found during virus infection (2). Upon coexpression with NP, VP40-WT was partially redistributed into these inclusion bodies, although peripheral clusters containing VP40, but no NP, were still apparent. Similarly, VP40-WEA could be found colocalized with NP in inclusion bodies, suggesting that oligomerization-deficient VP40 is still able to interact with NP. Again, cells expressing VP40-WEA and NP did not show peripheral VP40 clusters.
FIG. 5.
Interaction of VP40 and NP. (A) Colocalization of VP40 and NP. myc-tagged NP and VP40 (wild type [WT] or mutant WEA, as indicated) were coexpressed in HUH-7 cells. Twenty-one hours after transfection, cells were fixed in 4% paraformaldehyde, permeabilized, and stained using a monoclonal mouse-anti-VP40 antibody and a polyclonal rabbit-anti-myc antiserum. As secondary antibodies, rhodamine-coupled anti-mouse and fluorescein isothiocyanate-coupled anti-rabbit antibodies were used. Nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI). (B) Coimmunoprecipitation of NP and VP40. Flag-tagged VP40 (VP40-WT or VP40-WA, -EA, and -WEA mutants, as indicated) and NP were expressed in 293 cells. Forty-eight hours after transfection, cells were lysed and the lysate was subjected to coimmunoprecipitation using anti-Flag agarose. NP and VP40 in both the cell lysate and the immunoprecipitate were detected by Western blotting using a monoclonal mouse-anti-NP antibody and a polyclonal guinea pig-anti-VP40 antiserum. As secondary antibodies, IRDye-680 anti-mouse (shown in green) and IRDye-800 anti-guinea pig (shown in red) antibodies were used. (C) Quantification of coimmunoprecipitation. Experiments were performed as described for panel B. Signals in the cell lysate and the immunoprecipitate were quantified using an Odyssey infrared imaging system and are graphed as the ratio between immunoprecipitate and cell lysate, with VP40-WT set to 1. The average and standard deviation of 3 independent experiments are shown.
In order to further confirm that the interaction of NP and VP40 is independent of VP40 oligomerization, coimmunoprecipitation experiments were performed using Flag-tagged VP40-WT, VP40-WA, VP40-EA, and VP40-WEA. As previously shown (34), VP40-WT was able to interact with NP in this assay (Fig. 5B). Also, all VP40-mutants were able to coimmunoprecipitate NP. Quantification showed no significant differences in the amount of precipitated NP (Fig. 5C), confirming the results of the coimmunofluorescence analysis.
Oligomerization of VP40 is important for the regulation of viral transcription, but not viral replication.
We have recently reported that VP40 is involved in the regulation of viral genome replication as well as viral transcription, which are novel functions for VP40 (17). In order to assess the role of VP40 oligomerization in the regulation of these processes, a minigenome assay was performed in the presence or absence of VP40-WT or the oligomerization-deficient VP40-WEA mutant. The minigenome assay used in this approach resembles an iVLP assay; however, typically only the nucleocapsid proteins and a minigenome are expressed. Thus, no iVLPs are formed, and reporter activity in this assay only reflects viral genome replication and transcription.
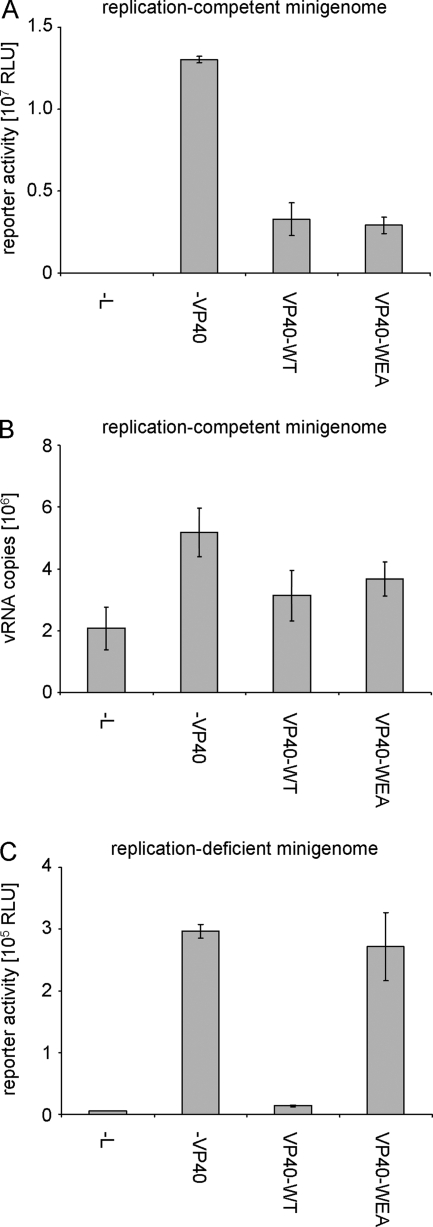
As previously reported, VP40-WT induced an approximately 80% reduction in reporter activity in a minigenome assay compared to the control without VP40 (Fig. 6A). This inhibition has been previously shown to be independent of the budding activity of VP40 (17). Interestingly, VP40-WEA showed an almost identical reduction in reporter activity, compared to VP40-WT. Reporter activity in minigenome assays is dependent on both viral genome replication and subsequent viral genome transcription, but does not allow the differentiation of these two processes. Therefore, vRNA levels in cells transfected with the minigenome assay components and VP40-WT or VP40-WEA were compared by quantitative RT-PCR in order to assess the impact of VP40 oligomerization on the regulation of viral genome replication alone. As previously reported, in the absence of the polymerase some vRNA copies were still detected. These reflect the initial expression of vRNA molecules from the minigenome plasmid (Fig. 6B) (17). In the presence of L, replication of these initial minigenome copies was observed in the form of an increase in the number of vRNA copies. Upon coexpression of either VP40-WT or VP40-WEA, replication was significantly reduced (Fig. 6B), consistent with the observed overall reduction in reporter activity (Fig. 6A). Both VP40-WT and VP40-WEA were able to suppress replication to a similar extent, showing that VP40 oligomerization is not required for the regulation of viral genome replication by VP40.
FIG. 6.
Influence of VP40 homooligomerization on viral minigenome replication and transcription. (A) Role in a classical minigenome assay. 293 cells were transfected with a T7-driven minigenome encoding Renilla luciferase, and expression plasmids for the viral proteins NP, L, VP35, VP30, and VP40 (wild type [VP40-WT] or mutant VP40-WEA, as indicated) and accessory plasmids as described in Materials and Methods. Forty-eight hours after transfection, cells were lysed and reporter activity, reflecting both viral replication and transcription, was measured. The average and standard deviation of 3 independent experiments are shown. RLU, relative light units. (B) Influence of VP40 homooligomerization on minigenome replication. 293 cells were transfected with minigenome assay components as described for panel A. Forty-eight hours after transfection, total RNA from these cells was isolated and subjected to a strand-specific quantitative RT-PCR detecting only negative-sense vRNA copies. The average vRNA copy number and standard deviation from 2 independent experiments are shown. (C) Influence of VP40 homooligomerization on transcription of a replication-deficient minigenome. 293 cells were transfected with minigenome assay components as described for panel A; however, instead of a classical minigenome, a replication-deficient minigenome was used. Forty-eight hours after transfection, cells were lysed and reporter activity, reflecting only viral transcription, was measured. The average and standard deviation of 3 independent experiments are shown.
In order to analyze the effect of VP40 oligomerization on the regulation of viral genome transcription, a replication-deficient minigenome was used (17). This minigenome contains a 55-base deletion at the extreme 5′ end of the trailer. We have previously shown that this deletion prevents replication of this minigenome but does not affect its transcription (17). Interestingly, due to the abolished replication, absolute levels of reporter activity from this construct were about 50-fold lower than in a classical minigenome system, emphasizing the importance of viral replication in increasing the pool of vRNAs available for transcription. Using this replication-deficient minigenome, we could readily observe reporter activity reflecting only viral transcription, which was measured at about 70-fold above background activity (Fig. 6C, cf. −VP40 and −L). Upon coexpression of VP40-WT, reporter activity dramatically dropped, consistent with the previously reported impact of VP40 on viral genome transcription (Fig. 6C) (17). Surprisingly, in the presence of oligomerization-deficient VP40-WEA no reduction in reporter activity compared to the control sample without VP40 was observed, suggesting that VP40 oligomerization is still involved in the regulation of viral transcription, although it does not influence viral replication. This result also indicates that the observed effect of VP40 on transcription is not due to impaired initial encapsidation of minigenome RNA, as there are no differences in the interaction of oligomerization-deficient VP40 and competent VP40 with NP. Since NP is responsible for minigenome encapsidation, any effect of VP40 on this process would then also be independent of the oligomerization ability of VP40, which is clearly not the case.
DISCUSSION
While oligomerization of EBOV VP40 has long been known to occur, and its importance for the virus life cycle has been demonstrated, the functions of the different oligomeric forms of VP40 remain largely unknown. Among other NNSVs, oligomerization of the matrix protein has also been shown to occur. Bornavirus M was shown to form tetramers, which have the tendency to assemble into larger two-dimensional lattices (26). The matrix proteins of both measles virus and Nipah virus have been shown to oligomerize as well (5, 38); however, similar to the filoviruses, the function of this oligomerization remains unclear. Therefore, in order to better understand the function of matrix protein oligomerization, we have generated oligomerization-deficient mutants of EBOV VP40 and compared their function to that of wild-type VP40.
Previously, we have shown that octameric VP40, while essential for virus rescue, is not important for VLP formation, suggesting that VP40 octamers are not involved in particle formation (19). These experiments were performed using the VP40 R134A point mutant (VP40-RA), in which the “octameric” interdimeric (dimer-dimer) interface was destabilized. This interdimeric interface has been predicted to differ between hexamers and octamers, while the intradimeric monomer-monomer interface was predicted to be similar, with amino acids W95 and E160 involved in its stabilization (11, 33, 44) (Fig. 1A and B). Therefore, while octamerization of VP40-RA was abolished, hexamerization of this mutant should still be possible. Thus, in order to impair formation of all higher-order oligomers, we now have destabilized the intradimeric interface by mutating W95 and E160. Using both mammalian two-hybrid assays and in vitro cross-linking, we could show that these two amino acids are indeed required for in vitro self-interaction of the oligomerization domain of VP40 and efficient formation of any higher-order oligomers of VP40.
Interestingly, in the mammalian two-hybrid assay, we were unable to detect any self-interaction of wild-type VP40, despite overwhelming evidence that direct VP40-VP40 interactions do occur (11, 19, 37, 40, 42, 44). It has been shown that membrane-associated VP40 is predominantly in an oligomeric state, whereas soluble protein is monomeric (37, 42), and it has been suggested that membrane interaction triggers oligomerization (15). Thus, we suggest that the membrane association of oligomeric VP40 fusion proteins prevents their import into the nucleus, which is required for a functional mammalian two-hybrid assay. Consistent with this, once the C-terminal membrane binding domain was removed, self-interaction of the N-terminal oligomerization domain was readily detectable. Mutation of W95 and E160 to alanine in this context then abolished VP40-VP40 interaction, a finding that was then reproduced in the context of full-length VP40 by in vitro cross-linking of bacterially expressed recombinant protein.
When analyzing the different VP40 mutants in an iVLP assay, which models the viral life cycle under biosafety level 2 conditions (18), the completely oligomerization-incompetent VP40-WEA led to a strongly reduced infection of target cells. This lack of infection was due to the fact that no VLPs were produced by the oligomerization-deficient VP40-WEA. In contrast the single mutants, which still show some higher-order oligomerization, showed no significant difference in comparison to wild-type VP40. We have previously shown that octamerization-deficient VP40 is still able to produce VLPs (19), and no difference is observed between wild-type VP40 and octamerization-deficient VP40 in iVLP assays (data not shown). This suggests that a lack of VP40 hexamerization is responsible for the observed inability of our oligomerization-incompetent VP40 mutant to produce VLPs. This interpretation is consistent with a report by Noda et al., which showed that nucleocapsid-like structures are incorporated into VLPs induced by either wild-type VP40 or octamerization- and RNA-binding-deficient VP40 (36). Taken together with our data, this clearly speaks against a role of VP40 octamerization in morphogenesis and budding but suggests a role of VP40 hexamerization in these processes.
When further analyzing the oligomerization-deficient mutant VP40-WEA, it became apparent that both its transport to the cell surface and membrane binding of this mutant were disturbed. This observation complements previous observations indicating that membrane-bound VP40 is mostly oligomeric (37, 42) and provides experimental evidence that VP40 oligomerization is a requirement for stable membrane binding. Interestingly, it has been previously suggested that binding to membranes is a trigger for oligomerization (7, 42). However, these two observations are not mutually exclusive, since it is possible that monomeric VP40 transiently binds to cellular membranes, thus triggering oligomerization. This could in turn stabilize the membrane association, possibly via additional membrane interactions through the C-terminal domains of the newly added VP40 subunits (Fig. 7A). In contrast, oligomerization-deficient VP40 would not be able to bind to further VP40 subunits and would rapidly dissociate from the membrane (Fig. 7B).
FIG. 7.
Hypothetical model of membrane binding by VP40. (A) Oligomerization-competent VP40. Monomeric VP40 binds to membranes, which leads to a displacement of the C-terminal domain relative to the N-terminal domain and exposure of the oligomerization interface within the N-terminal domain (depicted as a dark gray box). Oligomerization leads to the recruitment of additional membrane-binding domains, stabilizing the interaction of VP40 with the membrane. For simplicity, only two subunits of VP40 oligomers are depicted. (B) Oligomerization-deficient VP40. Oligomerization-deficient VP40 is not able to recruit additional membrane binding domains and quickly dissociates from cellular membranes.
The idea that oligomerization and membrane binding of VP40 are important for particle formation is supported by a number of studies. Oligomeric VP40 can be found in lipid raft microdomains, which have been suggested to serve as budding platforms for EBOV (37). Further, only oligomeric VP40 interacts strongly with Nedd4 (45), a cellular ubiquitin ligase involved in the budding of EBOV (48). Interestingly, McCarthy et al. have reported that mutations in the C-terminal domain of VP40 lead to a budding-incompetent phenotype (30). These mutants showed an altered intracellular localization and, interestingly, a much stronger tendency to oligomerize—possibly due to the fact that the interface between the N-terminal domain and the C-terminal domain was destabilized by the introduced mutations, thus exposing the oligomerization interface in the N-terminal domain. Also, a recent study has shown that the deletion or mutation of amino acids 96 to 101 abolished the budding activity of EBOV VP40 and led to an altered intracellular distribution similar to the one observed here for oligomerization-deficient VP40 (28). The proximity between the region analyzed in this study and W95, which is involved in the formation of the intradimeric interface, is striking, and indeed these mutants also show an altered oligomerization pattern. However, the authors of this study observed a much stronger formation of high-molecular-weight forms of VP40 for their mutants, with no remaining monomeric VP40. Therefore, it is likely that while oligomerization is necessary for efficient particle formation, the balance between monomeric and oligomeric forms of VP40 also plays an important role in this process.
One obvious problem with studies involving mutated proteins is the possibility that an observed loss of function is due to an overall misfolding of the protein. In order to exclude this possibility, we sought to identify biological functions of VP40 that are independent of its ability to oligomerize. VP40 has previously been shown to interact with NP (34). We analyzed this interaction using coimmunofluorescense analysis, which has been previously used to identify interaction partners for NP (1, 3, 12), and with coimmunoprecipitation studies. We could show that in both assays, the interaction of VP40 and NP is independent of the ability of VP40 to oligomerize and can be detected for both VP40-WT and VP40-WEA. Also, we have recently identified VP40 as a negative regulator of viral genome replication and transcription (17). When comparing the influence of wild-type VP40 and oligomerization-deficient VP40 on these processes, we observed that there was no apparent difference between VP40-WT and VP40-WEA in the reduction of reporter activity in a classical minigenome assay or the reduction of vRNA copies, which reflects the impact of VP40 on viral genome replication. Therefore, we have shown two biological functions of VP40 that are not impaired by the introduced point mutations, suggesting that the observed effects on membrane binding, transport, and subsequent particle formation are indeed due to the abolished oligomerization ability rather than an overall misfolding of oligomerization-deficient VP40.
Surprisingly, when analyzing the regulation of viral transcription by VP40, we observed that this regulation was strongly dependent on the oligomerization ability of VP40. While negative regulation of viral transcription by matrix proteins has been known for many years, the dependence of this regulation on oligomerization is a new finding. For rhabdoviruses, it was reported that the matrix protein M inhibits viral transcription of purified RNP complexes (4). This effect has been suggested to be linked to the ability of M to condense RNP complexes into tight structures that are no longer able to serve as templates for transcription or replication (6). It was also shown that even prior to condensation of RNP complexes, M shifts the balance between replication and transcription toward replication (10) and that this function can be genetically separated from other functions of M (9). For paramyxoviruses, it has been recently reported that M inhibits viral replication and/or transcription by interacting with the nucleoprotein N (21). Similarly, the matrix protein Z of Tacaribe virus inhibits replication and/or transcription in a minigenome assay (29), and this inhibition is dependent on an interaction between Z and the viral polymerase L (22). Interestingly, we have previously shown that the negative regulatory effect of VP40 on viral transcription is independent of both its RNA-binding and octamerization functions (17). Together with our present finding, that oligomerization is required for this regulatory effect, this suggests that it is VP40 hexamers that are involved in this process. Also, the fact that oligomerization is important for the regulation of viral transcription, but not viral genome replication, suggests that these two regulatory processes are mechanistically distinct. The mechanistic details of negative regulation of viral genome transcription and replication will be the subject of future studies.
In summary, our results show that oligomerization of the EBOV matrix protein is essential for the membrane binding of VP40, its transport to the cell surface and, subsequently, for the production of virus particles. The amino acids W95 and E160 are critical for VP40 oligomerization, as suggested by the crystal structure of VP40 octamers. Analysis of oligomerization-deficient mutants showed that oligomerization of VP40 is not necessary for its interaction with NP. Interestingly, negative regulation of viral genome replication by VP40, a novel VP40 function that we have recently identified, is independent of its ability to oligomerize, whereas the negative regulation of viral transcription seems to be dependent on VP40 oligomerization. This is the first time that oligomerization of an NNSV viral matrix protein has been shown to be essential for particle release. Further analysis of the identified oligomerization-deficient VP40 mutants will allow elucidation of the molecular details of the role of VP40 in morphogenesis and the regulation of viral transcription and replication in the future.
Acknowledgments
We thank Astrid Herwig and Katharina Kowalski for their technical support. We are further very grateful to Markus Eickmann for his help establishing the quantitative RT-PCR and to Hosam Shams-Eldin for his help generating the guinea pig antiserum.
Financial support came from the German Chemical Industry Association VCI (T.H.), the Natural Sciences and Engineering Research Council of Canada (T.H.), the Schering Foundation (T.H. and N.B.), the Canadian Institutes of Health Research (A.G.), the German National Academic Foundation (S.J.), and the German Research Foundation DFG (SPP1175 and SFB593; S.B).
Footnotes
Published ahead of print on 12 May 2010.
REFERENCES
- 1.Becker, S., C. Rinne, U. Hofsass, H. D. Klenk, and E. Muhlberger. 1998. Interactions of Marburg virus nucleocapsid proteins. Virology 249:406-417. [DOI] [PubMed] [Google Scholar]
- 2.Bjorndal, A. S., L. Szekely, and F. Elgh. 2003. Ebola virus infection inversely correlates with the overall expression levels of promyelocytic leukaemia (PML) protein in cultured cells. BMC Microbiol. 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehmann, Y., S. Enterlein, A. Randolf, and E. Muhlberger. 2005. A reconstituted replication and transcription system for Ebola virus Reston and comparison with Ebola virus Zaire. Virology 332:406-417. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, A. R., and R. R. Wagner. 1979. Role of the membrane (M) protein in endogenous inhibition of in vitro transcription by vesicular stomatitis virus. J. Virol. 29:134-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciancanelli, M. J., and C. F. Basler. 2006. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J. Virol. 80:12070-12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De, B. P., G. B. Thornton, D. Luk, and A. K. Banerjee. 1982. Purified matrix protein of vesicular stomatitis virus blocks viral transcription in vitro. Proc. Natl. Acad. Sci. U. S. A. 79:7137-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dessen, A., V. Volchkov, O. Dolnik, H. D. Klenk, and W. Weissenhorn. 2000. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 19:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmann, H., V. E. Volchkov, V. A. Volchkova, and H. D. Klenk. 1999. The glycoproteins of Marburg and Ebola virus and their potential roles in pathogenesis. Arch. Virol. Suppl. 15:159-169. [DOI] [PubMed] [Google Scholar]
- 9.Finke, S., and K. K. Conzelmann. 2003. Dissociation of rabies virus matrix protein functions in regulation of viral RNA synthesis and virus assembly. J. Virol. 77:12074-12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finke, S., R. Mueller-Waldeck, and K. K. Conzelmann. 2003. Rabies virus matrix protein regulates the balance of virus transcription and replication. J. Gen. Virol. 84:1613-1621. [DOI] [PubMed] [Google Scholar]
- 11.Gomis-Ruth, F. X., A. Dessen, J. Timmins, A. Bracher, L. Kolesnikowa, S. Becker, H. D. Klenk, and W. Weissenhorn. 2003. The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure (Camb.) 11:423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groseth, A., J. E. Charton, M. Sauerborn, F. Feldmann, S. M. Jones, T. Hoenen, and H. Feldmann. 2009. The Ebola virus ribonucleoprotein complex: a novel VP30-L interaction identified. Virus Res. 140:8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 14.Hartlieb, B., T. Muziol, W. Weissenhorn, and S. Becker. 2007. Crystal structure of the C-terminal domain of Ebola virus VP30 reveals a role in transcription and nucleocapsid association. Proc. Natl. Acad. Sci. U. S. A. 104:624-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartlieb, B., and W. Weissenhorn. 2006. Filovirus assembly and budding. Virology 344:64-70. [DOI] [PubMed] [Google Scholar]
- 16.Hoenen, T., A. Groseth, L. Kolesnikova, S. Theriault, H. Ebihara, B. Hartlieb, S. Bamberg, U. Stroher, H. Feldmann, and S. Becker. 2006. Infection of naive target cells with virus-like particles: implications for the function of Ebola virus VP24. J. Virol. 80:7260-7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoenen, T., S. Jung, A. Herwig, A. Groseth, and S. Becker. 3 May 2010, posting date. Both matrix proteins of Ebola virus contribute to the regulation of viral replication and transcription. Virology [Epub ahead of print.] doi: 10.1016/j.virol.2010.04.002. [DOI] [PubMed]
- 18.Hoenen, T., L. Kolesnikova, and S. Becker. 2007. Recent advances in filovirus- and arenavirus-like particles. Future Virol. 2:193-203. [Google Scholar]
- 19.Hoenen, T., V. Volchkov, L. Kolesnikova, E. Mittler, J. Timmins, M. Ottmann, O. Reynard, S. Becker, and W. Weissenhorn. 2005. VP40 octamers are essential for Ebola virus replication. J. Virol. 79:1898-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, Y., L. Xu, Y. Sun, and G. J. Nabel. 2002. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 10:307-316. [DOI] [PubMed] [Google Scholar]
- 21.Iwasaki, M., M. Takeda, Y. Shirogane, Y. Nakatsu, T. Nakamura, and Y. Yanagi. 2009. The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J. Virol. 83:10374-10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacamo, R., N. Lopez, M. Wilda, and M. T. Franze-Fernandez. 2003. Tacaribe virus Z protein interacts with the L polymerase protein to inhibit viral RNA synthesis. J. Virol. 77:10383-10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jasenosky, L. D., and Y. Kawaoka. 2004. Filovirus budding. Virus Res. 106:181-188. [DOI] [PubMed] [Google Scholar]
- 24.Jasenosky, L. D., G. Neumann, I. Lukashevich, and Y. Kawaoka. 2001. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 75:5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayakar, H. R., E. Jeetendra, and M. A. Whitt. 2004. Rhabdovirus assembly and budding. Virus Res. 106:117-132. [DOI] [PubMed] [Google Scholar]
- 26.Kraus, I., E. Bogner, H. Lilie, M. Eickmann, and W. Garten. 2005. Oligomerization and assembly of the matrix protein of Borna disease virus. FEBS Lett. 579:2686-2692. [DOI] [PubMed] [Google Scholar]
- 27.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, Y., L. Cocka, A. Okumura, Y. A. Zhang, J. O. Sunyer, and R. N. Harty. 2010. Conserved motifs within Ebola and Marburg virus VP40 proteins are important for stability, localization, and subsequent budding of virus-like particles. J. Virol. 84:2294-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez, N., R. Jacamo, and M. T. Franze-Fernandez. 2001. Transcription and RNA replication of Tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 75:12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarthy, S. E., R. F. Johnson, Y. A. Zhang, J. O. Sunyer, and R. N. Harty. 2007. Role for amino acids 212KLR214 of Ebola virus VP40 in assembly and budding. J. Virol. 81:11452-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhlberger, E., M. Weik, V. E. Volchkov, H. D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumann, G., H. Ebihara, A. Takada, T. Noda, D. Kobasa, L. D. Jasenosky, S. Watanabe, J. H. Kim, H. Feldmann, and Y. Kawaoka. 2005. Ebola virus VP40 late domains are not essential for viral replication in cell culture. J. Virol. 79:10300-10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen, T. L., G. Schoehn, W. Weissenhorn, A. R. Hermone, J. C. Burnett, R. G. Panchal, C. McGrath, D. W. Zaharevitz, M. J. Aman, R. Gussio, and S. Bavari. 2005. An all-atom model of the pore-like structure of hexameric VP40 from Ebola: structural insights into the monomer-hexamer transition. J. Struct. Biol. 151:30-40. [DOI] [PubMed] [Google Scholar]
- 34.Noda, T., H. Ebihara, Y. Muramoto, K. Fujii, A. Takada, H. Sagara, J. H. Kim, H. Kida, H. Feldmann, and Y. Kawaoka. 2006. Assembly and budding of Ebolavirus. PLoS Pathog. 2:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noda, T., H. Sagara, E. Suzuki, A. Takada, H. Kida, and Y. Kawaoka. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76:4855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noda, T., S. Watanabe, H. Sagara, and Y. Kawaoka. 2007. Mapping of the VP40-binding regions of the nucleoprotein of Ebola virus. J. Virol. 81:3554-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panchal, R. G., G. Ruthel, T. A. Kenny, G. H. Kallstrom, D. Lane, S. S. Badie, L. Li, S. Bavari, and M. J. Aman. 2003. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc. Natl. Acad. Sci. U. S. A. 100:15936-15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pohl, C., W. P. Duprex, G. Krohne, B. K. Rima, and S. Schneider-Schaulies. 2007. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J. Gen. Virol. 88:1243-1250. [DOI] [PubMed] [Google Scholar]
- 39.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin alpha 1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruigrok, R. W., G. Schoehn, A. Dessen, E. Forest, V. Volchkov, O. Dolnik, H. D. Klenk, and W. Weissenhorn. 2000. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J. Mol. Biol. 300:103-112. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez, A., T. Geisbert, and H. Feldmann. 2007. Filoviridae: Marburg and Ebola viruses, p. 1410-1448. In D. Knipe (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 42.Scianimanico, S., G. Schoehn, J. Timmins, R. H. Ruigrok, H. D. Klenk, and W. Weissenhorn. 2000. Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 19:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takimoto, T., and A. Portner. 2004. Molecular mechanism of paramyxovirus budding. Virus Res. 106:133-145. [DOI] [PubMed] [Google Scholar]
- 44.Timmins, J., G. Schoehn, C. Kohlhaas, H. D. Klenk, R. W. Ruigrok, and W. Weissenhorn. 2003. Oligomerization and polymerization of the filovirus matrix protein VP40. Virology 312:359-368. [DOI] [PubMed] [Google Scholar]
- 45.Timmins, J., G. Schoehn, S. Ricard-Blum, S. Scianimanico, T. Vernet, R. W. Ruigrok, and W. Weissenhorn. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326:493-502. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe, S., T. Noda, P. Halfmann, L. Jasenosky, and Y. Kawaoka. 2007. Ebola virus (EBOV) VP24 inhibits transcription and replication of the EBOV genome. J. Infect. Dis. 196(Suppl. 2):S284-S290. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe, S., T. Watanabe, T. Noda, A. Takada, H. Feldmann, L. D. Jasenosky, and Y. Kawaoka. 2004. Production of novel Ebola virus-like particles from cDNAs: an alternative to Ebola virus generation by reverse genetics. J. Virol. 78:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yasuda, J., M. Nakao, Y. Kawaoka, and H. Shida. 2003. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 77:9987-9992. [DOI] [PMC free article] [PubMed] [Google Scholar]