Abstract
Endogenous plant pararetroviruses (EPRVs) are viral sequences of the family Caulimoviridae integrated into the nuclear genome of numerous plant species. The ability of some endogenous sequences of Banana streak viruses (eBSVs) in the genome of banana (Musa sp.) to induce infections just like the virus itself was recently demonstrated (P. Gayral et al., J. Virol. 83:6697-6710, 2008). Although eBSVs probably arose from accidental events, infectious eBSVs constitute an extreme case of parasitism, as well as a newly described strategy for vertical virus transmission in plants. We investigated the early evolutionary stages of infectious eBSV for two distinct BSV species—GF (BSGFV) and Imové (BSImV)—through the study of their distribution, insertion polymorphism, and structure evolution among selected banana genotypes representative of the diversity of 60 wild Musa species and genotypes. To do so, the historical frame of host evolution was analyzed by inferring banana phylogeny from two chloroplast regions—matK and trnL-trnF—as well as from the nuclear genome, using 19 microsatellite loci. We demonstrated that both BSV species integrated recently in banana evolution, circa 640,000 years ago. The two infectious eBSVs were subjected to different selective pressures and showed distinct levels of rearrangement within their final structure. In addition, the molecular phylogenies of integrated and nonintegrated BSVs enabled us to establish the phylogenetic origins of eBSGFV and eBSImV.
Members of the Caulimoviridae are double-stranded DNA viruses replicating their 7- to 8-kbp genome by reverse transcription (30, 31). Also called plant pararetroviruses, they are phylogenetically related to Metaviridae (Ty3-Gypsy elements) (43). Although the viral replication cycle has no obligatory integration step, members of the Caulimoviridae family exist as both episomal (i.e., nonintegrated) viruses and as endogenous (i.e., integrated) sequences (endogenous pararetrovirus [EPRV]) in the host plant genome (62). To date, EPRVs have been described in nine distantly related mono- and dicotyledonous plant families. Each originated from independent integration events from five of the six genera of the Caulimoviridae family (49, 63, 64). Although most EPRVs are probably eliminated from the plant genome, they can also be retained through an endogenization process. Viral sequences first integrate into the germinal cells to become part of the plant genome. EPRVs are then fixed in plant populations by evolutionary forces such as natural selection and/or genetic drift (28, 31). The integration mechanism is thought to involve illegitimate recombination between the plant and viral genomes (63).
Despite their accidental origin, the presence of EPRVs has major consequences for host plants. First, EPRVs may contribute to genome size modification, induce changes in the methylation status of the host genome, and also act as genomic reorganizers by inducing chromosomal rearrangements (28), much like transposable elements (1, 35). In petunia (Petunia sp.) and tobacco (Nicotiana sp.) plants, for instance, EPRVs occur in several hundreds to thousands of copies, respectively (22, 34, 53), found mainly in heterochromatin. This amplification was probably the result of integration mediated by transposable elements found embedded or close to EPRVs (44, 63).
In addition, EPRVs are thought to serve host functions. A low level of EPRV transcription and subsequent small interfering RNA (siRNA) production was observed in petunia (Petunia hybrida) and tomato (Solanum lycopersicum) (48, 61). It is assumed that EPRV-induced homology-dependent gene silencing targeted the counterpart nonintegrated viruses: Petunia vein clearing virus (PVCV; genus Petuvirus) in petunia plants and Tobacco vein clearing virus (TVCV; genus Cavemovirus) in tomato plants. This mechanism is also proposed for Banana streak virus (BSV; genus Badnavirus) in wild diploid Musa balbisiana resistant to both endogenous BSV (eBSV) and BSV (28). Resistance mechanisms against episomal and endogenous forms of members of the Caulimoviridae family are seen as plant counteradaptations in response to the presence of EPRVs in their genomes. This gene silencing mechanism is based on the release from EPRVs of double-stranded RNA, which is likely produced by the transcription of antisense viral sequences. This could constitute a selective force acting to maintain the tandem repeat structures frequently observed in EPRVs (18, 47, 53).
Finally, some EPRVs are identified as infectious by the release of functional full-length viral genomes, from which viral multiplication and plant infection can arise. To date, infectious EPRVs have been observed in three pathosystems: BSV, banana; PVCV, petunia; and TVCV, tobacco. Interspecific hybridization and new genome combinations in plants are associated with EPRV activation in all pathosystems (39, 42, 53). Cultures under stress conditions, such as wounding for petunia, or in vitro culture for banana, trigger activation of PCVC (53) and BSV (9), respectively. In banana plants, genetic hybridization is another context to study the activation of infectious eBSV for triploid interspecific hybrids (AAB genome) resulting from genetic crosses between the two diploids Musa acuminata cv. IDN1104x (AAAA genome) and Musa balbisiana cv. Pisang Klutuk Wulung (PKW) (BB genome), since infectious eBSV reported to be present in the B genome is only transmitted by the diploid BB parent. Two molecular mechanisms are proposed to enable activation of infectious EPRVs: (i) homologous recombination between repeat regions surrounding them, resulting in the excision of a circular viral genome (47), and (ii) transcription of EPRVs leading to a viral pregenomic RNA (48, 53).
The presence of infectious EPRVs also impacts the evolution of the virus itself. Infectious EPRVs are an extreme case of parasitism and represent a newly described strategy of viral transmission among plant viruses. However, the endogenous state may be disadvantageous for the virus. EPRVs can become defective since they accumulate deleterious mutations with time, proportionately to the substitution rate of the Musa genome.
BSV naturally infects banana (Musa sp.) and is found in all banana-producing areas worldwide (41). The error-prone viral reverse transcriptase of badnaviruses would in part account for the genetic diversity and species richness observed in this genus (12, 13). BSV is indeed the generic name of several species showing up to 30% nucleotidic divergence but provoking the same disease in banana plants. Based on the RT/RNase H region of ORFIII, three major phylogenetic groups (BSV-1 to BSV-3) are observed (25). Banana streak Imové virus (BSImV) and Banana streak GF virus (BSGFV) belong to group BSV-1, which also encompasses at least five other fully described BSV species. Groups BSV-2 and BSV-3 each contain dozens of putative species (2, 17, 25), which remain to be fully characterized with whole-genome data and electronic microscopy.
The BSV-banana pathosystem is a model well adapted to investigate the evolution of infectious EPRVs. First, BSV phylogeny has been actively investigated, and a good picture of its genetic diversity and of the BSV integrations existing in the Musa genome is now available (2, 17, 20). Second, unlike EPRVs in Solanaceae, which can reach several hundreds to thousands of copies per genome (22, 34, 53, 61), the integrations of BSImV (eBSImV) and of BSGFV (eBSGFV) are simpler models present as single copies in the Musa balbisiana cv. PKW genome (18; F. C. Baurens et al., unpublished data). eBSImV (15.8 kbp) is monoallelic integration and eBSGFV is a diallelic integration (alleles eaBSGFV-7 [13.3 kpb] and eBSGFV-9 [15.6 kbp]); only eaBSGFV-7 is infectious (eaBSGFV-7 for endogenous and activatable BSGFV-7, according to the nomenclature of EPRVs [62]). Third, the biology of infectious integrants has been investigated: eBSImV and eBSGFV are able to reconstitute infectious viruses in interspecific hybrids between M. balbisiana cv. PKW and M. acuminata cv. ‘IDN 110 4x’ (33, 39). Genetic and environmental factors triggering eBSV activation (9, 39), and the mechanisms involved in their activation (P. Gayral et al., unpublished data), have also been studied. However, the evolutionary history explaining the presence and distribution of infectious eBSV in the Musa genus, and the phylogeny of M. balbisiana, the host plant species, are poorly documented. The few published studies concerning the genetic diversity of M. balbisiana focused on region-specific sampling rather than on exhaustive genetic diversity (19, 69). Consequently, no comparative study connecting EPRV distribution to host phylogeny has been performed. However, information on the evolution of this unusual biological model would help explain why infectious EPRVs are maintained in plants.
In the present study, we aimed to delineate the early history and evolutionary fate of infectious eBSImV and eBSGFV in relation to their banana (Musa sp.) host plant. Are the integrations of BSImV and BSGFV ancient or recent and were they contemporaneous? What was their evolutionary fate: conservation or degradation, fixation or loss? Did eBSImV and eBSGFV evolve in the same way? To answer these questions, we sampled 60 banana accessions representative of the genetic diversity of the Musa genus. Each accession was screened with five and eight PCR markers specific for eBSImV and eBSGFV, respectively. This provided data on both the polymorphism of insertion (through analysis of the distribution of each eBSV) and the evolution of their structure (through the presence or absence of PCR markers). These results were then interpreted in the light of a phylogenetic framework of Musa evolution. To do so, we chose 31 representative accessions from the initial sample of 60 and reconstructed Musa phylogeny using 19 microsatellite loci and chloroplast sequences. Scenarios of eBSGFV evolution were refined by estimating the age of the integration and the timing of evolution from sequence divergence analysis. Furthermore, we used molecular phylogenies to determine the origins and nearest ancestors of both eBSVs.
MATERIALS AND METHODS
Plant material and DNA extraction.
The 60 banana accessions listed in Table 1 represent the four sections of the genus Musa. Accessions of wild Musa balbisiana (BB genome) were predominantly sampled. Ensete ventricosum belongs to the second genus of the family Musaceae and was included as an outgroup taxon. Fresh leaf samples were kindly supplied by the CIRAD collection in Guadeloupe and from the International Institute of Tropical Agriculture (IITA) in Nigeria; dried leaf samples and plantlets from in vitro culture were supplied from the INIBAP Transit Center (ITC) in Leuven, Belgium.
TABLE 1.
Musa accession numbers and screen of EPRV from plants used in this study
| Section | Species, group, and hybrid | Phylogenyb | Subspecies | Genomec | Named | Abbreviationd | Accession no.e | EPRV amplificationf |
|
|---|---|---|---|---|---|---|---|---|---|
| BSImV | BSGFV | ||||||||
| Ensetea | E. glaucum (Roxb) Cheesm. | Cp | Vudu Vudu | ITC0775 | −−−−− | −−−−−−−− | |||
| Australimusa | M. textilis Née | TT | NEU0001 | −−−−− | −−−−−−−− | ||||
| Callimusa | M. beccarii Simmonds | TT | NEU0005 | −−−−− | −−−−−−−− | ||||
| M. coccinea Andr. | NEU0003 | −−−−− | −−−−−−−− | ||||||
| Rhodochlamys | M. laterita E. E. Cheesm. | Cp/Ms | NEU0008 | −−−−− | −−−−−−−− | ||||
| M. ornata Roxb. | Cp/Ms | NEU0007 | −−−−− | −−−−−−−− | |||||
| M. mannii H.Wendl | Cp/Ms | NEU0011 | −−−−− | −−−−−−−− | |||||
| M. velutina H.Wendl & Drude | Cp/Ms | ITC0638 | −−−−− | −−−−−−−− | |||||
| M. sanguinea | Cp/Ms | NEU0010 | −−−−− | −−−−−−−− | |||||
| Eumusa | M. basjoo | Cp/Ms | NEU0060 | −−−−− | −−−−−−−− | ||||
| M. itinerans E. E. cheesm. | Cp/Ms | −−−−− | −−−−−−−− | ||||||
| M. schizocarpa Simmonds | SS | M. schizocarpa N1 | ITC0599 | −−−−− | −−−−−−−− | ||||
| M. schizocarpa Simmonds | SS | M. schizocarpa | ITC0852 | −−−−− | −−−−−−−− | ||||
| M. schizocarpa Simmonds | SS | M. schizocarpa | ITC0890 | −−−−− | −−−−−−−− | ||||
| M. schizocarpa Simmonds | SS | M. schizocarpa | ITC0926 | −−−−− | −−−−−−−− | ||||
| M. schizocarpa Simmonds | SS | M. schizocarpa | ITC1002 | −−−−− | −−−−−−−− | ||||
| M. schizocarpa Simmonds | SS | M. schizocarpa | ITC1024 | −−−−− | −−−−−−−− | ||||
| M. schizocarpa Simmonds | Cp/Ms | SS | M. schizocarpa | ITC0846 | −−−−− | −−−−−−−− | |||
| M. schizocarpa Simmonds | SS | M. schizocarpa | ITC0856 | −−−−− | −−−−−−−− | ||||
| M. acuminata Colla | errans Blanco | AAw | Agutay | NEU0033 | −−−−− | −−−−−−−− | |||
| M. acuminata Colla | microcarpa Simmonds | AAw | Bornéo | NEU0028 | −−−−− | −−−−−−−− | |||
| M. acuminata Colla | truncata (Ridl.) Shepherd | AAw | Truncata | NEU0027 | −−−−− | −−−−−−−− | |||
| M. acuminata Colla | siamea Simmonds | AAw | Khae (Phrae) | NEU0025 | −−−−− | −−−−−−−− | |||
| M. acuminata Colla | siamea Simmonds | AAw | Pa (Rayong) | NEU0024 | −−−−− | −−−−−−−− | |||
| M. acuminata Colla | Cp/Ms | burmanicoides (De Langhe & Devreux) | AAw | Kolkata 4 | BUR | NEU0017 | −−−−− | −−−−−−−− | |
| M. acuminata Colla | burmanica Simmonds | AAw | Long Tavoy | NEU0016 | −−−−− | −−−−−−−− | |||
| M. acuminata Colla | Cp/Ms | malaccencis Simmonds | AAw | Pahang | MAL | NEU0013 | −−−−− | −−−−−−−− | |
| M. acuminata Colla | malaccencis Simmonds | AAw | Malaccensis | NEU0012 | −−−−− | −−−−−−−− | |||
| M. acuminata Colla | malaccencis Simmonds | AAw | P. cici (Bresil) | NEU0015 | −−−−− | −−−−−−−− | |||
| M. acuminata Colla | Cp/Ms | zebrina nom. nud. | AAw | Zebrina | ZEB | NEU0029 | −−−−− | −−−−−−−− | |
| M. acuminata Colla | banksii (F. Muell) Simmonds | AAw | Madang | ITC0254 | −−−−− | −−−−−−−− | |||
| M. acuminata Colla | AAw | Pa (Songkhla) | NEU0043 | −−−−− | −−−−−−−− | ||||
| M. acuminata Colla | AAw | THA018 | NEU0034 | −−−−− | −−−−−−−− | ||||
| M. acuminata Colla | AAw | Rung Hoa Xoan | ITC1432 | −−−−− | −−−−−−−− | ||||
| M. acuminata Colla | AAcv | Sa | NEU0149 | −−−−− | −−−−−−−− | ||||
| M. acuminata Colla | AAcv | Idn 110 d | NEU0137 | −−−−− | −−−−−−−− | ||||
| Banksii | AAcv | Wikago | NEU0102 | −−−−− | −−−−−−−− | ||||
| Desert banana | AAcv | Ta | NEU0096 | −−−−− | −−−−−−−− | ||||
| Rio | AAAcv | Leite | NEU0226 | −−−−− | −−−−−−−− | ||||
| Cavendish | Cp | AAAcv | Grande Naine | CAV | NEU0172 | −−−−− | −−−−−−−− | ||
| M. balbisiana Colla | Cp/Ms | BBw | Pisang Klutuk Wulung | M. balbisiana PKW | NEU0054 | +++++ | ++++++++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | Pisang Klutuk | M. balbisiana PKL | NEU0056 | +++++ | ++++++++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | Pisang Batu | M. balbisiana PBA | NEU0055 | +++++ | ++++++++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | Klue Tani | M. balbisiana KTA | NEU0053 | +++++ | ++++++++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | Honduras | M. balbisiana HDN | NEU0049 | −−−−− | ++++++++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | Balbisiana (10852) | M. balbisiana 852 | ITC0094/ONN0149 | +++++ | ++++++++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | M. balbisiana 545 | ITC0545 | +++++ | ++++++++ | |||
| M. balbisiana Colla | Cp/Ms | BBw | Cameroun | M. balbisiana CAM | NEU0050 | +++++ | +++++−++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | Lal Velchi | M. balbisiana LVE | NEU0051 | ++−−+ | +++++−++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | Singapuri | M. balbisiana SIN | NEU0052 | ++−−+ | +++++−++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | Los Banos | M. balbisiana LBA | ONN0151 | +++++ | +++++−++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | Montpellier | M. balbisiana MPL | ONN0152 | ++−++ | +++++−++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | I 63 | M. balbisiana I 63 | ONN0154 | −−−−− | +++++−++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | Eti Kehel | M. balbisiana EKE | ONN | +++++ | +++++−++ | ||
| M. balbisiana Colla | Cp/Ms | BBw | M. balbisiana HDN−211 | ITC0211 | +++++ | +++++−++ | |||
| M. balbisiana Colla | Cp/Ms | BBw | M. balbisiana I63−080 | ITC0080 | −−−−− | +++++−++ | |||
| M. balbisiana Colla | Cp/Ms | BBw | M. balbisiana LBA−342 | ITC0342 | +++++ | +++++−++ | |||
| M. balbisiana Colla | Cp/Ms | BBw | M. balbisiana 626 | ITC0626 | +++++ | +++++−++ | |||
| M. balbisiana Colla | Cp/Ms | BBw | M. balbisiana 1016 | ITC1016 | +++++ | ++++++++ | |||
| Hyb. M. balbisiana | Cp/Ms | BB | Butuhan | M. balbisiana BUT | ITC0565 | +++++ | +++++−++ | ||
E. ventricosum is in the genus Ensete.
Used in this study for microsatellites (Ms) and chloroplast molecular phylogeny (Cp).
w, wild type; cv, cultivar.
The names and abbreviations are specified if different from the species name.
Collections: ITC, INIBAP Transit Center; NEU, CIRAD-Neufchâteau; ONNE, IITA.
+, Presence of PCR amplification at the expected size; -, absence of amplification. The order of PCR markers for BSImV EPRV is Musa/F2, F1/F3, F3/F4, F4/F5, F5/Musa; the order of PCR markers for BSGFV EPRV is VM1, VV1, VV2, VV3, VV4, VV5, VV6, VM2.
Total genomic DNA was extracted from banana leaf tissue by the method of Gawel and Jarret (16). The quality of DNA was assessed visually under UV light after migration of 5 μl of DNA sample in a 0.8% agarose gel in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA [pH 8]), stained with ethidium bromide, and by PCR amplification of the housekeeping Musa actin gene (see below).
Nucleotide sequences.
Two chloroplast loci were sequenced for the phylogenetic analysis of Musa genus. A newly designed pair of primers—MatKHB1Musa (5′-ATGGAAGAATTACAAGGATATTTAG-3′) and MatK1326RMusa (5′-AGCACACGAAAGTCGAAGT-3′)—with 1.5 mM MgCl2 was used to amplify 1,250 bp of the 5′ trnK intron and partial matK gene (GenBank accession no. GQ374836 to GQ374868). These new primers were adapted from the primers MatKHB1 (http://www.plantbio.ohiou.edu/epb/faculty/faculty/heb.htm) and MatK1326R (8), respectively, after alignment with matK sequences from Musa acuminata (gb EU016987), M. basjoo (EMBL accession no. AJ581437), M. beccari (GenBank accession no. AF434869), and M. rosea (Emb AM114725). The primers trnL-F C (5′-CGAAATCGGTAGACGCTACG-3′) and trnF F (5′-ATTTGAACTGGTGACACGAG-3′) (63) amplified 800 to 900 bp of the tRNA-Leu(UAA) intron and tRNA-Leu-tRNA-Phe(GAA) intergenic spacer (PCR with 3 mM MgCl2) (GenBank accession no. GQ374803 to GQ374835).
Phylogenetic analysis of BSGFV was performed using sequences amplified with the primers PhyloEPRV-Gf-F (5′-CAGCTCCAGGAGATTGGAAA-3′) and PhyloEPRV-Gf-R (5′-GGAGGAATCTATCCCATGGAC-3′) with 2 mM MgCl2. This primer pair amplified 1,313 bp containing the RT/RNase H domains frequently used in Badnavirus phylogeny studies (gb GQ374869 to GQ374888).
The following thermal cycling profiles were used: (i) for the trnK-matK region, 5 min of denaturation at 94°C, 30 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 58°C, and 1 min 15 s of extension at 72°C, and then 10 min for final extension; (ii) for the trnL-trnF region, 2 min of denaturation at 94°C, 30 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 52°C, and 3 min of extension at 72°C, and then 10 min for final extension; and (iii) for PhyloEPRV-Gf, 5 min of denaturation at 95°C, 35 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 50°C, and 1 min 30 s of extension at 72°C, and then 10 min at 72°C for final extension. See below for details of the PCR mixture. Sequencing was performed by Cogenics Genome Express SA (Grenoble, France) on the reverse strand for trnL-trnF and on both strands for PhyloEPRV-Gf and matK.
Phylogenetic inference.
Sequences were aligned by using CLUSTAL W (66) implemented in Bioedit (24) and corrected manually when necessary. Primer sequences were discarded from the alignments. The software DAMBE version 4.5.20 (72) was used to detect substitution saturation in each of the six alignments according to a previously described method (73). For this purpose, the percentage of invariant sites was first estimated by PhyML v2.4.4 software (23) using a GTR+I substitution model with eight categories of the gamma parameter. The expected saturation index for a symmetric tree topology was calculated.
Trees topologies were computed with MrBayes 3.1.2 (29) by running five chains and 106 generations using the default priors of the GTR model. Bayesian posterior probabilities were calculated from majority-rule consensus trees sampled every 20 generations once the Markov chains had become stationary (determined by empirical checking of likelihood values).
Maximum likelihood (ML) phylogenies were inferred with PhyML. A GTR model with eight categories of the gamma parameter and with a fixed proportion of invariable sites (0.01%) was used; 1,000 bootstrap iterations were performed to assess the robustness of tree topologies.
Sequences of the RT/RNase H region of ORFIII of BSV species BSImV and BSGFV were retrieved from public databanks. Episomal BSImV virus (BSImVUg) clones 10.1 (EMBL accession no. AJ968444), 20.3 (EMBL accession no. AJ968445), 21.6 (EMBL accession no. AJ968447), 39.1 (EMBL accession no. AJ968449), 49.2 (EMBL accession no. AJ968450) and 49.6 (EMBL accession no. AJ968451) and BSGFV virus clones GFUg50.1 (EMBL accession no. AJ968435), 54.4 (EMBL accession no. AJ968439), 54.6 (EMBL accession no. AJ968441), 54.8 (EMBL accession no. AJ968442), 54.2 (EMBL accession no. AJ968438), and 54.5 (EMBL accession no. AJ968440) originated from BSV epidemics in Uganda (25). BSGFV episomal clones Col20 (GenBank accession no. EU076416), Col23 (GenBank accession no. EU076417), Col28 (GenBank accession no. EU076418), Col30 (GenBank accession no. EU076420), Col31 (GenBank accession no. EU076421), Col32 (GenBank accession no. EU076422), and Col34 (GenBank accession no. EU076423) originated from Colombian epidemics. eBSImV clones Bat27 (GenBank accession no. AY189426) and KT32 (GenBank accession no. AY452264) were amplified from healthy Musa balbisiana cv. Pisang Batu (genotype BB) and from banana cv. ‘Klue Tiparot’ (genotype ABB), respectively (20). Complete BSV genomic sequences of BSAcVNV (GenBank accession no. AY750155) (40) and of BSGFV (GenBank accession no. AY493509) (18) were used to retrieve the RT/RNase H region. Episomal BSImV sequence (gb GQ374801) and eBSImV sequence (GenBank accession no. GQ374802) originating from a BSImV-infected M. acuminata, and a BSV-free M. balbisiana cv. PKW, respectively, were also used.
Microsatellite genotyping.
The 19 microsatellite loci used in the present study were independent (I. Hippolyte et al., unpublished data). They were developed in M. acuminata cv. ‘Gobusik’ (7, 37) and M. balbisiana cv. PKW (Table 2). Genotyping was performed by using a PCR with fluorescently labeled primers. PCR was carried out in 25 μl of a mixture containing 25 ng of DNA, 0.14 μM reverse primer, 0.12 μM M13-tailed forward primer, 1.25 U of GoTaq Flexi DNA Polymerase (Promega, Madison, WI), 0.1 mM concentrations of each deoxynucleoside triphosphate (dNTP), 1.5 mM MgCl2, 5 μl of 5× Colorless GoTaq Flexi buffer, and 0.16 μM M13 primer fluorescently labeled with hexachlorocarboxyfluorescein (HEX) and carboxyfluorescein (6-FAM) (Eurogentec, Maastricht, Netherlands). An initial denaturing step of 2 min at 94°C was followed by 40 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min, and then by 10 min at 72°C. The PCR products were diluted (1/5; depending on their concentration). The sizes of the amplified fragments were measured by using a MegaBACE capillary sequencing machine (Amersham Biosciences, Freiburg, Germany). Alleles were scored by using MegaBACE Genetic Profiler v1.0 software (Amersham Biosciences). Alleles included in final consensus genotypes were observed at least twice. Two samples with known genotypes served as positive controls and were included in each run of 96 PCRs to standardize genotyping across experiments.
TABLE 2.
Description of the microsatellite loci used and summary of the allelic variation
| Locusa | GenBank accession no. | Repeat motif | No. of alleles sampled |
Allele size range (nt) | Heb |
Hoc | |||
|---|---|---|---|---|---|---|---|---|---|
| M. balbisiana | Other Musa spp. | Total | M. balbisiana | Other Musa spp. | |||||
| mMaCIR08† | X87264 | (TC)6N24(TC)7 | 5 | 11 | 13 | 251-287 | 0.573 | 0.964 | 0.608 |
| mMaCIR307‡ | AM950533 | (CA)6 | 1 | 4 | 4 | 162-170 | 0.000 | 0.727 | 0.045 |
| mMaCIR07† | X87258 | (GA)13 | 6 | 11 | 14 | 148-188 | 0.504 | 0.945 | 0.514 |
| mMaCIR13† | X90745 | (GA)16N76(GA)8 | 6 | 9 | 13 | 266-307 | 0.676 | 0.933 | 0.550 |
| Ma3-90* | NAd | NAa | 4 | 9 | 13 | 142-177 | 0.554 | 0.918 | 0.429 |
| mMaCIR01† | X87262 | (GA)20 | 6 | 9 | 14 | 248-329 | 0.609 | 0.933 | 0.572 |
| mMaCIR39† | Z85970 | (CA)5GATA(GA)5 | 6 | 9 | 12 | 295-388 | 0.681 | 0.918 | 0.634 |
| mMaCIR260‡ | AM950515 | (TG)8 | 3 | 6 | 7 | 208-256 | 0.608 | 0.855 | 0.332 |
| mMaCIR03† | X87263 | (GA)10 | 2 | 6 | 7 | 116-148 | 0.516 | 0.782 | 0.382 |
| mMaCIR40† | Z85977 | (GA)13 | 5 | 10 | 14 | 170-232 | 0.750 | 0.955 | 0.500 |
| mMaCIR150‡ | AM950440 | (CA)10 | 3 | 6 | 7 | 253-271 | 0.495 | 0.819 | 0.436 |
| mMaCIR214‡ | AM950480 | (AC)7 | 3 | 2 | 4 | 122-130 | 0.611 | 0.527 | 0.420 |
| mMaCIR264‡ | AM950519 | (CT)17 | 1 | 8 | 9 | 240-280 | 0.000 | 0.927 | 0.136 |
| mMaCIR152‡ | AM950442 | (CTT)18 | 6 | 6 | 11 | 160-196 | 0.564 | 0.844 | 0.450 |
| mMaCIR164‡ | AM950454 | (AC)14 | 4 | 9 | 13 | 300-436 | 0.716 | 0.964 | 0.300 |
| mMaCIR196‡ | AM950462 | (TA)4(TC)17TG(TC)3 | 4 | 8 | 11 | 162-198 | 0.737 | 0.933 | 0.503 |
| mMaCIR231‡ | AM950497 | (TC)10 | 2 | 10 | 12 | 240-280 | 0.523 | 0.909 | 0.325 |
| mMaCIR45† | Z85968 | (TA)4CA(CTCGA)4 | 2 | 8 | 9 | 278-296 | 0.329 | 0.936 | 0.241 |
| mMaCIR24† | Z85972 | (TC)7 | 4 | 7 | 9 | 239-289 | 0.655 | 0.767 | 0.400 |
Genetic distances and microsatellite genetic variation.
The interindividual microsatellite genetic distance “Simple Matching” implemented in DARwin v5.0.155 software (X. Perrier and J. P. Jacquemoud-Collet [http://darwin.cirad.fr/darwin]) was used. This distance, also called the “1-Dps” distance, takes into account the proportion of shared alleles. The experimental data suggested that the proportion of shared alleles was effective in obtaining correct genealogical relationships (26, 46). A total of 1,000 bootstrap replicates were performed by using DARwin software. Distance matrices were then used to construct dendrograms with the neighbor-joining (NJ) algorithm implemented in DARwin software. The program FSTAT v.2.9.3.2 (21) (http://www2.unil.ch/popgen/softwares/fstat.htm) was used to compute the values of the standard genetic diversity indices.
PCR screen of eBSVs.
Details on the primers used for eBSV screen by PCR are shown in the supplemental material at http://southgreen.cirad.fr/sites/all/files/uploads/supplementary_material_article_no_jvi0401_10.doc. Figure 1 shows the amplified eBSV fragments (see also Results).
FIG. 1.
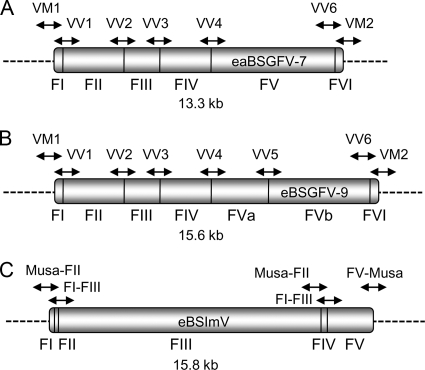
Schematic representation of eaBSGFV-7 (A), eBSGFV-9 (B), and eBSImV (C). Horizontal lines indicate junctions between two rearranged viral fragments. Fragments are numbered below eBSVs. Dashed lines represent the flanking Musa genome. Arrows above the fragments show the region amplified by PCR for eBSV detection.
All PCRs were performed with 5 to 20 ng of DNA, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 100 mM concentrations of each dNTP, 1.5 mM MgCl2, 10 pmol of each primer, and 1 U of Taq DNA polymerase (Eurogentech, Seraing, Belgium) in a total reaction volume of 25 μl. PCR conditions were as follows: 1 cycle at 94°C for 5 min, followed by 30 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and then one elongation cycle at 72°C for 10 min. PCR products were visualized under UV light after migration of 10 μl of PCR products on a 1.5% agarose gel in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA, [pH 8]) stained with ethidium bromide.
For the housekeeping actin gene amplification, the following primers and conditions were used: Actine1F (5′-TCCTTTCGCTCTATGCCAGT-3′), Actine1R (5′-GCCCATCGGGAAGTTCATAG-3′), and a Tm of 58°C for 25 cycles with 1.5 mM MgCl2.
PCR products were cloned into TOPO-TA (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
RESULTS
Classification of bananas based on morphological and genetic characters divided the Musa genus into four sections: Eumusa, Rhodochlamys, Australimusa, and Callimusa (5, 60, 66). The initial sample comprised 59 Musa sp. accessions and Ensete glaucum as an outgroup. This sample emphasized the section Eumusa, to which Musa balbisiana belongs (Table 1). The PCR-based screen was performed on all 60 accessions. Chloroplast-based molecular phylogeny and microsatellite-based phylogeny encompassed 33 and 31 accessions of the initial sample, respectively.
PCR screen of eBSVs.
The annotated structure and biological properties of eBSGFV from M. balbisiana cv. PKW were described in Gayral et al. (18). The structures of eBSGFV and eBSImV are shown in Fig. 1. These eBSVs are independent integrations of two distinct BSV species—BSGFV and BSImV—but they show a similar organization in M. balbisiana cv. PKW. They are both composed of juxtaposed viral fragments in both orientations. Analysis of the distribution of eBSGFV and eBSImV among the genus Musa was performed by PCR amplification of the eight and five characterized junctions of eBSGFV and eBSImV in PKW (unpublished data). Therefore, each set of markers produced a unique signature for each eBSV (Fig. 1).
For eBSGFV, VV primers (for virus-virus junction) amplify the five internal junctions of the six fragments present in both alleles (Fig. 1A and B). Primer pair VV5 only amplifies the junction involved in the additional fragment of the allele eBSGFV-9. Primers VM (for virus-Musa junction) amplify the 5′ and 3′ flanking regions of eBSGFV and ensure detection of the integration locus in the Musa genome. Two additional VM primers amplify those of eBSGFV-9. The same approach was used for the homozygous BSImV integrant, which is composed of five fragments in cv. PKW (Fig. 1C). Since several fragments were very short (30 and 27 nucleotides for fragments I and II, respectively), it was not possible to amplify each junction separately. Three PCR markers (primers F1/F3, F3F4, and F4/F5) specific to the internal fragments of this integration were designed and used in the present study. Two additional PCR markers (Musa/F2 and F5/Musa) amplifying 5′ and 3′ boundaries of BSImV EPRV were specific for the integration locus in the Musa genome. Two additional PCR markers (Musa/F2 and F5/Musa), amplifying the 5′ and 3′ flanking regions of eBSImV were specific for the integration locus in the Musa genome.
PCR-based genotyping errors resulting in no amplified PCR product were lowered by two complementary approaches. First, we detected low DNA quantity or problems with the quality of extracted DNA by performing a parallel PCR amplification of the housekeeping actin gene in all samples (data not shown). PCRs or DNA extraction was repeated if no acceptable amplification of the actin gene was observed. Next, to ensure that the absence of amplification did not result from SNPs, PCR with a set of primers designed to be external to the first set on the same DNA fragment was performed (labeled “bis” in the supplemental material [http://southgreen.cirad.fr/sites/all/files/uploads/supplementary_material_article_no_jvi0401_10.doc]). Genotyping errors due to unspecific PCR amplification were also reduced. We first ensured that none of these PCR primers cross-amplified with the episomal form of the corresponding BSGFV and BSImV viruses, which would have resulted in false-positives. Then, the few PCR products showing unexpected product sizes were systematically cloned, sequenced, and then discarded if they were not BSV-related sequences (data not shown).
Detection of substitution saturation.
Substitution saturation, i.e., when substitutions occurred repeatedly at the same position in a nucleotide sequence, can create a bias in phylogenetic reconstruction (51). We tested whether some or all sequences in the data set have already lost phylogenetic information due to substitution saturation. It is assumed that phylogenetic information is basically lost when the observed saturation index is equal to, or larger than, half of the full substitution saturation (71). We estimated the expected saturation indices, assuming half of the full substitution saturation, and compared them to the observed saturation indices. Substitution saturation is detected when the observed indices are higher than the expected indices. No saturation was detected in any of the three alignments used in the present study, i.e., the concatenation of the matK gene and the TrnL-F region (see below) and the RT/RNase H region of BSGFV and BSImV (P < 10−4) (Table 3).
TABLE 3.
Detection of saturation substitution with the program DAMBE
| Alignment | Observed saturation index (Iss) | Expected saturation index (Iss.cSym) | Ta | dfb | Pc |
|---|---|---|---|---|---|
| Concat-matK_trnL-F | 0.126 | 0.810 | 36.75 | 2,158 | 0.0000 |
| BSGVF | 0.083 | 0.717 | 40.80 | 342 | 0.0000 |
| BSImV | 0.086 | 0.734 | 59.28 | 460 | 0.0000 |
Test statistic (see reference 72).
T, number of degrees of freedom (df) of the model.
The statistical significance of the difference between observed and expected saturation indexes was assessed with a two-tailed test.
Chloroplast molecular phylogeny of Musa.
All phylogenetic reconstructions presented here were based on Bayesian and maximum-likelihood (ML) methods. These methods are supported by probabilistic models known to be more sophisticated and more robust and that introduce less bias than maximum-parsimony or distance-based methods (55). Figure 2 shows the Bayesian chloroplast phylogeny of Musa inferred from a combined alignment of 2,159 nucleotides made from concatenation of matK and trnL-trnF alignments. When used separately, matK phylogeny and trnL-trnF phylogeny were congruent with the combined analysis (data not shown). ML analysis using the combined alignment produced a tree with very similar topology (data not shown), and bootstraps of the corresponding nodes are indicated in Fig. 2.
FIG. 2.
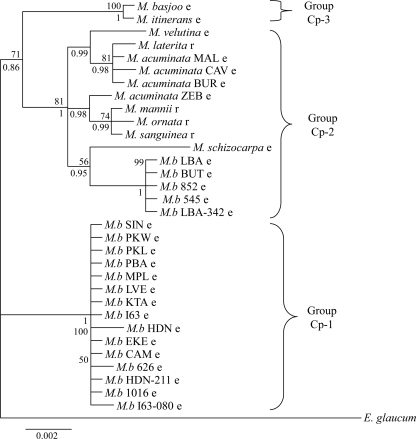
Bayesian tree of Musa based on the matk gene and trnL-trnF region concatenated into a single alignment of 2.1 kbp. Posterior probabilities are given below branches. Bootstrap values (over 50%) of the corresponding nodes in the ML tree are given above branches. Lowercase letters after species or accession names indicate the Eumusa (e) or Rhodochlamys (r) section, respectively. Ensete glaucum was used as outgroup. M.b, M. balbisiana.
This phylogeny separated the accessions into three, well-supported groups: Cp-1 to Cp-3 (Cp for chloroplast). Group Cp-1 contains most of the Musa balbisiana genotypes displaying very low diversity. The remaining accessions of M. balbisiana species (acc. ‘Butuhan’ [BUT], ‘Los Banos’ [LBA], LBA-342, -545, and -852) clustered in group Cp-2. This latter group also encompassed species belonging to the Rhodochlamys section, as well as the Eumusa species M. acuminata and M. schizocarpa. The phylogeny of group Cp-2 suggests close relationships between M. acuminata (section Eumusa) and species of the section Rhodochlamys. Our phylogenetic analysis suggested that Rhodochlamys is not a valid taxonomic group. Group 3 encompassed two closely related Eumusa species: M. itinerans and M. basjoo.
We further analyzed insertion/deletion mutations (indels) from matK and trnL-trnF, and their distribution in the chloroplast phylogeny. Four indels were informative, i.e., found in more than one Musa species and accessions (Table 4). Indel 1 was found in all M. balbisiana accessions of group Cp-2, as well as three species (M. mannii, M. sanguinea, and M. schizocarpa) that are closely related in the chloroplast phylogeny. Indel 2 was found specifically in all M. balbisiana accessions of group Cp-1. The last two indels were found in sister species according to the chloroplast phylogeny: M. itinerans and M. basjoo for indel 3 and M. mannii, M. ornata, and M. sanguinea for indel 4. The topology of the chloroplast phylogeny and the clustering of M. balbisiana were thus confirmed by indel data.
TABLE 4.
Shared indels from the chloroplast matK gene and the trnL-trnF region
| Indel no. | Locus | Positiona |
Size (nt) | Musa species (accessions concerned) | |
|---|---|---|---|---|---|
| Start | End | ||||
| 1 | matK | 326 | 340 | 15 | M. mannii, M. sanguinea, M. schizocarpa, M. balbisiana (LBA-342, 545, 852, BUT, LBA) |
| 2 | trnL-trnF | 247 | 261 | 15 | M. balbisiana (I63-080, 1016, HDN-211, 626, CAM, EKE, HDN, I63, KTA, LVE, MPL, PBA, PKL, PKW, SIN) |
| 3 | trnL-trnF | 341 | 365 | 25 | M. basjoo, M. itinerans |
| 4 | trnL-trnF | 813 | 832 | 20 | M. mannii, M. ornata, M. sanguinea |
Position refers to single-gene alignments (i.e., not concatenated).
Microsatellite phylogeny.
We performed a microsatellite-based analysis to assess the phylogeny of wild diploids M. balbisiana harboring eBSImV and eBSGFV. This provided a robust framework for the analysis of the distribution of both integrants among our Musa sampling. 31 diploid Musa accessions used in the present study (Table 1) were genotyped at 19 loci. Figure 3 represents the NJ tree inferred from microsatellite data and rooted by M. ornata (section Rhodochlamys) that clustered outside the Eumusa/Rhodochlamys group in studies based on morphological characters (60) and amplified fragment length polymorphism markers (67).
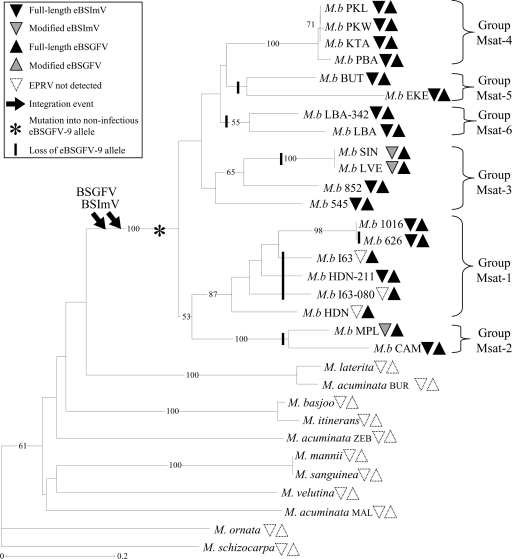
FIG. 3.
NJ tree of wild Musa species, reconstructed from the “Simple Matching” genetic distance from 19 microsatellite loci. Bootstrap values over 50% (percentage from 1,000 replicates) are shown to the left of the nodes. Distribution of eBSVs was investigated with eight PCR markers for eBSGFV and five PCR markers for eBSImV. Full-length eBSVs are represented by black triangles, modified BSVs are represented by shaded triangles, and the absence of eBSV is represented by empty triangles (see Table 1 for details). Downward arrows indicate inferred BSGFV and BSImV integration events. The presumed appearance of the noninfectious eBSGFV-9 allele and its loss in Musa genomes are represented by an asterisk and vertical lines, respectively. The tree was rooted with the Musa ornata sequence.
The 20 accessions of M. balbisiana species formed a monophyletic group (i.e., forming a clade that consists of all descendants belonging to a common ancestor) with a bootstrap value of 100% for the supporting node. Furthermore, the analysis of microsatellite variation (Table 2) revealed the presence of genetic diversity in this species. All microsatellites were polymorphic, with 10.3 different alleles per locus, on average. We observed high values of gene diversity He in M. balbisiana (average, 0.53; range, 0 to 0.75) and in the other Musa species (average, 0.87; range, 0.57 to 0.97). Furthermore, there were 54 private alleles (alleles detected only in one sample), 4 of which were specific for M. balbisiana species and 18 of which were specific for M. acuminata.
M. balbisiana species were structured into six groups. Groups Msat-1, Msat-2, and Msat-4 were supported by strong bootstrap value (87, 100, and 100%, respectively); the remaining groups were less supported. The microsatellite topology was congruent with the RFLP-based study of Carreel et al. (4), that included some of the M. balbisiana accessions used in the present work. We therefore numbered the M. balbisiana groups accordingly.
Distribution of eBSImV and eBSGFV among the Musa genus.
Detailed distribution of eBSV markers after genotyping of the 60 Musa accessions are presented in Table 1, and the eBSV distribution mapped on Musa microsatellite phylogeny is summarized in Fig. 3. To interpret eBSV distribution, we postulated that independent integrations would result in a distinct organization of the BSV fragments comprising them. This scenario would have been revealed by no amplification of eBSV markers during PCR and/or by amplification of eBSV markers of an unexpected size, which was never observed. The presence of a full-length eBSV in distantly related accessions is therefore interpreted as a shared character state. Conversely, we observed six modified eBSVs revealed by one or several missing markers (Fig. 3). These mutations likely correspond to derived, rather than shared, character states, since distinct fragments of eBSV were modified.
eBSImV and eBSGFV show the same distribution within the Musa genus since both are restricted to the species M. balbisiana. The other 13 species failed to amplify any eBSV markers. Since all six genetic groups of M. balbisiana harbor eBSImV and eBSGFV, these two integrations probably took place after M. acuminate and M. balbisiana speciation but before M. balbisiana diversification.
BSImV integration exhibited large structural polymorphism. Among the 20 M. balbisiana accessions, 3 lacked a variable number of eBSV markers at different positions within their integration (Table 1), attesting to modification or truncation of the integration. Such modified eBSVs were observed in several groups (Msat-2 and Msat-3), suggesting that the degradation of eBSImV was repeated during M. balbisiana evolution. Three other accessions found in group Msat-1 (I63, I-63-080, and HDN) completely lacked eBSImV, as evidenced by the absence of amplification of the 5 eBSV markers. This result suggested that a single event of loss of eBSImV occurred in the common ancestor of these three accessions. Note that the low resolution in the tips of the tree could explain why the monophyly of this group is not observed in Fig. 3.
In comparison, the structure of eBSGFV remained remarkably stable. The only observed polymorphism concerned the differentiation in two alleles found in cv. PKW: eaBSGFV-7 (infectious) and eBSGFV-9 (not infectious). Comparison of these two alleles suggested that eBSGFV-9 was derived from eaBSGFV-7 by accumulation of deleterious mutations in ORFs, together with an internal tandem duplication of 2.3 kbp (18). The distribution of the two alleles is listed in Table 1, and the results are summarized in the microsatellite phylogeny shown in Fig. 3. Three groups (Msat-2, -5, and -6) lacked the noninfectious eBSGFV-9 allele. In addition, the microsatellite tree showed no particular structure in the distribution of the eBSGFV-9 allele. Instead, the tree topology showed that eBSGFV-9 is present in all groups and corroborated the view that eBSGFV-9 appeared early, prior to the divergence into six groups. Interestingly, eBSGFV-9 is not associated with the release of BSV particles. We thus expected it to have followed the same evolutionary dynamics as advantageous alleles, i.e., its fixation during the diversification of M. balbisiana. On the contrary, we observed that independent loss of this allele occurred five times during M. balbisiana evolution (Fig. 3).
Age of integration and allelic divergence of eBSGFV.
In a previous study, we showed that eBSGFV was embedded within a Ty3/Gypsy retroelement. One hypothesis to explain this situation was a recombination event between the retroelement during its retrotransposition, and a BSGFV pregenomic RNA originating from a BSGFV infection in M. balbisiana. The chimera formed would have then retrotransposed into the M. balbisiana genome (18). The divergence between the two long terminal repeats (LTRs) sequences of this retroelement, estimated with ML using the HKY model implemented in PhyloWin (15), was used to determine the approximate date of integration. For eBSGFV, the substitution rate along the 351 bp of LTRs was estimated at 0.0058 substitutions per site. Applying an average synonymous substitution rate of 4.5 per 109 years for nuclear genes in the order Zingiberales, to which Musaceae belongs (38), eBSGFV would have integrated approximately 0.64 million years ago (MYA) [i.e., 0.0058 substitutions per site/(2 × 4.5 × 10−9 substitutions per site per year)]. The divergence time between the eaBSGFV-7 and eBSGFV-9 alleles was estimated by using the same approach. From analysis of a 13,280-bp alignment, their divergence was estimated at 0.0022 substitution per site, suggesting that they diverged approximately 0.24 MYA [0.0022 substitutions per site/(2 × 4.5 × 10−9 substitutions per site per year)].
Origin of BSGFV and BSImV integrants.
Phylogenetic relationships between endogenous and episomal BSGFV and BSImV sequences were investigated using ML phylogeny with 550 bp of the RT/RNase H region present in the ORFIII of the virus. We retrieved all publicly available sequences of episomal and endogenous BSImV and BSGFV and then sequenced the RT/RNase H region of eBSGFV from 20 M. balbisiana accessions.
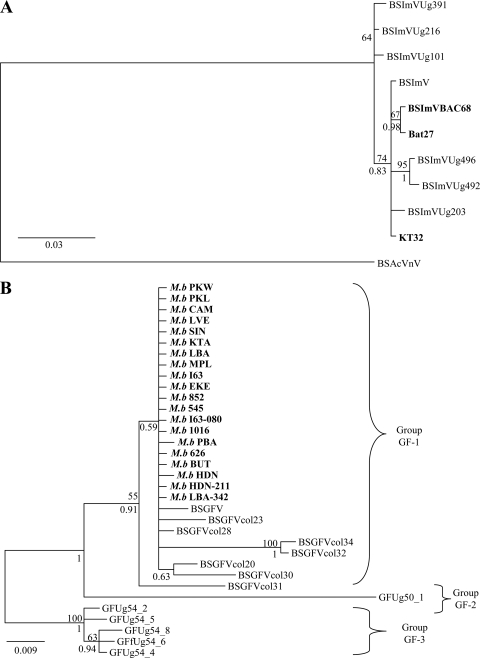
Episomal BSImV showed little sequence polymorphism, and endogenous sequences did not form a divergent cluster (Fig. 4A). The situation was different for BSGFV (Fig. 4B). This species was more polymorphic, and episomal sequences formed three distinct groups (named GF-1 to GF-3). Our results showed that eBSVs derived from group GF-1 viruses. In this group, we observed that the episomal BSGFV and eBSGFVs originated from the same node, suggesting a recent common ancestor. Furthermore, the branches leading to eBSVs were particularly short (<0.002 substitution/site on average), whereas those of the episomal sequence (BSGFV) were slightly longer (0.006 substitution/site). This result indicated a slower rate of evolution of eBSGFV compared to that of the corresponding episomal sequences.
FIG. 4.
Bayesian tree based on the RT/RNase H region (550 bp). Endogenous sequences found in the M. balbisiana genome are in boldface, episomal sequences are in normal font. Posterior probabilities are given below branches. Bootstrap values (>50%) of the corresponding nodes in ML tree are given above the branches. (A) Phylogeny of BSImV. The tree is rooted with the closely related species BSAcVNV. (B) Phylogeny of BSGFV. Phylogenetic groups among this viral species are indicated with brackets.
DISCUSSION
The aim of this study was to investigate the origin and evolution of infectious integrants of Banana streak virus (eBSV) for Imové- and GF-BSV species in the Musa genus. Using a PCR-based approach, we monitored the distribution of infectious eBSImV and eBSGFV in the genome of 60 accessions, using 13 specific eBSV markers. By focusing on diploid M. balbisiana genotypes and several closely related banana species, we were able to set BSV integrations in a host phylogenetic context, an essential step toward dissecting the early evolution stages of these integration events.
Nearly 2,000 accessions exist in Musa germplasm collections. Accordingly, banana phylogenetic studies have thus far been aimed at organizing the huge amount of genetic resources needed for agronomical purposes. Previous studies have focused mostly on domesticated genotypes, such as diploid or triploid M. acuminata-derived cultivars (AA or AAA), or polyploid interspecific cultivars (AB, AAB, ABB, and AAAB) rather than on wild species (10, 27, 50). In addition, very few studies have included a representative sample of the genetic diversity of M. balbisiana species (68, 70) in which infectious eBSVs have been discovered. To circumvent this problem, we constructed a phylogeny of wild Musa species with an emphasis on the diploid M. balbisiana species, using two chloroplast genes and 19 nuclear microsatellites.
Mainly due to the low level of polymorphism, the chloroplast phylogeny did not achieve reconstruction of the phylogenetic relationships among M. balbisiana accessions. However, it provided valuable markers for observing the gene flows of the chloroplast genome, which is inherited maternally in banana plants (14). In the chloroplast phylogeny, five M. balbisiana species clustered apart from the rest, near Rhodochlamys species (group Cp-2 in Fig. 2). This discrepancy was confirmed by both molecular phylogeny and the analysis of informative indels. This result strongly suggests ancient chloroplast gene flows between M. balbisiana and other species. Based on RFLP markers on cpDNA, Carreel et al. (4) showed that the M. balbisiana accession ‘Butuhan’ (BUT) differed in its chloroplast pattern and was closer to that of Australimusa species. BUT was suspected to be a backcross of M. balbisiana with an interspecies hybrid (M. textilis × M. balbisiana). The introgression of the chloroplast genome from M. textilis could thus explain the intermediary position of M. balbisiana accessions within group Cp-2, as observed in the chloroplast phylogeny. We hypothesize that hybridization between Musa species might have allowed the spread of eBSVs other than BSGFV or BSImV throughout the genus Musa. To test this hypothesis, as well as to gain a better understanding of the evolutionary dynamics of eBSV in the Musa genome, we must now take into account the huge biodiversity of BSV species, since dozens of such species are integrated in the Musa genome (2, 17, 20).
In the last decade, microsatellite analysis has been used successfully to infer accurate phylogenetic relationships (58). The phylogeny proposed here is based on a microsatellite analysis of 19 polymorphic loci, since several recent studies using between 5 and 20 loci provided strong phylogenetic signals and accurate phylogenetic reconstructions in both plant and vertebrates species (6, 46, 52, 54). Several lines of evidence suggest that microsatellite-based phylogeny is robust and can be trusted. Despite the presence of homoplasy (i.e., similarities due to convergent evolution) in the allele size occurring between divergent species, the use of microsatellites is possible at the intraspecific level (58), such as among M. balbisiana genotypes and the closely related Musa species. Furthermore, several observations indicated that this phylogeny produced accurate results among closely related species. First, as expected between distant species, the basal branching order was poorly resolved, and the nodes displayed low bootstrap values (<50%), whereas the topology was well resolved and robust among the M. balbisiana genotypes. Second, the closely related species detected in chloroplast phylogeny such as M. laterita and M. acuminata subsp. burmanica, M. basjoo and M. itinerans, or M. manii and M. sanguinea, remained also closely related in microsatellite phylogeny. Third, all M. balbisiana accessions clustered in a single group. Fourth, previous studies on Musa genetic diversity found close genetic relationships between the four M. balbisiana accessions PKL, PKW, KTA, and PBA, between the two accessions SIN and LVE (4), and between the two accessions BUT and EKE (67). These accessions remained closely related in the microsatellite phylogeny. Fifth, based on chloroplast and mitochondrial DNA RFLPs, Carreel et al. (4) described genetic groups from nine accessions of M. balbisiana species, and a very good agreement of accession clustering was also observed in the present study using the 20 accessions available. Carreel et al. (4) observed that HDN, CAM, and BUT belong to three distinct groups, LVE and SIN to a fourth group, and PKL, PKW, KTA, and PBA to a fifth group. By reflecting the evolution of the whole nuclear genome, microsatellite phylogeny revealed an important polymorphism of M. balbisiana and confirmed the monophyly of the species. The study of the evolution of infectious eBSGFV and eBSImV within M. balbisiana species was based on this phylogenetic framework.
The structure of eBSV is complex, and it is thought to have arisen randomly via illegitimate recombinations (28, 63). We assumed that each independent eBSV (i.e., those not arising by duplication) has a unique structure. To detect the structure of eBSGFV and eBSImV and their locus of integration in the Musa genome, we designed PCR markers specific for each internal rearrangement and for the two flanking regions of the plant genome. Recording of all of these markers gives a signature for each eBSV. As a result, the presence of the same signature in different banana genomes revealed orthologous eBSVs sharing a common ancestor. The analysis of their distribution in our Musa sampling allowed us to begin to unravel the origin of BSV integration.
The distribution patterns of infectious eBSImV and eBSGFV in Musa were very similar. They were both restricted to the M. balbisiana genome and absent from the other wild Musa species. eBSImV and eBSGFV are composed of repeated homologous regions, a characteristic common to all known eBSV (18, 47). Such repeats are good templates for intra-eBSV recombination. We therefore expected a rapid evolution of their structure. This expectation was confirmed for eBSImV only, since the eBSGFV structure remained unaltered. Further studies of the evolutionary dynamics of the genome at the integration site, and of the type of selection acting on the integrant itself, are now required to better understand the consequence of these factors on the fate of eBSV.
The two allelic forms of eBSGFV, the infectious eaBSGFV-7 and the noninfectious eBSGFV-9, are present in the M. balbisiana genome. Our study enabled us to follow the emergence, fixation, and loss of both alleles in M. balbisiana species. Analysis of the mutation pattern suggested that the infectious allele appeared first, followed by its mutation into eBSGFV-9 (18). The molecular detection of eBSGFV-9 was based on the presence of a specific indel, suggesting that all of the eBSGFV-9 detected originated from a single event. eBSGFV-9 is present in half of the genetic groups (groups Msat-1, -3, and -4) of M. balbisiana. As a nondeleterious allele, we expected that eBSGFV-9 would have invaded wild M. balbisiana populations and eventually replaced the deleterious eaBSGFV-7 allele. However, we observed the opposite. The infectious allele was retained, and the noninfectious allele was lost several times independently. We hypothesize that infectious eaBSGFV-7 was conserved in the M. balbisiana genome because it plays, or has played, some beneficial role in the evolution of the host. Such integrants are indeed suspected to induce a resistance against viral multiplication whatever its origin, whether cognate episomal virus or eBSVs (28, 32, 45, 63). M. balbisiana cv. PKW, and probably other accessions of this species, have evolved resistance against infectious eBSV and BSV infection (33). Only interspecific hybrids with M. acuminata are sensitive to infectious eBSV. Infectious eBSV would therefore be beneficial for M. balbisiana plants but deleterious for the interspecific hybrids that occur naturally in the sympatric area of Southeast Asia (50, 59). Synthetic hybrids from contemporaneous M. balbisiana × M. acuminata are polyploid and poorly fertile, and inbreeding depression would have prevented large introgression of nuclear DNA between these two species in natural populations (59). Infectious eBSVs might also have contributed to reinforcing speciation of M. balbisiana and M. acuminata species by producing BSV-infected hybrids with lower fitness.
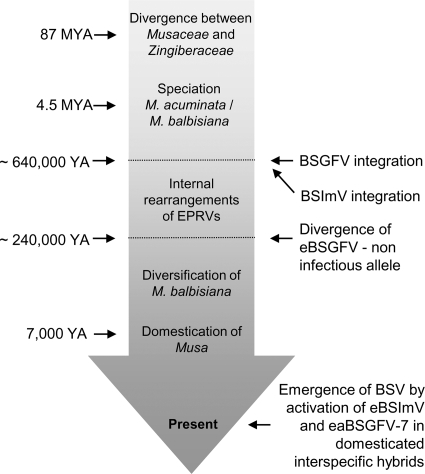
Large-scale phylogenetic analysis conducted on different episomal and integrated BSV species has previously suggested that the majority of integrations in the Musa genomes occurred after speciation between M. acuminata and M. balbisiana (17, 20). The distribution analysis in the present study firmly confirms that BSGFV and BSImV integrated after this Musa speciation, which occurred ca. 4.5 MYA (38). Analysis of the substitution rate at the eBSGFV locus indicated that the origin of integrations was even more recent, ca. 0.63 MYA. Allelic divergence between infectious and noninfectious eBSGFV followed chronologically and arose circa 0.23 MYA ago. The evolutionary history of eBSVs and their Musa host are summarized in Fig. 5. Even though the divergence dates are approximate, based on rough estimates of minimum divergence times, this approach has been used successfully for transposable elements in other plants (57). Since divergence is likely to be diminished by gene conversion occurring between homologous sequences of eBSVs, this method is conservative and tends to underestimate divergence time. eBSVs are young elements in the banana genome and are still infectious elements displaying functional coding and regulatory viral sequences.
FIG. 5.
Dates and events associating the evolutionary history of eBSVs and their host. Schematic diagram illustrating key events in the evolutionary history of BSImV and BSGFV described in the present study in association with estimated dates during the evolution of the Musa genus. Estimation of the age of the most recent common ancestor for the Musacaceae and Zingiberaceae is taken from (3, 36, 56) and between M. acuminata and M. balbisiana from (38). The date of the emergence of agriculture and domestication of the banana is from reference 11.
Our study has finally allowed us to retrace the phylogenetic relationships between integrated sequences and episomal viruses in domesticated bananas carrying the B genome. We found little variation between BSImV sequences, from either an integrated or an episomal origin. This could indicate that episomal BSImV sampled in Colombian and Ugandan banana crops both derive from recent activation of the same eBSV. However, additional BSImV sequence data are now required to confirm that episomal BSImV sequences truly display low polymorphism and to show that insufficient sampling is not an alternative causal factor. Concerning BSGFV species, we confirmed the presence of three distinct genetic groups as previously described (25), and showed that eBSV is derived from group 1 viruses. The episomal “BSGFV” sequence used in the present study came from first complete BSGFV genome sequenced, isolated primarily from an interspecific hybrid containing the B genome. Interestingly, this episomal virus clustered very close to an eBSV clade. This suggests that episomal virus could originate from activation of infectious EPRVs. We furthermore expected that episomal virus would accumulate more mutations than eBSV sequences, because BSV genomes evolve faster than those of Musa (17). This trend was confirmed by observing longer branches for episomal sequences than in endogenous sequences.
Our study has shown that endogenous BSGFV and BSImV sequences are useful in retracing the recent history of infectious eBSVs. Despite the fact that eBSImV and eBSGFV lineages are approximately the same age, their evolutionary fates were distinct. The structure of eBSGFV was conserved, whereas that of eBSImV was relatively altered. Further studies will be required to link the polymorphism of eBSVs and their ability to reconstitute infectious virus. Since both eBSVs are still able to produce infectious episomal virus, they have undeniably played an important role in the evolution of the banana-BSV interaction, as a reservoir protecting viral populations from local extinction.
Acknowledgments
P.G. was supported by Ph.D. grant CIRAD-Région Languedoc Roussillon.
We thank Serge Galzi, Nathalie Laboreau, Marie Umber, and Pierre Olivier Duroy for technical assistance; Liying Zhang and Benham E. L. Lockhart for providing the two BSGFV clones; Andrew Geering for sequencing the BSImV genome; and the curators of Musa collections for providing plant material (Christophe Jenny [Station de Recherches Fruitières de Neufchâteau, CIRAD], Ines Van Den Houwe, and A. M. Ayodele [INIBAP Transit Center-ITC, Katholieke Universiteit Leuven], and Perpetua Udu and A. Tenkouano [International Institute of Tropical Agriculture, Onne Station]). We are also very grateful to Elisabeth Fournier, Didier Tharreau, and one anonymous reviewer for critical reading of the manuscript.
Footnotes
Published ahead of print on 28 April 2010.
REFERENCES
- 1.Bennetzen, J. L. 2000. Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42:251-269. [PubMed] [Google Scholar]
- 2.Bousalem, M., E. J. P. Douzery, and S. E. Seal. 2008. Taxonomy, molecular phylogeny, and evolution of plant reverse transcribing viruses (family Caulimoviridae) inferred from full-length genome and reverse transcriptase sequences. Arch. Virol. 153:1085-1102. [DOI] [PubMed] [Google Scholar]
- 3.Bremer, K. 2000. Early cretaceous lineages of monocot flowering plants. Proc. Natl. Acad. Sci. U. S. A. 97:4707-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carreel, F., D. G. de Leon, P. Lagoda, C. Lanaud, C. Jenny, J. P. Horry, and H. T. du Montcel. 2002. Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome 45:679-692. [DOI] [PubMed] [Google Scholar]
- 5.Cheesman, E. E. 1947. Classification of the banana. Kew Bull. 2:97-117. [Google Scholar]
- 6.Chirhart, S. E., R. L. Honeycutt, and I. F. Greenbaum. 2005. Microsatellite variation and evolution in the Peromyscus maniculatus species group. Mol. Phylogenet. Evol. 34:408-415. [DOI] [PubMed] [Google Scholar]
- 7.Crouch, H. K., J. H. Crouch, R. L. Jarret, P. B. Cregan, and R. Ortiz. 1998. Segregation at microsatellite loci in haploid and diploid gametes of Musa. Crop Sci. 38:211-217. [Google Scholar]
- 8.Cuenoud, P., V. Savolainen, L. W. Chatrou, M. Powell, R. J. Grayer, and M. W. Chase. 2002. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 89:132-144. [DOI] [PubMed] [Google Scholar]
- 9.Dallot, S., P. Acuna, C. Rivera, P. Ramirez, F. Cote, B. E. L. Lockhart, and M. L. Caruana. 2001. Evidence that the proliferation stage of micropropagation procedure is determinant in the expression of Banana streak virus integrated into the genome of the FHIA 21 hybrid (Musa AAAB). Arch. Virol. 146:2179-2190. [DOI] [PubMed] [Google Scholar]
- 10.De Langhe, E., L. Vrydaghs, P. de Maret, X. Perrier, and T. Denham. 2009. Why bananas matter: an introduction to the history of banana domestication. Ethnobot. Res. Applications 7:165-177. [Google Scholar]
- 11.Denham, T. P., S. G. Haberle, C. Lentfer, R. Fullagar, J. Field, M. Therin, N. Porch, and B. Winsborough. 2003. Origins of agriculture at Kuk Swamp in the highlands of New Guinea. Ann. Bot. 301:189-193. [DOI] [PubMed] [Google Scholar]
- 12.Duffy, S., L. A. Shackelton, and E. C. Holmes. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9:267-276. [DOI] [PubMed] [Google Scholar]
- 13.Fargette, D., G. Konate, C. Fauquet, E. Muller, M. Peterschmitt, and J. M. Thresh. 2006. Molecular ecology and emergence of tropical plant viruses. Annu. Rev. Phytopathol. 44:235-260. [DOI] [PubMed] [Google Scholar]
- 14.Faure, S., J. L. Noyer, F. Carreel, J. P. Horry, F. Bakry, and C. Lanaud. 1994. Maternal inheritance of chloroplast genome and paternal inheritance of mitochondrial genome in bananas (Musa acuminata). Curr. Genet. 25:265-269. [DOI] [PubMed] [Google Scholar]
- 15.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 16.Gawel, N. J., and R. L. Jarret. 1991. A modified CTAB DNA extraction procedure for Musa and Ipomea. Plant Mol. Biol. Rep. 9:262-266. [Google Scholar]
- 17.Gayral, P., and M. L. Iskra-Caruana. 2009. Phylogeny of banana streak virus reveals recent and repetitive endogenization in the genome of its banana host (Musa sp.). J. Mol. Evol. 69:65-80. [DOI] [PubMed] [Google Scholar]
- 18.Gayral, P., J.-C. Noa-Carrazana, M. Lescot, F. Lheureux, B. E. L. Lockhart, T. Matsumoto, P. Piffanelli, and M.-L. Iskra-Caruana. 2008. A single Banana streak virus integration event in the banana genome as the origin of infectious endogenous pararetrovirus. J. Virol. 82:6697-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge, X. J., M. H. Liu, W. K. Wang, B. A. Schaal, and T. Y. Chiang. 2005. Population structure of wild bananas, Musa balbisiana, in China determined by SSR fingerprinting and cpDNA PCR-RFLP. Mol. Ecol. 14:933-944. [DOI] [PubMed] [Google Scholar]
- 20.Geering, A. D. W., N. E. Olszewski, G. Harper, B. E. L. Lockhart, R. Hull, and J. E. Thomas. 2005. Banana contains a diverse array of endogenous badnaviruses. J. Gen. Virol. 86:511-520. [DOI] [PubMed] [Google Scholar]
- 21.Goudet, J. 1995. FSTAT (version 1.2): a computer program to calculate F-statistics. J. Hered. 86:485-486. [Google Scholar]
- 22.Gregor, W., M. F. Mette, C. Staginnus, M. A. Matzke, and A. J. M. Matzke. 2004. A distinct endogenous pararetrovirus family in Nicotiana tomentosiformis, a diploid progenitor of polyploid tobacco. Plant Physiol. 134:1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 24.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 25.Harper, G., D. Hart, S. Moult, R. Hull, A. Geering, and J. Thomas. 2005. The diversity of Banana streak virus isolates in Uganda. Arch. Virol. 12:2407-2420. [DOI] [PubMed] [Google Scholar]
- 26.Harr, B., S. Weiss, J. R. David, G. Brem, and C. Schlötterer. 1998. A microsatellite-based multilocus phylogeny of the Drosophila melanogaster species complex. Curr. Biol. 8:1183-1187. [DOI] [PubMed] [Google Scholar]
- 27.Heslop-Harrison, J. S., and T. Schwarzacher. 2007. Domestication, genomics and the future for banana. Ann. Bot. 100:1073-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hohn, T., K. R. Richert-Pöggeler, G. Harper, T. Schwarzacher, C. H. Teo, P. Y. Techeney, M. L. Iskra-Caruana, and R. Hull. 2008. Evolution of integrated plant viruses, p. 58-81. In M. Roossinck (ed.), Plant virus evolution. Springer, Heidelberg, Germany.
- 29.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 30.Hull, R. 1999. Classification of reverse transcribing elements: a discussion document. Arch. Virol. 144:209-214. [DOI] [PubMed] [Google Scholar]
- 31.Hull, R., and S. N. Covey. 1995. Retroelements: propagation and adaptation. Virus Genes 11:105-118. [DOI] [PubMed] [Google Scholar]
- 32.Hull, R., G. Harper, and B. Lockhart. 2000. Viral sequences integrated into plant genomes. Trends Plant Sci. 5:362-365. [DOI] [PubMed] [Google Scholar]
- 33.Iskra-Caruana, M. L., F. Lheureux, J. C. Noa-Carrazana, P. Piffanelli, F. Carreel, C. Jenny, N. Laboureau, and B. E. L. Lockhart. 2003. Unstable balance of relation between pararetrovirus and its host plant: the BSV-EPRV banana pathosystem, p. 8. EMBO Workshop: Genomic Approaches in Plant Virology, Keszthely, Hungary.
- 34.Jakowitsch, J., M. F. Mette, J. van der Winden, M. A. Matzke, and A. J. M. Matzke. 1999. Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants. Proc. Natl. Acad. Sci. U. S. A. 96:13241-13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kidwell, M. G., and D. R. Lisch. 2000. Transposable elements and host genome evolution. Trends Ecol. Evol. 15:95-99. [DOI] [PubMed] [Google Scholar]
- 36.Kress, W. J., L. M. Prince, W. J. Hahn, and E. A. Zimmer. 2001. Unraveling the evolutionary radiation of the families of the Zingiberales using morphological and molecular evidence. Syst. Biol. 50:926-944. [DOI] [PubMed] [Google Scholar]
- 37.Lagoda, P. J., J. L. Noyer, D. Dambier, F. C. Baurens, A. Grapin, and C. Lanaud. 1998. Sequence-tagged microsatellite site (STMS) markers in the Musaceae. Mol. Ecol. 7:659-663. [PubMed] [Google Scholar]
- 38.Lescot, M., P. Piffanelli, A. Y. Ciampi, M. Ruiz, G. Blanc, J. Leebens-Mack, F. R. da Silva, C. M. Santos, A. D'Hont, O. Garsmeur, A. D. Vilarinhos, H. Kanamori, T. Matsumoto, C. M. Ronning, F. Cheung, B. J. Haas, R. Althoff, T. Arbogast, E. Hine, G. J. Pappas, T. Sasaki, M. T. Souza, R. N. Miller, J. C. Glaszmann, and C. D. Town. 2008. Insights into the Musa genome: syntenic relationships to rice and between Musa species. BMC Genomics 9:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lheureux, F., F. Carreel, C. Jenny, B. Lockhart, and M. Iskra-Caruana. 2003. Identification of genetic markers linked to banana streak disease expression in inter-specific Musa hybrids. Theor. Appl. Genet. 106:594-598. [DOI] [PubMed] [Google Scholar]
- 40.Lheureux, F., N. Laboureau, E. Muller, B. E. Lockhart, and M. L. Iskra-Caruana. 2007. Molecular characterization of Banana streak acuminata Vietnam virus isolated from Musa acuminata siamea (banana cultivar). Arch. Virol. 152:1409-1416. [DOI] [PubMed] [Google Scholar]
- 41.Lockhart, B., and D. Jones. 2000. Banana streak, p. 263-274. In D. R. Jones (ed.), Diseases of banana, abaca, and enset. CAB International, Wallingford, United Kingdom.
- 42.Lockhart, B. E., J. Menke, G. Dahal, and N. E. Olszewski. 2000. Characterization and genomic analysis of Tobacco vein clearing virus, a plant pararetrovirus that is transmitted vertically and related to sequences integrated in the host genome. J. Gen. Virol. 81:1579-1585. [DOI] [PubMed] [Google Scholar]
- 43.Malik, H. S., and T. H. Eickbush. 2001. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 11:1187-1197. [DOI] [PubMed] [Google Scholar]
- 44.Matzke, M., W. Gregor, M. F. Mette, W. Aufstatz, T. Kanno, J. Jakowitsch, and A. J. M. Matzke. 2004. Endogenous pararetroviruses of allotetraploid Nicotiana tabacum and its diploid progenitors, N. sylvestris and N. tomentosiformis. Biol. J. Linn. Soc. 82:627-638. [Google Scholar]
- 45.Mette, M. F., T. Kanno, W. Aufsatz, J. Jakowitsch, J. van der Winden, M. A. Matzke, and A. J. M. Matzke. 2002. Endogenous viral sequences and their potential contribution to heritable virus resistance in plants. EMBO J. 21:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muir, G., C. C. Fleming, and C. Schlotterer. 2000. Species status of hybridizing oaks. Nature 405:1016. [DOI] [PubMed] [Google Scholar]
- 47.Ndowora, T., G. Dahal, D. LaFleur, G. Harper, R. Hull, N. E. Olszewski, and B. Lockhart. 1999. Evidence that Badnavirus infection in Musa can originate from integrated pararetroviral sequences. Virology 255:214-220. [DOI] [PubMed] [Google Scholar]
- 48.Noreen, F., R. Akbergenov, T. Hohn, and K. R. Richert-Pöggeler. 2007. Distinct expression of endogenous Petunia vein clearing virus and the DNA transposon dTph1 in two Petunia hybrida lines is correlated with differences in histone modification and siRNA production. Plant J. 50:219-229. [DOI] [PubMed] [Google Scholar]
- 49.Pahalawatta, V., K. Druffel, and H. Pappu. 2008. A new and distinct species in the genus Caulimovirus exists as an endogenous plant pararetroviral sequence in its host, Dahlia variabilis. Virology 376:253-257. [DOI] [PubMed] [Google Scholar]
- 50.Perrier, X., F. Bakry, F. Carreel, C. Jenny, J.-P. Horry, V. Lebot, and I. Hippolyte. 2009. Combining biological approaches to shed light on the evolution of edible bananas. Ethnobot. Res. Applications 7:199-216. [Google Scholar]
- 51.Philippe, H., and P. Forterre. 1999. The rooting of the universal tree of life is not reliable. J. Mol. Evol. 49:509-523. [DOI] [PubMed] [Google Scholar]
- 52.Richard, M., and R. S. Thorpe. 2001. Can. microsatellites be used to infer phylogenies? Evidence from population affinities of the Western Canary Island lizard (Gallotia galloti). Mol. Phylogenet. Evol. 20:351-360. [DOI] [PubMed] [Google Scholar]
- 53.Richert-Pöggeler, K. R., F. Noreen, T. Schwarzacher, G. Harper, and T. Hohn. 2003. Induction of infectious petunia vein clearing (pararetro) virus from endogenous provirus in petunia. EMBO J. 22:4836-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritz, L. R., M. L. Glowatzki-Mullis, D. E. MacHugh, and C. Gaillard. 2000. Phylogenetic analysis of the tribe Bovini using microsatellites. Anim. Genet. 31:178-185. [DOI] [PubMed] [Google Scholar]
- 55.Salemi, M., and A. M. Vandamme (ed.). 2003. The phylogenetic handbook: a practical approach to DNA and protein phylogeny. Cambridge University Press, Cambridge, United Kingdom.
- 56.Sanderson, M. J., J. L. Thorne, N. Wikstrom, and K. Bremer. 2004. Molecular evidence on plant divergence times. Am. J. Bot. 91:1656-1665. [DOI] [PubMed] [Google Scholar]
- 57.SanMiguel, P., B. S. Gaut, A. Tikhonov, Y. Nakajima, and J. L. Bennetzen. 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20:43-45. [DOI] [PubMed] [Google Scholar]
- 58.Schlotterer, C. 2001. Genealogical inference of closely related species based on microsatellites. Genet. Res. 78:209-212. [DOI] [PubMed] [Google Scholar]
- 59.Simmonds, N. W. (ed.). 1962. The evolution of the bananas. Longmans Green, London, England.
- 60.Simmonds, N. W., and S. T. C. Weatherup. 1990. Numerical taxonomy of the wild bananas (Musa). New Phytol. 115:567-571. [DOI] [PubMed] [Google Scholar]
- 61.Staginnus, C., W. Gregor, M. F. Mette, C. H. Teo, E. G. Borroto-Fernandez, M. L. Machado, M. Matzke, and T. Schwarzacher. 2007. Endogenous pararetroviral sequences in tomato (Solanum lycopersicum) and related species. BMC Plant Biol. 7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Staginnus, C., M. L. Iskra-Caruana, B. Lockhart, T. Hohn, and K. R. Richert-Poggeler. 2009. Suggestions for a nomenclature of endogenous pararetroviral sequences in plants. Arch. Virol. 154:1189-1193. [DOI] [PubMed] [Google Scholar]
- 63.Staginnus, C., and K. R. Richert-Pöggeler. 2006. Endogenous pararetroviruses: two-faced travelers in the plant genome. Trends Plant Sci. 11:485-491. [DOI] [PubMed] [Google Scholar]
- 64.Su, L., S. Gao, Y. Huang, C. Ji, D. Wang, Y. Ma, R. Fang, and X. Chen. 2007. Complete genomic sequence of Dracaena mottle virus, a distinct Badnavirus. Virus Genes 35:423-429. [DOI] [PubMed] [Google Scholar]
- 65.Taberlet, P., L. Gielly, G. Pautou, and J. Bouvet. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17:1105-1109. [DOI] [PubMed] [Google Scholar]
- 66.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ude, G., M. Pillay, D. Nwakanma, and A. Tenkouano. 2002. Analysis of genetic diversity and sectional relationships in Musa using AFLP markers. Theor. Appl. Genet. 104:1239-1245. [DOI] [PubMed] [Google Scholar]
- 68.Ude, G., M. Pillay, D. Nwakanma, and A. Tenkouano. 2002. Genetic diversity in Musa acuminata Colla and Musa balbisiana Colla and some of their natural hybrids using AFLP markers. Theor. Appl. Genet. 104:1246-1252. [DOI] [PubMed] [Google Scholar]
- 69.Uma, S., S. A. Siva, M. S. Saraswathi, P. Durai, T. Sharma, D. B. Singh, R. Selvarajan, and S. Sathiamoorthy. 2005. Studies on the origin and diversification of Indian wild banana (Musa balbisiana) using arbitrarily amplified DNA markers. J. Hortic. Sci. Biotech. 80:575-580. [Google Scholar]
- 70.Wong, C., R. Kiew, G. Argent, O. Set, S. K. Lee, and Y. Y. Gan. 2002. Assessment of the validity of the sections in Musa (Musaceae) using AFLP. Ann. Bot. London 90:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xia, X. 1999. DAMBE (Software Package for Data Analysis in Molecular Biology and Evolution) user manual. Department of Ecology and Biodiversity, University of Hong Kong.
- 72.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]
- 73.Xia, X., Z. Xie, M. Salemi, L. Chen, and Y. Wang. 2003. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 26:1-7. [DOI] [PubMed] [Google Scholar]