Abstract
The human immunodeficiency virus type 1 (HIV-1) protein Vif recruits the host E3 ubiquitin ligase, composed of cullin 5 (Cul5), Rbx2, Elongin B, and Elongin C (EloBC), to polyubiquitinate the antiviral protein APOBEC3G. Multiple regions in the C-terminal half of Vif interact with the E3 ligase. We have purified individual regions of Vif and investigated their thermodynamic contributions to the ligase assembly in vitro using isothermal titration calorimetry and fluorescence anisotropy. Our results quantify the high-affinity interactions between the Vif BC box and EloBC and between the Vif zinc finger and Cul5, as well as the modest interaction between the Vif cullin box and Cul5. Our purified Vif constructs also provide direct biochemical evidence that the Vif cullin box, containing the PPLP region, leads to the dimerization of Vif-EloBC complexes but not Cul5-Vif-EloBC complexes.
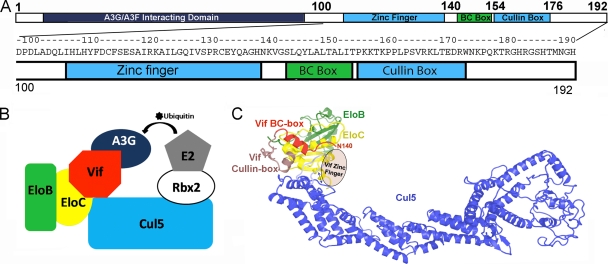
HIV Vif antagonizes the human antiviral protein APOBEC3G by hijacking the human Elongin B/C (EloBC)-cullin-SOCS box (ECS)-type E3 ubiquitin ligase, resulting in the polyubiquitination of APOBEC3G and subsequently its proteasomal degradation. Canonical ECS-type ubiquitin ligases consist of a cullin scaffold protein to which adaptor and substrate receptor proteins bind at the N terminus. HIV Vif serves as a substrate receptor protein—its N terminus recruits APOBEC3G, while multiple C-terminal regions assemble with the E3 ligase (9, 13, 24). The E3 ligase interacting regions include a zinc finger (residues 100 to 140), a BC box (residues 141 to 154), and a cullin box (residues 155 to 176) (Fig. 1).
FIG. 1.
(A) A sequence schematic of Vif showing the regions that interact with A3G, A3F, EloBC, and Cul5. (B) An illustration of the assembly of the Vif-E3 ubiquitin ligase. (C) A homology model of Vif-Cul5-EloBC, where the Vif BC box-EloBC is actual structural data (PDB ID 3DCG).
Vif binds the cullin adaptor proteins EloB and EloC through the BC-box region (24). The BC box is a loop-helix motif with the consensus sequence (T/S)LxxxCxxx(V/L/I) (7), and it also exists in cellular proteins that interact with EloBC. While Vif does not fit this consensus perfectly, it still binds EloBC with high affinity, and this interaction is lost upon mutation or deletion of consensus BC-box residues (10, 24, 25). This interaction has been described previously for the cellular proteins VHL (15), SOCS2 (3), SOCS3 (1), SOCS4 (4), and recently HIV Vif (14).
Both the Vif zinc finger and cullin box interact with the E3 ligase scaffold protein cullin 5 (Cul5) (11, 12, 20, 21). It has been established that the zinc finger is required for Vif to bind Cul5. Mutation of critical histidine or cysteine residues in this region or the addition of the zinc chelator N,N,N′,N′-tetrakis(2-pyridylmethyl)-ethylenediamine (TPEN) abolishes the Vif-Cul5 interaction (8, 11, 25). The sequence of the Vif cullin box is not as conserved as those of cellular SOCS-box proteins, which have a defined structure and determine the specificities of their respective cullins (6). The role of the Vif cullin box is not clear, but it has been suggested to promote dimerization of Vif, involving the conserved PPLP region (22, 23), and has recently been implicated in APOBEC3G binding (5, 17). While its importance in Cul5 binding has been demonstrated in coimmunoprecipitation experiments (14), experimental data also exist showing that the Vif zinc finger alone still immunoprecipitates Cul5 (11, 21).
To dissect the assembly of the Vif-E3 ubiquitin ligase, we quantified the binding interactions between various C-terminal Vif constructs, EloBC, and Cul5 by isothermal titration calorimetry (ITC) and fluorescence polarization (FP). We additionally probed the effects of the cullin box on Vif dimerization.
MATERIALS AND METHODS
Cloning, expression, and purification.
Vif constructs were cloned into pETDuet-1 (Novagen) with an N-terminal His tag and TEV protease cleavage site. EloB and EloC (residues 17 to 112) were expressed from the pACYCDuet plasmid (a gift from Alex Bullock, University of Oxford, Oxford, United Kingdom). The Vif BC-box mutations were made by site-directed mutagenesis. The proteins were overexpressed in Escherichia coli BL21(DE3) cells overnight at 18°C by induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Purification consisted of nickel affinity, anion exchange, His tag removal, and gel filtration. Murine Cul5 N-terminal domain (NTD) was purified as previously described (1). Briefly, Cul5 NTD was expressed in E. coli BL21(DE3) cells from pGEX-4T-1 overnight at 18°C by induction with 0.5 mM IPTG. Purification consisted of glutathione S-transferase (GST) affinity, GST tag removal, and gel filtration chromatography.
Isothermal titration calorimetry.
ITC experiments were performed using a Microcal iTC200 microcalorimeter. Prior to each experiment, proteins were dialyzed overnight against 1 liter of 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 0.1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in 3,500-molecular-weight-cutoff (MWCo) minidialysis units (Thermo). All experiments were performed at 25°C with a 10-min pretitration delay and 1,000-rpm stirring speed. Data analyses for ITC experiments were performed using the Origin 7.0 software package. Calorimetric data curves were fit to a single-site binding model. Control experiments were performed by injecting protein into buffer to correct for the heat of dilution during titration. Each ITC experiment was conducted three times to ensure the reproducibility and reliability of the results, except in one case (binding of Vif139-192-EloBC to Cul5), where it was performed twice due to the limitation of the large amount of materials required. For the wild-type (WT) Vif experiments, a competition assay was employed. The affinity of Vif139-192 L148A to EloBC was determined, and then WT Vif139-192 was titrated into the Vif139-192-EloBC complex. The WT Vif139-192-EloBC affinity was then calculated using the equation Kapp = K/(1 + Kmutant[Vif139-192 L148A]) (16), where Kapp, K, and Kmutant are the dissociation constants for the competition assay, WT Vif, and the Vif mutant, respectively.
Fluorescence polarization.
Increasing amounts of EloBC (0 to 100 μM) were titrated into 1 nM fluorescein isothiocyanate (FITC)-labeled Vif139-154 (synthesized by Yale University Keck Facility) in buffer (25 mM Tris-HCl [pH 7.5], 100 mM NaCl, and 0.1 mM TCEP). Samples were excited at 480 nm with polarized light, and emissions at 535 nm were monitored. The depolarization signal generated upon the formation of the FITC-Vif139-154-EloBC complex was calculated (ΔmP). Data were fit to the following binding equation: ΔmP = ΔmPmax[EloBC]/(Kd + [EloBC]), where Kd is the dissociation constant.
Multiangle laser light-scattering and size exclusion chromatography.
Each Vif complex sample was concentrated to 50 μM, and 500 μl was loaded onto a Superdex 75 (10/300 GL) column (GE), which had been calibrated using cytochrome c (molecular weight, 12,400), carbonic anhydrase (29,000), ovalbumin (44,000), and bovine serum albumin (BSA) (67,000). Multiangle laser light-scattering experiments were performed in buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCL, and 0.1 mM TCEP) at room temperature. Light-scattering data were collected on a Dawn Eos spectrometer (Wyatt Technology) coupled to an Optilab Dsp (Wyatt Technologies) interferometric refractometer. Samples (100 μl) were injected and run over a Superdex 200 (10/300 GL) gel filtration column (GE) at a flow rate of 0.5 ml min−1. Multiangle laser light scattering (690 nm), absorbance (280 nm), and the refractive index were monitored after elution. Before samples were run, the system was calibrated and normalized to monomeric bovine serum albumin. The dn/dc value (change in solution refractive index with respect to protein concentration) is relatively constant for proteins (18), and the value for all experiments and analysis reported was set to 0.19. Data were processed using the software ASTRA as previously described (19).
RESULTS
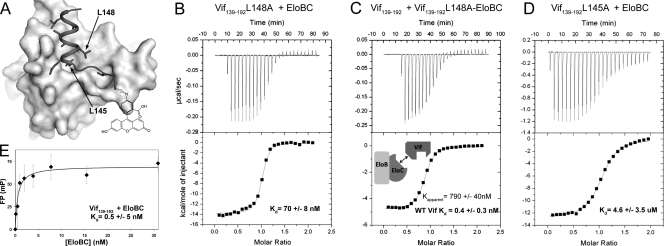
The Vif BC-box binds EloBC with subnanomolar affinity (Kd of ∼0.4 nM) through hydrophobic interactions (Fig. 2).
FIG. 2.
(A) A surface representation of EloBC bound to FITC-Vif140-154 (ribbon) (14). (B to D) ITC analysis of WT and mutant Vif139-192 titrated into EloBC. (E) FP binding curve of FITC-Vif140-154-EloBC.
A competition assay (16) was employed to accurately measure the high-affinity interaction by ITC. We first measured that the Vif mutant, Vif139-192 L148A, binds to EloBC at 70.2 ± 8.4 nM (Fig. 2B). We then titrated WT Vif139-192 into Vif139-192 L148A-EloBC complex to obtain an apparent affinity of 780 ± 21 nM (Fig. 2C). We calculate the WT Vif139-192-EloBC affinity as 0.4 ± 0.3 nM. To ascertain that the binding affinity is contributed only from the BC box, rather than the cullin box also present in the Vif constructs, we used an FP assay to measure the binding of the Vif BC box (Vif139-154) to EloBC (Fig. 2E). The FP assay suggests an affinity of ∼0.5 ± 5 nM, in excellent agreement with the ITC measurement, and confirms that the binding is achieved mainly through the BC-box region. We further measured the binding of Vif139-192 L145A to EloBC. The single point mutation of this important residue, which binds to a deep hydrophobic pocket on the EloC surface, dropped the binding affinity by nearly 104-fold to 4.6 μM (Fig. 2D).
The Vif zinc finger binds Cul5 with high affinity.
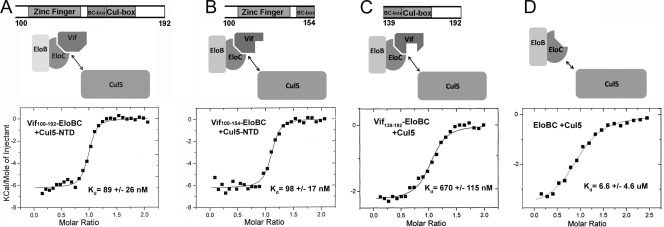
The C terminus of Vif (Vif100-192) binds to Cul5 with high affinity (Kd of ∼90 nM) (Fig. 3), and the interaction is achieved mostly through the Vif zinc finger domain. To finely map the Cul5-binding domains on Vif, we titrated Cul5 into several Vif-EloBC complexes containing Vif100-154, Vif100-176, or Vif100-192. All of these complexes bind Cul5 with approximately the same affinity of ∼90 nM (Fig. 3). We used Vif-EloBC complexes because the insolubility of Vif100-192 on its own prevented us from directly measuring the interaction between Vif and Cul5. Nonetheless, as demonstrated below, the binding is mostly attributed to the zinc finger domain rather than the BC box, the cullin box, or EloBC, which are also in the constructs.
FIG. 3.
(A to C) ITC analysis of Cul5 titrated into the Vif100-192-EloBC, Vif100-154-EloBC, and Vif139-192-EloBC complexes, respectively. (D) ITC analysis of Cul5 titrated into EloBC. The upper panels show the Vif domains used in the experiments and schematics of the E3 ligase assembly.
The Vif cullin box binds Cul5 weakly.
To clarify the contribution of the Vif cullin box to the binding of Cul5, we measured binding of Vif139-192-EloB/EloC to Cul5 and obtained a Kd of ∼670 nM (Fig. 3C). We further confirmed that the Vif BC box does not contribute to Cul5 binding by titrating Cul5 into Vif139-154-EloBC and finding that the two do not interact. A Vif L163S mutation has been implicated to affect the function and conformation of the PPLP loop in the Vif cullin box (14). We tested the binding of Vif100-192 L163S-EloBC to Cul5 and obtained the same affinity as that of WT Vif100-192-EloBC (data not shown). These results confirm that the Vif zinc finger mainly contributes to the interaction between Vif and Cul5, while the cullin box does interact with Cul5, with an affinity ∼10-fold lower.
EloBC binds Cul5 with very low affinity.
Because Vif forms a high-affinity complex with EloBC, which may also contribute to binding of the Vif-EloBC subcomplex to Cul5, we next asked whether the binding of EloBC to Cul5 plays a major role in assembly of the Vif-containing E3 ligase. Purified EloBC was titrated into Cul5. While EloBC does have an affinity for Cul5 on its own, it was found to be much weaker (6.6 ± 4.6 μM) than that of the Vif-EloBC constructs with the Vif zinc finger domain (Fig. 3D). The weak interaction between Cul5 and EloBC suggests that EloBC does not have a significant contribution to the recruitment of Cul5 to assemble the Vif-containing E3 ubiquitin ligase.
Vif C-terminal domain binds to the E3 ligase with 1:1 stoichiometry.
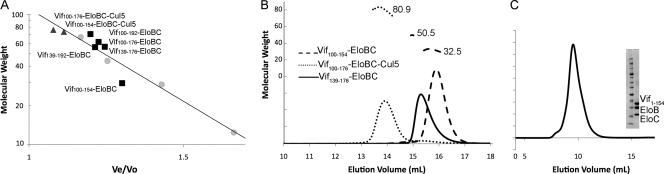
In addition to thermodynamic parameters, ITC experiments also provide information about the stoichiometry of the interactions. All of the Vif-EloBC constructs we tested demonstrated 1:1 stoichiometry with Cul5 in our ITC experiments (Fig. 2 and 3). We confirmed this using size exclusion chromatography (Fig. 4 A) and multiangle laser light scattering experiments (Fig. 4B), both of which determined a molecular weight of each Vif-EloBC-Cul5 complex consistent with a Vif/EloB/EloC/Cul5 ratio of 1:1:1:1.
FIG. 4.
(A) The relationship of molecular weight and elution volume for various Vif-EloBC and Vif-Cul5-EloBC complexes on a Superdex 75 (10/300 GL) size exclusion column. The expected molecular weight (in thousands) is plotted against predicted positions in the line calibrated using molecular weight standards. The Vif100-154-EloBC is plotted as a monomer, and all other complexes are plotted as dimers. (B) Multiangle laser light scattering results of the molecular weights of Cul5-Vif100-176-EloBC, Vif139-176-EloBC, and Vif100-154-EloBC. The measured molecular weights of Cul5-Vif100-176-EloBC, Vif139-176-EloBC, and Vif100-154-EloBC (80,900, 50,500, and 32,500, respectively) match the theoretical values of monomers for Cul5-Vif100-176-EloBC (76,200) and Vif100-154-EloBC (29,700) and the theoretical value for a dimer of Vif139-176-EloBC (56,500). (C) Size exclusion chromatogram of Vif1-154-EloBC and an SDS-PAGE gel of the peak fraction showing a pure, monomeric complex.
Vif dimerizes through interactions involving the PPLP region in the cullin box.
We further investigated the oligomerization state of the Vif complexes, since Vif has been implicated in dimerization (22, 23). We used two methods, size exclusion chromatography and multiangle laser light scattering, to demonstrate that the cullin-box region of Vif, containing the PPLP loop, leads to the dimerization of Vif in the absence of Cul5 (Fig. 4). While size exclusion chromatography provides useful information about the size of the molecule, it is shape dependent, and a protein with a more extended conformation can elute at a position with a larger apparent molecular weight. In contrast, light scattering is shape independent and provides a more accurate measurement of molecular weight. Our results show that all constructs containing the PPLP region, such as Vif100-176-EloBC, Vif139-192-EloBC, Vif139-176-EloBC, and Vif100-192-EloBC, are dimers, while Vif100-154-EloBC without the PPLP region is monomeric (Fig. 4). Vif100-154-EloBC appears to be slightly larger than its predicted molecular weight of 29,700 as judged by its elution volume from the size exclusion column (Fig. 4A); however, light scattering unambiguously shows the correct molecular weight of a monomeric Vif100-154-EloBC complex (Fig. 4B). The difference in the Vif100-154-EloBC molecular weight measured by the two techniques suggests that the Vif construct containing the zinc finger but without the cullin box adopts a more extended conformation such that its shape distorts the size exclusion chromatography results.
To probe the oligomerization state of full-length Vif, we attempted expression of the entire protein; however, our attempts to express full-length Vif in the presence or absence of Cul5 and EloBC yielded aggregated complexes, since full-length Vif is known to be prone to aggregation and insolubility. We did succeed, however, in purifying a near-full-length construct of Vif1-154 when it is coexpressed in complex with EloBC. This construct lacks the PPLP region needed for dimerization and was purified as a monomer (Fig. 4C). This result indicates that the N-terminal domain of Vif is not sufficient to promote Vif self-association. In addition, since the Vif1-154-EloBC and Vif100-154-EloBC (above) complexes do not dimerize, we exclude the possibility that the EloBC component in the complexes is responsible for dimer formation, confirming that the Vif PPLP region is the principal Vif dimerization site.
DISCUSSION
In this study, we quantified the binding thermodynamics of various Vif C-terminal regions for the human E3 ligase components EloBC and Cul5 (Table 1). The results show that the Vif C-terminal domain is capable of forming a high-affinity complex with the E3 ligase with 1:1 stoichiometry. Our data agree with the available biochemical data that HIV Vif uses two critical regions to assemble with the human E3 ubiquitin ligase: the Vif BC box interacts with EloBC (0.4 nM affinity), and the Vif zinc finger interacts with Cul5 (90 nM affinity). The Vif cullin box is plausible as an additional Cul5 recruiting site but with an affinity an order of magnitude lower (670 nM affinity). The interaction between EloBC and Cul5 is another order of magnitude lower (6.6 μM affinity). In addition, the amino acids beyond the Vif cullin box are dispensable for Cul5 binding. These data provide a more detailed picture of how HIV Vif hijacks the human E3 ubiquitin ligase machinery to evade the cellular innate immune system.
TABLE 1.
Vif-EloBC-Cul5 binding thermodynamicsa
| ITC syringe | ITC cell | ΔH (kcal/mol) | Kd | ΔG (kcal/mol) | −TΔS (kcal/mol) |
|---|---|---|---|---|---|
| WT Vif139-192 | EloB/EloC | −20.2 ± 2.0 | 0.4 ± 0.3 nM | −12.9 ± 0.6 | 7.3 ± 1.5 |
| Vif139-192 L145A | EloB/EloC | −10.7 ± 3.6 | 4.6 ± 3.5 μM | −7.4 ± 0.4 | 3.3 ± 3.1 |
| Vif139-192 L148A | EloB/EloC | −14.0 ± 0.1 | 70.2 ± 8 nM | −9.8 ± 0.1 | 4.3 ± 0.1 |
| Cul5 NTD | Vif139-192-EloB/EloC | −3.1 ± 1.1 | 670 ± 115 nM | −8.4 ± 0.1 | −5.3 ± 1.0 |
| Cul5 NTD | Vif100-154-EloB/EloC | −6.4 ± 0.5 | 97.5 ± 17 nM | −9.6 ± 0.1 | −3.2 ± 0.6 |
| Cul5 NTD | Vif100-192-EloB/EloC | −6.5 ± 0.3 | 89.0 ± 26 nM | −9.6 ± 0.2 | −3.1 ± 0.4 |
| Cul5 NTD | EloB/EloC | 6.6 ± 4.6 μM | −7.2 ± 0.4 |
ΔH, change in observed enthalpy; T, experimental temperature; ΔS, change in entropy.
We have found that Vif binds EloBC with subnanomolar affinity and that the mutation of two key hydrophobic residues at the interface drastically reduces the affinity. The importance of L145 and L148 is evident from the crystal structure of the Vif BC box-EloBC complex (14), and our ITC and FP results confirm that this interaction is mainly driven by hydrophobic interactions. These data also corroborate coimmunoprecipitation studies where L145A results in the loss of Vif and EloBC binding (25). The remarkably high affinity between Vif and EloBC shows that HIV has evolved with a very effective strategy to highjack the host E3 ubiquitin ligase to degrade APOBEC3G.
The interaction between Vif and Cul5 is mediated mainly by the zinc finger of Vif, with a small contribution by the cullin box. Our results show that the Vif cullin box has a small but noticeable effect on the assembly of the E3 ligase, with an affinity for Cul5 ∼10-fold weaker than that of the zinc finger. This is supported by earlier homology modeling results that the Vif cullin box makes modest contact with Cul5 (14) (Fig. 1C). The weak interaction explains the lack of an apparent binding effect from the cullin box when it was included in Vif constructs also containing the zinc finger in our ITC experiments and why the Vif zinc finger alone still immunoprecipitates Cul5 (11, 21). Nonetheless, the binding effect can still be noticeable under certain experimental conditions and offers a possible explanation for the observed importance of the Vif cullin box in coimmunoprecipitation experiments with the L163S or L169S cullin box mutant (14).
Our experiments focus on the recruitment of the E3 ubiquitin ligase by the Vif C-terminal domain (CTD), since all current data show the Vif CTD is responsible for the interactions with the ligase components. Our attempt to include full-length Vif in our study did not yield soluble complexes amenable to biochemical studies. According to our data on various Vif truncation constructs, full-length Vif is likely to dimerize in the EloBC complex and to form a monomer complex with EloBC and Cul5. Nonetheless, the results for the Vif CTD show that it is sufficient to recruit the E3 ligase and that the affinity compares well with those of similar cellular substrate receptor components in the ligase (2). It is possible that the Vif N-terminal domain can still contribute to the binding, which would further enhance the ability of Vif to hijack the host ubiquitination machinery.
The cullin box also plays a key role in the dimerization of Vif through the PPLP region. Our gel filtration and multiangle laser light scattering experiments provide direct biochemical evidence that the PPLP loop of Vif is responsible for the formation of Vif dimers. Vif constructs containing only the N-terminal domain, the zinc finger, and the BC box are monomeric, while all Vif-EloBC constructs containing the cullin box are dimers. Interestingly, the addition of Cul5 to all Vif-EloBC complexes results in monomeric Vif-EloBC-Cul5 complexes (Fig. 4A and C). This may be a result of Cul5 interacting with the cullin box of Vif and therefore blocking the PPLP dimerization site. This is consistent with earlier homology modeling that the Vif cullin box makes contacts with Cul5 (14). These results indicate that the assembly of the E3 ligase does not involve Vif dimerization. The functional importance of Vif dimerization through the PPLP region perhaps lies in other cellular functions of Vif. One possibility is to aid the recruitment of APOBEC3G, since it was reported recently that the Vif PPLP region is required for APOBEC3G binding (5, 17).
Acknowledgments
We thank Alex Bullock for providing the Elongin BC plasmid, Bryan Cullen for providing Vif HXB3 DNA, Jeffery Babon for the murine Cul5 clone, Michael Salcius for help with FP data analysis, and Ann Valentine and Jean Gaffney for discussions of ITC analysis.
This work is supported in part by a Smith Family New Investigator Award and a grant from the NIH (AI078831) to Y.X.
Footnotes
Published ahead of print on 12 May 2010.
REFERENCES
- 1.Babon, J. J., J. K. Sabo, A. Soetopo, S. Yao, M. F. Bailey, J. G. Zhang, N. A. Nicola, and R. S. Norton. 2008. The SOCS box domain of SOCS3: structure and interaction with the elonginBC-cullin5 ubiquitin ligase. J. Mol. Biol. 381:928-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babon, J. J., J. K. Sabo, J. G. Zhang, N. A. Nicola, and R. S. Norton. 2009. The SOCS box encodes a hierarchy of affinities for Cullin5: implications for ubiquitin ligase formation and cytokine signalling suppression. J. Mol. Biol. 387:162-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullock, A. N., J. E. Debreczeni, A. M. Edwards, M. Sundstrom, and S. Knapp. 2006. Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 103:7637-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullock, A. N., M. C. Rodriguez, J. E. Debreczeni, Z. Songyang, and S. Knapp. 2007. Structure of the SOCS4-ElonginB/C complex reveals a distinct SOCS box interface and the molecular basis for SOCS-dependent EGFR degradation. Structure 15:1493-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donahue, J. P., M. L. Vetter, N. A. Mukhtar, and R. T. D'Aquila. 2008. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology 377:49-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamura, T., K. Maenaka, S. Kotoshiba, M. Matsumoto, D. Kohda, R. C. Conaway, J. W. Conaway, and K. I. Nakayama. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18:3055-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamura, T., S. Sato, D. Haque, L. Liu, W. G. Kaelin, Jr., R. C. Conaway, and J. W. Conaway. 1998. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 12:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo, K., Z. Xiao, E. Ehrlich, Y. Yu, B. Liu, S. Zheng, and X. F. Yu. 2005. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. U. S. A. 102:11444-11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 10.Mehle, A., J. Goncalves, M. Santa-Marta, M. McPike, and D. Gabuzda. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18:2861-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehle, A., E. R. Thomas, K. S. Rajendran, and D. Gabuzda. 2006. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 281:17259-17265. [DOI] [PubMed] [Google Scholar]
- 12.Paul, I., J. Cui, and E. L. Maynard. 2006. Zinc binding to the HCCH motif of HIV-1 virion infectivity factor induces a conformational change that mediates protein-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 103:18475-18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell, R. A., and V. K. Pathak. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81:8201-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanley, B. J., E. S. Ehrlich, L. Short, Y. Yu, Z. Xiao, X. F. Yu, and Y. Xiong. 2008. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J. Virol. 82:8656-8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stebbins, C. E., W. G. Kaelin, Jr., and N. P. Pavletich. 1999. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science 284:455-461. [DOI] [PubMed] [Google Scholar]
- 16.Velazquez-Campoy, A., Y. Kiso, and E. Freire. 2001. The binding energetics of first- and second-generation HIV-1 protease inhibitors: implications for drug design. Arch. Biochem. Biophys. 390:169-175. [DOI] [PubMed] [Google Scholar]
- 17.Walker, R. C., Jr., M. A. Khan, S. Kao, R. Goila-Gaur, E. Miyagi, and K. Strebel. 2010. Identification of dominant negative human immunodeficiency virus type 1 Vif mutants that interfere with the functional inactivation of APOBEC3G by virus-encoded Vif. J. Virol. 84:5201-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen, J., T. Arakawa, and J. S. Philo. 1996. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal. Biochem. 240:155-166. [DOI] [PubMed] [Google Scholar]
- 19.Wyatt, P. J. 1993. Light-scattering and the absolute characterization of macromolecules. Anal. Chim. Acta 272:1-40. [Google Scholar]
- 20.Xiao, Z., E. Ehrlich, K. Luo, Y. Xiong, and X. F. Yu. 2007. Zinc chelation inhibits HIV Vif activity and liberates antiviral function of the cytidine deaminase APOBEC3G. FASEB J. 21:217-222. [DOI] [PubMed] [Google Scholar]
- 21.Xiao, Z., E. Ehrlich, Y. Yu, K. Luo, T. Wang, C. Tian, and X. F. Yu. 2006. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology 349:290-299. [DOI] [PubMed] [Google Scholar]
- 22.Yang, B., L. Gao, L. Li, Z. Lu, X. Fan, C. A. Patel, R. J. Pomerantz, G. C. DuBois, and H. Zhang. 2003. Potent suppression of viral infectivity by the peptides that inhibit multimerization of human immunodeficiency virus type 1 (HIV-1) Vif proteins. J. Biol. Chem. 278:6596-6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang, S., Y. Sun, and H. Zhang. 2001. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J. Biol. Chem. 276:4889-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 25.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-Elongin B-Elongin C E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]