Abstract
Coronavirus (CoV) 3C-like proteinase (3CLpro), located in nonstructural protein 5 (nsp5), processes the replicase polyproteins 1a and 1ab (pp1a and pp1ab) at 11 specific sites to produce 12 mature nonstructural proteins (nsp5 to nsp16). Structural and biochemical studies suggest that a conserved Gln residue at the P1 position is absolutely required for efficient cleavage. Here, we investigate the effects of amino acid substitution at the P1 position of 3CLpro cleavage sites of infectious bronchitis virus (IBV) on the cleavage efficiency and viral replication by in vitro cleavage assays and reverse genetic approaches. Our results demonstrated that a P1-Asn substitution at the nsp4-5/Q2779, nsp5-6/Q3086, nsp7-8/Q3462, nsp8-9/Q3672, and nsp9-10/Q3783 sites, a P1-Glu substitution at the nsp8-9/Q3672 site, and a P1-His substitution at the nsp15-16/Q6327 site were tolerated and allowed recovery of infectious mutant viruses, albeit with variable degrees of growth defects. In contrast, a P1-Asn substitution at the nsp6-7/Q3379, nsp12-13/Q4868, nsp13-14/Q5468, and nsp14-15/Q5989 sites, as well as a P1-Pro substitution at the nsp15-16/Q6327 site, abolished 3CLpro-mediated cleavage at the corresponding position and blocked the recovery of infectious viruses. Analysis of the effects of these lethal mutations on RNA synthesis suggested that processing intermediates, such as the nsp6-7, nsp12-13, nsp13-14, nsp14-15, and nsp15-16 precursors, may function in negative-stranded genomic RNA replication, whereas mature proteins may be required for subgenomic RNA (sgRNA) transcription. More interestingly, a mutant 3CLpro with either a P166S or P166L mutation was selected when an IBV infectious cDNA clone carrying the Q6327N mutation at the nsp15-16 site was introduced into cells. Either of the two mutations was proved to enhance significantly the 3CLpro-mediated cleavage efficiency at the nsp15-16 site with a P1-Asn substitution and compensate for the detrimental effects on recovery of infectious virus.
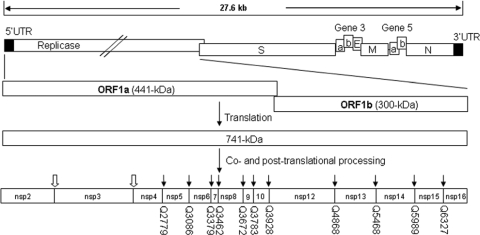
Coronavirus (CoV)-encoded 3C-like proteinase (3CLpro), together with one or two papain-like proteinases (PLpro), mediates the extensive proteolytic processing of two large replicase polyproteins (pp), pp1a and pp1ab, and yields multiple mature nonstructural proteins (nsp) essential for the assembly and function of the viral replication complex. PLpro cleaves the N-terminal regions of the polyproteins at two or three sites, and 3CLpro is responsible for the processing of all downstream parts of the replicase polyproteins at 11 conserved cleavage sites (50, 55, 72, 74). Like its homologs in other coronaviruses, 3CLpro of avian infectious bronchitis virus (IBV) is encoded by open reading frame 1a (ORF1a) and resides in nsp5. This proteinase specifically cleaves polyproteins 1a and 1ab at 11 sites to produce 12 mature products (nsp5 to nsp16) (24, 36-39, 41-43, 64) (Fig. 1).
FIG. 1.
IBV genome organization and proteolytic processing of the replicase polyproteins. Cleavage sites and the processed products of IBV replicase polyproteins pp1a (nsp2 to nsp10) and pp1ab (nsp2 to nsp10 and nsp12 to nsp16) are shown. The cleavage sites of 3CLpro are indicated with black arrows, and the conserved glutamine (Q) residue at the P1 position of each cleavage site is indicated. The cleavage sites of papain-like proteinase (PLpro) are indicated with block arrows. UTR, untranslated region; S, spike protein; E, envelope protein; M, membrane protein; N, nucleocapsid protein; a, 3a or 5a protein; b, 3b or 5b protein.
The structures of many nonstructural proteins have been determined, and their functions are partially elucidated. nsp7 and nsp8 form a hexadecameric structure able to encircle and bind RNA (70); nsp8 alone is described as a second RdRp of severe acute respiratory syndrome CoV (SARS-CoV) (28). nsp9 is a single-stranded RNA-binding protein that may stabilize nascent and template RNA strands during replication and transcription and may also be involved in RNA processing (9, 16, 53). nsp10 forms a dodecamer structure, and two zinc fingers have been identified in the monomer, implying that it may function in the RNA synthesis machinery (32, 52). More recently, murine hepatitis virus (MHV) nsp10 has been shown to be a critical regulator of viral RNA synthesis and to play an important role in polyprotein processing (13, 14). nsp14 is a bifunctional protein: a 3′-to-5′ exonuclease (ExoN) (40) with a role in maintaining the fidelity of RNA transcription (15) and a cap N7 methyltransferase (10) involved in RNA cap formation. nsp15 is a poly(U)-specific endoribonuclease (NendoU) (6, 8, 25, 33). nsp16, an S-adenosylmethionine-dependent ribo-2′-O-methyltransferase (2′O-MTase), has also been thought to be involved in cap formation (11, 50, 60).
Because of its pivotal role in the viral life cycle and the conservation in structure, 3CLpro becomes the most promising target for drug design against coronaviruses (23, 63, 67, 69). The substrate specificity of coronavirus 3CLpro is determined mainly by the P1, P2, and P1′ positions. Comparative analyses of 77 3CLpro cleavage sites from seven coronaviruses suggest amino acid conservation at the P2 (Leu, Val, Ile, Phe, and Met), P1 (Gln), and P1′ (Ser, Ala, Gly, Asn, and Cys) positions. Among these, the P1 position is exclusively occupied by Gln, and the bulky hydrophobic residues (mainly Leu) are dominant at the P2 position (22, 26, 56, 74). Insights into the catalytic activity and substrate specificity of 3CLpros obtained by the recently determined crystal structures of five 3CLpros from transmissible gastroenteritis virus (TGEV), human coronavirus 229E (HCoV-229E), SARS-CoV, IBV, and HCoV HKU1 further confirm the necessity of a conserved Gln residue at the P1 position (2, 3, 66, 68, 71).
The importance of coronavirus 3CLpro-mediated processing of replicase polyproteins in viral replication was verified by proteinase inhibitors (35, 66, 67), but data on the necessity of coronavirus 3CLpro-mediated individual cleavages are very limited. Reverse genetics studies indicated recently that proteolytic processing at the nsp7-8, nsp8-9, and nsp10-11/12 cleavage sites of MHV was essential for viral replication, while cleavage at the nsp9-10 site of MHV and at the nsp10-11/12 site of IBV was dispensable for viral replication (12, 20). In this report, we investigate the effects of amino acid substitutions at the P1 position of 10 IBV 3CLpro cleavage sites, except the nsp10-11/12 site, on the cleavage efficiency and viral replication by in vitro cleavage assays and IBV reverse genetics. Our results demonstrated that mutation of Gln to Asn at the P1 position of the nsp4-5/Q2779, nsp5-6/Q3086, nsp7-8/Q3462, nsp8-9/Q3672, and nsp9-10/Q3783 sites, P1-Glu substitution at the nsp8-9/Q3672 site, and P1-His substitution at the nsp15-16/Q6327 site were tolerated, since each of these mutations allowed the recovery of infectious virus, albeit with various degrees of defects in viral growth. Our data also showed that proteolytic processing at the nsp6-7/Q3379, nsp12-13/Q4868, nsp13-14/Q5468, nsp14-15/Q5989, and nsp15-16/Q6327 cleavage sites was required for IBV replication. In addition, a distal mutation, P166S or P166L, in 3CLpro was identified when a point mutation, nsp15-16/Q6327N, representing a P1-Asn substitution at the nsp15-16 site, was introduced into the IBV infectious cDNA clone. Either of the two mutations was proved to enhance significantly the 3CLpro-mediated cleavage efficiency at the nsp15-16 site with a P1-Asn substitution and compensate for the detrimental effects on recovery of infectious virus. This study provides further insights into the substrate specificity of coronavirus 3CLpro and the regulatory mechanism of viral RNA synthesis as well as helpful information on the drug design against coronaviruses.
MATERIALS AND METHODS
Cells and virus.
Vero cells and H1299 cells were cultured at 37°C, respectively, in minimal essential medium (MEM) and RPMI supplemented with 10% fetal bovine serum (FBS), penicillin (100 units/ml), and streptomycin (100 μg/ml). A recombinant IBV (rIBV) generated from an infectious clone (19) was used as the wild-type control. All mutant IBVs were propagated in Vero cells in FBS-free MEM.
Transient expression and processing in mammalian cells.
ORFs placed under the control of a T7 promoter were expressed transiently in mammalian cells using a vaccinia virus-T7 expression system (21). Briefly, confluent H1299 cells in 6-well plates were inoculated with a recombinant vaccinia virus (vTF7-3) expressing T7 RNA polymerase. At 1 h postinfection, medium was removed and plasmids carrying wild-type or mutant cleavage sites of IBV 3CLpro together with pIBV3C containing the full-length IBV 3CLpro sequence (39) were transfected into cells with Lipofectamine 2000 transfection reagent according to the manufacturer's instructions (Invitrogen). Cells were harvested after incubation at 37°C for 16 to 20 h.
SDS-PAGE and Western blot analysis.
Transfected cells were lysed with 2× SDS loading buffer containing 100 mM dithiothreitol (DTT) and 5 mM iodoacetamide, boiled at 100°C for 5 min, and clarified. The proteins were separated by 15% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Stratagene). The membrane was blocked overnight at 4°C or for 2 h at room temperature in blocking buffer (5% fat-free milk powder in phosphate-buffered saline [PBS] buffer containing 0.1% Tween 20 [PBST]) and then was incubated with diluted primary antibodies in blocking buffer for 2 h at room temperature. After the membrane was washed three times with PBST, it was incubated with 1:2,000 diluted anti-mouse or anti-rabbit IgG antibodies conjugated with horseradish peroxidase (Dako) in blocking buffer for 1 h at room temperature. After the membrane was washed three times with PBST, the polypeptides were detected with a chemiluminescence detection kit (ECL kit; Amersham Biosciences) according to the manufacturer's instructions. The films were exposed and developed.
Generation of mutant viruses.
The DNA fragment containing the nsp4-5/Q2779, nsp5-6/Q3086, nsp6-7/Q3379, nsp7-8/Q3462, nsp8-9/Q3672, nsp9-10/Q3783, and nsp12-13/Q4868 cleavage sites was mapped to plasmid pXL-IBVC, and the fragment containing the nsp13-14/Q5468, nsp14-15/Q5989, and nsp15-16/Q6327 cleavage sites to pGEM-IBVD (19, 54). Constructs containing a single Gln-to-Asn/Glu/Pro/His substitution at the P1 position were produced by using a QuikChange site-directed mutagenesis kit (Stratagene). The full-length infectious clone was assembled as previously described (19, 54) by replacing the corresponding fragment with the mutant fragment. The mutations were verified by automated nucleotide sequencing.
Full-length transcripts were generated in vitro using the mMessage mMachine T7 kit (Ambion, Austin, TX) and electroporated into Vero cells with one pulse at 450 V and 50 μF with a Bio-Rad Gene Pulser II electroporator. The transfected Vero cells were cultured overnight in 1% FBS-containing MEM and further cultured in MEM without FBS (19, 54). The transfected cells were monitored daily for formation of cytopathic effect (CPE). Recovered viruses were plaque purified and passaged on Vero cells. For the mutants that failed to produce observable CPE, three independent experiments were conducted.
Northern hybridization.
Vero cells were infected with rIBV and recovered mutant viruses at a multiplicity of infection (MOI) of ∼1 PFU/cell, and 10 μg of total RNA extracted from the infected cells was added to a mixture of 1× MOPS, 37% formaldehyde, and formamide and incubated at 65°C for 20 min prior to gel electrophoresis. The separated RNA bands were transferred onto a Hybond N+ membrane (Amersham Biosciences) via capillary action overnight and fixed by UV cross-linking (Stratalinker). Hybridization of digoxigenin (Dig)-labeled DNA probes corresponding to the IBV 3′ untranslated region (nucleotides [nt] 27104 to 27510) was carried out at 50°C in a hybridization oven overnight. Membranes were washed three times for 15 min each with the stringency buffers, and signals were detected with CDPStar (Roche) according to the manufacturer's instructions.
Viral growth assays.
A confluent monolayer of Vero cells on six-well plates was infected with recovered viruses at the indicated MOI and harvested at different times postinfection. Viral stocks were prepared by freeze-thawing the cells three times. The 50% tissue culture infection dose (TCID50) of each sample was determined by infecting five wells of Vero cells on 96-well plates with a 10-fold serial dilution of each viral stock and using Reed-Muench calculation.
RT-PCR analysis.
Total RNAs were prepared from the electroporated or infected Vero cells with TRI reagent (Molecular Research Center, Inc.). For the mutations that did not produce the infectious virus, the in vitro transcripts prior to electroporation and the extracted RNAs were treated with DNase I and subjected to phenol-chloroform extraction and ethanol precipitation to completely rule out possible DNA contamination. Reverse transcription (RT) was performed with Expand reverse transcriptase (Roche) according to the manufacturer's instructions using the sense primer IBV leader (5′-26CTATTACACTAGCCTTGCGCT46-3′) for the detection of negative-stranded subgenomic RNA (sgRNA) and the antisense primer IBV24803-R (5′-24803CTCTGGATCC AATAACCTAC24784-3′) for the detection of positive-stranded sgRNA. The two primers were then used for PCR. If transcription of subgenomic mRNAs did occur, a 415-bp PCR product corresponding to the 5′-terminal region of subgenomic mRNA 4 and a 1,010-bp fragment corresponding to the 5′-terminal region of subgenomic mRNA 3 would be expected. Similarly, RT was carried out with the sense primer IBV14931-F (5′-14931GCTTATCCACTAGTACATC14949-3′) for the detection of negative-stranded genomic RNA, and IBV14931-F and the antisense primer IBV15600-R (5′-15600CTTCTCGCACTTCTGCACTAGCA15578-3′) were used for PCR. If replication of viral RNA occurred, a 670-bp PCR fragment would be expected.
Construction of plasmids.
Plasmid pXL-IBV-C (nt 8689 to 15532) or pGEM-IBV-D (nt 15511 to 20930) (19) was used to construct a series of T7-driven expression plasmids. A DNA fragment covering each of 10 wild-type cleavage sites on IBV polyproteins was amplified with Turbo Pfu DNA polymerase (Stratagene), and the purified PstI-digested PCR product was inserted in frame into EcoRV/PstI-digested vector pFlag. The resulting plasmid allowed the expression of an N-terminal Flag-tagged protein under the control of the T7 promoter. As templates, these plasmids were used to make the corresponding mutant plasmids containing different single mutations in place of the conserved Gln residue at the P1 position of every cleavage site by using the QuikChange site-directed mutagenesis kit (Stratagene). Plasmids and the primers used for PCR are listed in Table 1 .
TABLE 1.
Plasmids and primers
| Plasmid | Siteb | Insert (nt) | Primera |
|---|---|---|---|
| pFlag-Q2779 | nsp4-5 | 8569-9069 | 5′-ATCCACTCGTAAGCTGTATGATGGC |
| 5′-CGGCTGCAGTTATTGAGTTGTAACTTCAAACTC | |||
| pFlag-Q2779N | 5′-AGTAGATTAAACTCTGGTTTTAAG | ||
| pFlag-Q2779E | 5′-AGTAGATTAGAATCTGGTTTTAAG | ||
| pFlag-Q3086 | nsp5-6 | 9490-9981 | 5′-ATCCGCAATTATTAGTGTTAAGGAG |
| 5′-CGGCTGCAGTTAAGTATCCATATATGCCATAAC | |||
| pFlag-Q3086N | 5′-GGTGTTAGATTAAACTCTTCTTTT | ||
| pFlag-Q3086E | 5′-GGTGTTAGATTAGAATCTTCTTTT | ||
| pFlag-Q3379 | nsp6-7 | 10531-10860 | 5′-ATCCGTTTCAGTAGATCAATATAGG |
| 5′-CGGCTGCAGTTAAGTAGAATCTATACAAAATAG | |||
| pFlag-Q3379N | 5′-GCTACAGTTAACGCTAAATTGAGT | ||
| pFlag-Q3379E | 5′-GCTACAGTTGAAGCTAAATTGAGT | ||
| pFlag-Q3462 | nsp7-8 | 10666-11544 | 5′-ATCCGCTAAATTGAGTGATGTAAAG |
| 5′-CGCTGCAGTTATTGCAAAACAACATCAACCTT | |||
| pFlag-Q3462N | 5′-ACTGTATTAAACTCGGTTACTCAA | ||
| pFlag-Q3462E | 5′-ACTGTATTAGAATCGGTTACTCAA | ||
| pFlag-Q3672 | nsp8-9 | 10915-11877 | 5′-ATCCTCGGTTACTCAAGAATTCTCA |
| 5′-CGCTGCAGTTACTGTAAGACAACAACATTAGA | |||
| pFlag-Q3672N | 5′-GTTTTGAACAATAATGAGCTTATG | ||
| pFlag-Q3672E | 5′-GTTTTGGAAAATAATGAGCTTATG | ||
| pFlag-Q3783 | nsp9-10 | 11590-12120 | 5′-ATCCGCAGGTGTAGATCAAGCACAT |
| 5′-CGGCTGCAGTTAGTGTGCTCTACAATAGAGACA | |||
| pFlag-Q3783N | 5′-GTTGTCTTAAACTCTAAAGGGCAT | ||
| pFlag-Q3783E | 5′-GTTGTCTTAGAGTCTAAAGGGCAT | ||
| pFlag-Q4868 | nsp12-13 | 14979-15479 | 5′-ATCCTTTGTTCTCCTTGCATATATC |
| 5′-CGGCTGCAGTTAAATGGACCAATTAGTAGTAGC | |||
| pFlag-Q4868N | 5′-ACGACTTTAAACTCTTGTGGCGTT | ||
| pFlag-Q4868E | 5′-ACGACTTTAGAATCTTGTGGCGTT | ||
| pFlag-Q5468 | nsp13-14 | 16785-17159 | 5′-ATCCGCAGATTCGCAGCATGCACTG |
| 5′-CGGCTGCAGTTAATCACGTGTTATAAACATGTT | |||
| pFlag-Q5468N | 5′-GAAACAAGTCTGAACGGTACAGGT | ||
| pFlag-Q5468E | 5′-GAAACAAGTCTGGAAGGTACAGGT | ||
| pFlag-Q5989 | nsp14-15 | 18219-18719 | 5′-ATCCAGCTTCCGCAATTTGAAAGCT |
| 5′-CGGCTGCAGTTATAAACCTTTCAAAATACGGTT | |||
| pFlag-Q5989N | 5′-TCAGCTCTCAACTCTATCGACAAT | ||
| pFlag-Q5989E | 5′-TCAGCTCTCGAGTCTATCGACAAT | ||
| pFlag-Q6327 | nsp15-16 | 19299-19799 | 5′-ATCCTTTGTATTGTCGGACAATGGT |
| 5′-CGGCTGCAGTTAATCATTATCGACAAGGAGTGT | |||
| pFlag-Q6327N | 5′-CCACAGCTTAACTCAGCATGGACG | ||
| pFlag-Q6327E | 5′-CCACAGCTTGAATCAGCATGGACG | ||
| pFlag-Q6327H | 5′-CCACAGCTTCACTCAGCATGGACG | ||
| pFlag-Q6327P | 5′-CCACAGCTTCCATCAGCATGGACG |
Sequences are based on the IBV strain Beaudette P65 (DQ001339.1). The underlined type indicates a mutated codon. Only the forward primer for each mutant is shown, and the reverse primer is completely complementary with it.
The location of IBV 3CLpro cleavage sites.
A DNA fragment encoding full-length 3CLpro of the SARS-CoV Sin2774 strain (GenBank accession no. AY283798) was amplified by PCR with primers 5′-CGGGATCCACCATG9969AGTGGTTTTAGGAAAATGGCA-3′ and 5′-CGGAATTCTTA10886TTGGAAGGTAACACCAGAGCA-3′. PCR products were digested with BamHI and EcoRI and cloned into BglII- and EcoRI-digested vector pKTO to get plasmid pSARS-3C.
RESULTS
Differential effects of amino acid substitutions at the P1 position of individual cleavage sites on processing efficiency.
To examine whether IBV 3CLpro can tolerate other amino acids instead of a Q residue at the P1 position, a set of expression plasmids, containing IBV sequences flanking either wild-type or mutant cleavage sites with mutation of P1-Q to N or E, was constructed (Fig. 2 A). These plasmids and the corresponding primers used are listed in Table 1. To facilitate detection of the full-length and cleavage products, a Flag tag was fused to the N termini of the expressed IBV peptides. The calculated molecular masses of the Flag-tagged uncleaved (U) and cleaved (C) products are illustrated in Fig. 2A. Wild-type and mutant substrates were coexpressed with IBV 3CLpro, and processing was analyzed by Western blotting with an anti-Flag antibody. The effect of the mutations on the cleavage efficiency was evaluated by measuring the relative density of the U and C bands using the QUANTITY ONE program (Bio-Rad) and calculating the relative cleavage efficiency (RCE) [RCE = C/(U + C) × 100%]. In this expression system, IBV 3CLpro effectively cleaved the substrates at all 10 wild-type cleavage sites, albeit with different cleavage efficiencies (Fig. 2B to D). Notably, nearly complete cleavage at the nsp8-9 site, a so-called noncanonical site with an N residue instead of S, A, or G at the P1′ position, was observed (Fig. 2C). In contrast, its counterparts in HCoV-229E, TGEV, MHV, and SARS-CoV were found to be hydrolyzed with far less efficiency (18, 26). The reasons for this discrepancy remain unclear but are likely due to different experimental approaches and substrates used.
FIG. 2.
Proteolytic processing of IBV polyproteins by IBV and SARS-CoV 3CLpros. (A) Diagram showing the constructs containing sequences flanking IBV cleavage sites and the 3CLpro-mediated processing in the present study. Uncleaved (U) and cleaved (C) products are represented with straight lines. The predicted sizes of the Flag-tagged (black box) uncleaved and cleaved products are indicated. Western blot analyses of the 3CLpro-mediated processing of wild-type and mutant substrates flanking nsp4-5/Q2779 (B), nsp5-6/Q3086 (B), nsp6-7/Q3379 (B), nsp7-8/Q3462 (C), nsp8-9/Q3672 (C), nsp9-10/Q3783 (C), and nsp12-13/Q4868 (C) and nsp13-14/Q5468 (D), nsp14-15/Q5989 (D), and nsp15-16/Q6327 (D) in IBV 1a and 1ab polyproteins, respectively. The Flag-tagged wild-type and P1-mutant substrates were coexpressed with IBV (I-3C) or SARS-CoV (S-3C) 3CLpro in H1299 cells using the vaccinia virus-T7 system. The transfected cell lysates were resolved on an SDS-15 to 20% polyacrylamide gel and subjected to Western blotting with anti-Flag antibodies. Q, E, H, N, and P represent constructs containing the corresponding amino acid residue at the P1 position. As a control, the amount of expressed 3CLpro was probed with IBV or SARS-CoV anti-3CLpro antibodies. The uncleaved (U) and cleaved (C) products and the relative cleavage efficiency (RCE) are indicated.
Compared to wild-type sites, mutation of P1-Q to N showed variable effects on the cleavage efficiency, depending on the location of the site. Complete abolishment of cleavage was observed for the nsp6-7/Q3379N, nsp12-13/Q4868N, nsp13-14/Q5468N, and nsp14-15/Q5989N P1-N mutants, while no inhibitory effect for the nsp4-5/Q2279N mutant was observed (Fig. 2B to D). A minor or moderate reduction in the cleavage efficiency for the nsp7-8/Q3462N, nsp5-6/Q3086N, and nsp8-9/Q3672N mutants (from 9% to 31%), as well as a more dramatic decrease for the nsp9-10/Q3783N and nsp15-16/Q6327N mutants (58% and 69%, respectively), was detected (Fig. 2B to D). Mutation of Q to E showed effects similar to those of the Q-to-N substitution in most cases, except at the nsp13-14 and nsp15-16 sites. Cleavage at the nsp13-14/Q5468E site showed low efficiency (17%), but no cleavage was observed at the nsp13-14/Q5468N site (0%) (Fig. 2D). To the contrary, higher cleavage efficiency was observed at the nsp15-16/Q6327E site (36%) than at the nsp15-16/Q6327N site (5%) (Fig. 2D). Replacement of the P1-Q residues with H and P at the nsp15-16 site was also analyzed. Mutation of P1-Q to H at the nsp15-16 site maintained a relatively high level of cleavage efficiency (44%), but no cleavage was observed when P1-Q was replaced by P (0%) (Fig. 2D). Taken together, these findings show that IBV 3CLpro was able to tolerate mutation of Q to other amino acids, such as N, E, or H, at several sites. The requirement for a conserved P1-Q residue at cleavage sites in the 1b region was more stringent than at those in the 1a region, except the nsp6-7 site.
In previous in vitro cleavage assays, synthetic peptides flanking the N-terminal autocleavage site of a certain coronavirus 3CLpro could be cleaved by 3CLpros from all three groups of coronaviruses (3, 26, 67), probably due to the structural similarity among these proteinases (3, 68). To test if this low-stringency requirement for enzyme substrate specificity could be extended to all other sites, cleavage of substrates with either wild-type or mutant sites by a heterogeneous 3CLpro from SARS-CoV was analyzed. Cleavage with various efficiencies (from 45% to 100%) was observed at the nsp4-5, nsp5-6, nsp7-8, nsp8-9, nsp9-10, and nsp15-16 wild-type sites, whereas no detectable cleavage was observed at other sites (Fig. 2B to D). Moreover, SARS-CoV 3CLpro could also tolerate the mutation of P1-Q to N or E at the two sites (nsp4-5/Q2279 and nsp5-6/Q3086) flanking IBV 3CLpro, although the processing efficiency of the mutant N-terminal site was far higher than that of the mutant C-terminal site (Fig. 2B), in contrast with the previous results showing that cleavage was completely abolished when the P1-N or P1-E substitution was introduced into the N-terminal self-cleavage site of SARS-CoV (17). In addition, a low level (8%) of cleavage at the nsp15-16 site with a P1-H substitution was also detected (Fig. 2D).
Introduction of Q-N/E substitution into a full-length IBV clone and recovery of mutant viruses from IBV clones containing cleavable Q-N/E substitution.
To evaluate the impact of Q-N point mutations at the P1 positions on virus replication, full-length cDNA clones containing each of the mutant cleavage sites were assembled. In vitro transcripts were synthesized and electroporated into Vero cells. For mutations that allowed efficient proteolytic processing, including nsp4-5/Q2779N, nsp5-6/Q3086N, nsp7-8/Q3462N, nsp8-9/Q3672N, and nsp9-10/Q3783N, the corresponding mutant virus was recovered at 2 or 3 days postelectroporation. In contrast, mutations which completely abolished the cleavage activity, including nsp6-7/Q3379N, nsp12-13/Q4868N, nsp13-14/Q5468N, and nsp14-15/Q5989N, blocked the generation of infectious viruses. In cells electroporated with nsp15-16/Q6327N mutant transcripts, formation of several small syncytial cells was observed at 3 days postelectroporation. These syncytia disappeared after prolonged incubation. Following 6 rounds of successive reinfection of fresh Vero cells with freeze-thawed cell preparations, sgRNA was still detectable by RT-PCR, but no syncytium formation was observed (data not shown).
As cleavage was also observed at sites between nsp4 and nsp5, nsp5 and nsp6, nsp8 and nsp9, nsp9 and nsp10, and nsp15 and nsp16 when Q was replaced with E, this point mutation at the site between nsp8 and nsp9 (nsp8-9/Q3672E) was introduced into the infectious IBV clone, and infectious virus was recovered from cells electroporated with the mutant transcripts.
Growth properties and genetic stability of the recovered mutant viruses.
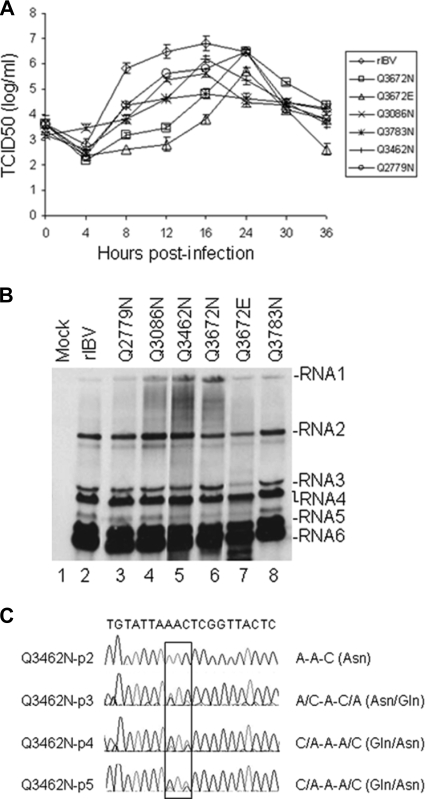
The five recovered mutant viruses, the nsp4-5/Q2779N, nsp5-6/Q3086N, nsp7-8/Q3462N, nsp8-9/Q3672N, and nsp9-10/Q3783N mutants, were plaque purified, and their growth properties were characterized by TCID50 analysis of passage 2 of each mutant virus. The results showed that the nsp7-8/Q3462N, nsp5-6/Q3086N, and nsp9-10/Q3783N mutants exhibited reduced growth phenotypes, decreasing about 2, 1, and 0.5 log10 of the peak TCID50 value at 16 h postinfection compared to wild-type rIBV, while the nsp4-5/Q2779N and nsp8-9/Q3672N mutants displayed delayed and slightly reduced growth phenotypes (Fig. 3 A). The TCID50 values of these two viruses reached the peak at 24 h postinfection, which was 8 h later than that for rIBV (Fig. 3A). Analysis of the growth properties of the nsp8-9/Q3672E mutant virus by a TCID50 assay showed that the virus exhibited a delayed and reduced growth phenotype compared to rIBV, reaching the peak titer at 24 h postinfection, which was 8 h later than for the wild-type virus (Fig. 3A). Moreover, the peak titer was approximately 10-fold less than that for rIBV (Fig. 3A). Compared to the nsp8-9/Q3672N mutant virus, the nsp8-9/Q3672E mutant virus showed similar growth kinetics, but its peak titer was approximately 5-fold lower (Fig. 3A).
FIG. 3.
Characterization of the growth properties and genetic stability of mutant viruses. (A) Growth properties of rIBV and mutant viruses. Vero cells were infected with wild-type and mutant viruses at an MOI of 1 PFU/cell and harvested at 0, 4, 8, 12, 16, 24, and 36 h postinoculation. Viral stocks were prepared by freeze-thawing the cells three times, and the TCID50 of each viral stock was determined by infecting five wells of Vero cells on 96-well plates in triplicate with a 10-fold serial dilution of each viral stock. Error bar shows standard error of the mean. (B) Northern blot analysis of the genomic and subgenomic RNAs in cells infected with wild-type and mutant viruses. Vero cells infected with wild-type and mutant viruses at an MOI of 1 PFU/cell. Total RNA (10 μg) extracted from the infected cells at 12 h postinfection was separated on 1% agarose gel and transferred to a Hybond N+ membrane. Viral RNAs were probed with a Dig-labeled DNA probe corresponding to the 3′-end 400 nucleotides of the IBV genome. Numbers on the right indicate the genomic and subgenomic RNA species of IBV. (C) Partial reversion of Asn to Gln at amino acid position 3462 during passage of the Q3462N mutant virus. The plaque-purified Q3462N mutant virus was successively passaged on Vero cells using supernatants from the cells infected with this mutant virus at an MOI of 1 PFU/cell for 16 h. Total RNA was extracted from the infected cells, and RT-PCR was carried out. Sequencing the gel-purified RT-PCR products displayed an increasing trend reverting AAC to CAA, causing partial reversion of Asn to wild-type Gln from passage 2 to 5.
Northern blot analysis was performed to analyze the total RNA extracted from cells infected with the wild type and the recovered mutant viruses. The results showed that most mutant viruses displayed patterns similar to those of the wild-type virus, except that an additional, relatively strong band migrating below sgRNA6 and a relatively reduced sgRNA3 were simultaneously detected in cells infected with the Q3672E mutant virus (Fig. 3B). As the complete sequence of this mutant virus was not determined, it was not clear at the moment if the detection of these aberrant bands was due to the presence of additional mutations in the genome. Further studies are required to clarify this issue.
The genetic stability of these mutant viruses was investigated by successive passage of plaque-purified mutant viruses on fresh Vero cells up to at least passage 5. Total RNA was extracted from infected cells, and the specific regions flanking cleavage sites were amplified by RT-PCR. Sequence analysis of the RT-PCR products confirmed the presence of the introduced mutations in the nsp4-5/Q2779N, nsp5-6/Q3086N, nsp8-9/Q3672N, nsp8-9/Q3672E, and nsp9-10/Q3783N mutant viruses, suggesting that these mutant viruses were stable in cultured cells. No amino acid change in the nsp5 coding region was identified by sequencing the entire region. Since the recovered mutant viruses were not fully sequenced, additional mutations in other regions could not be ruled out. Nevertheless, these results demonstrated that no other mutations in 3CLpro may be involved in the recovery of these mutant viruses. Interestingly, an increasing trend of reverting to the wild-type sequence was observed during passage of the nsp7-8/Q3462N mutant virus. This reversion (AAC→CAA) was initially detected in passage 3 despite the predominance of the mutant AAC codon (Fig. 3C). Subsequently, the wild-type codon (CAA) became more dominant (Fig. 3C). The RT-PCR products from passage 5 were cloned and sequenced. Eight of ten clones contained the wild-type codon (CAA), while the other two clones contained the mutant codon (AAC). Similarly, no mutations were found in 3CLpro.
Detrimental effects of Q3379N, Q4868N, Q5468N, Q5989N, and Q6327N mutations on subgenomic RNA transcription.
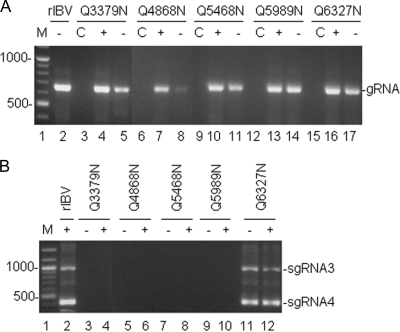
The lethality of the nsp6-7/Q3379N, nsp12-13/Q4868N, nsp13-14/Q5468N, and nsp14-15/Q5989N substitutions to the recovery of infectious virus may be due to the detrimental effects of these mutations on RNA synthesis. To examine this possibility, RT-PCR was carried out to amplify the genomic and subgenomic RNAs. Vero cells were electroporated with either wild-type or mutant RNA transcripts and cultured at 37°C for 48 h. Total RNA was extracted from the electroporated cells for reverse transcription. RT-PCR products from nucleotides 14931 to 15600, representing both positive and negative strands of the genomic RNA, were detected from cells electroporated with all four full-length mutant transcripts (Fig. 4 A, lanes 4, 5, 7, 8, 10, 11, 13, and 14). Analysis of the same RNA extracts for the detection of subgenomic RNA species 3 and 4 (sgRNA3 and 4) by RT-PCR showed no detectable RT-PCR products (Fig. 4B, lanes 3 to 10). These results suggested that mutations at these positions allowed the replication of the negative-stranded genomic RNA but disrupted the transcription of the subgenomic RNA.
FIG. 4.
Analysis of IBV genomic RNA replication and subgenomic RNA transcription in cells electroporated with full-length mutant transcripts. (A) Detection of positive- and negative-strand genomic RNA (gRNA) replication in cells electroporated with wild-type and mutant full-length transcripts. Total RNA was prepared from Vero cells electroporated with in vitro-synthesized full-length transcripts at 48 h postelectroporation. The region corresponding to nucleotides 14931 to 15600 of the positive (+)- and negative (−)-sense IBV genomic RNA was amplified by RT-PCR and analyzed on 1.2% agarose gel. Lane 1 shows DNA markers, and lanes C show negative control. (B) Detection of the negative- and positive-strand subgenomic RNA synthesis in cells electroporated with wild-type and mutant transcripts. Total RNA was prepared from Vero cells electroporated with in vitro synthesized full-length transcripts at 48 h postelectroporation. Regions corresponding to the 5′-terminal 415 and 1010 nucleotides of subgenomic RNA 4 and 3, respectively, were amplified by RT-PCR and analyzed on 1.2% agarose gel. Lane 1 shows DNA markers. The sizes of the molecular weight markers are indicated on the left.
Interestingly, substitution of nsp15-16/Q6327 with an N significantly decreased the cleavage efficiency but allowed the synthesis of both negative genomic RNA and sgRNA in cells electroporated with the nsp15-16/Q6327N mutant transcripts (Fig. 4A, lanes 16 and 17; Fig. 4B, lanes 11 and 12). These results suggest that minimal levels of viral RNA replication and transcription occurred in these cells and prompt more detailed characterization of the site.
Further characterization of the cleavage site between nsp15 and nsp16.
As sgRNA was still detected after six rounds of successive incubation of fresh cells with the nsp15-16/Q6327N mutant, but no apparent syncytium formation was observed, the possibility of additional mutations occurring in the spike protein was checked. Total RNA was extracted from the infected cells, and the RT-PCR products covering the full-length spike protein gene were obtained and sequenced. Sequence comparison illustrated that no amino acid change was found. Also, no additional mutation was detected in sequences flanking nsp15-16, except the introduced mutation. Western blotting showed no detectable expression of the structural proteins (data not shown). These data support the argument that only a minimal amount of infectious virus was produced in cells infected with this mutant virus.
To investigate whether this phenotype is associated with a nucleotide sequence across this site that may affect viral RNA packaging, a mutant construct containing six silent mutations at nucleotides 19499 (A→G), 19502 (G→A), 19505 (T→C), 19508 (A→G), 19511 (A→G), and 19514 (A→T) was made and introduced into the infectious clone. Infectious virus carrying six silent mutations was recovered, suggesting that this region is not part of the IBV packaging signal.
Mutation of P1-Q to H allowed the production of viable virus (nsp15-16/Q6327H) with a delayed growth phenotype (Fig. 5 A). Sequencing the RT-PCR products from infected Vero cells confirmed the presence of this mutation. It was reported that a putative cleavage site in which the P1 position is occupied by the H residue instead of the regular Q was predicted in human coronavirus NL63 (HCoV-NL63) (58), HCoV-HKU1 (61), and bat coronavirus HKU5 (62) based on sequence alignment. In addition, Goetz et al. reported that IBV, MHV, and SARS-CoV 3CLpros exhibit a strong preference for P1-H-containing substrates and both H and Q can adopt the same binding mode in the S1 pocket (23). Furthermore, a recently resolved structure of HCoV-HKU1 and enzyme activity assays confirmed the existence of such a cleavage event in vitro (71). This result provided the first biological evidence that IBV 3CLpro can recognize the P1-H-containing nsp15-16 site.
FIG. 5.
Effects of amino acid substitutions at Q6327 on IBV replication. (A) Growth properties of the Q6327H mutant virus. Vero cells were infected with wild-type and mutant viruses at an MOI of 1 PFU/cell and harvested at 0, 4, 8, 12, 16, 24, and 36 h postinoculation. Viral stocks were prepared by freeze-thawing the cells three times, and the TCID50 of each viral stock was determined by infecting five wells of Vero cells on 96-well plates in triplicate with a 10-fold serial dilution of each viral stock. Error bar shows standard error of the mean. (B) Detection of genomic and subgenomic RNA synthesis in cells electroporated with full-length Q6327P mutant transcripts. Total RNA was prepared from Vero cells electroporated with in vitro-synthesized full-length transcripts at 48 h postelectroporation. Regions corresponding to nucleotides 14931 to 15600 of the positive (+)- and negative (−)-sense IBV genomic RNA, and the 5′-terminal 415 and 1010 nucleotides of subgenomic RNA 4 and 3, respectively, were amplified by RT-PCR and analyzed on 1.2% agarose gel. Lanes 1 and 9 show DNA markers. The sizes of the molecular weight markers are indicated on the left.
Replacement of nsp15-16/Q6327 with P abolished completely proteolytic processing as well as virus recovery. Analysis of RNA synthesis in cells electroporated with the mutant transcripts showed that nsp15-16/Q6327P allowed the synthesis of negative-strand genomic RNA but disrupted the sgRNA transcription (Fig. 5B), suggesting that proteolytic processing at the nsp15-16 site is essential for viral replication.
Selection of mutant 3CLpros that compensate for the sublethal Q6327N mutation.
Coincidently, in a repeated experiment using double amounts of the in vitro synthesized full-length nsp15-16/Q6327N transcripts for electroporation, mutant virus was recovered. After propagation of the recovered virus for 9 passages, five RT-PCR products covering the entire IBV genome were obtained and directly sequenced, showing mixed peaks at two positions (C-to-T changes at nucleotide positions 9361 and 9362) (Fig. 6A), besides the presence of the introduced Q6327N mutation. To trace the source of the mutations, the corresponding regions of the early virus passages P1 and P3 were sequenced, revealing the presence of the two mutations (Fig. 6A). In contrast, only the wild-type sequence was identified in passage 6 of the Q6327N virus which did not produce CPE (Fig. 6A, top). Similarly, only wild-type sequences were detected by sequencing the RT-PCR products from in vitro transcripts prior to electroporation, ruling out the possibility that the mutations were introduced during in vitro transcription. To further confirm the mutations, 11 plaque-purified isolates were randomly selected from passage 9 for sequence analysis, showing that 7 out of 11 isolates carried C9362T, while the other four isolates possessed C9361T. These mutations result in amino acid changes from P to L and from P to S, respectively, at amino acid position 166 of 3CLpro. No wild-type sequences were identified in the same region.
FIG. 6.
Effects of a single amino acid mutation (P166S or P166L) in 3CLpro on the cleavage efficiency for the nsp15-16/Q6327N mutant substrate and IBV replication. (A) Existence of mixed nucleotides at nucleotide positions 9361 and 9362 in passage 1, 3, and 9 of the recovered double mutant viruses. Wild-type sequence in passage 6 of the recovered Q6327N mutant virus, which did not show syncytium formation, is shown on top. Viral RNA was extracted, and RT-PCR was performed. The purified RT-PCR products were directly sequenced. (B) Growth properties of mutant viruses. Vero cells were infected with the indicated viruses at an MOI of 1 PFU/cell and harvested at 0, 4, 8, 12, 16, and 24 h postinoculation. Viral stocks were prepared by freeze-thawing the cells three times, and the TCID50 of each viral stock was determined by infecting five wells of Vero cells on 96-well plates in triplicate with a 10-fold serial dilution of each viral stock. Error bar shows standard error of the mean. (C) Comparison of the cleavage efficiencies of wild-type (wt) and mutant 3CLpro at the nsp15-16 site containing the Q6327N mutation. Wild-type and mutant substrates were coexpressed with wild-type and mutant 3CLpro in H1299 cells using the vaccinia virus-T7 system. The transfected cell lysates were resolved on a 15% SDS-polyacrylamide gel and subjected to Western blotting with anti-Flag antibody. As a control, the amount of expressed 3CLpro was probed with anti-3CLpro antibody. The relative cleavage efficiency (RCE) is shown.
It was speculated that P166S and P166L mutations in 3CLpro may compensate for the detrimental effects on viral replication caused by the nsp15-16/Q6327N mutation. To confirm this possibility, P166S, P166L, P166S/Q6327N, and P166L/Q6327N were introduced individually into the IBV genome. As expected, viable mutant viruses were recovered from all four constructs, and TCID50 assays showed that all four mutant viruses (P166L, P166S, P166L/Q6327N, and P166S/Q6327N) exhibited a slightly reduced growth phenotype, decreasing about 0.4 to 1.0 log unit of the peak TCID50 value at 16 h postinfection, compared to rIBV (Fig. 6B).
To determine whether 3CLpro carrying the P166S or P166L mutation affects cleavage at the mutant nsp15-16 site, processing of wild-type and mutant substrates by wild-type or mutant 3CLpro was tested. Two expression plasmids containing P166S or P166L in 3CLpro were constructed. Upon cotransfection of wild-type substrate with wild-type 3CLpro and each of the mutant 3CLpros, nearly complete cleavage was observed (Fig. 6C). Upon cotransfection of nsp15-16/Q6327N mutant substrate with wild-type 3CLpro, a trace amount of the cleaved product was detected besides the uncleaved peptide (Fig. 6C). In contrast, much more efficient cleavage of the mutant substrate was observed when either P166S or P166L mutant 3CLpro was coexpressed (Fig. 6C), confirming that the mutant 3CLpros could strongly enhance the cleavage efficiency of the mutant substrates flanking the nsp15-16 site.
Attempts to recover mutant viruses carrying double mutations P166S/Q5468N, P166L/Q5468N, P166S/Q5989N, and P166L/Q5989N failed. It was also found that neither 3CLpro with P166S nor 3CLpro with P166L was able to process P1-N-containing substrates flanking the nsp13-14 and nsp14-15 sites, suggesting that this enhancement effect is specific for the P1-N-containing nsp15-16 site.
DISCUSSION
Among nidoviral 3CLpros, arterivirus and torovirus 3CLpros are serine proteinases, while ronivirus and coronavirus 3CLpros are cysteine proteinases (49, 51, 73, 74). With respect to substrate specificity, arterivirus and ronivirus 3CLpros prefer the Glu residue at the P1 position of the substrates, while coronavirus and torovirus 3CLpros require the Gln residue (49, 51, 73, 74). This differential preference at the P1 position has been thought to be determined by the S1 substrate-binding pocket (2, 3, 5, 66, 68). Among all identified 3CLpro cleavage sites of arterivirus, one site contains P1-Gln, which can also tolerate Gln-to-Asn/Glu substitution at the P1 position (59). The catalytic site Cys-to-Ser substitution in both IBV and SARS-CoV 3CLpro yields an active enzyme (27, 37, 57); conversely, Ser-to-Cys substitution in equine torovirus retains the enzymatic activity (49). In this report, we show that replacement of P1-Gln with Asn at the nsp4-5, nsp5-6, nsp7-8, nsp8-9, and nsp9-10 sites, Glu at the nsp8-9 site, and His at the nsp15-16 site of IBV allowed the recovery of the corresponding infectious mutant virus and the 3CLpro-mediated cleavage of these mutant substrates, suggesting that IBV 3CLpro is able to accept Asn, Glu, and His instead of the conserved Gln residue at the P1 position of several cleavage sites (Table 2). Further confirmation of the effect of these mutations on cleavage at the individual mutant sites would be required for cells infected with the corresponding recovered mutant viruses. However, due to the lack of specific antibodies, these studies cannot be performed. On the other hand, although the nsp4-5/Q2779N, nsp5-6/Q3086N, nsp7-8/Q3462N, nsp8-9/Q3672N, nsp8-9/Q3672E, and nsp9-10/Q3783N mutant viruses were recovered, all of them displayed reduced and/or delayed growth to a certain extent. Moreover, a reversion of Asn to Gln was observed in the nsp7-8/Q3462N mutant virus, indicating that IBV 3CLpro has a strong preference for the Gln residue at the P1 position, though it can tolerate the P1-Asn or -Glu substitution in these sites. The flexibility for substrate specificity displayed by IBV and arterivirus 3CLpros may reflect the evolutionary relationships between virus-encoded 3C-like serine and 3C-like cysteine proteinases, implying that the two proteinases may derive from a common ancestor.
TABLE 2.
Summary of the effects of mutations at the P1 position on 3CLpro-mediated cleavage and virus replication
| Mutation | Site | Cleavage in vitroa | Virus recovery | Virus growth | RNA synthesisb |
|
|---|---|---|---|---|---|---|
| gRNA | sgRNA | |||||
| Q2779N | nsp4-5 | +++ | + | Delayed and reduced | + | + |
| Q3086N | nsp5-6 | ++ | + | Reduced | + | + |
| Q3379N | nsp6-7 | − | − | + | − | |
| Q3462N | nsp7-8 | ++ | + | Reduced | + | + |
| Q3672N | nsp8-9 | ++ | + | Delayed and reduced | + | + |
| Q3672E | nsp8-9 | ++ | + | Delayed and reduced | + | + |
| Q3783N | nsp9-10 | + | + | Reduced | + | + |
| Q4868N | nsp12-13 | − | − | + | − | |
| Q5468N | nsp13-14 | − | − | + | − | |
| Q5989N | nsp14-15 | − | − | + | − | |
| Q6327N | nsp15-16 | + | + | Noncytopathic infection | + | + |
| Q6327H | nsp15-16 | ++ | + | Delayed | + | + |
| Q6327P | nsp15-16 | − | − | + | − | |
+++, complete cleavage; ++, moderate cleavage efficiency; +, low cleavage efficiency; −, no detectable cleavage.
gRNA, genomic RNA.
Complete abolishment of proteolysis and failure to recover infectious virus caused by mutations nsp6-7/Q3379N, nsp12-13/Q4868N, nsp13-14/Q5468N, nsp14-15/Q5989N, and nsp15-16/Q6327P showed that cleavage at the nsp6-7, nsp12-13, nsp13-14, nsp14-15, and nsp15-16 sites is essential for IBV replication. nsp6 contains multiple transmembrane regions and is believed to be involved in the assembly of the replicase complex (4, 44). nsp7 and nsp8 form a hexadecameric structure with RNA binding activity (70). It is possible that the unprocessed nsp6-7 intermediate may affect the formation of a functional replicase complex as well as the nsp7-nsp8 supercomplex and thus impede the initiation of sgRNA synthesis.
Considering the significance and requirement of the processed products of the ORF1b-encoded polyprotein, including RdRp, helicase, and three highly conserved RNA-processing enzymes (NendoU, ExoN, and 2′-O-MTase) for virus replication (1, 7, 10, 11, 15, 29-31, 34, 40, 45, 55, 56), the lethality caused by these mutations may be due to the loss of related enzymatic and other accessory activities. It is likely that the nsp12-nsp13, nsp13-nsp14, nsp14-nsp15, or nsp15-nsp16 fusion protein results in the change in the conformation or structure of proteins, or disruption of the protein-protein interactions associated with sgRNA transcription. Coronavirus nsp14 contains an ExoN domain with the conserved residues DE-D-D (50). Alanine substitutions for the DE-D-D residues of SARS-CoV nsp14 nearly abolished its nucleolytic activities in vitro (40). Interestingly, alanine substitutions for these active-site residues were lethal to HCoV-229E replication (40), while viable mutant MHV viruses with growth and RNA synthesis defects were recovered in the context of an infectious cDNA system (15). The reason for this marked difference is unclear, but it would be interesting to investigate whether ExoN mutants of IBV are viable and whether the nsp13-14 or nsp14-15 intermediate affects ExoN activity. nsp15 exhibits endoribonuclease activity (6, 29, 34, 65), and its critical role in coronavirus replication was established with a SARS-CoV replicon (1) and reverse-genetics-based mutagenesis studies of HCoV-229E and MHV (29, 34). The biochemical and structural data reveal an active site with a highly conserved H-H-K triad as putative catalytic residues and the requirement of nsp15 hexamerization for optimal enzymatic activity (25, 29, 33, 34, 45, 65). Mutation of either or both of the two catalytic H residues in MHV nsp15 resulted in greatly reduced but detectable endoribonuclease activity in vitro and the recovery of mutant viruses with only modest defects in viral RNA synthesis, suggesting that the nsp15 endoribonuclease is needed for optimal MHV replication (34). In contrast, alanine substitutions for a noncatalytic residue, D324, in MHV nsp15 and the corresponding D6408 in HCoV-229E nsp15 were lethal to virus production (29, 34). Expression of MHV nsp15 containing the D324A mutation and corresponding SARS-CoV nsp15 carrying D296A in bacteria resulted in formation of insoluble proteins (25, 34). Structurally, this Asp residue is highly conserved in the nsp15 orthologs of all coronaviruses and is involved in an extensive hydrogen bond network (45). The D296A mutation was believed to have structural effects on nsp15 that indirectly impair the function of this protein, suggesting that the structural integrity of nsp15 is essential for viral replication (45). In the present study, the nsp14-15 and nsp15-16 intermediates resulting from nsp14-15/Q5989N and nsp15-16/Q6327P may block the hexamer formation of IBV nsp15, completely abolishing the enzymatic activity.
Some stable intermediate cleavage products, such as the IBV nsp12-13 (160 kDa) and nsp14-15-16 (132 kDa) intermediates and MHV nsp4-10/11 (150 kDa) intermediates, were identified in virus-infected cells and were thought to play a role in virus replication (48, 64). A recent model suggests that unprocessed intermediates, such as MHV nsp4-10/11 (P150), function as a polyprotein involved in negative-stranded RNA synthesis, while mature proteins are required for positive-stranded RNA synthesis (46, 47). The results from this report revealed an intimate correlation between 3CLpro-mediated cleavage and regulation of RNA synthesis (Table 2). Mutation of P1-Gln to Asn at the nsp6-7, nsp12-13, nsp13-14, and nsp14-15 sites, as well as mutation of P1-Gln to Pro at the nsp15-16 site, which resulted in the complete abolishment of proteolytic processing, allowed negative-stranded genomic RNA replication but blocked sgRNA transcription. In contrast, mutation of P1-Gln to Asn at the nsp15-16 site allowed the synthesis of both genomic and sgRNA, even though the amount of cleaved products was extremely low due to significantly reduced processing efficiency. These findings suggested that unprocessed intermediates, such as the nsp6-7, nsp12-13, nsp13-14, nsp14-15, and nsp15-16 precursors, may function in genomic RNA replication, while mature proteins may be involved in sgRNA transcription. More extensive studies of cells infected with the recovered mutant viruses and transfected with full-length transcripts carrying the mutations would be required to support this assumption.
Coronavirus nsp15 is an endoribonuclease involved in RNA processing (6, 8, 29, 65), and nsp16 possesses 2′O-methyltransferase activity, thought to be responsible for RNA capping (11, 50, 60). Both proteins were proven to be essential for RNA synthesis (1, 34). In this study, low-level, noncytopathic infection was observed with cells infected with Q6327N mutant virus, due to inefficient transcription of positive-stranded sgRNA. It suggests that one of the main functions of mature nsp15 and nsp16 is at this step of the viral replication cycle. However, attempts to rescue the phenotype by coexpression of individual nsp15 and nsp16 with the full-length mutant transcripts in cells were unsuccessful. Further biochemical and functional characterization of the two proteins would be required to address this issue.
Perhaps the most intriguing result is the selection of 3CLpro carrying the P166S or P166L mutation. Both the P166S and P166L mutants specifically enhance the cleavage efficiency at the nsp15-16 site containing the P1-Asn substitution. Structurally, Pro-166 in IBV 3CLpro, corresponding to Pro-168 in SARS-CoV 3CLpro, is not conserved in the 3CLpros of MHV, TGEV, and HCoV-229E where a Ser or a Gly residue is located (66). Residues 164 to 168 form an antiparallel sheet with residues P6 to P1 of a substrate, but Pro-166 is not a required determinant for substrate specificity (66). It is possible that the P166S or P166L mutation may alter the interaction between amino acid residues in the substrate-binding pocket, leading to the increased proteolytic cleavage activity at the mutant nsp15-16 site and promoting virus replication. Alternatively, interaction between the mutant 3CLpro and the mutant nsp15-16 intermediate may induce conformational change of the substrate to facilitate 3CLpro-mediated cleavage. As this enhancement effect of the mutant 3CLpros is observed only with the mutant nsp15-16 site containing the P1-Asn substitution, the substrate that induces the emergence of the mutant enzyme, further structural studies would reveal novel insights into the enzyme substrate specificity and catalytic mechanisms of this important proteinase.
Footnotes
Published ahead of print on 5 May 2010.
REFERENCES
- 1.Almazán, F., M. L. Dediego, C. Galan, D. Escors, E. Alvarez, J. Ortego, I. Sola, S. Zuniga, S. Alonso, J. L. Moreno, A. Nogales, C. Capiscol, and L. Enjuanes. 2006. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 80:10900-10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, K., G. J. Palm, J. R. Mesters, S. G. Siddell, J. Ziebuhr, and R. Hilgenfeld. 2002. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 21:3213-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 4.Baliji, S., S. A. Cammer, B. Sobral, and S. C. Baker. 2009. Detection of nonstructural protein 6 in murine coronavirus-infected cells and analysis of the transmembrane topology by using bioinformatics and molecular approaches. J. Virol. 83:6957-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrette-Ng, I. H., K. K. Ng, B. L. Mark, D. Van Aken, M. M. Cherney, C. Garen, Y. Kolodenko, A. E. Gorbalenya, E. J. Snijder, and M. N. James. 2002. Structure of arterivirus nsp4. The smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J. Biol. Chem. 277:39960-39966. [DOI] [PubMed] [Google Scholar]
- 6.Bhardwaj, K., L. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78:12218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhardwaj, K., S. Palaninathan, J. M. Alcantara, L. L. Yi, L. Guarino, J. C. Sacchettini, and C. C. Kao. 2008. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J. Biol. Chem. 283:3655-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhardwaj, K., J. Sun, A. Holzenburg, L. A. Guarino, and C. C. Kao. 2006. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 361:243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, B., S. Fang, J. P. Tam, and D. X. Liu. 2009. Formation of stable homodimer via the C-terminal alpha-helical domain of coronavirus nonstructural protein 9 is critical for its function in viral replication. Virology 383:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Y., H. Cai, J. Pan, N. Xiang, P. Tien, T. Ahola, and D. Guo. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 106:3484-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decroly, E., I. Imbert, B. Coutard, M. Bouvet, B. Selisko, K. Alvarez, A. E. Gorbalenya, E. J. Snijder, and B. Canard. 2008. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 82:8071-8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deming, D. J., R. L. Graham, M. R. Denison, and R. S. Baric. 2007. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J. Virol. 81:10280-10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donaldson, E. F., R. L. Graham, A. C. Sims, M. R. Denison, and R. S. Baric. 2007. Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing. J. Virol. 81:7086-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donaldson, E. F., A. C. Sims, R. L. Graham, M. R. Denison, and R. S. Baric. 2007. Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J. Virol. 81:6356-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckerle, L. D., X. Lu, S. M. Sperry, L. Choi, and M. R. Denison. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 81:12135-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egloff, M. P., F. Ferron, V. Campanacci, S. Longhi, C. Rancurel, H. Dutartre, E. J. Snijder, A. E. Gorbalenya, C. Cambillau, and B. Canard. 2004. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. U. S. A. 101:3792-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan, K., L. Ma, X. Han, H. Liang, P. Wei, Y. Liu, and L. Lai. 2005. The substrate specificity of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 329:934-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan, K., P. Wei, Q. Feng, S. Chen, C. Huang, L. Ma, B. Lai, J. Pei, Y. Liu, J. Chen, and L. Lai. 2004. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 279:1637-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang, S., B. Chen, F. P. Tay, B. S. Ng, and D. X. Liu. 2007. An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology 358:136-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang, S. G., H. Shen, J. Wang, F. P. Tay, and D. X. Liu. 2008. Proteolytic processing of polyproteins 1a and 1ab between non-structural proteins 10 and 11/12 of Coronavirus infectious bronchitis virus is dispensable for viral replication in cultured cells. Virology 379:175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, F., H. Y. Ou, L. L. Chen, W. X. Zheng, and C. T. Zhang. 2003. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 553:451-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goetz, D. H., Y. Choe, E. Hansell, Y. T. Chen, M. McDowell, C. B. Jonsson, W. R. Roush, J. McKerrow, and C. S. Craik. 2007. Substrate specificity profiling and identification of a new class of inhibitor for the major protease of the SARS coronavirus. Biochemistry 46:8744-8752. [DOI] [PubMed] [Google Scholar]
- 24.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guarino, L. A., K. Bhardwaj, W. Dong, J. Sun, A. Holzenburg, and C. Kao. 2005. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 353:1106-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegyi, A., and J. Ziebuhr. 2002. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 83:595-599. [DOI] [PubMed] [Google Scholar]
- 27.Huang, C., P. Wei, K. Fan, Y. Liu, and L. Lai. 2004. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry 43:4568-4574. [DOI] [PubMed] [Google Scholar]
- 28.Imbert, I., J. C. Guillemot, J. M. Bourhis, C. Bussetta, B. Coutard, M. P. Egloff, F. Ferron, A. E. Gorbalenya, and B. Canard. 2006. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 25:4933-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U. S. A. 101:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov, K. A., and J. Ziebuhr. 2004. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 78:7833-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joseph, J. S., K. S. Saikatendu, V. Subramanian, B. W. Neuman, A. Brooun, M. Griffith, K. Moy, M. K. Yadav, J. Velasquez, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2006. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J. Virol. 80:7894-7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joseph, J. S., K. S. Saikatendu, V. Subramanian, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2007. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 81:6700-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang, H., K. Bhardwaj, Y. Li, S. Palaninathan, J. Sacchettini, L. Guarino, J. L. Leibowitz, and C. C. Kao. 2007. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 81:13587-13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, J. C., R. A. Spence, P. F. Currier, X. Lu, and M. R. Denison. 1995. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology 208:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, D. X., I. Brierley, K. W. Tibbles, and T. D. Brown. 1994. A 100-kilodalton polypeptide encoded by open reading frame (ORF) 1b of the coronavirus infectious bronchitis virus is processed by ORF 1a products. J. Virol. 68:5772-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, D. X., and T. D. Brown. 1995. Characterisation and mutational analysis of an ORF 1a-encoding proteinase domain responsible for proteolytic processing of the infectious bronchitis virus 1a/1b polyprotein. Virology 209:420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, D. X., S. Shen, H. Y. Xu, and S. F. Wang. 1998. Proteolytic mapping of the coronavirus infectious bronchitis virus 1b polyprotein: evidence for the presence of four cleavage sites of the 3C-like proteinase and identification of two novel cleavage products. Virology 246:288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, D. X., H. Y. Xu, and T. D. Brown. 1997. Proteolytic processing of the coronavirus infectious bronchitis virus 1a polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J. Virol. 71:1814-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minskaia, E., T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard, and J. Ziebuhr. 2006. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 103:5108-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng, L. F., and D. X. Liu. 2000. Further characterization of the coronavirus infectious bronchitis virus 3C-like proteinase and determination of a new cleavage site. Virology 272:27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng, L. F., and D. X. Liu. 1998. Identification of a 24-kDa polypeptide processed from the coronavirus infectious bronchitis virus 1a polyprotein by the 3C-like proteinase and determination of its cleavage sites. Virology 243:388-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng, L. F., and D. X. Liu. 2002. Membrane association and dimerization of a cysteine-rich, 16-kilodalton polypeptide released from the C-terminal region of the coronavirus infectious bronchitis virus 1a polyprotein. J. Virol. 76:6257-6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oostra, M., M. C. Hagemeijer, M. van Gent, C. P. Bekker, E. G. te Lintelo, P. J. Rottier, and C. A. de Haan. 2008. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 82:12392-12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ricagno, S., M. P. Egloff, R. Ulferts, B. Coutard, D. Nurizzo, V. Campanacci, C. Cambillau, J. Ziebuhr, and B. Canard. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. U. S. A. 103:11892-11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawicki, S. G., D. L. Sawicki, and S. G. Siddell. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawicki, S. G., D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schiller, J. J., A. Kanjanahaluethai, and S. C. Baker. 1998. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smits, S. L., E. J. Snijder, and R. J. de Groot. 2006. Characterization of a torovirus main proteinase. J. Virol. 80:4157-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snijder, E. J., A. L. Wassenaar, L. C. van Dinten, W. J. Spaan, and A. E. Gorbalenya. 1996. The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases. J. Biol. Chem. 271:4864-4871. [DOI] [PubMed] [Google Scholar]
- 52.Su, D., Z. Lou, F. Sun, Y. Zhai, H. Yang, R. Zhang, A. Joachimiak, X. C. Zhang, M. Bartlam, and Z. Rao. 2006. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J. Virol. 80:7902-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sutton, G., E. Fry, L. Carter, S. Sainsbury, T. Walter, J. Nettleship, N. Berrow, R. Owens, R. Gilbert, A. Davidson, S. Siddell, L. L. Poon, J. Diprose, D. Alderton, M. Walsh, J. M. Grimes, and D. I. Stuart. 2004. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 12:341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan, Y. W., S. Fang, H. Fan, J. Lescar, and D. X. Liu. 2006. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 34:4816-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 75:6676-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 57.Tibbles, K. W., I. Brierley, D. Cavanagh, and T. D. Brown. 1996. Characterization in vitro of an autocatalytic processing activity associated with the predicted 3C-like proteinase domain of the coronavirus avian infectious bronchitis virus. J. Virol. 70:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Hoek, L., K. Pyrc, and B. Berkhout. 2006. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol. Rev. 30:760-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Dinten, L. C., S. Rensen, A. E. Gorbalenya, and E. J. Snijder. 1999. Proteolytic processing of the open reading frame 1b-encoded part of arterivirus replicase is mediated by nsp4 serine protease and is essential for virus replication. J. Virol. 73:2027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Grotthuss, M., L. S. Wyrwicz, and L. Rychlewski. 2003. mRNA cap-1 methyltransferase in the SARS genome. Cell 113:701-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woo, P. C., Y. Huang, S. K. Lau, H. W. Tsoi, and K. Y. Yuen. 2005. In silico analysis of ORF1ab in coronavirus HKU1 genome reveals a unique putative cleavage site of coronavirus HKU1 3C-like protease. Microbiol. Immunol. 49:899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woo, P. C., M. Wang, S. K. Lau, H. Xu, R. W. Poon, R. Guo, B. H. Wong, K. Gao, H. W. Tsoi, Y. Huang, K. S. Li, C. S. Lam, K. H. Chan, B. J. Zheng, and K. Y. Yuen. 2007. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 81:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, C. Y., J. T. Jan, S. H. Ma, C. J. Kuo, H. F. Juan, Y. S. Cheng, H. H. Hsu, H. C. Huang, D. Wu, A. Brik, F. S. Liang, R. S. Liu, J. M. Fang, S. T. Chen, P. H. Liang, and C. H. Wong. 2004. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U. S. A. 101:10012-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu, H. Y., K. P. Lim, S. Shen, and D. X. Liu. 2001. Further identification and characterization of novel intermediate and mature cleavage products released from the ORF 1b region of the avian coronavirus infectious bronchitis virus 1a/1b polyprotein. Virology 288:212-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu, X., Y. Zhai, F. Sun, Z. Lou, D. Su, Y. Xu, R. Zhang, A. Joachimiak, X. C. Zhang, M. Bartlam, and Z. Rao. 2006. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J. Virol. 80:7909-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xue, X., H. Yu, H. Yang, F. Xue, Z. Wu, W. Shen, J. Li, Z. Zhou, Y. Ding, Q. Zhao, X. C. Zhang, M. Liao, M. Bartlam, and Z. Rao. 2008. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 82:2515-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, H., W. Xie, X. Xue, K. Yang, J. Ma, W. Liang, Q. Zhao, Z. Zhou, D. Pei, J. Ziebuhr, R. Hilgenfeld, K. Y. Yuen, L. Wong, G. Gao, S. Chen, Z. Chen, D. Ma, M. Bartlam, and Z. Rao. 2005. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 3:e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, H., M. Yang, Y. Ding, Y. Liu, Z. Lou, Z. Zhou, L. Sun, L. Mo, S. Ye, H. Pang, G. F. Gao, K. Anand, M. Bartlam, R. Hilgenfeld, and Z. Rao. 2003. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U. S. A. 100:13190-13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang, S., S. J. Chen, M. F. Hsu, J. D. Wu, C. T. Tseng, Y. F. Liu, H. C. Chen, C. W. Kuo, C. S. Wu, L. W. Chang, W. C. Chen, S. Y. Liao, T. Y. Chang, H. H. Hung, H. L. Shr, C. Y. Liu, Y. A. Huang, L. Y. Chang, J. C. Hsu, C. J. Peters, A. H. Wang, and M. C. Hsu. 2006. Synthesis, crystal structure, structure-activity relationships, and antiviral activity of a potent SARS coronavirus 3CL protease inhibitor. J. Med. Chem. 49:4971-4980. [DOI] [PubMed] [Google Scholar]
- 70.Zhai, Y., F. Sun, X. Li, H. Pang, X. Xu, M. Bartlam, and Z. Rao. 2005. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 12:980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao, Q., S. Li, F. Xue, Y. Zou, C. Chen, M. Bartlam, and Z. Rao. 2008. Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. J. Virol. 82:8647-8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 287:57-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ziebuhr, J., S. Bayer, J. A. Cowley, and A. E. Gorbalenya. 2003. The 3C-like proteinase of an invertebrate nidovirus links coronavirus and potyvirus homologs. J. Virol. 77:1415-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]