Abstract
Human T-lymphotropic virus 1 (HTLV-1) causes an aggressive malignancy of T lymphocytes called adult T-cell leukemia/lymphoma (ATLL), and expression of HTLV-1 Tax influences cell survival, proliferation, and genomic stability in the infected T lymphocytes. Cyclin-dependent kinase inhibitor 1A (CDKN1A/p21waf1/Cip1) is upregulated by Tax, without perturbation of cell cycle control. During an analysis of the gene expression profiles of ATLL cells, we found very low expression of CDKN1A in ATLL-derived cell lines and ATLL cells from patient samples, and epigenetic abnormalities including promoter methylation are one of the mechanisms for the low CDKN1A expression in ATLL cells. Three HTLV-1-infected cell lines showed high levels of expression of both CDKN1A and Tax, but expression of CDKN1A was detected in only two of six ATLL-derived cell lines. In both the HTLV-1-infected and ATLL cell lines, we found that activated Akt phosphorylates CDKN1A at threonine 145 (T145), leading to cytoplasmic localization of CDKNIA. In HTLV-1-infected cell lines, cytoplasmic CDKN1A did not inhibit the cell cycle after UV irradiation; however, following treatment with LY294002, a PI3K inhibitor, CDKN1A was dephosphorylated and relocalized to the nucleus, resulting in suppression of the cell cycle. In the ATLL cell lines, treatment with LY294002 did not inhibit the cell cycle but induced apoptosis with the cytoplasmic localization. Therefore, the low CDKN1A expression in ATLL cells may be a key player in ATLL leukemogenesis, and the abnormal genomic methylation may influence the expression of not only HTLV-1 Tax but also CDKN1A during long-term development of ATLL from the HTLV-1-infected T lymphocytes.
Human T-cell lymphotropic virus type 1 (HTLV-1) is the etiologic agent of adult T-cell leukemia/lymphoma (ATLL), a fatal CD4+ leukemia (20, 21, 38). At present, an estimated 10 to 20 million people worldwide are infected with HTLV-1. The HTLV-1 infection is endemic in southwestern Japan, Africa, the Caribbean Islands, and South America. The prognosis of patients with aggressive ATLL remains poor, with a median survival time of less than 1 year despite advances in both chemotherapy and supportive care (28, 29, 37). The viral determinant critical for the progression to T-cell malignancy in HTLV-1 carriers is thought to be the HTLV-1 transactivator/oncoprotein Tax (1). Tax is a 40-kDa protein that functions as a transactivator of viral gene expression and is considered a key component of the leukemogenic process that results from HTLV-1 infection (12). Tax interacts with multiple transcription factors, such as cyclic AMP-responsive element binding protein (CREB), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) family members, TATA-binding protein (TBP), and transcription factor IIA (TFIIA). Tax also stimulates the transcription of many genes, including interleukin-2 (IL-2), IL-2Rα, PCNA, and PTHrP, as well as the proto-oncogenes c-fos and c-sis (17). Intriguingly, Tax increases the levels of cyclin-dependent kinase 1A (CDKN1A/p21waf1) expression in HTLV-1-infected cell lines (5, 13). The CDKN1A gene product was originally thought to be purely a cell cycle inhibitor; however, HTLV-1-transformed T cells grow and proliferate normally, despite abundant CDKN1A expression. On the other hand, the majority of ATLL cells do not produce a large amount of Tax protein in vivo since methylation and deletion of HTLV-1 genomic DNA are frequently found in ATLL cells (14, 30, 32). Therefore, many important differences may exist between the intracellular environments of ATLL cells and HTLV-1-infected cells because several types of transformation events must be accumulated in order for ATLL to develop.
Recently, we reported that tumor suppressor in lung cancer 1 (TSLC1/IgSF4/CADM1) is overexpressed in acute-type ATLL cells in a DNA microarray-based survey of gene expression (24). Expression of a cell adhesion molecule, TSLC1, plays an important role in the organ infiltration of ATLL cells (6). In this report, we examined the expression profile of ATLL cells, focusing on genes regulated by HTLV-1 infection. Within the Tax-regulated genes, we found that CDKN1A was specifically downregulated in ATLL cells compared with CD4+ T lymphocytes, while CDKN1A was upregulated in the HTLV-1-infected cell lines. Compared with HTLV-1-infected cell lines, a majority of ATLL-derived cell lines and primary ATLL cells showed DNA methylation of the CDKN1A promoter region, with low or no expression of CDKN1A and Tax; however, no DNA methylation in CDKN1A was found in the three HTLV-1-infected cell lines that showed high levels of CDKN1A and Tax. Interestingly, CDKN1A was mainly localized in the cytoplasm in HTLV-1-infected cell lines and ATLL cell lines and was also phosphorylated at threonine 145 (T145) by activated Akt in both cell lines. After treatment with the phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002, Akt was dephosphorylated, and CDKN1A was dephosphorylated at T145 and relocalized to the nucleus in HTLV-1-infected cell lines, which increased the percentage of G1-arrested cells. However, dephosphorylated CDKN1A remained in the cytoplasm in ATLL cells to induce apoptosis after LY294002 treatment; the reason for this is unknown. Therefore, we found that CDKN1A showed different expression patterns and different functions in HTLV-1-infected and ATLL-derived cell lines. Since many kinds of genetic and epigenetic abnormalities are accumulated in the genomes of HTLV-1-infected cells during the course of development to ATLL cells, different expression patterns and functions of CDKN1A in both cell lines might reflect the developmental changes from HTLV-1-infected T cells to ATLL cells.
MATERIALS AND METHODS
Patient samples.
All samples were collected at the time of admission to the hospital before the patients started chemotherapy. The diagnosis of ATLL was based on clinical features, hematologic characteristics, and the presence of anti-HTLV-1 antibodies in patient sera. The integration of monoclonal HTLV-1 provirus into the DNA of leukemic cells was confirmed by Southern blot hybridization. Peripheral blood mononuclear cells (PBMCs) from patients with ATLL were isolated using Histopaque (Sigma, Saint Louis, MO) by density gradient centrifugation. Each patient had more than 90% leukemic cells in the blood at the time of analysis. The study was approved by the Institutional Review Board (IRB) of the Faculty of Medicine, University of Miyazaki. Informed consent was obtained from all blood and tissue donors according to the Helsinki Declaration.
Cell lines.
Human T-cell acute lymphoblastic leukemia (T-ALL) cell lines (Jurkat, MOLT4, and MKB-1), HTLV-1-infected T-cell lines (MT-2, MT-4 and HUT102), and ATLL-derived cell lines (ED, Su9T01, and S1T) were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. The interleukin-2 (IL-2)-dependent ATLL-derived cell lines KOB, SO4, and KK1 were maintained in complete RPMI 1640 medium supplemented with 100 Japan reference units (JRU)/ml of recombinant human IL-2 (Takeda Chemical Industries, Osaka, Japan).
Oligonucleotide microarray and data analyses.
Five micrograms of total RNA was hybridized to the HU95A microarray (Affymetrix, Santa Clara, CA). The standard protocol used for sample preparation and microarray processing is available from Affymetrix and has been described previously (24). Expression data were analyzed using Microarray Suite, version 5.0 (Affymetrix).
RT-PCR.
Total RNA was extracted using Trizol (Life Technologies, Carlsbad, CA) and converted to cDNA using an RNA-PCR kit (Takara Shuzo, Kyoto, Japan) according to the manufacturer's protocol. The following primers (forward and reverse, respectively) were used: for CDKN1A, 5′-ATGTCAGAACCGGCTGGGGAT-3′ and 5′-TAGGGCTTCCTCTTGGAGAAG-3′ (annealing temperature of 55°C); for HTLV-1 Tax, 5′-CCGGCGCTGCTCTCATCCCGAT-3′ and 5′-GGCCGAACATAGTCCCCCAGAG-3′ (60°C); for p53, 5′-CCAAGCTTATGGAGGAGCCGCAGTCAGATCCTAGCG-3′ and 5′-TCACAACCTCCGTCATGTGC-3′ (55°C); for β-actin, 5′-TCCTTCTGCATCCTGTCGGCT-3′ and 5′-CCAGAGATGGCCACGGCTGCT-3′ (55°C). The reverse transcription-PCR (RT-PCR) products were run on agarose gels and visualized by ethidium bromide staining.
Western blot analysis.
Cells were lysed with sodium dodecyl sulfate (SDS) sample buffer (0.05 M Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 0.01% bromophenol blue, 0.6% 2-mercaptoethanol), subjected to SDS polyacrylamide gel electrophoresis, and then transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5% nonfat dried milk in Tris-HCl-buffered saline plus 0.1% Tween 20 and incubated with anti-p21 (C-19; Santa Cruz Biotechnology, Santa Cruz, CA), anti-p27 (C-19; Santa Cruz), anti-Tax (MI73; a kind gift of M. Matsuoka, Institute for Virus Research, Kyoto University), anti-p53 (Ab-6; EMD Biosciences, San Diego, CA), anti-Akt (9272; Cell signaling), anti-phospho-Akt (Ser473; 9271, Cell signaling Technology, Beverly, MA), anti-phospho-CDKN1A (Thr145; sc-20220-R, Santa Cruz), or anti-β-actin (AC-15; Sigma) antibodies overnight at 4°C and then incubated with horseradish peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG for 2 h at room temperature. Immunoblots were then reacted with Lumi-Light Plus reagent (Roche Diagnostics, Indianapolis, IN). Can Get Signal (Toyobo, Osaka, Japan) was used to enhance antigenic signals.
Assay for HTLV-1 proviral copy numbers.
The primers and the probe for the pX gene region of HTLV-1 provirus were as follows: the forward primer (pX2-S, 5′-CGGATACCCAGTCTACGTGTT-3′; positions 7359 to 7379), the reverse primer (pX2-AS, 5′-CAGTAGGGCGTGACGATGTA-3′; positions 7458 to 7439), and the 6-carboxyfluorescein (FAM)-labeled probe (5′-FAM-CTGTGTACAAGGCGACTGGTGCC-TAMRA-3′, where TAMRA is 6-carboxytetramethylrhodamine) (31). The nucleotide position numbers of HTLV-1 provirus are according to the published reports (25). RNase P control reagent (Applied Biosystems, Foster City, CA) was used for the primers and the probe for the human RNase P DNA gene as an internal control.
Cell growth analysis.
Cells were seeded in six-well plates at 1 × 106 cells/ml and treated with UV radiation (20 J/m2) and/or LY294002 (20 μM). Rates of proliferation were determined by counting the number of cells every 24 h using the trypan blue exclusion method.
Real-time quantitative RT-PCR.
Real-time RT-PCR was performed on an ABI Prism 7700 SDS using a predeveloped TaqMan RT-PCR kit (Applied Biosystems). The expression levels of CDKN1A mRNA and the internal reference β-actin were measured following the manufacturer's instructions. The primers and probes were purchased from Applied Biosystems as TaqMan Gene Expression Assays.
MSP with bisulfite treatment.
One microgram of genomic DNA was treated with sodium bisulfite as described previously (22). Methylation-specific PCR (MSP) primers for CDKN1A were designed according to published literature (40). The following primer sets were used: 5′-GTTGTTTGTTGGAATTCGGTTAG-3′ and 5′-CGACGAATCCGCGCC-3′ for the methylated CDKN1A sequence, located at −182 to +48 upstream of the transcription start site (230 bp), and 5′-GTTTGTTGGAATTTGGTTAG-3′ and 5′-CAACAAATCCACACCCAA-3′ for the unmethylated CDKN1A sequence (230 bp). Each MSP reaction mixture incorporated approximately 10 ng of bisulfite-treated DNA, 20 pmol of each primer, 100 pmol of the deoxynucleoside triphosphates (dNTPs), 2 μl of 10× PCR buffer, 0.2 μl of ExTaq Polymerase (Takara, Tokyo, Japan), and 0.2 μl of Anti-ExTaq monoclonal antibody (Takara), in a final reaction volume of 20 μl. Cycle conditions were the following: 1 cycle of 5 min at 95°C; 35 cycles of 30 s at 95°C, 30 s at 50°C, and 30 s at 72°C; and a final cycle of 5 min at 72°C. PCR products were run on 2% agarose gels and visualized by ethidium bromide staining.
Immunofluorescence staining.
Cells were fixed with 1% formaldehyde, permeabilized with 0.5% Triton X-100, stained with antibodies against CDKN1A or CDKN1B followed by Alexa Fluor 488 green-conjugated second antibody (LifeTechnologies), and then observed under a confocal microscope (Olympus IX81 fluorescence microscope; Olympus, Tokyo, Japan). Cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) to visualize nuclei.
Cell cycle and apoptosis analyses.
Serum-starved cells (2 × 106) were treated with UV (20 J/m2) and/or LY294002 (20 μM) for the time periods indicated in the figure legends and stained with propidium iodide (PI). The cells were then subjected to fluorescence-activated cell sorter (FACS) analysis. The percentage of cells in each phase of the cell cycle was computed by using the ModFit LT software program (Becton Dickinson, San Jose, CA). For apoptosis analysis, cells were harvested and stained using an annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (MBL Nagoya, Japan) according to the manufacturer's protocol.
Statistical analysis.
For the cell growth shown in Fig. 3A and 4C, statistical analyses of the associations between treated and control cells were performed by a Student's t test with significance set at P values of <0.05 or <0.001, and all data are expressed as the means ± standard deviations of the values as analyzed by the t test.
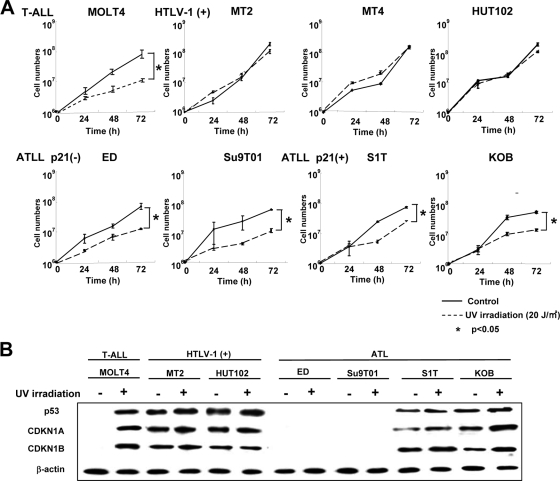
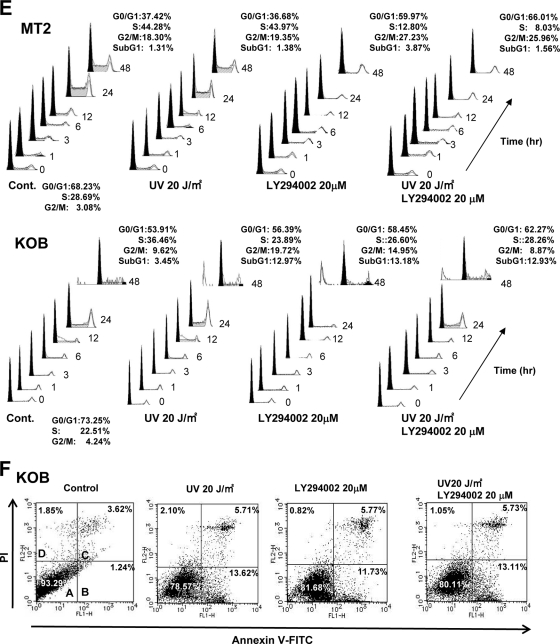
FIG. 3.
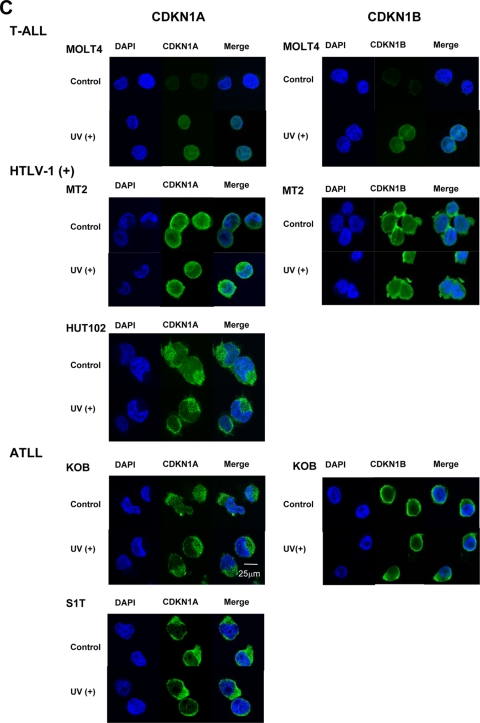
Loss of functional CDKN1A response to UV irradiation in HTLV-1-infected and ATLL cell lines. (A) Cell growth curves of an HTLV-1-negative T-ALL cell line (MOLT4), three HTLV-1-infected cell lines (MT-2, MT-4, and HUT102), and four ATLL-derived cell lines (ED, Su9T01, S1T, and KOB) before and after UV irradiation. Viable cells were counted using the trypan blue exclusion method at the indicated times after 20 J/m2 UV irradiation. Samples at 0 h were nonirradiated controls. A Student's t test was used for statistical analysis. (B) Western blot analyses of p53, CDKN1A, and CDKN1B following UV irradiation. Each cell line, either untreated (−) or UV irradiated (+; 20 J/m2),was cultured for 72 h to examine protein expression. (C) Subcellular localization of CDKN1A and CDKN1B with or without (control) UV irradiation. Localization of CDKN1A and CDKN1B was detected by confocal immunofluorescence analysis. DAPI stain was used to visualize the nuclei. Magnification, ×400; bar, 25 μm.
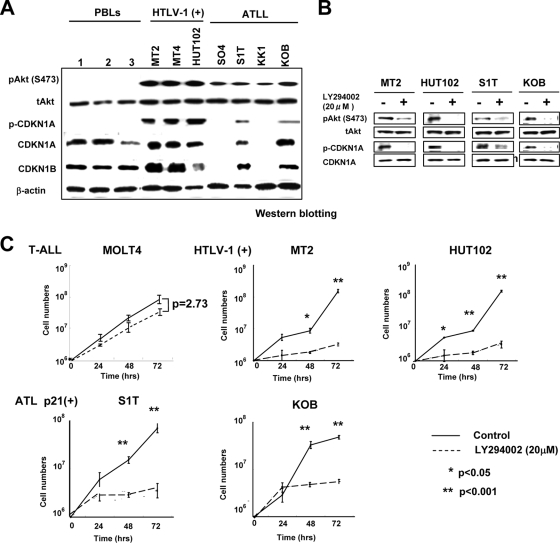
FIG. 4.
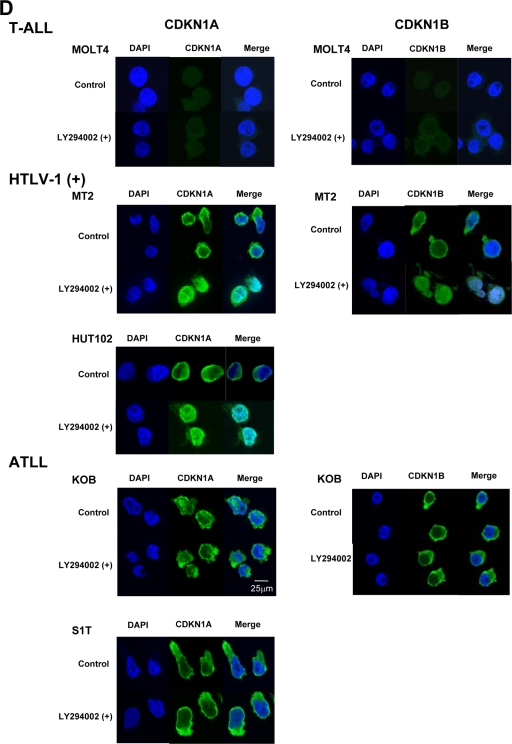
Cytoplasmic sequestration and functional inactivation of CDKN1A in HTLV-1-infected and ATLL cell lines. (A) Expression of Akt, phospho-Akt (Ser473), CDKN1A, phospho-CDKN1A (Thr145), and CDKN1B in PBLs and HTLV-1-infected and ATLL cell lines was examined by Western blotting. (B) HTLV-1-infected cell lines and ATLL cell lines were treated with 20 μM LY294002 for 2 h, and Western blot analyses were performed for phospho-Akt, Akt, phospho-CDKN1A, and CDKN1A. (C) Survival curves of various types of T-cell leukemia cell lines after treatment with the PI3K inhibitor LY294002. Cells were treated with 20 μM LY294002 for each indicated time point, and their cell numbers were measured by trypan blue exclusion method (0 h represents the untreated controls). A Student's t test was used for statistical analysis. (D) The subcellular localization of CDKN1A and CDKN1B with or without LY294002 treatment was detected by immunofluorescence in various types of T-cell leukemia cell lines. DAPI stain was used to visualize the nuclei. Magnification, ×400; bar, 25 μm. (E) MT-2 and KOB cells were grown under serum-free conditions and either left untreated (control) or treated with UV radiation (20 J/m2), LY294002 (20 μM), or both UV radiation and LY294002 and cultured for the indicated times. Cell cycle progression and apoptosis were analyzed by FACS analysis upon PI staining. The percentage of cells in each fraction at 0 h and 48 h is presented. (F) KOB ATLL cells were treated with UV radiation and/or LY294002 as described for panel E. The percentage of cells undergoing apoptosis was quantitated by staining with annexin V-FITC and PI, and the distribution of cells is presented in each quadrant by flow cytometric analysis.
RESULTS
Low expression of CDKN1A in ATLL cells.
In a previous study, we reported that 192 genes were specifically upregulated in acute-type adult T-cell leukemia/lymphoma (ATLL) cells compared with CD4+ T lymphocytes using gene expression profiles of a DNA microarray system (24). On the other hand, many genes were induced by HTLV-1 Tax, including IL-2Rα, CDKN1A, and others (4, 9, 18). Previously, the Tax-induced genes were reported by comparing the expression profiles of HTLV-1-infected cell lines with those of normal activated peripheral blood lymphocytes (PBLs) using Affymetrix Hu6800 arrays (19). In the present study we determined the expression patterns of a series of Tax-induced and Tax-suppressed genes in comparison with the expression profiles of acute-type ATLL cells and CD4+ T lymphocytes using Affymetrix HU95 arrays. We detected 108 genes that were upregulated at least 2-fold (77 genes) or suppressed more than 0.5-fold (31 genes) in the HTLV-1-infected cells (see Table S1 in the supplemental material). Of those 108 genes, 22 (including IL-2Rα) were expressed at least 2-fold higher, and 28 genes were suppressed by at least 0.5-fold in ATLL cells compared with CD4+ lymphocytes (Fig. 1A). Interestingly, 18 out of 28 genes upregulated by HTLV-1 infection were downregulated, with under 0.5-fold expression, in ATLL cells (see Table S1-d in the supplemental material). Cyclin-dependent kinase inhibitor p21waf1 (CDKN1A), an important gene induced by HTLV-1 infection, was one of the 18 oppositely regulated genes.
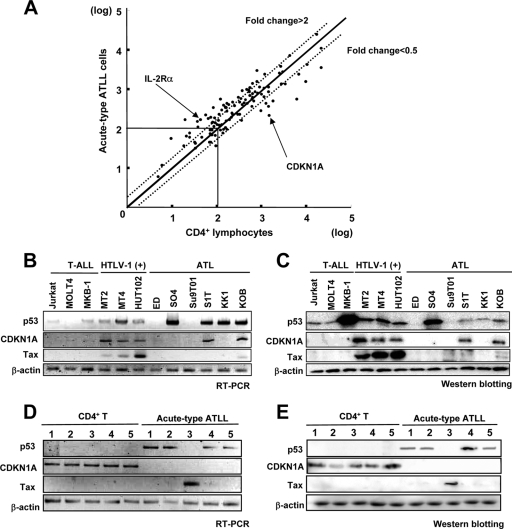
FIG. 1.
Downregulation of CDKN1A in ATLL cells. (A) cDNA microarray analysis of gene expression changes in acute-type ATLL. The scatter plot of DNA microarray signal intensities of ATLL cells from acute-type ATLL patients plotted against the signal intensities of CD4+ T lymphocytes of healthy volunteers is shown. Each dot shows a series of genes which were regulated by HTLV-1 infection (19). The flanking diagonal lines indicate a 2-fold change in expression. Data points (marked with arrows) corresponding to differentially expressed genes IL-2Rα and CDKN1A are also shown. Names of the genes that were regulated by HTLV-1 infection are listed in Table S1 in the supplemental material. (B and C) Expression of mRNA (B) and protein (C) for p53, CDKN1A, and Tax in three HTLV-1-uninfected T-ALL cell lines, three HTLV-1-infected cell lines [HTLV-1 (+)], and six ATLL-derived cell lines was examined by semiquantitative RT-PCR and Western blotting, respectively. The expression of β-actin is shown at the bottom as a control. (D and E) Expression of mRNA (D) and protein (E) for p53, CDKN1A, and Tax in five patients with acute-type ATLL and in five healthy controls (CD4+ T).
To confirm the downregulation of CDKN1A expression in ATLL cells, we initially determined expression of p53, CDKN1A, and Tax in three T-ALL cell lines (Jurkat, MOLT4, and MKB-1), the three HTLV-1-infected cell lines (MT-2, MT-4, and HUT102), and the six ATLL-derived cell lines (ED, SO4, Su9T01, S1T, KK1, and KOB) by semiquantitative RT-PCR and Western blotting (Fig. 1B and C). Three HTLV-1-infected cell lines highly expressed CDKN1A and Tax while only two of six ATLL cell lines expressed CDKN1A with low levels of Tax (S1T and KOB), but every ATLL cell line had minimal expression of Tax. Therefore, the CDKN1A expression in S1T and KOB is probably derived from the Tax expression. For reference, proviral DNA copy numbers in each cell line were calculated by the pX gene region of HTLV-1 DNA per RNase P DNA copy as an internal control (Fig. 2B). Three HTLV-1-infected cell lines and four out of six ATLL cell lines expressed various levels of p53 mRNA and protein. Among them, MKB-1 showed a high level of p53 protein expression with a low level of p53 mRNA expression, suggesting that the p53 protein degradation system might be inhibited in MKB-1. We next examined the expression of p53, CDKN1A, and Tax in CD4+ T lymphocytes and leukemia cells from the patients with acute-type ATLL (Fig. 1D and E). In CD4+ single-positive (SP) T lymphocytes, mRNA and protein of CDKN1A were clearly expressed, but p53 and Tax were not expressed in any of the five samples. In ATLL cells, mRNA and protein of CDKN1A were not detected in any of the five samples. However, four out of five ATLL samples showed expression of p53 mRNA and protein but not Tax. Tax was detected in one sample, but the same sample lacked p53. Compared with the CDKN1A expression levels in CD4+ SP T lymphocytes and HTLV-1-infected cell lines (Fig. 2B), CDKN1A was highly expressed in HTLV-1-infected cell lines, but there was minimal expression of CDKN1A in ATLL-derived cell lines and primary ATLL cells and no expression of CDKN1A in primary CD4+ lymphocytes from acute-type ATLL compared to CD4+ lymphocytes from healthy volunteers. We next determined the mechanism for the low CDKN1A expression in ATLL cells because downregulation of CDKN1A expression in leukemia is considered one of the major events required for leukemogenesis.
FIG. 2.
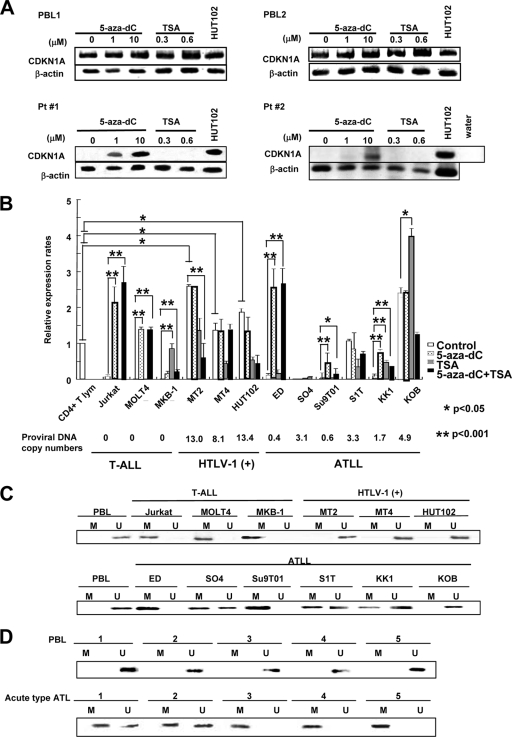
Promoter methylation of CDKN1A is involved in suppression of CDKN1A expression in ATLL cells. (A) Semiquantitative RT-PCR of CDKN1A expression was performed with mRNA isolated from PBLs from two healthy volunteers and leukemia cells from two acute-type ATLL patients (Pt 1 and 2) after treatment with 10 μM 5-aza-dC for 72 h or with 1.2 μM TSA for 24 h. The HUT102 cell line was used as a control. (B) Quantitative RT-PCR of CDKN1A expression was performed with mRNA isolated from various T-leukemia cell lines treated with 10 μM 5-aza-dC for 72 h, with 1.2 μM TSA for 24 h, or with 1.2 μM TSA for 24 h followed by 10 μM 5-aza-dC for 48 h. CDKN1A mRNA expression levels were normalized to β-actin mRNA expression and are expressed relative to the mRNA level in the normal CD4+ T lymphocytes. Proviral DNA copy numbers for HTLV-1 were measured in each cell line. Statistical analysis was done by a Student's t test. (C and D) MSP analysis of the CDKN1A gene promoter. DNAs from various T-leukemia cell lines (C) and primary ATLL cells (D) were treated with bisulfite and subjected to MSP. A product in the U gel indicates that the CDKN1A gene is unmethylated; a product in the M gel indicates that the gene is methylated. CD4+ T lymphocytes of healthy volunteers were used as a control.
Downregulation of CDKN1A due to methylation of its promoter region.
Genomic mutations and allelic loss of the CDKN1A gene are rarely detected in many kinds of cancer cells, but epigenetic alteration of the CDKN1A locus is reported in select cases (3, 7, 11, 27). We initially determined the genetic alterations at the CDKN1A locus on chromosome 6p21.2 by spectral karyotyping of acute-type ATLL cells from 61 patients (10) and by a comparative genomic hybridization method using a high-density DNA microarray of 10 patient samples and eight ATLL cell lines. We found that no translocations or genetic deletions at the CDKN1A locus were found in any of the ATLL samples (data not shown). Since hypermethylation at the promoter region of CDKN1A was reported in acute lymphocytic leukemia in children (23), we further investigated epigenetic modifications in the promoter region of CDKN1A in leukemia cells from acute-type ATLL patients.
Leukemic cells from eight patients with acute-type ATLL and PBLs from two healthy volunteers were cultured with various concentrations of the DNA-demethylating agent 5-aza-2-deoxycytidine (5-aza-dC) or the histone deacetylase inhibitor trichostatin A (TSA) for 2 to 4 days. After the treatments, we found that levels of CDKN1A mRNA were increased in five of eight ATLL samples by 5-aza-dC but not by TSA although expression levels of CDKN1A did not change in PBLs from two healthy volunteers (Fig. 2A and Table 1). Along with the patient samples, various kinds of T-cell leukemia cell lines (three T-ALL, three HTLV-1-infected, and six ATLL-derived) were treated with 5-aza-dC and/or TSA, and the expression of CDKN1A was determined by quantitative RT-PCR (Fig. 2B). CDKN1A was highly expressed in all three HTLV-1-infected cell lines and in two of six ATLL cell lines (S1T and KOB), but it was expressed at lower levels in all three T-ALL and four of six ATLL cell lines (ED, SO4, Su9T01, and KK1). After the 5-aza-dC treatment, upregulation of CDKN1A expression was observed in two of three T-ALL cell lines (Jurkat and MOLT4) and three of six ATLL cell lines (ED, Su9T01, and KK1). After the TSA treatment, upregulation of CDKN1A expression was observed in one T-ALL (MKB-1) and one ATLL (KK1) cell line, suggesting that downregulation of CDKN1A in T-ALL and ATLL cell lines was frequently due to DNA methylation of the CDKN1A promoter region.
TABLE 1.
Induction of the CDKN1A gene transcription in acute-type ATLL cells or PBLs after treatment with 5-aza-dC or TSAa
| Treatment |
CDKN1A transcription (no. of samples) |
|||
|---|---|---|---|---|
| Unchanged |
Upregulated |
|||
| ATLL cells | PBLs | ATLL cells | PBLs | |
| Control | 8 | 2 | 0 | 0 |
| 5-Aza-dC | 3 | 2 | 5 | 0 |
| TSA | 8 | 2 | 0 | 0 |
Leukemic cells from eight patients with acute-type ATLL and PBLs from two healthy volunteers were cultured with or without 5-aza-dC or TSA for 2 to 4 days. After the treatment, expression of CDKN1A was determined by RT-PCR (Fig. 2A).
We next examined the methylation status of the promoter region of CDKN1A in various types of T-cell leukemia cell lines and primary ATLL cells by methylation-specific PCR (MSP). Since the basal expression of CDKN1A is reportedly due to six tandem Sp1 binding sites in the CDKN1A promoter region adjacent to exon 1 (Materials and Methods), a 230-bp fragment of the CDKN1A promoter region containing Sp1 binding sites in CpG islands was treated with bisulfite and amplified by a pair of methylation- or nonmethylation-specific primers. As shown in Fig. 2C, the promoter region in all three T-ALL cell lines (Jurkat, MOLT4, and MKB-1) was methylated, but it was unmethylated in all three HTLV-1-infected cell lines (MT-2, MT-4, and HUT102). In ATLL cell lines, the promoter region was methylated in two cell lines (ED and Su9T01), partially methylated in three cell lines with lower expression of CDKN1A (SO4, S1T, and KK1), and not methylated in one cell line with high expression of CDKN1A (KOB) (Fig. 2B and C). On the other hand, all five DNA samples from lymphocytes in peripheral blood from healthy volunteers were not methylated and showed high expression levels of CDKN1A; three samples from ATLL leukemia cells were partially methylated, and two samples were methylated, with minimal expression of CDKN1A (Fig. 1D and 2D). After 23 DNA samples from primary acute-type ATLL cells were examined for promoter methylation by MSP analysis, we found that the promoter region of CDKN1A was methylated in 22 of 23 samples, with partial methylation of 17 samples (Table 2). Therefore, the promoter region of CDKN1A was methylated in most of the ATLL cells with lower CDKN1A expression levels.
TABLE 2.
Methylation status of the promoter region of CDKN1A as determined by MSP
| Methylation status of CDKN1A promoter region | No. of samples by cell linea |
No. of primary samples by typeb |
|||
|---|---|---|---|---|---|
| T-ALL | HTLV-1-infected | ATLL | PBL | Acute-type ATLL | |
| Methylated | 3 | 0 | 2 | 0 | 6 |
| Partially methylated | 0 | 0 | 3 | 0 | 17 |
| Unmethylated | 0 | 3 | 1 | 5 | 1 |
| Total (% of methylation rates) | 3 (100) | 3 (0) | 6 (82) | 5 (0) | 24 (96) |
Cell lines used were three T-ALL cell lines (Jurkat, MOLT4, and MKB-1), three HTLV-1-infected T-cell lines (MT-2, MT-4, and HUT102), and six ATLL-derived cell lines (ED, SO4, Su9T01, S1T, KK1, and KOB).
For primary samples, PBLs were obtained from five healthy volunteers, and ATLL cells were from 24 patients with acute-type ATLL.
CDKN1A does not contribute to G1 arrest in response to UV irradiation in HTLV-1-infected and ATLL-derived cell lines.
To examine the functional differences between CDKN1A expression in HTLV-1-infected and ATLL cell lines, we investigated the p53-mediated DNA repair responses to UV irradiation (Fig. 3A ), with the MOLT4 T-ALL cell line as a control. Three HTLV-1-infected cell lines with high expression levels of CDKN1A (MT-2, MT-4, and HUT102) did not change their rates of growth after high-dose UV irradiation (20 J/m2); however, MOLT4 and ATLL-derived cell lines either without (ED and Su9T01) or with low (S1T and KOB) CDKN1A expression significantly decreased their rates of growth after UV irradiation. To further investigate the differences between the responses of HTLV-1-infected and ATLL-derived cell lines to UV irradiation, protein expression levels of p53, CDKN1A, and CDKN1B (p27/KIP1) were examined before and after UV irradiation since both CDKN1A and CDKN1B induce cell cycle arrest following DNA damage. After UV irradiation, the expression level of CDKN1A in KOB cells increased 24 h after UV irradiation and was mostly sustained until 72 h (data not shown). In MOLT4 cells, the levels of p53, CDKN1A, and CDKN1B markedly increased up to 72 h postirradiation (data not shown). On the other hand, there was no significant change in any of these proteins at any of the time points after the UV irradiation in the MT-2 cells (data not shown). In two of the HTLV-1-infected cell lines (MT-2 and HUT102), the expression levels of p53, CDKN1A, and CDKN1B were similar before and 72 h after UV irradiation (Fig. 3B). On the other hand, ATLL-derived ED and Su9T01 cells did not express either p53, CDKN1A, or CDKN1B while in the S1T and KOB cell lines the levels of p53, CDKN1A, and CDKN1B were slightly increased after UV irradiation. The HTLV-1-infected cell lines did not respond to UV irradiation with the p53-mediated DNA repair pathway, suggesting that the overexpression of CDKN1A in HTLV-1-infected cells did not contribute to the cell cycle arrest. Therefore, we next determined the subcellular localization of CDKN1A and CDKN1B in these leukemia cell lines, with MOLT4 cells used as a control, by immunofluorescence staining (Fig. 3C). In MOLT4 cells, both CDKN1A and CDKN1B were weakly expressed and localized in the nucleus and cytoplasm. After UV irradiation, both CDKN1A and CDKN1B were significantly induced and mostly localized in the nucleus of MOLT4 cells. Interestingly, both CDKN1A and CDKN1B proteins mainly localized in the cytoplasm in both HTLV-1-infected and ATLL-derived cell lines before and after UV irradiation. Therefore, cytoplasmic CDKN1A and CDKN1B may contribute to cellular responses other than cell cycle arrest in response to UV irradiation in HTLV-1-infected and ATLL-derived cell lines.
Cytoplasmic CDKN1A induced by phosphorylated Akt in HTLV-1-infected and ATLL-derived cells.
Since recent reports show that cytoplasmic CDKN1A is phosphorylated at threonine 145 (T145) of CDKN1A by phosphorylated Akt (p-Akt) (39), we next examined the phosphorylation status of CDKN1A and the activation of the PI3K/Akt signaling pathway in HTLV-1-infected and ATLL cell lines. As shown in Fig. 4A, a high degree of p-Akt was detected in three HTLV-1-infected cell lines (MT-2, MT-4, and HUT102) and four ATLL cell lines (SO4, S1T, KK1, and KOB), compared with low expression of p-Akt in three peripheral lymphocytes from healthy volunteers. The expression levels of CDKN1A and CDKN1B were higher in three HTLV-1 cell lines than in lymphocytes and two ATLL cell lines (S1T and KOB) (data not shown). CDKN1A was highly phosphorylated at T145 in three HTLV-1-infected cell lines and two ATLL cell lines (S1T and KOB), and these cell lines also exhibited activated PI3K/Akt signaling, as evidenced with p-Akt. These data suggest that the phosphorylation of CDKN1A was probably derived from activation of PI3K/Akt signaling.
To investigate the relationship between activation of PI3K/Akt signaling and CDKN1A phosphorylation, the expression level of p-Akt was determined in these cell lines after treatment with the PI3K inhibitor LY294002. After the treatment with LY294002, the expression levels of p-Akt and p-CDKN1A were significantly decreased in two HTLV-1-infected cell lines (MT-2 and HUT102) and two ATLL cell lines (S1T and KOB) in parallel (Fig. 4B). The LY294002 treatment did not inhibit the growth rate of the control MOLT4 T-ALL cell line, with low Akt phosphorylation (data not shown), but significantly inhibited the growth rates of HTLV-1-infected cell lines and ATLL cell lines (Fig. 4C). The treatment with LY294002 also induced cytoplasmic CDKN1A and CDKN1B to relocalize to the nucleus in both HTLV-1-infected cell lines, but CDKN1A and CDKN1B did not relocalize to the nucleus in ATLL cell lines (Fig. 4D).
To determine the difference of cellular responsiveness after LY294002 treatment between HTLV-1-infected and ATLL cell lines, the cell cycle progression of MT-2 HTLV-1-infected cells and KOB ATLL cells was examined before and after UV irradiation and/or LY294002 treatment. In MT-2 HTLV-1-infected cells, approximately 60% of the cells were arrested in the G1 phase after LY294002 with or without UV irradiation (Fig. 4E). However, ∼13% of the KOB ATLL cells were in apoptotic cell death after LY294002 and/or UV irradiation (Fig. 4E and F). In the HTLV-1-infected cell lines, the relocalization of dephosphorylated CDKN1A to the nucleus was correlated with G1 cell cycle arrest after PI3K/Akt inactivation. On the other hand, T145-dephosphorylated CDKN1A remained in the cytoplasm but could not prevent apoptotic cell death after treatment with LY294002 and/or UV irradiation in ATLL cells. Therefore, along with the phosphorylation of CDKN1A at T145, the lower expression of CDKN1A in ATLL cells may contribute to the dysregulation of the p53-dependent cell cycle arrest.
DISCUSSION
In this study, we showed that CDKN1A was frequently downregulated in ATLL cells and that the mechanism of the transcriptional repression primarily involved promoter methylation, in spite of the upregulation of CDKN1A in HTLV-1-infected cell lines by Tax. Because severe methylation of the CDKN1A promoter region has been reported in acute lymphoblastic leukemia and was associated with an unfavorable clinical outcome (23), the downregulation of CDKN1A in ATLL cells by promoter methylation is probably one of the important events for ATLL leukemogenesis. Low expression of CDKN1A was detected in primary acute-type ATLL cells and a few ATLL cell lines with higher promoter methylation, but CDKN1A was expressed in some ATLL cell lines with partial promoter methylation. Since CDKN1A in HTLV-1-infected and ATLL cell lines was mostly localized to the cytoplasm when CDKN1A was phosphorylated at T145 by p-Akt, CDKN1A did not contribute to p53-dependent G1 arrest after UV irradiation in either cell line. Therefore, downregulation of CDKN1A transcription in ATLL cells may contribute to ATLL leukemogenesis by permitting progression of the cell cycle.
Earlier results have indicated that the loss of CDKN1B (p27KIP1) plays a critical role in T-cell transformation following HTLV-1 infection (2). However, the expression levels of CDKN1B were not different between three samples of peripheral blood lymphocytes and three HTLV-1-infected cell lines (Fig. 4A). Moreover, the expression levels of CDKN1B were not significantly different between CD4+ lymphocytes and acute-type ATLL cells (data not shown). On the other hand, recent findings from several labs have also shown that HTLV-1 Tax and HTLV-1 infection drastically upregulate expression levels of both CDKN1A and CDKN1B to cause cell cycle arrest or cellular senescence just after the infection (15, 33). However, many reports also showed that the HTLV-1-infected cell lines could grow well and proliferate intensely, despite abundant CDKN1A expression (5). Notably, we observed the cytoplasmic localization of CDKN1A protein phosphorylated at T145 by Akt in HTLV-1-infected and ATLL cell lines. It is reported that activation of PI3K/Akt signaling contributes to the phosphorylation of CDKN1A at T145 and of CDKN1B at T157 with their cytoplasmic localization (15, 26, 34, 35, 36, 39). In HTLV-1-infected cell lines, Tax is reported to promote Akt phosphorylation by directly binding to PI3K (18) or by downregulating PTEN transcription through NF-κB signaling (8). In this study, LY294002-mediated dephosphorylation of Akt induced relocation of CDKN1A and CDKN1B into the nucleus, with accumulation of cells in the G1 phase, suggesting that cytoplasmic localization of CDKN1A is an important factor in the antiapoptotic effect in HTLV-1-infected cell lines. However, most of the CDKN1A and CDKN1B still remained in the cytoplasm of ATLL cells after LY294002 treatment, suggesting that mechanisms other than phosphorylation may contribute to the subcellular localization of CDKN1A in ATLL cells, and we further speculate that the nuclear import system of CDKN1A may be disturbed in ATLL cells for unknown reasons.
Neither UV irradiation nor LY294002 treatment induced apoptotic cell death in HTLV-1-infected cell lines. Tax could account, at least in part, for the antiapoptotic property of HTLV-1-infected cells, through, for instance, activation of the PI3K/Akt pathway, costimulatory receptor signaling (OX40/OX40L), and enhanced expression of various antiapoptotic proteins (c-Flip, Bcl-xL, Bfl-1, and Hiap-1) (12). On the other hand, apoptotic cell death was induced in ATLL cells after UV irradiation or LY294002 treatment. Since inactivation of Tax was observed in over 70% of ATLL cells because of genetic abnormalities and DNA methylation in the HTLV-1 genome, apoptotic cell death induced after UV irradiation or LY294002 treatment may be due to a lack of Tax-mediated antiapoptotic effects. As the promoter region of CDKN1A was also methylated in the majority of the ATLL patients, the accumulation of abnormal DNA methylation in ATLL cells may be involved in leukemogenesis by the downregulation of Tax and CDKN1A during the progression from HTLV-1-infected cells to ATLL cells. Recently, the HTLV-1 bZIP factor gene (HBZ) within unmethylated or undeleted regions of the 3′ long terminal repeat (LTR) has been shown to be consistently expressed and to promote the cell growth of ATLL cells, suggesting that the HBZ gene likely is critical for pathogenesis of ATL cells along with the downregulation of Tax and CDKN1A.
Supplementary Material
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research of Priority Area from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by a research fund from the Miyazaki Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence, JST.
Footnotes
Published ahead of print on 5 May 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Akagi, T., H. Ono, and K. Shimotohno. 1995. Characterization of T cells immortalized by Tax1 of human T-cell leukemia virus type 1. Blood 86:4243-4249. [PubMed] [Google Scholar]
- 2.Cereseto, A., R. Washington Parks, E. Rivadeneira, and G. Franchini. 1999. Limiting amounts of p27Kip1 correlates with constitutive activation of cyclin E-CDK2 complex in HTLV-I-transformed T-cells. Oncogene 18:2441-2450. [DOI] [PubMed] [Google Scholar]
- 3.Claus, R., and M. Lubbert. 2003. Epigenetic targets in hematopoietic malignancies. Oncogene 22:6489-6496. [DOI] [PubMed] [Google Scholar]
- 4.de La Fuente, C., L. Deng, F. Santiago, L. Arce, L. Wang, and F. Kashanchi. 2000. Gene expression array of HTLV type 1-infected T cells: Up-regulation of transcription factors and cell cycle genes. AIDS Res. Hum. Retroviruses 16:1695-1700. [DOI] [PubMed] [Google Scholar]
- 5.de La Fuente, C., F. Santiago, S. Y. Chong, L. Deng, T. Mayhood, P. Fu, D. Stein, T. Denny, F. Coffman, N. Azimi, R. Mahieux, and F. Kashanchi. 2000. Overexpression of p21waf1 in human T-cell lymphotropic virus type 1-infected cells and its association with cyclin A/Cdk2. J. Virol. 74:7270-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dewan, M. Z., N. Takamatsu, T. Hidaka, K. Hatakeyama, S. Nakahata, J. Fujisawa, H. Katano, N. Yamamoto, and K. Morishita. 2008. Critical role for TSLC1 expression in the growth and organ infiltration of adult T-cell leukemia cells in vivo. J. Virol. 82:11958-11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang, J. Y., and Y. Y. Lu. 2002. Effects of histone acetylation and DNA methylation on p21 (WAF1) regulation. World J. Gastroenterol. 8:400-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda, R. I., K. Tsuchiya, K. Suzuki, K. Itoh, J. Fujita, A. Utsunomiya, and T. Tsuji. 2009. Human T-cell leukemia virus type I Tax down-regulates the expression of phosphatidylinositol 3,4,5-trisphosphate inositol phosphatases via the NF-κB pathway. J. Biol. Chem. 284:2680-2689. [DOI] [PubMed] [Google Scholar]
- 9.Harhaj, E. W., L. Good, G. Xiao, and S. C. Sun. 1999. Gene expression profiles in HTLV-I-immortalized T cells: deregulated expression of genes involved in apoptosis regulation. Oncogene 18:1341-1349. [DOI] [PubMed] [Google Scholar]
- 10.Hidaka, T., S. Nakahata, K. Hatakeyama, M. Hamasaki, K. Yamashita, T. Kohno, Y. Arai, T. Taki, K. Nishida, A. Okayama, Y. Asada, R. Yamaguchi, H. Tsubouchi, J. Yokota, M. Taniwaki, Y. Higashi, and K. Morishita. 2008. Down-regulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood 112:383-393. [DOI] [PubMed] [Google Scholar]
- 11.Jiemjit, A., T. E. Fandy, H. Carraway, K. A. Bailey, S. Baylin, J. G. Herman, and S. D. Gore. 2008. p21(WAF1/CIP1) induction by 5-azacytosine nucleosides requires DNA damage. Oncogene 27:3615-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katrin, S., and G. Ralph. 2007. Human T cell leukemia virus type 1 Tax-induced signals in cell survival, proliferation, and transformation. Signal Transduct. 7:34-52. [Google Scholar]
- 13.Kehn, K., L. Deng, C. de la Fuente, K. Strouss, K. Wu, A. Maddukuri, S. Baylor, R. Rufner, A. Pumfery, M. E. Bottazzi, and F. Kashanchi. 2004. The role of cyclin D2 and p21/waf1 in human T-cell leukemia virus type 1 infected cells. Retrovirology 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koiwa, T., A. Hamano-Usami, T. Ishida, A. Okayama, K. Yamaguchi, S. Kamihira, and T. Watanabe. 2002. 5′-Long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 76:9389-9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang, J., J. Zubovitz, T. Petrocelli, R. Kotchetkov, M. K. Connor, K. Han, J. H. Lee, S. Ciarallo, C. Catzavelos, R. Beniston, E. Franssen, and J. M. Slingerland. 2002. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat. Med. 8:1153-1160. [DOI] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17.Matsuoka, M., and K. T. Jeang. 2007. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer. 7:270-280. [DOI] [PubMed] [Google Scholar]
- 18.Peloponese, J. M., Jr., and K. T. Jeang. 2006. Role for Akt/protein kinase B and activator protein-1 in cellular proliferation induced by the human T-cell leukemia virus type 1 tax oncoprotein. J. Biol. Chem. 281:8927-8938. [DOI] [PubMed] [Google Scholar]
- 19.Pise-Masison, C. A., M. Radonovich, R. Mahieux, P. Chatterjee, C. Whiteford, J. Duvall, C. Guillerm, A. Gessain, and J. N. Brady. 2002. Transcription profile of cells infected with human T-cell leukemia virus type I compared with activated lymphocytes. Cancer Res. 62:3562-3571. [PubMed] [Google Scholar]
- 20.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Proietti, F. A., A. B. Carneiro-Proietti, B. C. Catalan-Soares, and E. L. Murphy. 2005. Global epidemiology of HTLV-I infection and associated diseases. Oncogene 24:6058-6068. [DOI] [PubMed] [Google Scholar]
- 22.Raizis, A. M., F. Schmitt, and J. P. Jost. 1995. A bisulfite method of 5-methylcytosine mapping that minimizes template degradation. Anal. Biochem. 226:161-166. [DOI] [PubMed] [Google Scholar]
- 23.Roman-Gomez, J., J. A. Castillejo, A. Jimenez, M. G. Gonzalez, F. Moreno, C. Rodriguez Mdel, M. Barrios, J. Maldonado, and A. Torres. 2002. 5′ CpG island hypermethylation is associated with transcriptional silencing of the p21(CIP1/WAF1/SDI1) gene and confers poor prognosis in acute lymphoblastic leukemia. Blood 99:2291-2296. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki, H., I. Nishikata, T. Shiraga, E. Akamatsu, T. Fukami, T. Hidaka, Y. Kubuki, A. Okayama, K. Hamada, H. Okabe, Y. Murakami, H. Tsubouchi, and K. Morishita. 2005. Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute-type adult T-cell leukemia. Blood 105:1204-1213. [DOI] [PubMed] [Google Scholar]
- 25.Seiki, M., S. Hattori, Y. Hirayama, and M. Yoshida. 1983. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc. Natl. Acad. Sci. U. S. A. 80:3618-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin, I., F. M. Yakes, F. Rojo, N. Y. Shin, A. V. Bakin, J. Baselga, and C. L. Arteaga. 2002. PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat. Med. 8:1145-1152. [DOI] [PubMed] [Google Scholar]
- 27.Shin, J. Y., H. S. Kim, J. Park, J. B. Park, and J. Y. Lee. 2000. Mechanism for inactivation of the KIP family cyclin-dependent kinase inhibitor genes in gastric cancer cells. Cancer Res. 60:262-265. [PubMed] [Google Scholar]
- 28.Siegel, R. S., R. B. Gartenhaus, and T. M. Kuzel. 2001. Human T-cell lymphotropic-I-associated leukemia/lymphoma. Curr. Treat. Options Oncol. 2:291-300. [DOI] [PubMed] [Google Scholar]
- 29.Tajima, K. 1990. The 4th nation-wide study of adult T-cell leukemia/lymphoma (ATL) in Japan: estimates of risk of ATL and its geographical and clinical features. The T- and B-cell Malignancy Study Group. Int. J. Cancer 45:237-243. [DOI] [PubMed] [Google Scholar]
- 30.Takeda, S., M. Maeda, S. Morikawa, Y. Taniguchi, J. Yasunaga, K. Nosaka, Y. Tanaka, and M. Matsuoka. 2004. Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int. J. Cancer. 109:559-567. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka, G., A. Okayama, T. Watanabe, S. Aizawa, S. Stuver, N. Mueller, C. C. Hsieh, and H. Tsubouchi. 2005. The clonal expansion of human T lymphotropic virus type 1-infected T cells: a comparison between seroconverters and long-term carriers. J. Infect. Dis. 191:1140-1147. [DOI] [PubMed] [Google Scholar]
- 32.Taniguchi, Y., K. Nosaka, J. Yasunaga, M. Maeda, N. Mueller, A. Okayama, and M. Matsuoka. 2005. Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology 2:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tripp, A., P. Banerjee, M. Sieburg, V. Planelles, F. Li, and G. Feuer. 2005. Induction of cell cycle arrest by human T-cell lymphotropic virus type 1 Tax in hematopoietic progenitor (CD34+) cells: modulation of p21cip1/waf1 and p27kip1 expression. J. Virol. 79:14069-14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viglietto, G., M. L. Motti, P. Bruni, R. M. Melillo, A. D'Alessio, D. Califano, F. Vinci, G. Chiappetta, P. Tsichlis, A. Bellacosa, A. Fusco, and M. Santoro. 2002. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat. Med. 8:1136-1144. [DOI] [PubMed] [Google Scholar]
- 35.Winters, Z. E., R. D. Leek, M. J. Bradburn, C. J. Norbury, and A. L. Harris. 2003. Cytoplasmic p21WAF1/CIP1 expression is correlated with HER-2/ neu in breast cancer and is an independent predictor of prognosis. Breast Cancer Res. 5:R242-R249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia, W., J. S. Chen, X. Zhou, P. R. Sun, D. F. Lee, Y. Liao, B. P. Zhou, and M. C. Hung. 2004. Phosphorylation/cytoplasmic localization of p21Cip1/WAF1 is associated with HER2/neu overexpression and provides a novel combination predictor for poor prognosis in breast cancer patients. Clin. Cancer Res. 10:3815-3824. [DOI] [PubMed] [Google Scholar]
- 37.Yamada, Y., M. Tomonaga, H. Fukuda, S. Hanada, A. Utsunomiya, M. Tara, M. Sano, S. Ikeda, K. Takatsuki, M. Kozuru, K. Araki, F. Kawano, M. Niimi, K. Tobinai, T. Hotta, and M. Shimoyama. 2001. A new G-CSF-supported combination chemotherapy, LSG15, for adult T-cell leukaemia-lymphoma: Japan Clinical Oncology Group Study 9303. Br. J. Haematol. 113:375-382. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida, M., I. Miyoshi, and Y. Hinuma. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. U. S. A. 79:2031-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, B. P., Y. Liao, W. Xia, B. Spohn, M. H. Lee, and M. C. Hung. 2001. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat. Cell Biol. 3:245-252. [DOI] [PubMed] [Google Scholar]
- 40.Zhu, W. G., K. Srinivasan, Z. Dai, W. Duan, L. J. Druhan, H. Ding, L. Yee, M. A. Villalona-Calero, C. Plass, and G. A. Otterson. 2003. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21 (Cip1) promoter. Mol. Cell. Biol. 23:4056-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.