Abstract
During human adenovirus 5 infection, a temporal cascade of gene expression leads ultimately to the production of large amounts of the proteins needed to construct progeny virions. However, the mechanism for the activation of the major late gene that encodes these viral structural proteins has not been well understood. We show here that two key positive regulators of the major late gene, L4-22K and L4-33K, previously thought to be expressed under the control of the major late promoter itself, initially are expressed from a novel promoter that is embedded within the major late gene and dedicated to their expression. This L4 promoter is required for late gene expression and is activated by a combination of viral protein activators produced during the infection, including E1A, E4 Orf3, and the intermediate-phase protein IVa2, and also by viral genome replication. This new understanding redraws the long-established view of how adenoviral gene expression patterns are controlled and offers new ways to manipulate that gene expression cascade for adenovirus vector applications.
Although years of study have produced a detailed understanding of most molecular events during human adenovirus type 5 (Ad5) infection (4, 27), how the transition in viral gene expression from the early to the late phase is controlled has remained poorly defined. This control is crucial, since it determines the activity of the genes that encode virion proteins and, hence, the productivity of the infection. Residual activity from these genes is a confounding factor in the utility of E1-deleted Ad5 vectors for long-term gene delivery (51).
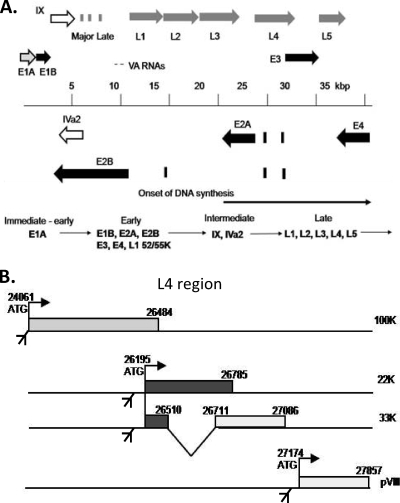
The initial expression of E1A from the linear Ad5 genome provides transcriptional activators that, with host proteins, turn on the expression of the remaining early genes E1B, E2, E3, and E4 (Fig. 1 A). The major-late transcription unit (MLTU) also is weakly active at this time, but only the most 5′-proximal L1 product is produced (1, 35, 41). Around the time of transition to the late phase of infection, when the replication of the viral genome also begins, the transcription of intermediate genes IX and IVa2 commences (13, 39, 48, 49), while major-late promoter (MLP) activity greatly increases, and its scope expands to direct the expression of a full set of around 15 MLTU products from regions L1 to L5 via alternative splicing and polyadenylation (35, 41). This transition in MLTU activity reflects transcriptional and posttranscriptional changes, both of which require proteins encoded by genes in the MLTU L4 region (Fig. 1B). L4-22K and L4-33K act posttranscriptionally to activate the production of the full set of MLTU mRNAs (16, 33, 44). At the same time, the MLP is further activated by IVa2 protein (30, 45) working with L4-22K and/or L4-33K (2, 33, 38).
FIG. 1.
(A) Ad5 transcription map showing immediate-early (light gray), early (black), intermediate (white), and major late transcription units (MLTU; dark gray arrows), which are expressed during infection in a temporal cascade (bottom). All transcription units except IX and IVa2 produce multiple mRNAs and protein products by alternative RNA processing. MLTU mRNAs each comprise the three exons of the tripartite leader (TPL) spliced to 1 of ∼15 possible acceptor sites within regions L1 to L5. (B) Organization of the L4 region of the MLTU showing L4-100K, L4-22K, L4-33K, and pVIII ORFs, with 3′ splice sites for their expression from the major late promoter. 22K and L4-33K share the same N-terminal sequence but have distinct C termini.
The essential role of L4-22K and L4-33K in producing full late-phase expression from the Ad5 MLTU creates a paradox since, according to the current model of Ad5 gene expression, their expression is achieved only as a consequence of this activation process. Here, we show that a novel Ad5 promoter expresses L4-22K and L4-33K independently from the MLP, resolving this paradox, and that this promoter is activated by a combination of viral proteins and viral DNA replication.
MATERIALS AND METHODS
Plasmids.
pTG3602-Ad5wt (pWT) is a clone of the complete wild-type (WT) Ad5 genome (11); pTG3602-L4-22K− (pL4-22K−) was derived from pWT and contains a premature stop codon within the C-terminal unique portion of the L4-22K open reading frame (ORF) (38). Linear genome was prepared from each of these plasmids by PacI digestion. Specific protein expression plasmids pCMV-IX (9); pMEPCMV-IVa2 (7); pCMV22KFLAG, pCMV33KFLAG, and pCMV100KFLAG (33); and pcDNA3.1Orf3 and pcDNA3.1Orf3 N82A (21) have been described previously. pE1A, provided by J. Logan, contains Ad5 bp 1 to 5788 cloned between the EcoRI and SalI sites of the pBR322 derivative, pML2, and with a deletion of the Ad5 SacI fragment (bp 1770 to 5644). pcDNA3.1Orf6 contains the Ad5 E4Orf6 sequence (bp 34089 to 33182), which was obtained by PCR and cloned at the EcoRI site of pcDNA3.1.
L4 luciferase reporter plasmids were generated by amplifying various fragments in the region of Ad5 positions 25887 to 26295 using primer pairs containing restriction recognition sites for KpnI (5′ primer) and NheI (3′ primer) and cloning into pGL3-Basic luciferase reporter plasmid (Promega) using these sites. pcDNA3.1HisLacZ (Invitrogen) was used as a transfection control. pA-22/33KFLAG was generated by amplifying the relevant sequence as an EcoRI fragment from Ad5 strain 300 wild-type viral DNA. pA-22KFLAG was generated from pA-22/33KFLAG by exchanging the HindIII/EcoRI 3′ fragment (Ad5 positions 26328 to 26785 and C-Terminal FLAG tag) with the equivalent fragment from pCMV22KFLAG (33).
pLoxPGFP was obtained by the modification of pBiEGFPPacI (16), with the insertion of a LoxP sequence between the promoter PBi-1 and the enhanced green fluorescent protein (EGFP) ORF and a zeocin resistance gene driven by the thymidine kinase promoter. L4 shuttle plasmids were constructed from pBiEGFPPacI (16) first by replacing the PBi-1 promoter and EGFP with a LoxP sequence and adding a hygromycin resistance cassette from pTK-Hyg (Clontech), and then inserting one of several L4 cassettes downstream of LoxP. These were (i) TPL-L4 cassettes, comprising the Ad5 tripartite leader (bp 6049 to 6089, 7111 to 7182, 9644 to 9733) and 109-bp intron sequence downstream of leader 3 (9734 to 9842), joined to L4 sequence from either 198 bp upstream of the 100K ORF or 177 bp upstream of the 22/33K ORF to a C-terminal FLAG tag on L4-33K, giving pShuttle100/22/33KFLAG and pShuttle22/33KFLAG, respectively; (ii) L4-only cassettes comprising the L4 components of the TPL-L4 cassettes, giving pShuttle26018-22KFLAG and pShuttle26018-22/33KFLAG; or (iii) the L4-22/33K ORF alone from its AUG at bp 26195, giving pShuttle22/33KFLAG ORF.
Production of L4P− genome.
pBR322ΔHindIII was generated by digesting pBR322 with HindIII, end-filling with DNA polI (Klenow fragment), and religating. The NdeI fragment from pWT was subcloned into pBR322ΔHindIII to generate pWTNdeI19548-31088. The core promoter (positions 26018 to 26098) was deleted from this subclone by a two-step PCR protocol using primers that incorporated sequence from each side of the deletion (Ad26003-26017/26099-26119f and Ad26112-26099/26017-25997r) with 3′- and 5′-flanking primers (Ad26511-26487r and Ad25198-26027f, respectively). The second-stage PCR product (positions 25198 to 26511) with the core promoter deleted was digested with AscI and HindIII at sites within the Ad5 sequence and used to replace the equivalent wild-type sequence, generating pWTNdeI19548-31088ΔP. The NdeI fragment from pWTNdeI19548-31088ΔP then was recloned into pWT to generate pL4P−, from which linear genome was prepared as above.
Cell culture.
293 and HeLa cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% newborn bovine serum. 293TETOFF cells (Clontech) and their derivatives were maintained in DMEM plus 10% fetal bovine serum on plates precoated with polylysine. Full details of the isolation of L4 cell lines will be presented elsewhere. Briefly, 293TETOFF cells, preselected for the stable expression of GFP following pLoxPGFP transfection and zeocin selection, were transfected with either pShuttle100/22/33KFLAG or pShuttle22/33KFLAG together with 500 ng Cre recombinase expression plasmid (pCre; Invitrogen), and hygromycin-resistant lines were isolated.
Transfection and inhibitors.
Transient transfections were carried out in 12-well cultures with cells plated at a density of 7 × 105 cells/well, using TransIT-LT1 (Cambridge Bioscience) at a ratio of 3 μl/μg DNA by following the manufacturers' protocols. For the inhibition of ATM/ATR signaling, cells were pretreated with 3 mM caffeine for 3 h, washed twice with serum-free medium, and then transfected as described above. Five hours posttransfection, medium was replaced with medium containing 3 mM caffeine until harvest at 48 h posttransfection. For the inhibition of viral DNA replication in genome-transfected cells, 5 h posttransfection media were replaced with medium containing 10 mM hydroxyurea until harvested 48 h posttransfection.
Luciferase reporter assays.
Assays were performed as previously described (33). β-Galactosidase activity was used to correct luciferase levels for differences in transfection efficiency. The effect of added E1A on β-galactosidase expression in HeLa cells was adjusted for by using the ratio of mean activity between cells expressing and not expressing E1A. Data are shown either as fold induction, where mean corrected luciferase expression from the specified basic or parental reporter plasmid is set as 1, or as the percentage of the activity of the defined L4 promoter reporter (bp 25887 to 26125). Values shown are the means from triplicate independent determinations within an experiment and are representative of multiple independent experiments.
Antibodies.
Proteins were detected using the following antibodies: anti-FLAG rabbit polyclonal serum (Sigma) at 1:10,000 for Western blotting and at 1:1,000 for immunofluorescence; AbJLB1 rabbit polyclonal serum to Ad5 late proteins at 1:10,000 (16); rabbit anti-L4-100K (W. C. Russell, University of St. Andrews) at 1:10,000 for Western blotting and 1:1,000 for immunofluorescence; rabbit anti-L4-33K at 1:1,000 (18); mouse anti-DNA binding protein monoclonal antibody (MAb) B6-8 at 1:10,000 (40); anti-E4-Orf3 (6A11) rat monoclonal antibody at 1:500 (34); rabbit anti-IVa2 at 1:10,000 (7); and anti-β-tubulin mouse MAb (Sigma) at 1:200. Secondary antibodies were goat-anti-mouse IgG-horseradish peroxidase (HRP) conjugate (Sigma) at 1:5,000, goat-anti-rabbit IgG-HRP (Santa Cruz) at 1:100,000, goat-anti-rat IgG-HRP (Chemicon) at 1:100,000, and Alexa Fluor 594 goat-anti-rabbit IgG (Invitrogen) at 1:500.
Protein expression detection.
Transfections used 500 ng pA-22KFLAG or pA-22/33KFLAG alone or cotransfected with either 1 μg linear viral genome or 500 ng various expression plasmids. All transfections were equalized for DNA content by the addition of either salmon sperm DNA or empty vector to account for the absence of genome or expression plasmid, respectively. Transfected cells were harvested 48 h posttransfection, and FLAG-tagged proteins were isolated using anti-FLAG (M2) agarose as described previously (21). Samples corresponding to 50% of the volume of immunoprecipitated proteins and 1 to 2% of the volume of cell lysates taken prior to immunoprecipitation were resolved through either 10% or 15% SDS-polyacrylamide gels as appropriate. Proteins were transferred to enhanced-chemiluminescence nitrocellulose membranes (GE Healthcare), and Western blot analysis was carried out as described previously (29) using horseradish peroxidase-conjugated secondary antibodies and detection via ECL-Advance (GE Healthcare). Immunofluorescence was performed as previously described (28).
RESULTS
Expression of Ad5 L4 22/33K and 100K proteins is separately regulated.
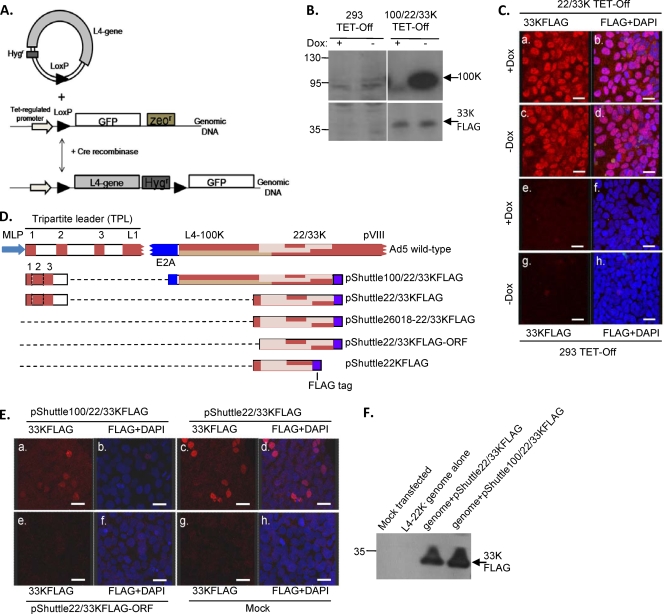
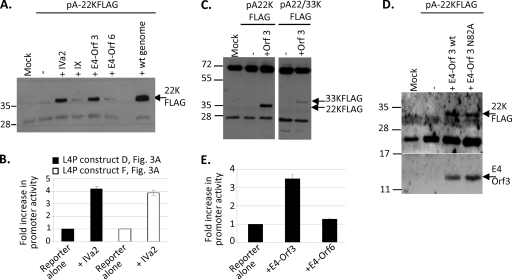
Early data showing that L4 mRNAs accumulated before those from L2, L3, and L5 during the onset of the late phase suggested that L4 expression is regulated differently from other MLTU regions, although transcription rate data did not suggest any additional promoter within the MLTU (25). L4 encodes several proteins (Fig. 1B). During attempts to produce stable cell lines expressing L4-100K, 22K, and 33K proteins from a shortened MLTU under the control of a tetracycline-regulated promoter (Fig. 2 A), cells were repeatedly obtained that showed correctly inducible 100K expression but the constitutive expression of 33K (Fig. 2B). Similar constructs designed to express just the 22K and 33K proteins also gave only constitutively expressing cell lines (Fig. 2C), while the shuttle plasmids (Fig. 2D) used to produce these cell lines produced readily detectable 33K in transient assays despite lacking any known promoter (Fig. 2E and F). The removal from these constructs of all Ad5 sequence upstream of the L4-33K reading frame abolished this expression (Fig. 2E). These results suggested that L4 22/33K could be expressed from a novel viral promoter independently of the Ad5 MLP.
FIG. 2.
Differences in control of L4-22K, L4-33K, and L4-100K expression. (A) Schematic representing the method for generating stable L4 protein cell lines. Target cells express GFP under doxycycline control, and this gene then is replaced with L4 sequences by Cre-mediated recombination with a promoterless shuttle plasmid. (B) 100/22/33KFLAG cells containing the L4 region linked to a tetracycline-regulated promoter or parental 293TETOFF cells were grown in the presence (+) or absence (−) of doxycycline for 3 days with daily medium changes. L4-100K and L4-33KFLAG in total cell lysates were detected by Western blot analysis. (C) 22/33KFLAG cells (a to d) or parental 293TETOFF cells (e to h) were grown in the presence (a, b, e, and f) or absence (c, d, g, and h) of doxycycline for 3 days with daily medium changes. Cells were fixed and stained for FLAG-tagged 33K (red) and for nuclear DNA (DAPI; blue), and images were collected sequentially using a Leica SP2 confocal microscope to avoid cross-talk between the fluors. FLAG and DAPI images were overlaid using Leica software (b, d, f, and h); scale bar, 20 μm. (D) Schematic representation of the Ad5 sequences present in pShuttle plasmids. Brown, MLTU exon sequences (rightward transcription); blue, E2A exon (leftward transcription); pale brown, L4 ORFs as indicated. (E) Transient expression from promoterless L4 shuttle plasmids. 293TETOFF cells were transfected with pShuttle100/22/33KFLAG (a and b), pShuttle22/33KFLAG (c and d), or pShuttle22/33KFLAG ORF (e and f), or they were mock transfected (g and h). Cells were fixed and stained 48 h later and were imaged as described for panel C; scale bar, 20 μm. (F) 293TETOFF cells were mock transfected or transfected with L4-22K− genome together with either pShuttle100/22/33KFLAG or pShuttle22/33KFLAG. 33KFLAG protein in total cell lysates was detected by Western blot analysis. Protein molecular mass markers migrated to the positions shown on the left of panels B and F (in kDa).
Ad5 L4 contains a promoter for 22K/33K expression.
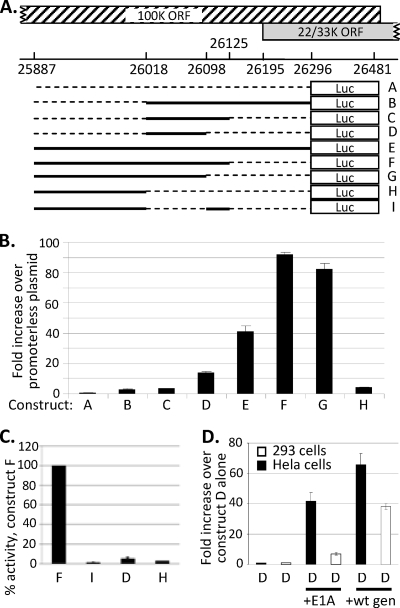
The promoterless shuttle plasmids, which nonetheless expressed L4-33K, retained the Ad5 DNA that encodes the tripartite leader (TPL), which is spliced onto all MLP-encoded mRNAs during Ad infection. However, this was not required for L4-22/33K expression, as plasmids lacking this TPL-encoding sequence were equally capable of expressing these proteins (data not shown). Attention therefore focused on the 177 bp of L4 DNA upstream of the 22/33K start codon also present in these constructs, which was hypothesized to contain a promoter for L4 22/33K expression. When this sequence plus 100 bp of the downstream 22/33K reading frame (Fig. 3 A, construct B) was placed in front of a promoterless luciferase reporter gene, it caused a modest 4-fold increase in activity (Fig. 3B). However, extending the sequence further upstream to position 25887 increased activation to 40-fold (Fig. 3B, construct E). A similar analysis of a series of constructs with different lengths of Ad sequence showed that an essential sequence for the L4 promoter was located between bp 26018 and 26098. This sequence alone activated luciferase expression more than 10-fold (Fig. 3B, construct D), and its deletion from the full promoter completely destroyed activity (Fig. 3C, construct I). Including sequence upstream of this essential sequence to position 25887 greatly increased promoter activity, while sequences downstream to 26125 were repressive; however, the presence of the upstream sequences overcame this repressive effect (Fig. 3B). The full L4 promoter (L4P), which gave a >80-fold increase in activity above the background, therefore was designated Ad5 position 25887 to 26125. The addition of further downstream sequence, to position 26296, gave lower activity than that of the full promoter, possibly because this sequence includes the L4 22/33K AUG, which would reduce translation from the luciferase start codon. Alternatively, this decrease in activity may be due to negative control elements within the extended region.
FIG. 3.
Defining sequences important for L4 promoter activity. (A) A schematic representation of the L4 sequences included in luciferase reporter constructs analyzed in panels B to D. (B and C) 293 cells were mock transfected or transfected with luciferase reporter constructs containing various lengths of L4 sequence as the promoter. Firefly luciferase activity, corrected for transfection efficiency using β-galactosidase expression from an independent control plasmid, is expressed as the fold difference from the activity of the promoterless reporter plasmid (pGL3Basic) (B) or as the percentage of the activity of the full L4 promoter, construct F (C). (D) HeLa cells (black bars) or 293 cells (white bars) were transfected with construct D plus either empty vector, E1A expression plasmid, or the Ad5 WT genome (wt gen). Firefly luciferase activity, corrected as described for panels B and C, is expressed as the fold difference from the activity of construct D in the presence of empty vector, which was set as 1. Each panel shows the mean values from biological triplicates within a single experiment (error bars indicate standard deviations) and is representative of at least three independent experiments.
Previously described Ad5 promoters are activated by the E1A proteins that are produced in the earliest stages of infection. These E1A proteins are constitutively expressed in 293 cells, where L4P activity was first detected (Fig. 3B). The importance of E1A to L4P activity therefore was tested in HeLa cells, in which the basal activity of the promoter was very low. Activity was increased 40-fold by E1A, whereas adding further E1A in 293 cells gave only a 6-fold enhancement of a much higher basal level (Fig. 3D). Thus, the L4P is responsive to E1A. The cotransfection of WT Ad5 genome also strongly activated the promoter (Fig. 3D); this activation is explored further below.
L4 promoter is functionally relevant and required for infection to enter late phase.
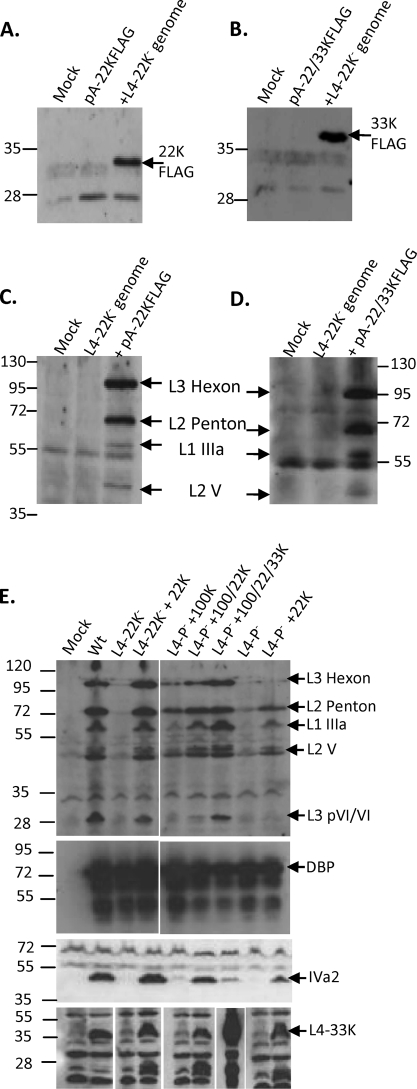
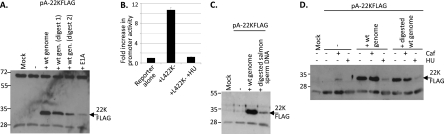
To determine whether L4P in its natural context directed the expression of L4 proteins, DNA from position 25887 to the 3′ end of the 22K coding sequence [FLAG tagged for detection and with a poly(A) site added] was cloned into a plasmid with no other viral or eukaryotic sequences present (pA-22KFLAG). This plasmid alone expressed little or no 22K-FLAG in 293 cells, but when cotransfected with 22K-deficient Ad5 genome, 22K-FLAG was readily detected (Fig. 4 A). Cotransfected genome also activated the expression of 33K from a similar construct capable of expressing both 22K (untagged) and 33K-FLAG (Fig. 4B). Ad5 superinfection also activated L4P in HeLa cells (data not shown). To test whether these amounts of 22K expressed from L4P were functionally significant, we made use of a complementation assay using the Ad5 22K− genome. Such genomes are substantially defective in the synthesis of all proteins encoded in the MLTU but can be complemented in trans with a 22K expression plasmid (33). Using this test, L4P directed the expression of functional amounts of 22K-FLAG, either from a 22K-only construct (Fig. 4C) or from a 22/33K construct (Fig. 4D). L4P therefore is active in its natural context, and the amount of 22K it produces is sufficient to promote the early-late transition in MLTU activity.
FIG. 4.
L4-22K expression from L4 promoter is functionally significant. (A to D) 293 cells were mock transfected or transfected with L4-22K− genome alone or with either pA-22KFLAG (A and C) or pA-22/33KFLAG (B and D). Immunoprecipitated FLAG-tagged 22K (A) or 33K (B), or Ad5 late proteins in total cell lysates (C and D), were detected by Western blot analysis. (E) Ad5 wild-type (Wt), L4-22K−, or L4P− genomes were transfected into 293 cells with combinations of expression plasmids for individual L4 proteins 100K, 22K, and 33K as indicated. Total cell extracts were analyzed as described for panels A and B for specific viral proteins indicated to the right of the panel. Where a blot with a given antibody is shown in segments, these all were taken from the same exposure of the same blot, with lanes rearranged for clarity of explanation. The positions to which proteins of known molecular mass migrated are indicated for all panels (in kDa).
To show the importance of L4P to the life cycle of the virus, the region bp 26018 to 26098 (which is essential for L4P activity) (Fig. 3C) was deleted within full-length wild-type genome plasmid to create the L4P− genome. This deletion also disrupts the essential L4-100K open reading frame; however, late proteins (except hexon) still are expressed from MLTU plasmids in the absence of 100K (16), meaning that the importance of L4P for gene expression from the MLTU could be tested using this mutant genome. The L4P− genome expressed amounts of DNA binding protein (DBP), a representative early protein, equivalent to those of the wild-type genome, but it essentially failed to express either MLTU-derived proteins or IVa2, reproducing the phenotype of the L4-22K− genome (Fig. 4E). The cotransfection of 22K expression plasmid with L4P− allowed the significant expression of IVa2 and MLTU proteins, except hexon, which accumulated when 100K was additionally expressed in trans. Thus, the L4P− genome is functionally deficient in L4-22K. Although the promoter deletion is some distance upstream of the major splice acceptor site by which L4-22K and 33K are expressed from the MLP (position 26158), its use could have been affected by the deletion, thus preventing 22K/33K expression. However, this is not the case, since L4-33K expression from the L4P− genome clearly was induced following complementation with 22K in trans (Fig. 4E). Thus, the gross defect in late gene expression from the L4P− genome is due to a lack of L4 promoter activity, which therefore is required to initiate late-phase gene expression. Surprisingly, the cotransfection of L4P− with 100K alone did cause some increase in certain late proteins, particularly penton base, although it was substantially less than that achieved by 22K complementation. This may reflect low levels of transcription from the MLP in the absence of L4-22K, from which translation can be enhanced by L4-100K. Alternatively, because, uniquely among the plasmids used here, the DNA present in the complementing L4-100K plasmid covers the L4P− deletion, some rescue of the mutation may be occurring by homologous recombination in this case only.
Defining activators of the L4 promoter.
The strong activation of the L4 promoter by the Ad5 genome in cells that already expressed E1A suggested that the genome supplied activators in addition to E1A. Full late-phase activation of the MLTU requires genome replication; given the expected timing of the activation of L4P at the early- to late-phase transition, we first tested whether or not the replication of the added genome was required for its effect. Genome DNA fragmented to prevent its replication by exhaustive digestion with BglII and HindIII (digest 1; 23 fragments) still significantly activated L4P (Fig. 5 A), suggesting that genome replication is not essential for L4P trans-activation. However, replication must contribute to the activation caused by intact genome, since (i) digest 1 has somewhat lower activity than intact genome, and (ii) hydroxyurea (HU), which blocks DNA synthesis among other effects, essentially blocked the stimulation of the promoter by genome (Fig. 5B and D).
FIG. 5.
Activation of L4P by Ad5 genome. (A) 293 cells were mock transfected or transfected with pA-22KFLAG together with undigested Ad5 WT genome (wt gen.), Ad5 WT genome digested with either BglII/HindIII (digest 1) or HaeII (digest 2), or an E1A expression plasmid, and 22KFLAG was detected as described for Fig. 4. (B) 293 cells were transfected with L4 promoter luciferase reporter construct D (Fig. 3A), with or without L4-22K− genome and/or treatment with 10 mM hydroxyurea (HU) from 5 h posttransfection. Luciferase expression, corrected for transfection efficiency, is expressed as the fold increase above the activity of core promoter alone, set as 1. Error bars show the standard deviations from three replicate determinations. The graph is representative of two experiments. (C) As described for panel A, except using digested salmon sperm DNA as an L4P activator in comparison with intact Ad5 genome. (D) 293 cells were mock treated or pretreated with 3 mM caffeine for 3 h and then were mock transfected or transfected with pA25887-22KFLAG together with intact Ad5 WT genome or digest 1. From 5 h posttransfection, cells were maintained in DMEM or DMEM supplemented with 3 mM caffeine (Caf) or 10 mM hydroxyurea (HU).
We considered the possibility that some component of L4P activation by complete or fragmented genome could be due to the expression of specific viral proteins. Ad5 genome digest 2 (HaeII; 77 fragments), which truncates or separates from their promoters all known open reading frames except protein IX, was a much less potent activator of the L4 promoter than genome digest 1, which could, in principle, express E1A, IX, IVa2, and E4 Orf1 from individual fragments (Fig. 5A). Since additional E1A contributed only modestly to 22K expression from L4P in 293 cells (Fig. 5A), while E4 Orf1 is expressed late in Ad5 infection (14) and Orf1 mutants express late proteins normally (8, 43), the involvement of these proteins in genome-mediated L4P activation in 293 cells was unlikely. In contrast, the production of IX and IVa2 begins around the early-late transition, making them plausible L4P activators. When tested, IVa2 but not IX activated L4P significantly (Fig. 6 A and B), although it was less potent than the intact genome.
FIG. 6.
Viral proteins activate the L4 promoter. (A, C, and D) 293 cells were mock transfected or transfected with pA-22KFLAG or pA-22/33KFLAG, together with viral protein expression plasmids as indicated, or with WT Ad5 genome. Immunoprecipitated 22KFLAG protein or proteins from unfractionated cell lysates were detected by Western blot analysis as indicated at the right of each panel. The positions of molecular mass marker proteins are shown on the left (in kDa). (B) 293 cells were mock transfected or transfected with L4 promoter luciferase reporter construct D or F (Fig. 3A), with or without IVa2 expression plasmid. Firefly luciferase activity, corrected for transfection efficiency, is expressed as the fold difference from the activity of the relevant promoter reporter alone, which was set as 1. Error bars show the standard deviations from three replicate determinations. (E) As described for panel B, except that cells were transfected with L4 promoter reporter construct D, with or without either E4-Orf3 or E4-Orf6 expression plasmid. The graphs are representative of two experiments.
Ad5 E4 Orf3 and E4 Orf6 also were tested for effects on L4 protein expression, initially because these proteins had been shown to affect MLTU splicing during Ad infection (36, 37), and it was conceivable that part of the induction of 22K by the genome was by altering the splicing balance between 22K and 33K expression. However, both 22K (from pA-22KFLAG) and 33K (from pA-22/33KFLAG) were induced by exogenous E4-Orf3 (Fig. 6C), suggesting that this was not the case. Moreover, Orf3 activated L4P while Orf6 did not (Fig. 6A and E). Thus, Orf3, like IVa2, is an activator of L4P. Furthermore, this activity was retained by mutant Orf3 N82A (Fig. 6D), a protein that is defective in other Orf3 activities.
The fact that highly fragmented genome (digest 2, 77 fragments) still was able to activate L4P to some extent (Fig. 5A), even though it was incapable of replicating or expressing any viral proteins other than potentially IX, which did not activate in trans, suggested that a general stress/DNA damage response, consequent upon the transfection of this fragmented DNA, was affecting the promoter. During infection, replicating linear adenovirus genome also activates this response. In support of this idea, a nonspecific fragmented DNA also activated L4P (Fig. 5C). The fragmentation of the DNA clearly was the critical factor in this activating response, since the same DNA was used unfragmented as the transfection control in all of these experiments and had no effect on the promoter. Cellular responses to double-strand DNA breaks are signaled via the kinase ATM, which is inhibited by caffeine. However, caffeine treatment did not inhibit L4P activation by digested genome (Fig. 5D). Thus, the mechanism by which L4P is activated by fragmented DNA remains unclear.
DISCUSSION
Our data show that L4-22K and 33K expression is driven by a previously undetected intermediate-phase promoter (L4P) embedded within the body of the Ad5 MLTU, and that this promoter triggers the progression of infection into the late phase. The ability of Ad5 to produce L4-22/33K proteins independently of the MLP resolves the paradox created by these otherwise MLP-derived proteins being required for their own production. This discovery therefore fills a significant gap in our understanding of how the Ad5 infectious cycle progresses to virus production.
As might have been expected from knowledge of other Ad5 promoters, L4P activity was strongly upregulated by several virus-derived factors. In particular, L4P was strongly activated by the presence and replication of viral genome. This stimulation was partly attributable to the expression of viral protein activators, including E1A, E4 Orf3, and IVa2, the synthesis of the latter itself being dependent on the onset of DNA replication (22). The role of E1A proteins was not surprising, since they activate several other Ad promoters (5, 23). Similarly, the role of IVa2 in L4P activation fits with its induction at the onset of replication and with its role as a transcription factor in activating the MLP (32, 45). Since we have shown recently that L4-22K increases the levels of IVa2, probably through protein-protein interaction and stabilization (33), there is a mutual enhancement relationship between these two proteins. This creates a molecular switch that can reinforce itself to drive the infection into the late phase.
The involvement of IVa2 in L4P activation may explain, at least in part, the increased activation of L4P that is observed when cotransfected genome is able to replicate, since the expression of IVa2 is itself activated by replication (22). IVa2 expression also may account for some of the difference in L4P trans-activation activity between digested genome preparations 1 and 2. Where the IVa2 gene remained intact (digest 1), the genome fragment preparation still was able to express low levels of IVa2 (data not shown), whereas equivalent amounts of digest 2 were, unsurprisingly, unable to express any IVa2 protein. However, there remains a component of activation by highly fragmented genome that cannot be accounted for by protein expression and that is not specific to the presence of Ad5 sequences. The basis of this component of L4P trans-activation remains to be determined.
The relevance to the L4P activation of known Orf3 activities, which either disrupt DNA damage responses through the mislocalization of key cellular proteins (42), disrupt promyelocytic leukemia (PML) nuclear bodies through direct interaction with PML protein (10, 15, 21) to overcome antiviral responses (46, 47), or bind the transcription factor TIF1α (52), can be discounted, since these activities are lost by the Orf3 N82A mutant, which we have shown retains full activity toward L4P. Two further Orf3 activities, the regulation of MLTU RNA splicing (36, 37) and the relief of the repression of cellular p53 activity that is imposed by other viral proteins during the early phase of infection (24), have not been tested for the N82A mutant; however, Orf3 activation of L4P was not diminished by a significant reduction in p53 levels (data not shown), suggesting that Orf3 does not act via this route. Thus, the basis for the activation of L4P by E4 Orf3 remains to be determined.
Differences were observed between the activity of L4P when incorporated into 293-based cell lines within an inducible expression cassette and when used transiently to drive L4-22K/33K expression. In the former case, the expression of L4 proteins was detectable (via their FLAG tag) without additional inducers, whereas the plasmid-based promoter required induction by one or more of several factors for its protein product to be detected. There are two potential reasons for this difference. First, the level of protein detected in cell lines reflects accumulation to steady state over a considerable time, whereas in a transient assay the level of protein observed is more dependent on its rate of synthesis. Second, the basal activity of L4P appears to be sensitive to the state of the cells since, in some transient assays, a low level of 22K was detected from L4P in the absence of genome and we have shown L4P induction by fragmented DNA, a known cell stressor. Because the cell lines were maintained in a cocktail of drugs to maintain the appropriate selection regimen, this also may have imposed a stress on the cells that served to activate L4P. There also were differences in the requirement for specific activators to observe detectable L4P activity between the natural context where L4P was driving the expression of L4-22K and the luciferase reporter context. In the natural context, activity was highly dependent on activators provided by the viral genome, while considerable luciferase reporter activity was seen in the absence of activation, although this was further increased by trans-activation. We believe this difference likely is due to the presence of negative regulatory elements downstream of our mapped promoter region, within the 22/33K coding sequence.
The DNA sequence within the mapped L4P contains potential binding sites for a large number of transcription factors, including known mediators of E1A activation, such as E2F and ATF, that may well be significant in the activity of L4P as well as numerous other factors, the significance of which cannot be predicted. It is notable that there is no obvious TATA box in the promoter region, and in this respect L4P is similar to the IVa2 promoter, which depends for its activity on an initiator element at the transcription start site (12). However, the functional significance of this lack of TATA box is uncertain, since TATA boxes, although once thought to be fairly ubiquitous features of eukaryotic promoters, more recently have been shown to be present in only around 25% of genes in genome-wide surveys (50).
L4P overlaps with the E2 late promoter (E2-L) on the opposite genome strand. Like L4P, E2-L becomes active during the intermediate phase of infection (3). However, the two promoters have different sequence requirements, with sequence from bp 25910 to 26065 providing E2-L full activity with correct temporal regulation within the Ad replication cycle (6), while the minimum L4 promoter was shown here to be bp 26018 to 26098. Moreover, the two promoters differ in their response to E1A, with L4P being activated by E1A while E2-L is insensitive (26) or repressed (20). Thus, L4P and E2-L are distinct and independent in their regulation.
The discovery of L4P creates a new understanding of how the full late phase of Ad5 infection is initiated, which is important in two ways. First, it creates a new opportunity to enhance the properties of Ad5-based gene delivery vectors. E1-deleted vectors have been intensively investigated for many applications, but in situations where the long-term persistence of the delivered gene is required, cellular immune responses to residual viral gene expression products are a significant confounding factor (51). While these problems can be avoided by removing all viral genes from the vector (31), such vectors are difficult to produce in quantity. Second, the importance of L4P can be speculatively linked to the ability of Ad5 to persist for extended periods in lymphoid cells in its natural host (17, 19). This property requires that Ad5 can regulate its infectious cycle to limit or prevent commitment to the late phase. The discovery of the essential role of L4P therefore presents further avenues through which to explore this aspect of Ad5 biology.
Acknowledgments
We thank D. Farley for the development of the promoterless shuttle plasmid system employed here and the following colleagues for their generous gifts of reagents: P. Hearing (pTG3602-L4-22K−, anti-33K antibody), S. J. Flint (pTG3602), T. Dobner (anti-Orf3 antibody), and W. C. Russell (anti-100K antibody).
This work was supported by the Biotechnology and Biological Sciences Research Council (grant numbers BBS/B/02169 and BB/E014550/1).
Footnotes
Published ahead of print on 5 May 2010.
REFERENCES
- 1.Akusjärvi, G., and H. Persson. 1981. Controls of RNA splicing and termination in the major late adenovirus transcription unit. Nature 292:420-426. [DOI] [PubMed] [Google Scholar]
- 2.Ali, H., G. LeRoy, G. Bridge, and S. J. Flint. 2007. The adenovirus L4 33-kilodalton protein binds to intragenic sequences of the major late promoter required for late phase-specific stimulation of transcription. J. Virol. 81:1327-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, C. C., and E. B. Ziff. 1981. Promoters and heterogeneous 5′ termini of the messenger-RNAs of adenovirus serotype-2. J. Mol. Biol. 149:189-221. [DOI] [PubMed] [Google Scholar]
- 4.Berk, A. J. 2006. Adenoviridae: the viruses and their replication, p. 63. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, and M. A. Martin (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams and Wilkins, New York, NY. [Google Scholar]
- 5.Berk, A. J., F. Lee, T. Harrison, J. Williams, and P. A. Sharp. 1979. Pre-early adenovirus-5 gene-product regulates synthesis of early viral messenger-RNAs. Cell 17:935-944. [DOI] [PubMed] [Google Scholar]
- 6.Bhat, G., L. Sivaraman, S. Murthy, P. Domer, and B. Thimmappaya. 1987. In vivo identification of multiple promoter domains of adenovirus EIIa-late promoter. EMBO J. 6:2045-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brey, S. 1999. Construction of IVa2-deficient adenovirus using complementing cell lines. Ph.D. thesis, University of Warwick, United Kingdom.
- 8.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caravokyri, C., and K. N. Leppard. 1995. Constitutive episomal expression of polypeptide IX (pIX) in a 293-based cell line complements the deficiency of pIX mutant adenovirus type 5. J. Virol. 69:6627-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho, T., J. S. Seeler, K. Öhman, P. Jordan, U. Pettersson, G. Akusjärvi, M. Carmo-Fonseca, and A. Dejean. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, H., and S. J. Flint. 1992. Mutational analysis of the adenovirus 2 IVa2 initiator and downstream elements. J. Biol. Chem. 267:25457-25465. [PubMed] [Google Scholar]
- 13.Chow, L. T., and T. R. Broker. 1978. The spliced structures of adenovirus 2 fiber message and other late mRNAs. Cell 15:497-510. [DOI] [PubMed] [Google Scholar]
- 14.Dix, I., and K. N. Leppard. 1993. Regulated splicing of adenovirus type 5 E4 transcripts and regulated cytoplasmic accumulation of E4 mRNA. J. Virol. 67:3226-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 16.Farley, D. C., J. L. Brown, and K. N. Leppard. 2004. Activation of the early-late switch in adenovirus type 5 major late transcription unit expression by L4 gene products. J. Virol. 78:1782-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox, J. P., C. D. Brandt, F. E. Wassermann, C. E. Hall, I. Spigland, A. Kogon, and L. R. Elveback. 1969. The virus watch program: a continuing surveillance of viral infections in metropolitan New York families. VI. Observations of adenovirus infections: virus excretion patterns, antibody responses, efficiency of surveillance, patterns of infections, and relation to illness. Am. J. Epidemiol. 89:25-50. [DOI] [PubMed] [Google Scholar]
- 18.Gambke, C., and W. Deppert. 1981. Late nonstructural 100,000- and 33,000-dalton proteins of adenovirus type 2. I. Subcellular localization during the course of infection. J. Virol. 40:585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnett, C. T., G. Talekar, J. A. Mahr, W. Huang, Y. Zhang, D. A. Ornelles, and L. R. Gooding. 2009. Latent species C adenoviruses in human tonsil tissues. J. Virol. 83:2417-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guilfoyle, R. A., W. P. Osheroff, and M. Rossini. 1985. Two functions encoded by adenovirus early region 1A are responsible for the activation and repression of the DNA-binding protein gene. EMBO J. 4:707-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoppe, A., S. J. Beech, J. Dimmock, and K. N. Leppard. 2006. Interaction of the adenovirus type 5 E4 Orf3 protein with promyelocytic leukemia protein isoform II is required for ND10 disruption. J. Virol. 80:3042-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iftode, C., and S. J. Flint. 2004. Viral DNA synthesis-dependent titration of a cellular repressor activates transcription of the human adenovirus type 2 IVa(2) gene. Proc. Natl. Acad. Sci. U. S. A. 101:17831-17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, N., and T. Shenk. 1979. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc. Natl. Acad. Sci. U. S. A. 76:3665-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.König, C., J. Roth, and M. Dobbelstein. 1999. Adenovirus type 5 E4orf3 protein relieves p53 inhibition by E1B-55-kilodalton protein. J. Virol. 73:2253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsson, S., C. Svensson, and G. Akusjärvi. 1992. Control of adenovirus major late gene-expression at multiple levels. J. Mol. Biol. 225:287-298. [DOI] [PubMed] [Google Scholar]
- 26.Leff, T., and P. Chambon. 1986. Sequence-specific activation of transcription by adenovirus E1a products is observed in HeLa cells but not in 293 cells. Mol. Cell. Biol. 6:201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leppard, K. N. 2008. Adenoviruses: molecular biology, p. 17-23. In B. W. J. Mahy and M. H. V. van Regenmortel (ed.), Encyclopedia of virology, vol. 1. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 28.Leppard, K. N., and R. D. Everett. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 80:997-1008. [DOI] [PubMed] [Google Scholar]
- 29.Lethbridge, K. J., G. E. Scott, and K. N. Leppard. 2003. Nuclear matrix localization and SUMO-1 modification of adenovirus type 5 E1b 55K protein are controlled by E4 Orf6 protein. J. Gen. Virol. 84:259-268. [DOI] [PubMed] [Google Scholar]
- 30.Lutz, P., and C. Kedinger. 1996. Properties of the adenovirus IVa2 gene product, an effector of late phase-dependent activation of the major late promoter. J. Virol. 70:1396-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCormack, W. M., M. P. Seiler, T. K. Bertin, K. Ubhayakar, D. J. Palmer, P. Ng, T. C. Nichols, and B. Lee. 2006. Helper-dependent adenoviral gene therapy mediates long-term correction of the clotting defect in the canine hemophilia A model. J. Thrombosis and Haemostasis. 4:1218-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mondesert, G., C. Tribouley, and C. Kedinger. 1992. Identification of a novel downstream binding protein implicated in late-phase-specific activation of the adenovirus major late promoter. Nucleic Acids Res. 20:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris, S. J., and K. N. Leppard. 2009. Adenovirus serotype 5 L4-22K and L4-33K proteins have distinct functions in regulating late gene expression. J. Virol. 83:3049-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nevels, M., B. Tauber, E. Kremmer, T. Spruss, H. Wolf, and T. Dobner. 1999. Transforming potential of the adenovirus type 5 E4orf3 protein. J. Virol. 73:1591-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nevins, J. R., and M. C. Wilson. 1981. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature 290:113-118. [DOI] [PubMed] [Google Scholar]
- 36.Nordqvist, K., K. Öhman, and G. Akusjärvi. 1994. Human adenovirus encodes 2 proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol. Cell. Biol. 14:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Öhman, K., K. Nordqvist, and G. Akusjärvi. 1993. Two adenovirus proteins with redundant activities in virus growth facilitate tripartite leader mRNA accumulation. Virology 194:50-58. [DOI] [PubMed] [Google Scholar]
- 38.Ostapchuk, P., M. E. Anderson, S. Chandrasekhar, and P. Hearing. 2006. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 80:6973-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Persson, H., U. Pettersson, and M. B. Mathews. 1978. Synthesis of a structural adenovirus polypeptide in the absence of viral DNA replication. Virology 90:67-79. [DOI] [PubMed] [Google Scholar]
- 40.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480-484. [DOI] [PubMed] [Google Scholar]
- 41.Shaw, A. R., and E. B. Ziff. 1980. Transcripts from the adenovirus-2 major late promoter yield a single early family of 3′ coterminal mRNAs and five late families. Cell 22:905-916. [DOI] [PubMed] [Google Scholar]
- 42.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, M. A., R. S. Broughton, F. D. Goodrum, and D. A. Ornelles. 2009. E4orf1 limits the oncolytic potential of the E1B-55K deletion mutant adenovirus. J. Virol. 83:2406-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Törmänen, H., E. Backström, A. Carlsson, and G. Akusjärvi. 2006. L4-33K, an adenovirus-encoded alternative RNA splicing factor. J. Biol. Chem. 281:36510-36517. [DOI] [PubMed] [Google Scholar]
- 45.Tribouley, C., P. Lutz, A. Staub, and C. Kedinger. 1994. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J. Virol. 68:4450-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ullman, A. J., and P. Hearing. 2008. Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J. Virol. 82:7325-7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ullman, A. J., N. C. Reich, and P. Hearing. 2007. Adenovirus E4 ORF3 protein inhibits the interferon-mediated antiviral response. J. Virol. 81:4744-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson, M. C., N. W. Fraser, and J. E. J. Darnell. 1979. Mapping of RNA initiation sites by high doses of uv irradiation: evidence for three independent promoters within the left 11% of the Ad-2 genome. Virology 94:175-184. [DOI] [PubMed] [Google Scholar]
- 49.Winter, N., and J.-C. d'Halluin. 1991. Regulation of the biosynthesis of subgroup C adenovirus protein IVa2. J. Virol. 65:5250-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, C., E. Bolotin, T. Jiang, F. M. Sladek, and E. Martinez. 2007. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 389:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, Y., F. A. Nunes, K. Berensci, E. E. Furth, E. Gonzol, and J. M. Wilson. 1994. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc. Natl. Acad. Sci. U. S. A. 91:4407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yondola, M. A., and P. Hearing. 2007. The adenovirus E4 ORF3 protein binds and reorganizes the TRIM family member transcriptional intermediary factor 1 alpha. J. Virol. 81:4264-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]