Abstract
A putative latency-associated transcript (LUNA) complementary to the human cytomegalovirus (HCMV) UL81-82 region previously identified in seropositive donors' monocytes is also expressed during lytic infection. Thus, the LUNA promoter is active during both lytic and latent infection. Consequently, the mechanisms regulating this promoter may provide further insight into factors that determine whether the outcome of HCMV infection is latent or lytic. By transfection, the LUNA promoter exhibited low but reproducible activity. Substantial activation by virus infection suggested that a viral factor was important for LUNA expression during lytic infection. IE72, a known transactivator of viral promoters, activated the LUNA promoter in cotransfection assays. Furthermore, coinfection with wild-type HCMV but not an IE72 deletion virus (CR208) also activated the LUNA promoter. Finally, diminished LUNA gene expression in CR208 virus-infected cells supported a role for IE72 in LUNA gene expression. The initial regulation of herpesvirus immediate-early gene expression is associated with proteins found at cellular nuclear domain 10 (ND10) bodies, such as PML, hDaxx, and ATRX. hDaxx transfection repressed LUNA promoter activity. Furthermore, we observed binding of hDaxx to the LUNA promoter, which was abrogated by IE72 gene expression via direct interaction. Finally, we show that small interfering RNA (siRNA) knockdown of the hDaxx interaction partner ATRX rescued LUNA gene expression in CR208-infected cells. Overall, these data show that hDaxx/ATRX-mediated repression of LUNA during lytic infection absolutely requires IE72 gene expression. It also suggests that the targeting of cellular factors by IE72 is important throughout the different phases of HCMV gene expression during productive infection.
Infection of healthy hosts with human cytomegalovirus (HCMV) is usually asymptomatic and results in a lifelong persistent infection, with up to 90% of some populations seropositive for the virus (23). In contrast to the outcome for healthy individuals, infection or reactivation in immunocompromised individuals is a significant cause of morbidity and mortality, and thus HCMV represents a major cause of disease in transplant patients, AIDS sufferers, and newborns (5, 13, 23, 51).
Like all herpesviruses, HCMV can undergo both lytic and latent life cycles, which show profound differences in patterns of viral gene expression. Viral gene expression during latent infection appears to be limited to a small subset of genes (6, 19, 28, 30), some of which are also expressed during lytic infection (6, 36, 55, 58). One of these genes, LUNA, is expressed from the opposite strand of the UL81-82 gene locus and can be detected during both lytic and latent infection (6).
Herpesvirus lytic infection is crucially dependent on the initial expression of viral immediate-early (IE) genes, but it is now clear that this itself is subject to regulation by cellular transcriptional regulators (39). For example, herpesvirus infection is profoundly affected by a number of cellular proteins that accumulate at subnuclear structures called nuclear domain 10 (ND10) bodies (1, 3, 15, 16, 26, 27, 31, 38, 57). Key constituents of these proteins include PML, hDaxx, and SP100, and it has been proposed that these structures represent a cellular antiviral strategy that the virus must overcome (9, 14, 42, 56). The well-studied herpes simplex virus type 1 (HSV-1) protein ICP0 has been shown to target the PML protein for degradation upon infection in a proteosome-dependent manner to facilitate a more efficient lytic infection (7, 10, 17). Similarly, HCMV also makes a concerted effort to target components of these structures. It has been well established, for instance, that one of the viral major IE proteins, IE72, contributes (along with pp71) to ND10 reorganization and disruption (3, 31, 59). Again, the effect of IE72 on ND10 is mediated via an interaction with PML, although in contrast to results for HSV, IE72 does not appear to promote the degradation of PML, just the relocalization of the protein away from ND10 bodies (32, 61).
It is becoming increasingly clear that the hDaxx component of ND10 is also a repressor of IE gene expression and inhibits the initiation of the lytic cycle (8, 47, 52, 53, 60). Thought to act specifically by recruitment of histone deacetylases (HDACs) (24, 33), hDaxx silences expression of the viral immediate-early genes by creating a repressive chromatin structure around the major immediate-early promoter (MIEP) (60). The repression is initially countered by the viral tegument protein pp71, which has been suggested to relieve hDaxx-mediated repression of the MIEP by degrading hDaxx in the nucleus (52, 53). The initial repression of the virus lytic cycle by hDaxx has, therefore, been suggested to play an important role in the establishment of latent infection (52), although this still remains contentious (22, 63) and requires further study.
HCMV IE72 and IE86 play important roles in regulating the characteristic temporal cascade of viral gene expression observed during HCMV lytic infection. A number of studies, including analyses of the IE72-deficient CR208 virus (20), have suggested that IE72 plays a pivotal role in the activation of early gene expression, although the growth defect of the IE72-deleted virus appears to be overcome at high multiplicities of infection (MOIs) (20, 40). Although there is no evidence that IE72 can bind to DNA directly, it can bind to a number of cellular proteins to mediate its effects. For instance, it has been suggested that IE72 can physically interact with class I histone deacetylases and that this interaction results in the sequestration of these enzymes from viral early promoters, resulting in their derepression (44). Consistent with this notion, the addition of the histone deacetylase inhibitor trichostatin A (TSA) to cells infected with IE72-deleted virus can compensate for the growth defect observed at low MOIs (44). Such observations, coupled with the increasing evidence that viral lytic gene expression is regulated by histone proteins at all phases of infection (12, 25, 41, 48), are consistent with IE72 expression acting to promote a nuclear environment conducive for early gene expression by modulating chromatin-mediated regulation of viral gene expression.
In this study, we show that the robust expression of the LUNA gene product during lytic infection is absolutely dependent on IE72. However, in contrast to previously published data for other IE72-dependent genes (44), the addition of TSA is not sufficient to rescue wild-type LUNA expression in CR208-infected cells. We next show that repression of the LUNA promoter during lytic infection is mediated by the recruitment of hDaxx to the LUNA promoter, that repression is abrogated by IE72, and that IE72 physically interacts with hDaxx. Taken together, these data suggest a mechanism by which IE72 promotes LUNA expression during lytic infection. Finally, we link hDaxx-mediated repression of LUNA with the binding of the ATRX protein to the LUNA promoter, suggesting that the hDaxx-mediated repression of LUNA gene expression at early stages of HCMV infection is ATRX dependent and that this needs to be overcome by IE72.
MATERIALS AND METHODS
Virus, cell lines, and culture.
The clinical isolate TB40/E, laboratory strain Towne, and CR208Q revertant virus were purified from infected fibroblasts on a sorbitol gradient. The IE72 deletion virus CR208 has been described previously (20) and was obtained, along with the repaired virus CR208Q, from Richard Greaves and Ed Mocarski. The K450R sumoylation-deficient IE72 virus and the complementary revertant were obtained from Michael Nevels and have been described previously (43). CR208 was propagated in the ihfie1.3 cell line, constitutively expressing IE72. Primary fibroblasts and the ihfie1.3 cell line were cultured in Dulbecco modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM l-glutamine.
Plasmids and transfection.
The putative LUNA promoter (nucleotides 117648 to 118306) (6) was amplified from viral DNA using 5′-ATA TTG GTA CCG ACA CAA CAA ACG-3′ and 5′-ATG AGA AGC TTT ATC ACG GTG TAG AAA-3′ primers, and the amplified product was cloned into pGL3-Basic luciferase via the introduction of KpnI and HindIII restriction sites. To generate MIEP-pGL3, the minimal AD169 IE promoter (positions −302 to +72) was excised from pUC19IE using EcoRI/SmaI digestion and cloned into pGL3. The IE72 expression vector (pON2203) was a kind gift of Ed Mocarski and was generated by excision of the IE72 Towne cDNA from clone pie1 and subsequent ligation into pME18S (29). The hDaxx expression vector was generated by insertion of the full-length hDaxx cDNA into pcDNA3 and has been described previously (60). The pHM 142 plasmid, encoding the luciferase gene under the control of the UL112/113 promoter, was a kind gift of Thomas Stamminger.
For transient transfection, approximately 2.5 × 105 cells were transfected using Fugene 6 (Roche) as described by the manufacturer. Briefly, 3 μl of Fugene was diluted into 97 μl of serum-free medium; after 5 min, 1 μg of luciferase reporter construct was added, and Lipofectamine-DNA complexes were allowed to form for 15 min. The reaction mix was then added directly to the cell culture medium and left for 24 h. For luciferase assays, cells were transferred 24 h posttransfection to a 96-well plate prior to any subsequent infection with Towne, CR208, or CR208Q. Luciferase activity was measured using Britelite plus (Perkins-Elmer). Briefly, medium was aspirated from the wells and replaced with 100 μl of phosphate-buffered saline (PBS); then, 100 μl of Britelite reagent was added to lyse cells, and luciferase was measured using a 96-well Costar machine. The results are averages from three independent experiments.
For small interfering RNA (siRNA) transfection, Lipofectamine 2000 was used to transfect fibroblasts with previously validated ATRX duplexes (ATRXHSS100879, 5′-GGC UGA UAU UAA GAA GGC UCA UCU U-3′ and 5′-AAG AUG AGC CUU UCU UAA UAU CAG GC-3′; ATRXHSS100880, 5′-GCG AUG GAU GCU GUA AAC AAA GAG A-3′ and 5′-UCU CUU UGU UUA CAG CAU CCA UCG C-3′ [BLOCKit; Invitrogen]) or the specific siRNA control (siRNA_control_121 and siRNA_control_157; Invitrogen). Briefly, 100 pmol of siRNA was diluted in 250 μl of Optimem reduced serum medium (Invitrogen). Concomitantly, 5 μl of Lipofectamine was diluted in 250 μl of Optimem. After 5 min, the incubation mixtures were combined, incubated at room temperature for 20 min, and then added to 105 fibroblasts plated in 2 ml of antibiotic-free medium (50% confluence). After 6 h, the medium was replaced with normal fibroblast growth medium. Knockdown of ATRX RNA and protein was observed by 24 h posttransfection.
Nucleic acid isolation, reverse transcription (RT), chromatin immunoprecipitation (ChIP), and PCR.
RNA was isolated from 105 cells using RNAeasy spin columns as described by the manufacturer (Qiagen). Following isolation, 10 μg of total RNA was incubated with DNase I (Promega) and then reverse transcribed using an ImpromII RT kit (Promega). DNA was harvested from infected cells using the sodium perchlorate method. Cells (106) were resuspended in 100 mM NaCl-5 mM EDTA (pH 8.0) before lysis with 10% SDS. Protein was then aggregated with the addition of 5 M sodium perchlorate before phenol-chloroform extraction of DNA and isopropanol precipitation.
ChIP assays were performed as previously described (48). Briefly, 105 cells either infected or transfected as described below were fixed in 1% formaldehyde (10 min) and lysed, and the DNA was sheared by sonication into 250- to 500-bp fragments. Immunoprecipitations were performed overnight at 4°C with mouse anti-IE protein (1:250; Chemicon), rabbit anti-hDaxx (1:100; Sigma), or goat anti-ATRX (clone C16, 1:100; Santa Cruz) or the corresponding mouse, rabbit, or goat IgG control (all from Sigma). A protein A/G bead mix was used to capture antibodies, and following cross-link reversal, the purified DNA was isolated by proteinase K (2 μg/ml) digestion and ethanol precipitated for downstream analysis.
Gene- and promoter-specific primers were then used to amplify target sequences by PCR using PCR 2× Mastermix (Promega), under the following cycling conditions: 95°C for 5 min, followed by 20 to 30 cycles of 94°C for 40 s, 55°C for 40 s (except where otherwise indicated), and 72°C for 60 s, and then a final extension at 72°C for 10 min. The following primers were used (MgCl2 concentrations are shown in parentheses): for LUNA (1.5 mM), 5′-ATG ACC TCT CCT CCA CAC C and 5′-GGA AAA ACA CGC GGG GGA; for IE72 (2 mM), 5′-CAT CCA CAT CTC CCG CTT AT and 5′-CAC GAC GTT CCT GCA GAC TAT G; for the IE protein (2.5 mM), 5′-CGT CCT TGA CAC GAT GGA GT and 5′-ATT CTT CGG CCA ACT CTG GA; for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (2.5 mM), 5′-GAG TCA ACG GAT TTG GTC GT and 5′-TTG ATT TTG GAG GGA TTC TCG; for actin (2.5 mM), 5′-GCT CCG GCA TGT GCA and 5′-AGG ATC TTC ATG AGG TAG T; for the LUNA promoter (1.5 mM), 5′-AGC GTC TAA AAG TCA CGT C and 5′-GGA GTA CAC ACG GTA GTA GTA G; for the MIEP (1.5 mM) (melting temperature [Tm] of 50°C): 5′-TGG GAC TTT CCT ACT TGG and 5′-CCA GGC GAT CTG ACG GTT; for ATRX (2 mM), 5′-CCA AAA GAA GAT GGG CTT CA and 5′-TCC ATT CCA TCT GAG TCA ACG; for pGL3 (1.5 mM), ACG CTC TCC ATC AAA ACA AAA and 5′-GCT TAC TTA AGA TCG CAG ATC TCG; and for UL44 (1.5 mM), 5′-GCT GTC GCT CTC CTC TTT CG and 5′-TCA CGG TCT TTC CTC CAA GG. PCRs were analyzed on 2% agarose gels using ethidium bromide detection.
Yeast two-hybrid analysis.
The Matchmaker LexA yeast two-hybrid system was used to detect interactions between IE72 and hDaxx essentially as described by the manufacturer (Clontech). Briefly, IE72 was expressed as a DNA-binding domain fusion in pLexA, and hDaxx was expressed as an activation domain fusion in B42AD. Transformants were selected for growth and subsequently analyzed for β-galactosidase activity by filter test experiments. The known interaction between the simian virus 40 (SV40) large T antigen and p53, achieved using LexA-SV40 large T antigen and B42AD-p53 fusions, was used as a positive control.
Immunoprecipitation and Western blotting.
Immunoprecipitation analyses were performed using U373 cells infected with laboratory strain AD169, Towne K450R, or the Towne K450R revertant at an MOI of 5. Briefly, a T175 flask was harvested in PBS at 16 h postinfection (hpi). Nuclear extracts were prepared essentially as previously described (4). Briefly, pellets were resuspended in 400 μl cold buffer A (10 mM HEPES-KOH, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 0.2 mM phenylmethylsulfonyl fluoride [PMSF]), incubated on ice for 10 min, and then vortexed for 10 s. Samples were centrifuged for 10 s, and the supernatant was discarded. The pellet was resuspended, lysed in 100 μl buffer B (20 mM HEPES-KOH, pH 7.9, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF), and incubated on ice for 20 min. Cell debris was cleared by centrifugation for 2 min at 4°C. Precleared lysates (50 μl of protein A-Sepharose beads incubated for 1 h at 4°C) were incubated with a mouse monoclonal anti-IE protein antibody (clone E13, 1:200; Argene Biosoft, France) or with a mouse IgG1 control (Sigma-Aldrich, United Kingdom) for a further 1 h at 4°C. Fifty microliters of protein A immobilized on Sepharose beads (diluted 1:1 in PBS) was added and incubated for a further 2 h at 4°C. Beads were then collected by centrifugation (5,000 rpm for 3 min), washed three times with 1 ml NETN (20 mM Tris [pH 8.0], 1 mM EDTA, 900 mM NaCl, 0.5% NP-40), and analyzed by Western blotting with the primary antibody goat anti-hDaxx (1:50; Santa Cruz Biotechnology) or rabbit anti-hDaxx (1:4,000; Sigma, Poole, United Kingdom) for 1 h at room temperature. After washing and incubation with an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (1:2,000; Santa Cruz Biotechnology), blots were exposed using enhanced chemiluminescence (Amersham Biosciences, United Kingdom). To detect ATRX expression in siRNA-transfected cells, protein from 105 cells was analyzed by Western blotting using goat anti-ATRX (C-16) antibody (diluted 1:200 and incubated overnight at 4°C) and HRP-conjugated rabbit anti-goat IgG (diluted 1:2,000 and incubated for 1 h at room temperature). The filter was then reprobed with an HRP-conjugated rabbit anti-GAPDH antibody (diluted 1:4,000 and incubated for 1 h at room temperature; Abcam) to confirm equivalent loading.
RESULTS
LUNA expression during productive infection requires viral gene expression.
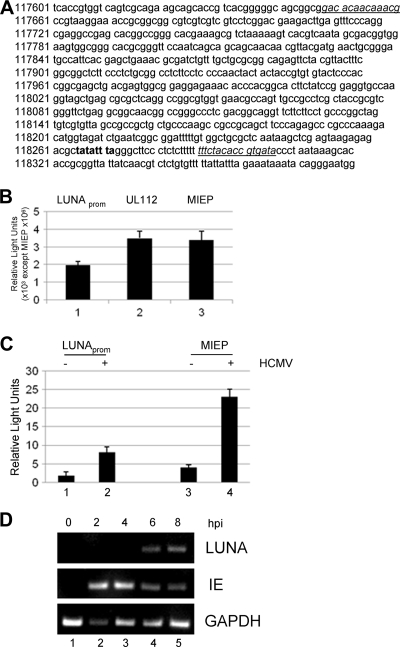
To understand the regulation of the LUNA promoter in HCMV-permissive cells, we generated a luciferase expression vector for analysis by transfection. The putative LUNA promoter (Fig. 1 A) was amplified from the clinical isolate TB40/E by use of the promoter-specific primers and then cloned into the luciferase reporter pGL3-basic. Positive clones were confirmed by sequencing.
FIG. 1.
HCMV infection activates the LUNA promoter in permissive cells. (A) The putative LUNA promoter was cloned into pGL3-Basic using the primer sequences underlined. The TATA box is highlighted in bold. (B) Primary fibroblasts transfected with pLUNA (LUNAprom), pUL112/113, or pMIEP were analyzed for luciferase gene expression. Results are from triplicate samples from 3 independent experiments. (C) Primary fibroblasts transfected for 24 h with pLUNA or pMIEP were either mock infected (−) or infected with Towne at an MOI of 5 (+) and analyzed for luciferase gene expression 24 h postinfection. Results are from triplicate samples from 3 independent experiments (results for the LUNA promoter, ×103; results for the MIEP, ×106). Error bars indicate standard deviations. (D) RT-PCR analysis was performed for LUNA, IE gene, and GAPDH RNA expression from HCMV-infected fibroblasts at 0 to 8 hpi.
To measure the basal activity of the LUNA promoter in comparison to that for the MIEP, each construct was transfected into fibroblasts and assayed for luciferase activity 48 h posttransfection. As expected, the MIEP exhibited high levels of luciferase activity (Fig. 1B, column 3). Transfection of LUNA promoter resulted in a reproducible but low level of transcriptional activity following transfection similar to the level of expression from the UL112/UL113 promoter (Fig. 1B, columns 1 and 2). We next tested the effect of HCMV infection on LUNA promoter activity. Fibroblasts were transfected followed by infection with HCMV at an MOI of 5 at 24 h posttransfection. HCMV infection resulted in a 5-fold increase in expression from both the LUNA promoter and the MIEP, suggesting that the LUNA promoter, similarly to the MIEP, could be activated by a virus infection (Fig. 1C).
To address the timing of LUNA activation during lytic infection, we next performed an RT-PCR analysis of HCMV-infected fibroblasts for both IE72 and LUNA gene expression. Consistent with previous data (6, 41), analysis of RNA isolated from infected fibroblasts between 0 and 8 hpi showed that LUNA gene expression was readily detectable at 6 h postinfection (Fig. 1D, lane 4). IE72 RNA expression was detected as early as 2 h postinfection (Fig. 1D, row 2, lane 2), and thus major IE gene expression was detectable prior to LUNA gene expression during lytic infection.
The IE72 protein activates LUNA gene expression during HCMV infection.
Both the transfection and the RT-PCR data suggested that an immediate-early function was responsible for activation of LUNA gene expression. IE72, the most abundant viral protein at IE times of infection, plays a pleiotropic role in HCMV infection, including the regulation of both viral and cellular gene expression. Consequently, we tested whether IE72 played any role in LUNA expression by both cotransfection and infection assays.
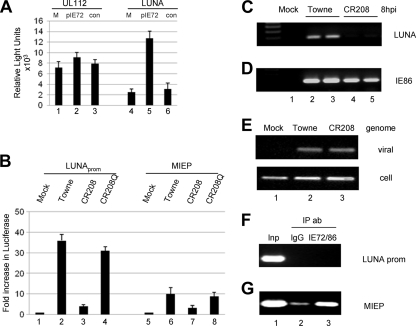
Fibroblasts were cotransfected with LUNA promoter or UL112/113 promoter in the presence or absence of IE72 and assayed for luciferase expression. Again, LUNA promoter exhibited low but reproducible activity in fibroblasts (Fig. 2 A, column 4) that was markedly increased upon cotransfection with pIE72 (Fig. 2A, column 5) but not with the empty-vector control (Fig. 2A, column 6). Interestingly, this effect of IE72 was not as marked on another early promoter, UL112/113 (Fig. 2A, columns 1 to 3), suggesting an important role for IE72 in LUNA gene expression. To confirm the dependence of LUNA activation on IE72, we next analyzed the transactivation of LUNA promoter in the context of virus infection in the presence or absence of IE72. Fibroblasts were transfected with LUNA promoter and then infected with the Towne, CR208 (IE72-deleted), or CR208Q (revertant) strain of HCMV at a high MOI (5 PFU/cell) and assayed 24 h postinfection. Consistent with the data presented in Fig. 1C, the Towne strain of HCMV activated the LUNA promoter (Fig. 2B, column 2). However, infection with the IE72-deficient CR208 virus severely abrogated viral activation of the LUNA promoter (Fig. 2B, column 3). To preclude the possibility that a defect elsewhere in CR208 was responsible for this inability of the virus to activate the LUNA promoter, we also analyzed the revertant virus, CR208Q, in which the deleted exon 4 of IE72 has been restored. This revertant activated pLUNA essentially to the same levels observed with the wild-type virus (Fig. 2B, column 4). Overall, these data suggest that LUNA gene expression in lytically infected cells is dependent upon IE72 gene expression.
FIG. 2.
IE72 activates the LUNA promoter but does not bind it directly. (A) Primary fibroblasts transfected with pUL112/113 or pLUNA were mock transfected (M) or cotransfected with pON2203 (pIE72) or pME18S (con [control]) and analyzed for luciferase gene expression 48 h posttransfection. (B) Primary fibroblasts transfected for 24 h with pLUNA or pMIEP were subsequently mock infected or infected with Towne, CR208, or CR208Q virus at an MOI of 5. Luciferase gene expression was quantified 24 h postinfection and is expressed as the fold increase over the level for mock-infected cells. (C and D) RT-PCR for LUNA and IE gene expression in mock-, Towne (MOI of 5)-, or CR208 (MOI of 5)-infected cells was performed 8 h after infection of fibroblasts. (E) Concomitant with the RT-PCR analysis, DNA isolated from mock-, Towne-, or CR208-infected fibroblasts was harvested 4 h postinfection and analyzed using IE gene (“viral”)- and GAPDH (“cell”)-specific PCRs. (F and G) ChIP assays were performed with Towne-infected fibroblasts at 12 h postinfection using an anti-IE protein antibody (ab) or a mouse IgG control. DNA purified before (input [Inp]) and after IP was amplified in LUNA promoter- and MIEP-specific PCRs. Error bars indicate standard deviations.
To confirm a role for IE72 in the context of virus infection, we analyzed LUNA expression in infected fibroblasts by using the CR208 (IE72-deleted) virus. RT-PCR analysis was performed on CR208- and Towne-infected cells for LUNA and IE gene expression at 8 h postinfection, the time at which LUNA gene expression is readily detectable (Fig. 1D). Consistent with the transfection data, LUNA gene expression was markedly decreased in CR208-infected cells (Fig. 2C, lanes 4 and 5) compared with that for the wild-type virus (Fig. 2C, lanes 2 and 3). RT-PCR for IE86 gene expression confirmed that both the CR208- and Towne-infected cells were lytically infected (Fig. 2D) and that, furthermore, the reduced LUNA gene expression was not due to an absence of the genome in the CR208-infected cells (Fig. 2E).
IE72-mediated activation of the LUNA promoter does not occur by direct binding to the LUNA promoter.
We next used ChIP analysis to determine whether IE72 mediates LUNA gene expression by direct binding to the LUNA promoter. Sheared chromatin from fixed, infected fibroblasts was immunoprecipitated with an anti-IE72/IE86 antibody or a mouse IgG control, and immunoprecipitated DNA was amplified using LUNA promoter- and MIEP-specific primers (Fig. 2F and G). We could not detect the immunoprecipitation of the LUNA promoter with an anti-IE protein antibody (Fig. 2F, lane 3), suggesting that IE72 (and/or IE86) does not directly bind to the LUNA promoter. In contrast, we did observe the immunoprecipitation of the MIEP by the IE protein antibody (Fig. 2G, lane 3), likely due to the known binding of IE86 to the cis-repression sequence (CRS) of the MIEP (11, 46). Thus, the inability to immunoprecipitate the LUNA promoter with an anti-IE protein antibody did not appear to be due to any intrinsic failure of the IP assay. Although IE72 is a well-established transcriptional activator, the failure to detect binding of IE72 to the LUNA promoter is consistent with previous evidence that IE72 mediates transcriptional activation indirectly, i.e., in the absence of direct binding to DNA.
TSA rescues LUNA promoter activity in transfection, but during infection, rescue is IE72 dependent.
At low multiplicities of infection, the IE72 protein plays a pivotal role in the efficient induction of viral early gene expression, and this has been associated with a functional interaction between IE72 and chromatin remodeling enzymes. For instance, a study by Nevels et al. (44) has shown that the addition of histone deacetylase inhibitors to CR208-infected cells results in levels of early and late gene expression approaching that observed in wild-type infections. Furthermore, this was not due to direct activation by IE72 per se but was likely due to the ability of IE72 to interact with histone deacetylases, sequestering them from viral promoters and relieving repression indirectly. If IE72 activates the LUNA promoter by a similar mechanism, we predict that the histone deacetylase inhibitor TSA increases LUNA promoter activity in transfection analyses. To test this, fibroblasts were incubated overnight with TSA or with dimethyl sulfoxide (DMSO) solvent control, transfected with either LUNA promoter or MIEP, and analyzed for luciferase activity at 48 h posttransfection. The MIEP was activated over 10-fold by the addition of TSA (Fig. 3 A, column 2), consistent with previous data showing that the MIEP is TSA responsive (48). Similarly, we also observed a 10-fold activation of the LUNA promoter in response to TSA addition (Fig. 3A, column 4), suggesting that in transfection assays, the LUNA promoter is subject to histone-mediated repression.
FIG. 3.
TSA does not rescue LUNA gene expression in CR208-infected cells. (A) Primary fibroblasts incubated with TSA (330 nM) or DMSO 24 h posttransfection were transfected with pMIEP or pLUNA. Luciferase activity was measured 24 h posttransfection, and the fold change in luciferase expression over the level observed with the DMSO control was calculated in the presence (+) of TSA. (B) Fibroblasts were preincubated overnight with 330 nM TSA (+) or DMSO (−) and then infected with Towne or CR208. RT-PCR analysis for IE gene, LUNA, UL44, and GAPDH expression was then performed. Error bars indicate standard deviations.
To confirm the transfection data in the context of virus infection, we repeated the experiments with Towne- or CR208-infected cells in the presence and absence of TSA and analyzed LUNA and IE gene expression by RT-PCR at 12 h postinfection. Consistent with the transfection data, the addition of TSA resulted in the upregulation of LUNA gene expression in Towne-infected cells (Fig. 3B, lane 2). Intriguingly, however, TSA did not induce LUNA gene expression in the CR208-infected cells (Fig. 3B, lane 4). However, addition of TSA did rescue the expression of another early gene, UL44, consistent with previously published data. Thus, these data suggest that TSA alone was not enough to rescue LUNA gene expression in the absence of the IE72 protein during infection. One likely explanation for the discrepancy between Towne and CR208 infection is that the addition of TSA, which is known to activate the MIEP (48), drives increased levels of IE72 expression (Fig. 3B) and thus that the increased LUNA gene expression in Towne-infected cells is a consequence of increased levels of IE72 gene expression, which would not occur in CR208 (IE72-deficient)-infected cells.
hDaxx, a known transcriptional repressor of viral IE gene expression, plays a role in the regulation of LUNA gene expression.
It is becoming increasingly clear that the regulation of herpesvirus gene expression by posttranslational modifications of histones is intimately associated with ND10, present in the nucleus (14, 37). The importance of these nuclear bodies in repression of herpesvirus lytic infection is perhaps underscored by the fact that both HCMV and herpes simplex virus type 1 (HSV-1) target these ND10 structures for dispersal during initial stages of infection. HSV-1 disrupts ND10 via ICP0-mediated degradation of PML, an essential component of ND10 formation (10). In contrast, IE72-mediated disruption of ND10 by HCMV occurs by the relocalization of PML away from ND10 bodies, a mechanism that does not involve PML degradation (61).
Another key component of ND10 bodies is the cellular protein hDaxx, and this too has been strongly implicated in the regulation of HCMV gene expression (37). Stable overexpression of the hDaxx protein makes normally permissive cells refractory for HCMV infection (60) and may be linked to the increased stability of the ND10 bodies in cells. To counter the effects of hDaxx, the tegument protein pp71 initially targets hDaxx for degradation, which advances lytic infection (53).
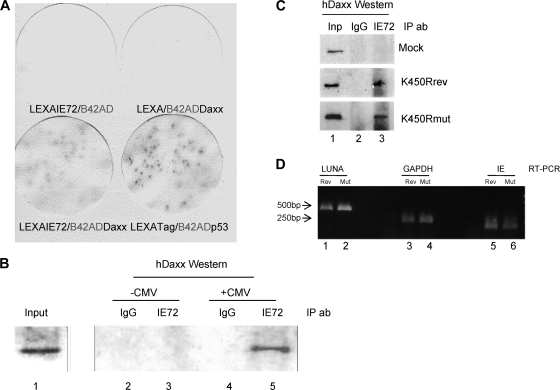
In a yeast two-hybrid screen to identify cellular proteins that could interact with IE72, we identified hDaxx as a potential interaction partner (data not shown). Figure 4 A confirms this interaction specifically between IE72 and hDaxx in filter lift experiments of transformed yeast. In order to confirm the interaction detected in the yeast two-hybrid assay, we next performed immunoprecipitation analyses of infected cells (Fig. 4B).
FIG. 4.
IE72 physically interacts with hDaxx in vitro and in vivo. (A) The LexA/B42AD system was used to screen for interactions between IE72 and hDaxx. Induction of β-galactosidase activity was determined by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining. The known interaction between the SV40 T antigen (LexATag) and p53 (B42ADp53) was used as a positive control. (B) Nuclear extracts from uninfected (−CMV) and infected (+CMV) U373 cells were harvested 18 hpi, immunoprecipitated with an anti-IE protein antibody (IE72) or an isotype-matched control antibody (IgG), and analyzed by Western blotting for IP of hDaxx. (C) Nuclear extracts from uninfected cells or cells infected with either the K450R virus (K450Rmut) or its revertant (K450Rrev) were harvested 18 hpi, immunoprecipitated with an anti-IE protein antibody or an isotype-matched control antibody, and analyzed by Western blotting for IP of hDaxx. (D) RT-PCR for LUNA, IE gene, and GAPDH expression was performed with K450Rmut (Mut)- and K450Rrev (Rev)-infected fibroblasts at 8 hpi.
Mock- and HCMV-infected U373 cells were lysed, and the nuclear extracts were immunoprecipitated with an anti-IE protein antibody or an isotype-matched control (Fig. 4B). Immunoprecipitates were then analyzed by Western blotting for hDaxx coimmunoprecipitation. These data clearly show that hDaxx complexes with IE72 in infected cells (Fig. 4B, lane 5). No such coimmunoprecipitation of hDaxx was observed with an isotype-matched control antibody (Fig. 4B, lane 4). As expected, the interaction of IE72 with hDaxx could be detected only in infected cells (Fig. 4B, lanes 4 and 5). Overall, these data suggest that during infection IE72 physically interacts with the hDaxx protein. Glutathione S-transferase (GST) pulldown analyses suggested that the interaction between IE72 and hDaxx was more efficient when IE72 was sumoylated (data not shown). To address this during infection, we infected cells with the IE72 sumoylation-deficient virus (K450R virus) or the revertant and performed the IP as described above (Fig. 4C). These data show that, during infection, IE72 sumoylation is not necessary for an interaction with hDaxx (Fig. 4C). Consistent with this, LUNA gene expression was not abrogated in K450R virus-infected cells (Fig. 4D).
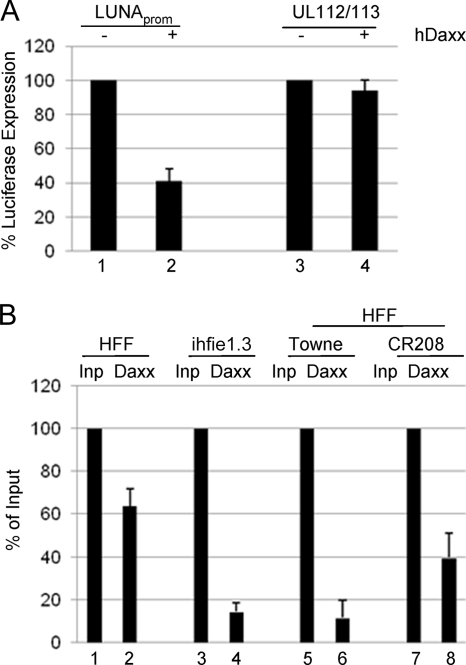
Having established that IE72 can physically interact with hDaxx, a known transcriptional regulator, we next tested whether hDaxx could repress the LUNA promoter. Transfection of LUNA promoter into permissive fibroblasts with either pcDNA3 or hDaxx showed that cotransfection with hDaxx specifically represses LUNA, but not UL112/113, promoter activity (Fig. 5 A), suggesting that hDaxx may also be involved in the repression of the LUNA promoter during lytic infection.
FIG. 5.
hDaxx binds and represses the LUNA promoter in transfection and infection assays. (A) Fibroblasts transfected with pLUNA or pUL112/113 were cotransfected with either pcDNA3 (−) or pcDNA-hDaxx (+) and were assayed 48 h later for luciferase gene expression. (B) Fibroblasts transfected with LUNA promoter for 24 h were mock infected (HFF) or infected with Towne or CR208 and then harvested for ChIP assays 24 h postinfection. Alternatively, an HFF line constitutively expressing IE72 was transfected with pLUNA in parallel (ihfie1.3). Following lysis, immunoprecipitation was performed with either anti-Daxx or a rabbit IgG serum control overnight, and then the purified DNA was amplified with LUNA promoter-specific primers. Band densitometry was performed on PCR products, and the corrected hDaxx samples were expressed as percentages of the input (Inp) controls. Error bars indicate standard deviations.
We next used ChIP analysis to determine whether hDaxx mediated repression by binding to the LUNA promoter. Cells transfected with pLUNA were immunoprecipitated with a specific anti-hDaxx antibody or a rabbit IgG control (Fig. 5B). Immunoprecipitated DNA was analyzed by PCR with pGL3 primers flanking the inserted LUNA promoter. For comparative analysis, the IP of the LUNA promoter with hDaxx was expressed as a percentage of the input after correction with the IgG isotype control. Following transfection into human foreskin fibroblasts (HFFs), IP of the LUNA promoter with hDaxx was observed (Fig. 5B, column 2), suggesting that hDaxx does indeed bind to the LUNA promoter. Knowing that IE72 can inhibit hDaxx-mediated repression of LUNA promoter activity, we also asked whether IE72 expression had any effect on the binding of hDaxx to the LUNA promoter in infected cells. First, the same analysis using the ihfie1.3 cell line (a cell line that constitutively expresses IE72) showed that the IP of the LUNA promoter was abrogated in the presence of IE72 (Fig. 5B, column 4). Similarly, infection of transfected HFFs with the Towne virus clearly resulted in a major decrease in hDaxx binding to the LUNA promoter (Fig. 5B, column 6), whereas infection in the absence of IE72 (with the CR208 virus) had only a minor effect on binding of hDaxx to the LUNA promoter (Fig. 5B, column 8). Again, these data support a role for IE72 preventing repression of LUNA expression by inhibiting promoter-specific binding of hDaxx to the LUNA promoter via a direct physical interaction.
hDaxx represses LUNA gene expression in an ATRX-dependent manner.
The repression of gene expression by hDaxx has been suggested to occur by a number of mechanisms. For example, hDaxx interacts with chromatin remodeling enzymes, such as histone deacetylases (24) (particularly HDAC-2), to mediate the repression of gene expression. However, Fig. 3B suggests that hDaxx-mediated repression of LUNA cannot be completely abrogated by TSA during infection. It has been reported that hDaxx also interacts with the transcriptional repressor ATRX (62), and a recent report from the Preston laboratory (35) has suggested that this interaction between hDaxx and the ATRX protein is important for the regulation of the HCMV MIEP at times immediately after virus entry. Consequently, we asked whether ATRX may play a role in the repression of the LUNA promoter by hDaxx and, specifically, whether siRNAs against ATRX rescue LUNA gene expression after infection in the absence of IE72.
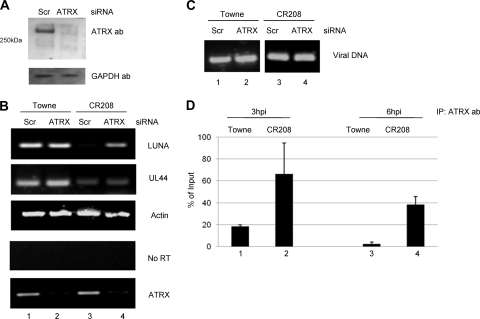
In order to knock down ATRX, we used a prevalidated BLOCKit siRNA cocktail (Invitrogen) that contains siRNAs to 3 regions of the ATRX gene which, as confirmed by Western blotting, knocked down ATRX expression by 48 h posttransfection (Fig. 6 A). To test the effect of ATRX knockdown on LUNA gene expression, cells were infected with either the Towne or the CR208 virus 48 h posttransfection and analyzed for LUNA gene expression 12 h postinfection (Fig. 6B). In siRNA control cells, no LUNA gene expression was detected after infection with CR208 (Fig. 6B, lane 3), which is consistent with data shown in Fig. 2C. In contrast, LUNA gene expression was detectable in the CR208-infected cells, in which levels of ATRX mRNA expression were clearly reduced (Fig. 6B, lane 4). An actin-specific RT-PCR (Fig. 6B), control samples with no prior cDNA synthesis (Fig. 6B), and an HCMV-specific DNA PCR (Fig. 6C) confirmed that the increase in LUNA gene expression was not due to differences in RNA level, DNA contamination, or level of HCMV infection in the cells but was specific to ATRX knockdown. Furthermore, we observed that the deletion of ATRX was specific for LUNA, as no effect on UL44 expression was observed. We observed little effect on LUNA expression after infection of ATRX knockdown cells with Towne virus (Fig. 6B, compare lanes 1 and 2). However, as IE72 expression occurs very quickly upon infection with wild-type virus, any potential effects on the activation of LUNA gene expression by ATRX depletion are likely to be masked by the robust IE72-mediated effects in the cells at the time of the analysis.
FIG. 6.
ATRX represses the LUNA promoter during infection by binding to the LUNA promoter. (A) Fibroblasts were transfected with a siRNA-ATRX cocktail (ATRX) or a mismatch control (Scr) and analyzed 24 h posttransfection by Western blotting for ATRX and GAPDH expression. (B and C) Fibroblasts were transfected with a siRNA-ATRX cocktail or a mismatch control; 24 h posttransfection, cells were infected (MOI of 5) with Towne or CR208, and RNA (B) and DNA (C) were harvested 12 h postinfection. RT-PCR was performed on the RNA using LUNA-, actin-, UL44-, and ATRX-specific primers. Alternatively, RNA was amplified in the LUNA-specific PCR without prior RT. An IE gene-specific PCR was used to amplify viral DNA to confirm equivalent numbers of genomes in the infected cells. (D) Fibroblasts infected with CR208 or Towne at an MOI of 3 were harvested at 3 and 6 h postinfection and analyzed by ChIP for ATRX binding. Sonicated cell lysates were immunoprecipitated with an anti-ATRX antibody or a goat IgG control serum, and the DNA was amplified in a LUNA promoter-specific PCR. The IgG control was used to correct the ATRX samples, which were then expressed as percentages of the input. Results from 2 ChIP experiments are shown. Error bars indicate standard deviations.
Finally, to further characterize the role of ATRX in repression of LUNA expression, we performed ChIP assays with Towne- and CR208-infected cells to analyze ATRX occupancy of the LUNA promoter at early times of infection (3 and 6 hpi) (Fig. 6D). Minimal binding of ATRX to the Towne LUNA promoter was detected at 3 or 6 h postinfection (Fig. 6D, columns 1 and 3). In contrast, ATRX binding to the LUNA promoter in CR208-infected cells was detected at both 3 and 6 h postinfection (Fig. 6D, columns 2 and 4). Consequently, the lack of LUNA gene expression observed in CR208-infected cells appears to correlate with the binding of ATRX to the LUNA promoter, suggesting that hDaxx-mediated repression of the LUNA promoter occurs in an ATRX-dependent manner.
DISCUSSION
The regulation of HCMV gene expression during lytic infection is a complicated interplay between viral and cellular factors. Virus infection promotes a strong antiviral response involving both immune and nonimmune pathways (54, 64). Part of the intrinsic response to virus infection is mediated through the action of ND10 bodies (14), nuclear structures that recruit a number of proteins that appear to antagonize viral gene expression (8, 32, 47, 53, 56, 60). In response, viruses make a concerted effort to abrogate these cellular responses to ultimately promote a lytic infection (2, 37, 53, 59, 61).
Of the proteins recruited to the ND10 bodies, those most intimately associated with HCMV gene expression are PML and hDaxx. PML, essential for ND10 formation, is targeted by the IE72 protein (3, 31), a function of IE72 that contributes to efficient lytic infection at low MOIs. Another target of IE72 is the class I histone deacetylases (44), which are also recruited to ND10 via their interactions with both PML and hDaxx proteins (24, 33). Indeed, increasing evidence suggests that viral gene expression is continuously regulated by chromatin-modifying enzymes and histone proteins throughout the course of lytic infection (12, 25, 41, 45, 48) as well as in latency (41, 50). Consistent with this, the activation of viral early promoters is thought to be enhanced by the sequestration of HDACs from viral promoters by IE72 (44).
In this study, we have analyzed the regulation of the LUNA promoter, an HCMV promoter predicted to be active in both lytic and latent infection (6). We show that LUNA gene expression becomes detectable between 4 and 6 h postinfection, consistent with previous data (6), and that LUNA expression is preceded by IE gene expression. The timing of LUNA expression was consistent with a role for IE72 in the activation of LUNA gene expression, and data from transfection assays as well as infection with the wild-type virus and the IE72-deleted virus (CR208) supported this prediction. IE72 alone was enough to activate the LUNA promoter, and transfection/infection studies showed that the CR208 virus was unable to activate the promoter efficiently; consistent with this, the CR208 virus showed markedly reduced levels of LUNA gene expression during lytic infection.
ChIP experiments confirmed that IE72 was not directly binding to DNA and was thus mediating its effect via an indirect mechanism. In contrast to previous data analyzing the regulation of other viral promoters with the CR208 virus (44), we observed that TSA could not rescue LUNA gene expression upon virus infection in the absence of IE72. It is worth noting that elevated LUNA expression was observed in wild-type-virus-infected cells expressing IE72 treated with TSA, but we believe that this is likely due to TSA-induced increases in IE72 expression. Thus, these observations are entirely consistent with the emerging role of histone posttranslational modification in the regulation of the MIEP in lytic infection (12, 21, 25, 41, 60) as well as in latency (34, 49, 50).
However, our results also suggest that IE72 uses a mechanism to activate LUNA gene expression different from the one proposed by Nevels et al. for other viral promoters, which appears to involve the sequestration of histone deacetylases (44). Interestingly, an early characterization of the CR208 virus reported that not all viral promoters were similarly affected by the IE72 deletion, suggesting that differing mechanisms are involved in viral gene expression (18). In our study, we show that IE72 can also bind to the cellular hDaxx protein. Previous data have shown that stable overexpression of the hDaxx protein is sufficient to block lytic infection (60). Consistent with the ability to titrate away this hDaxx-mediated repression of lytic infection with increasing amounts of virus, hDaxx has been shown to be a target for an incoming virion protein, pp71 (47, 53). The pp71 tegument protein promotes the degradation of hDaxx as a potential mechanism to prevent hDaxx-mediated repression of the MIEP. However, the data presented here suggest that the action of pp71 on hDaxx alone is not sufficient to rescue LUNA gene expression. Although hDaxx appears to repress the LUNA promoter, delivery of pp71 with the CR208 virus is not sufficient to activate LUNA gene expression to wild-type levels. However, it is worth noting that the transfection analysis of LUNA promoter activation (Fig. 2B) indicates that CR208 infection has a modest effect on the activity of the LUNA promoter, suggesting that other viral factors, such as pp71, may play a minor role in the activation of LUNA gene expression.
Having determined that hDaxx could repress the LUNA promoter by transfection, we then confirmed that hDaxx bound to the LUNA promoter and that this binding was abrogated by HCMV infection and, in particular, by IE72. As TSA did not rescue LUNA promoter activity during infection, we next asked whether hDaxx-mediated repression was effected via an interaction partner other than HDACs. Analysis of ATRX, a transcriptional repressor (54) which interacts with hDaxx and which has been suggested to be involved in hDaxx-mediated repression of the MIEP (35), showed that knockdown of ATRX rescued LUNA gene expression in CR208-infected cells. Furthermore, the ATRX protein could be detected on the LUNA promoter at 3 and 6 h postinfection in CR208-infected cells but not wild-type-virus-infected cells (expressing IE72).
Our findings support previous data suggesting that the IE72 protein is pivotal for the targeting of components of ND10 bodies within the cell. Our observations show that, as does PML, the IE72 protein interacts with the hDaxx protein, another constituent of ND10 bodies. Given the reports that the incoming pp71 tegument protein targets hDaxx for degradation, it may at first seem surprising that another viral protein (IE72) also targets hDaxx. However, the effects of pp71 are likely to be restricted to events immediately postentry, and although important for initial activation of the MIEP, we believe it is likely that this activity of pp71 wanes as infection progresses and that IE72 subsequently takes over this function of hDaxx/ND10 disruption.
In summary, the data presented here show that IE72 mediates the activation of the LUNA promoter during lytic infection and that this is likely achieved by a direct interaction between IE72 and hDaxx. As opposed to the activation of other viral promoters by IE72, the IE72-mediated activation of the LUNA promoter does not appear to be mediated solely by modification of histone acetylation on the LUNA promoter but appears to involve the hDaxx-interacting protein ATRX. Studies to determine the mechanism by which ATRX may functionally interact with hDaxx to regulate LUNA expression are in progress. Finally, as the interaction between IE72 and hDaxx is important for the regulation of viral lytic gene expression, it is possible that such an interaction between IE72 and hDaxx (and PML) may also be important in supporting the efficient reactivation of HCMV from latency.
Acknowledgments
We thank Joan Baillie, Linda Teague, and Verity Kew for technical assistance.
This work was funded by Novartis Institutes for Biomedical Research (M.R. and T.C.), a United Kingdom MRC studentship (D.W.), and a United Kingdom MRC program grant (J.S.).
Footnotes
Published ahead of print on 5 May 2010.
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39-55. [DOI] [PubMed] [Google Scholar]
- 3.Ahn, J. H., and G. S. Hayward. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arvin, A. M., P. Fast, M. Myers, S. Plotkin, and R. Rabinovich. 2004. Vaccine development to prevent cytomegalovirus disease: report from the National Vaccine Advisory Committee. Clin. Infect. Dis. 39:233-239. [DOI] [PubMed] [Google Scholar]
- 6.Bego, M., J. Maciejewski, S. Khaiboullina, G. Pari, and S. St. Jeor. 2005. Characterization of an antisense transcript spanning the UL81-82 locus of human cytomegalovirus. J. Virol. 79:11022-11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantrell, S. R., and W. A. Bresnahan. 2006. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 80:6188-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77:7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 11.Cherrington, J. M., E. L. Khoury, and E. S. Mocarski. 1991. Human cytomegalovirus ie2 negatively regulates alpha gene expression via a short target sequence near the transcription start site. J. Virol. 65:887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuevas-Bennett, C., and T. Shenk. 2008. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J. Virol. 82:9525-9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drew, W. L. 1988. Cytomegalovirus infection in patients with AIDS. J. Infect. Dis. 158:449-456. [DOI] [PubMed] [Google Scholar]
- 14.Everett, R. D., and M. K. Chelbi-Alix. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819-830. [DOI] [PubMed] [Google Scholar]
- 15.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:566-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D., C. Parada, P. Gripon, H. Sirma, and A. Orr. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82:2661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., S. Rechter, P. Papior, N. Tavalai, T. Stamminger, and A. Orr. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gawn, J. M., and R. F. Greaves. 2002. Absence of IE1 p72 protein function during low-multiplicity infection by human cytomegalovirus results in a broad block to viral delayed-early gene expression. J. Virol. 76:4441-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodrum, F., M. Reeves, J. Sinclair, K. High, and T. Shenk. 2007. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood 110:937-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groves, I. J., M. B. Reeves, and J. H. Sinclair. 2009. Lytic infection of permissive cells with human cytomegalovirus is regulated by an intrinsic ‘pre-immediate-early’ repression of viral gene expression mediated by histone post-translational modification. J. Gen. Virol. 90:2364-2374. [DOI] [PubMed] [Google Scholar]
- 22.Groves, I. J., and J. H. Sinclair. 2007. Knockdown of hDaxx in normally non-permissive undifferentiated cells does not permit human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 88:2935-2940. [DOI] [PubMed] [Google Scholar]
- 23.Ho, M. 1990. Epidemiology of cytomegalovirus infections. Rev. Infect. Dis. 12(Suppl. 7):S701-S710. [DOI] [PubMed] [Google Scholar]
- 24.Hollenbach, A. D., C. J. McPherson, E. J. Mientjes, R. Iyengar, and G. Grosveld. 2002. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 115:3319-3330. [DOI] [PubMed] [Google Scholar]
- 25.Ioudinkova, E., M. C. Arcangeletti, A. Rynditch, F. De Conto, F. Motta, S. Covan, F. Pinardi, S. V. Razin, and C. Chezzi. 2006. Control of human cytomegalovirus gene expression by differential histone modifications during lytic and latent infection of a monocytic cell line. Gene 384:120-128. [DOI] [PubMed] [Google Scholar]
- 26.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishov, A. M., O. V. Vladimirova, and G. G. Maul. 2002. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 76:7705-7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins, C., A. Abendroth, and B. Slobedman. 2004. A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J. Virol. 78:1440-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenkins, D. E., C. L. Martens, and E. S. Mocarski. 1994. Human cytomegalovirus late protein encoded by ie2: a trans-activator as well as a repressor of gene expression. J. Gen. Virol. 75:2337-2348. [DOI] [PubMed] [Google Scholar]
- 30.Kondo, K., J. Xu, and E. S. Mocarski. 1996. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc. Natl. Acad. Sci. U. S. A. 93:11137-11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korioth, F., G. G. Maul, B. Plachter, T. Stamminger, and J. Frey. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155-158. [DOI] [PubMed] [Google Scholar]
- 32.Lee, H. R., D. J. Kim, J. M. Lee, C. Y. Choi, B. Y. Ahn, G. S. Hayward, and J. H. Ahn. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, H., C. Leo, J. Zhu, X. Wu, J. O'Neil, E. J. Park, and J. D. Chen. 2000. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20:1784-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, X. F., S. Yan, M. Abecassis, and M. Hummel. 2008. Establishment of murine cytomegalovirus latency in vivo is associated with changes in histone modifications and recruitment of transcriptional repressors to the major immediate-early promoter. J. Virol. 82:10922-10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukashchuk, V., S. McFarlane, R. D. Everett, and C. M. Preston. 2008. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J. Virol. 82:12543-12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lunetta, J. M., and J. A. Wiedeman. 2000. Latency-associated sense transcripts are expressed during in vitro human cytomegalovirus productive infection. Virology 278:467-476. [DOI] [PubMed] [Google Scholar]
- 37.Maul, G. G. 2008. Initiation of cytomegalovirus infection at ND10. Curr. Top. Microbiol. Immunol. 325:117-132. [DOI] [PubMed] [Google Scholar]
- 38.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 39.Meier, J. L., and M. F. Stinski. 1996. Regulation of human cytomegalovirus immediate-early gene expression. Intervirology 39:331-342. [DOI] [PubMed] [Google Scholar]
- 40.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. U. S. A. 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy, J. C., W. Fischle, E. Verdin, and J. H. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Negorev, D. G., O. V. Vladimirova, A. Ivanov, F. Rauscher III, and G. G. Maul. 2006. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J. Virol. 80:8019-8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nevels, M., W. Brune, and T. Shenk. 2004. SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J. Virol. 78:7803-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nevels, M., C. Paulus, and T. Shenk. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. U. S. A. 101:17234-17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nitzsche, A., C. Paulus, and M. Nevels. 2008. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J. Virol. 82:11167-11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pizzorno, M. C., and G. S. Hayward. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J. Virol. 64:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preston, C. M., and M. J. Nicholl. 2006. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 87:1113-1121. [DOI] [PubMed] [Google Scholar]
- 48.Reeves, M., J. Murphy, R. Greaves, J. Fairley, A. Brehm, and J. Sinclair. 2006. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J. Virol. 80:9998-10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reeves, M. B., P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J. Gen. Virol. 86:2949-2954. [DOI] [PubMed] [Google Scholar]
- 50.Reeves, M. B., P. A. MacAry, P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. U. S. A. 102:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubin, R. H. 1990. Impact of cytomegalovirus infection on organ transplant recipients. Rev. Infect. Dis. 12(Suppl. 7):S754-S766. [DOI] [PubMed] [Google Scholar]
- 52.Saffert, R. T., and R. F. Kalejta. 2007. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J. Virol. 81:9109-9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saffert, R. T., and R. F. Kalejta. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinclair, J., and P. Sissons. 2006. Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 87:1763-1779. [DOI] [PubMed] [Google Scholar]
- 56.Tavalai, N., P. Papior, S. Rechter, M. Leis, and T. Stamminger. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tavalai, N., P. Papior, S. Rechter, and T. Stamminger. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82:126-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White, K. L., B. Slobedman, and E. S. Mocarski. 2000. Human cytomegalovirus latency-associated protein pORF94 is dispensable for productive and latent infection. J. Virol. 74:9333-9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilkinson, G. W., C. Kelly, J. H. Sinclair, and C. Rickards. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79:1233-1245. [DOI] [PubMed] [Google Scholar]
- 60.Woodhall, D. L., I. J. Groves, M. B. Reeves, G. Wilkinson, and J. H. Sinclair. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 281:37652-37660. [DOI] [PubMed] [Google Scholar]
- 61.Xu, Y., J. H. Ahn, M. Cheng, C. M. apRhys, C. J. Chiou, J. Zong, M. J. Matunis, and G. S. Hayward. 2001. Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression. J. Virol. 75:10683-10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xue, Y., R. Gibbons, Z. Yan, D. Yang, T. L. McDowell, S. Sechi, J. Qin, S. Zhou, D. Higgs, and W. Wang. 2003. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. U. S. A. 100:10635-10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan, J., X. Liu, A. W. Wu, P. W. McGonagill, M. J. Keller, C. S. Galle, and J. L. Meier. 2009. Breaking human cytomegalovirus major immediate-early gene silence by vasoactive intestinal peptide stimulation of the protein kinase A-CREB-TORC2 signaling cascade in human pluripotent embryonal NTera2 cells. J. Virol. 83:6391-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. U. S. A. 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]