Abstract
Determining the evolutionary basis of cross-species transmission and immune evasion is key to understanding the mechanisms that control the emergence of either new viruses or novel antigenic variants with pandemic potential. The hemagglutinin glycoprotein of influenza A viruses is a critical host range determinant and a major target of neutralizing antibodies. Equine influenza virus (EIV) is a significant pathogen of the horse that causes periodical outbreaks of disease even in populations with high vaccination coverage. EIV has also jumped the species barrier and emerged as a novel respiratory pathogen in dogs, canine influenza virus. We studied the dynamics of equine influenza virus evolution in horses at the intrahost level and how this evolutionary process is affected by interhost transmission in a natural setting. To this end, we performed clonal sequencing of the hemagglutinin 1 gene derived from individual animals at different times postinfection. Our results show that despite the population consensus sequence remaining invariant, genetically distinct subpopulations persist during the course of infection and are also transmitted, with some variants likely to change antigenicity. We also detected a natural case of mixed infection in an animal infected during an outbreak of equine influenza, raising the possibility of reassortment between different strains of virus. In sum, our data suggest that transmission bottlenecks may not be as narrow as originally perceived and that the genetic diversity required to adapt to new host species may be partially present in the donor host and potentially transmitted to the recipient host.
Influenza viruses inflict a persistent annual burden on human health, with the occasional appearance of strains with pandemic potential. A major research focus has been to determine the evolution of influenza A virus at the epidemic scale and across broad geographical areas (15, 22, 28, 30). However, little is known about the original source of viral genetic variation in natural animal hosts or how this variation impacts on the ability of the virus to adapt to new host species. RNA viruses are characterized by their ability to rapidly generate genetic variation, a consequence of their replication with highly error-prone RNA polymerases (8). Such mutational power, coupled with immense population sizes, is thought to be central to their classification as the most common agents of emerging disease (44). While the processes of mutation, segment reassortment, and natural selection are expected to shape the evolutionary dynamics of influenza viruses within an individual host, the nature, frequency, and interaction of these key evolutionary mechanisms have not been studied systematically in vivo in natural hosts or integrated into our understanding of viral evolution at the scale of global epidemics. Such data are also essential for addressing one of the most important questions in the cross-species transmission of pathogens: whether the genetic variation required for a virus to adapt to transmission in a new host species largely appears de novo in the recipient host or is seeded from the donor host (20). Similarly, the ability of natural selection to optimize such traits as host specificity is also in part dependent on the proportion of total genetic variability that is passed between hosts at transmission: the narrower the population bottleneck at transmission, the larger the stochastic component to viral evolution.
Influenza A viruses (IAVs) have their major reservoirs in wild aquatic birds (41), but periodically they transfer into humans and other mammals to cause epidemics or pandemics. Contemporary human influenza A viruses appear to have originated from a transfer that occurred shortly before the 1918 pandemic (39), and that virus subsequently exchanged three and then two of its genome segments with avian viruses in 1957 and 1968, respectively, to create new pandemic strains (31, 33). Phylogenetic analysis of recently emerged swine-origin H1N1 in humans suggests that this new pandemic virus is a reassortant between Eurasian and classic swine variants (11). While reassortment involves the transfer of entire genome segments, the more gradual process of mutation accumulation is required to facilitate both host adaptation and escape from immune responses and underlies the predictable seasonality of influenza in temperate regions (28, 30).
To determine the patterns and consequences of genetic variation in mammalian influenza viruses, we used equine influenza H3N8 virus as an experimental model system in horses. Equine influenza virus (EIV) of the H3N8 subtype was first isolated from horses in 1963 (40) and is apparently the only subtype currently circulating in the horse population. EIV has also recently jumped the species barrier and become established as a respiratory pathogen of dogs (5), so we are able to compare the variation of the virus in its established equine host, as well as in dogs which it initially infected at some point before 2004 (14). We undertook transmission experiments in naïve horses and examined the degree and composition of the within-host EIV genetic diversity of the hemagglutinin 1 (HA1) gene at different times postinfection. The HA1 gene encodes the major surface viral antigen and the receptor binding domain (42). We focused on the proportion of viral variability that persists during infection as well as that transmitted during natural chains of transmission, with the latter informing on the magnitude of any transmission bottleneck. To compare the variation observed within our experimental setting to that observed in the field, we also examined the variation in HA1 sequences of viruses recovered from natural cases of H3N8 influenza during an outbreak in the United Kingdom in 2003 (23). This allows us to link, for the first time, the process of viral evolution within and among hosts.
MATERIALS AND METHODS
Direct transmission experiments in naïve horses.
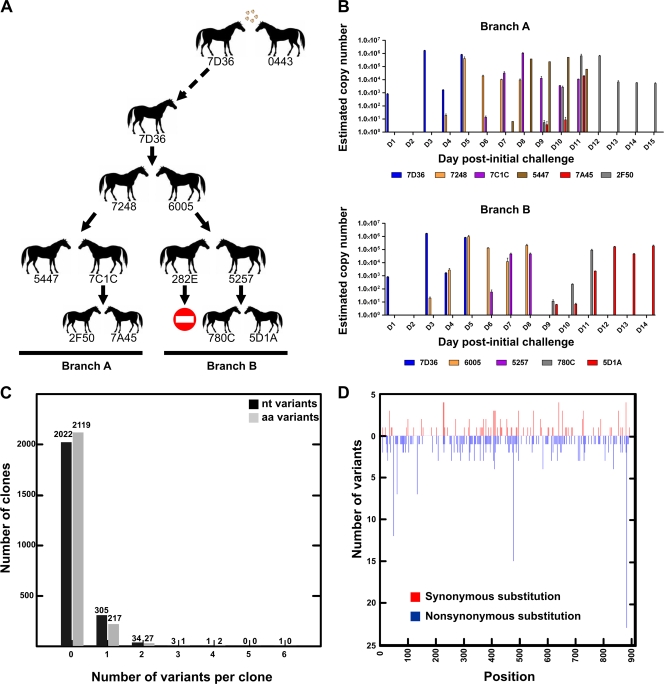
Two “transmitter” horses (7D36 and 0443) were nebulized with 20 ml of log10 106.3/ml 50% egg infective dose (EID50) of A/Equine/Newmarket/1/93 (21). On confirmation of infection using a rapid diagnostic enzyme-linked immunosorbent assay (ELISA) to detect influenza virus in swab extracts (4), one transmitter (7D36) was housed with two naïve horses (7248 and 6005) in the same stable. When horses 7248 and 6005 became infected, they were removed to clean separated stables, and each was housed with two other naïve horses (5447, 7C1C, 5257, and 282E) for another 72 h, when the procedure was repeated and horses 7C1C and 5257 were each housed as described above with two other horses (2F50, 7A45, 780C, and 5D1A) in individual clean stables (see Fig. 1). Nasal swabs were collected for 2 to 6 days after infection or contact, immersed in viral transport medium (5 ml), and stored at −80°C. All horses included in this experiment were considered naïve as they had no detectable antibodies against A/Equine/Newmarket/1/93 (measured by single radial hemolysis [43]). The animal work was done under Home Office license following full ethical approval.
FIG. 1.
Intrahost genetic variation of EIV in horses. (A) Layout of the transmission chain. Of the two horses experimentally inoculated with EIV, only one horse (7D36) was used to start the transmission chain (broken-line arrow). No virus was detected in nasal swabs from horse 282E (indicated with a stop sign). (B) Estimation of viral copy numbers from individual swabs by real-time PCR. (C) Number of clones with nucleotide (nt) and amino acid (aa) mutations. (D) Position and frequency of synonymous (red) and nonsynonymous (blue) mutations relative to the reference sequence (nucleotide level).
RNA extraction, real-time PCR, and PCR analysis.
Viral RNA from nasal swabs was isolated from 280-μl aliquots using the QIAamp viral RNA minikit (Qiagen) according to the manufacturer's instructions. Reverse transcription (RT), PCR, and real-time PCR (quantitative PCR [qPCR]) amplification were performed using a two-step reverse transcription-PCR protocol. cDNAs of the viral genomic M and HA genes were generated using Superscript III reverse transcriptase (Invitrogen) and primers Bm-M-1 and Bm-HA1 (14), respectively. RT was performed at 55°C for 90 min, followed by incubation at 70°C for 10 min. Viral copy numbers were estimated by a qPCR assay using the QuantiTect probe PCR kit (Qiagen) with fluorogenic hydrolysis type probes following the manufacturer's instructions. The primers and probe for qPCR were designed using Beacon designer (Premier Biosoft; sequences available from the authors upon request). Standard curves were generated using 10-fold dilutions of a plasmid containing the matrix segment (cloned from an egg-grown Equine/Newmarket/1/1993 isolate), ranging from 1 × 102 to 1 × 108 copies μl−1. For each run, all samples, no-template controls, plasmid standards, and positive and negative controls were run in triplicate and expressed as the mean number of viral RNA (vRNA) copies per microliter of cDNA. PCR amplification was performed using Platinum Pfx DNA polymerase (Invitrogen) and primers Bm-HA1 and EHA1007rw (5′TTGGGGCATTTTCCATATGT3′), spanning the region between nucleotide (nt) −43 (upstream of the HA1 start codon) and nt 1007. PCR amplification was performed for 40 cycles (94°C for 30 s, 55°C for 1 min, and 68°C for 1 min), followed by a final extension at 68°C for 10 min. PCR products were gel purified using the QIAquick gel extraction kit (Qiagen) and further cloned using the Zero-Blunt TOPO PCR cloning kit for sequencing (Invitrogen) following the manufacturer's instructions. Clones were sequenced at the Influenza Sequencing Pipeline established at the Wellcome Trust Sanger Institute.
Deep amplicon sequencing of HA1 from nasal swabs and controls.
Sequencing was performed using fluorescence sequencing chemistry and ABI 3730xl capillary sequencers. Forward and reverse sequencing reads from each clone were trimmed of vector sequence and poor-quality regions and assessed for quality; reads with an average Phred quality score (9, 10) of <20 were rejected. Overlapping forward and reverse reads were merged into a single contig with quality scores. Only contigs >900 bp in length were used for subsequent analyses. Contigs were aligned against the HA1 consensus sequence of an egg-grown isolate of A/Equine/Newmarket/1/93 using Ssaha2 (24), and high-quality variants were identified using the SNP_analysis.pm Perl module (http://sourceforge.net/projects/snpanalysis/) and our own Perl scripts. Nucleotide variants were considered real if their Phred score was >25. Nucleotides with a Phred score below that value were considered identical to the consensus nucleotide. Amino acid variants were considered real if all of the nucleotides in the codon had Phred scores of >25. Sequences containing high-quality insertions or deletions (indels) that altered the reading frame were counted and excluded from subsequent analyses.
Analysis of assay-related mutations.
To measure errors associated with the replication of plasmid DNA in bacteria and capillary sequencing, we sequenced 754 clones from a single plasmid clone. To assess for DNA polymerase errors during the PCR step, we amplified the HA1 segment from a single plasmid and cloned it into pCR4Blunt-TOPO as described above. This PCR product was cloned and sequenced as described above. To estimate mutations introduced during cDNA preparation, we performed in vitro transcription from a single plasmid clone template, using the Riboprobe in vitro transcription systems (Promega) following the manufacturers' instructions. In vitro-transcribed RNA was subject to RT-PCR, cloned, and sequenced as described above.
Evolutionary analysis.
A total of 2,366 intrahost EIV sequences were isolated from experimentally infected animals (EMBL data bank accession numbers FN398346 to FN400711), 372 intrahost EIV sequences isolated from natural cases (EMBL data bank accession numbers FN422006 to FN422377), and 158 epidemiological-scale sequences (see Table S1 in the supplemental material, obtained from the Influenza Virus Resource [http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html]) extending from the start codon of the HA open reading frame to nucleotide position 903 were collated. Sequence alignments were generated from the Ssaha2 output for the intrahost EIV sequences and using Se-Al (http://tree.bio.ed.ac.uk/software/seal/) for the EIV population sequences. Because of the very small genetic distances involved, the mean pairwise genetic diversity within each sample was calculated from the uncorrected pairwise distance matrix (p-distance) between taxa (available from the authors upon request). Maximum likelihood (ML) trees were estimated using the PAUP* 4.0b10 package (38) under the best-fit model of nucleotide substitution determined using Modeltest (27). For those data sets representing individual horses, expansive tree bisection and reconnection (TBR) branch swapping was used in all cases. However, for the extremely large data sets combining horses, simpler nearest-neighbor interchange (NNI) branch swapping was utilized to provide computational tractability. The mean numbers of nonsynonymous substitutions per site (dN) and synonymous substitutions per site (dS) (ratio dN/dS) were estimated using the single likelihood ancestor counting (SLAC) algorithm available in the Datamonkey web interface of the HyPhy software package (26). Minimum spanning trees allowing hypothetical intermediate nodes were calculated from the sequence data using Prim's algorithm in Bionumerics V4.5 (Applied Maths, Belgium) as described previously (29).
RESULTS
Transmission of equine influenza virus in naïve horses.
Our experimental system was designed to emulate natural transmission events of a virus in its natural host, and with two separate chains of transmission initiated from the first infected horse, internal comparison of any variation observed was also possible. Two naïve horses were experimentally infected with equine influenza virus A/Equine/Newmarket/1/93 (Fig. 1A), and then one of these two horses was housed with two other naïve horses, which were then separated and used to initiate two transmission chains (Fig. 1A). The aim of this first step was to readapt the egg-grown inocula to the horse. Of the 10 exposed animals, nine became infected, indicating that transmission under these conditions was efficient. Viral shedding was determined from copies of viral RNA in nasal swabs collected daily. The peak of excretion—in which millions of viral particles were detected in a single nasal swab—was usually observed around 72 h postcontact (hpc) (Fig. 1B). Thus, the sizes of the within-host viral populations are extremely large, particularly since nasal swabs capture only a very small proportion of the total population at a particular time point. In one exposure, only one of two recipients (horse 5257) became infected despite the fact that the donor displayed high levels of viral shedding (Fig. 1B). Horse 5257 subsequently exhibited lower levels of viral shedding (Fig. 1B), but it transmitted the virus to two contacts which appeared to shed less virus and showed a marked delay to reach the peak of viral shedding.
Intrahost genetic variation.
To determine the extent and structure of viral genetic diversity within infected horses, we examined the variation among multiple clones prepared from the RNA sequences of the first 903 nucleotides of the HA1 gene. Because nasal swabs displayed variable amounts of virus, we restricted our analysis to those days in which the viral populations were large enough to allow PCR amplification and further sequencing. Between 44 and 154 clones were prepared and sequenced from the virus collected in nasal swabs from each horse on each day examined, resulting in a total of 2,366 individual HA1 sequences. Table 1 summarizes the levels of sequence variability present in samples examined, as well as the relative numbers of synonymous and nonsynonymous substitutions per site as a measure of selection pressure.
TABLE 1.
Analysis of intrahost variation in EIV sequences from the transmission experiment
| Horse | Chaina | Dayb | No. of sequences | Total no. of nt mutationsc | Mean pairwise distance ± SE | Global dN/dS | No. of stop codons | No. of indelsd | No. of sites with >1 nonsyn. mutatione |
|---|---|---|---|---|---|---|---|---|---|
| 7D36f | A-1/B-1 | 3 | 152 | 29 | 0.00042 ± 0.00001 | 0.71 | 4 | 1 | 0 |
| 5 | 82 | 18 | 0.00048 ± 0.00002 | 1.04 | 0 | 0 | 1 | ||
| 7248 | A-2 | 3 | 154 | 15 | 0.00022 ± 0.00000 | 0.72 | 0 | 0 | 0 |
| 4 | 65 | 3 | 0.00010 ± 0.00001 | 0.20 | 0 | 0 | 0 | ||
| 5 | 154 | 24 | 0.00034 ± 0.00001 | 0.37 | 0 | 0 | 1 | ||
| 6 | 62 | 9 | 0.00032 ± 0.00001 | 0.37 | 0 | 0 | 1 | ||
| 5447 | A-3 | 3 | 69 | 4 | 0.00013 ± 0.00001 | 1.81 | 0 | 0 | 0 |
| 4 | 44 | 9 | 0.00045 ± 0.00003 | 1.25 | 0 | 0 | 0 | ||
| 5 | 50 | 12 | 0.00053 ± 0.00002 | 0.58 | 1 | 0 | 0 | ||
| 6 | 51 | 31 | 0.00123 ± 0.00003 | 1.11 | 2 | 0 | 2 | ||
| 7C1C | A-3 | 2 | 52 | 6 | 0.00026 ± 0.00001 | 0.56 | 0 | 0 | 0 |
| 3 | 54 | 8 | 0.00033 ± 0.00002 | 1.13 | 0 | 0 | 0 | ||
| 4 | 83 | 12 | 0.00032 ± 0.00001 | 0.90 | 0 | 0 | 2 | ||
| 6 | 75 | 14 | 0.00041 ± 0.00001 | 1.10 | 1 | 0 | 2 | ||
| 2F50 | A-4 | 3 | 49 | 4 | 0.00018 ± 0.00001 | 0.29 | 0 | 0 | 0 |
| 4 | 54 | 9 | 0.00041 ± 0.00002 | 0.13 | 0 | 0 | 0 | ||
| 5 | 80 | 13 | 0.00036 ± 0.00001 | 3.40 | 0 | 0 | 0 | ||
| 7 | 46 | 7 | 0.00033 ± 0.00002 | 0.34 | 0 | 0 | 0 | ||
| 7A45 | A-4 | 3 | 107 | 13 | 0.00027 ± 0.00001 | 0.58 | 0 | 0 | 0 |
| 6005 | B-2 | 3 | 67 | 9 | 0.00033 ± 0.00001 | N/Ag | 1 | 0 | 2 |
| 4 | 81 | 14 | 0.00038 ± 0.00001 | 3.49 | 1 | 1 | 1 | ||
| 5 | 52 | 7 | 0.00029 ± 0.00002 | N/A | 1 | 0 | 1 | ||
| 6 | 72 | 27 | 0.00073 ± 0.00002 | 1.98 | 0 | 0 | 3 | ||
| 5257 | B-3 | 2 | 112 | 16 | 0.00034 ± 0.00001 | 0.30 | 1 | 0 | 1 |
| 3 | 127 | 21 | 0.00038 ± 0.00001 | 0.40 | 3 | 0 | 0 | ||
| 780C | B-4 | 3 | 71 | 18 | 0.00056 ± 0.00002 | 0.85 | 0 | 0 | 0 |
| 5D1A | B-4 | 4 | 54 | 4 | 0.00016 ± 0.00001 | N/A | 0 | 0 | 0 |
| 5 | 81 | 12 | 0.00033 ± 0.00001 | 0.87 | 2 | 0 | 1 | ||
| 6 | 79 | 14 | 0.00039 ± 0.00001 | 1.60 | 0 | 0 | 3 | ||
| 0443f | 0 | 2 | 87 | 10 | 0.00026 ± 0.00001 | 0.85 | 0 | 0 | 0 |
Horses 7D36 and 0443 were experimentally infected, and the other horses were infected in a chain as described in Materials and Methods. The chain is indicated as follows: A or B for branch A or B. The number indicates the link in the chain (see Fig. 1A); for example, horse 7248 was the second horse to be infected from exposure to horse 7D36.
Day postinoculation (for inoculated animals) or day after initial contact with donor horse.
nt, nucleotide.
indels, insertions or deletions.
nonsyn., nonsynonymous.
Experimentally inoculated animals.
N/A, not available.
Overall, we observed a total of 392 mutations, which results in a mutation frequency of 1.8 × 10−4 mutations per nucleotide site. Each sample (i.e., nasal swab) contained a mixture of closely related genomes, with most clones being identical to each other, and a proportion (14.2%) containing 1 or sometimes 2 mutations (Fig. 1C). Only a few clones contained three, four, and six nucleotide mutations. Mutations were present at 246 nucleotide positions along the HA gene coding region examined. In most samples, the number of nonsynonymous mutations was greater than the number of synonymous mutations (Fig. 1D). Many clones shared a single mutation either in the same sample or in a different sample, and only two clones shared two mutations (C476T [Ala144Val] and A884G [Glu280Gly]), although these were derived from the same swab (animal 6005, day 6 postcontact).
We observed a gradual increase in the complexity of genetic diversity over the course of infection in individual animals, manifested as an increase in the number of synonymous and nonsynonymous mutations, the number of clones possessing a single mutation and multiple mutations, and the average pairwise divergence within samples (Table 1). Of note, the average pairwise distance is always significantly higher at the end of the infection period than at the beginning (P < 0.001 after correcting for multiple tests using Bonferroni's correction) in individual animals for which we had 3 or more samples. Some mutations were observed at the same site on multiple days from the same horse, and some of these mutations were also observed in other horses (Tables 2 and 3). Of the 392 detected mutations, 144 (distributed in 43 nucleotide sites) were observed in more than one of the 30 independent samples.
TABLE 2.
Intrahost EIV mutations present on multiple days
| Horse | Mutation (amino acid change) | Daya | No. of clones | Mutation present at the epidemiological scale? | Mutation variable at the epidemiological scale?d | Other horses (days) with the same mutatione |
|---|---|---|---|---|---|---|
| 7248 | A228Gb | 4 | 1 | 5257 (D3), 7C1C (D5) | ||
| 5 | 1 | |||||
| A855Gb | 3 | 1 | 5447 (D5) | |||
| 5 | 1 | |||||
| 2F50 | A231Gb | 4 | 2 | |||
| 7 | 2 | |||||
| 5447 | T29C (Leu10Proc) | 3 | 1 | No | No (L) | 7D36 (D5) |
| 5 | 1 | |||||
| G289A (Glu82Lys) | 4 | 1 | Yes | Yes (E, G, K) | ||
| 6 | 1 | |||||
| C407T (Thr121Ile) | 4 | 1 | Yes | Yes (I, M, T) | 5D1A (D5) | |
| A406C (Thr121Pro) | 6 | 1 | No | |||
| 6005 | C49A (Gln2Lys) | 4 | 1 | No | No (Q) | |
| 6 | 11 | |||||
| A61G (Ser6Gly) | 3 | 2 | Yes | Yes (D, G, N, S) | ||
| 4 | 4 | |||||
| 5 | 1 | |||||
| A884G (Glu280Gly) | 3 | 2 | No | Yes (E, K) | 0443 (D2), 7D36 (D5), 5257 (D2) | |
| 4 | 1 | |||||
| 5 | 3 | |||||
| 6 | 11 | |||||
| 5D1A | A413G (Glu123Gly) | 4 | 1 | No | No (E) | |
| 5 | 1 |
Day after initial contact with donor horse.
Synonymous mutations.
Signal peptide (absent in mature HA1).
If there was no variability, the invariable amino acid is shown in parentheses. If there was variability, the letters in parentheses list the alternate amino acids.
The number of the horse with the same mutation is shown. The day that the same mutation is shown in parentheses (e.g., D3, day 3).
TABLE 3.
Intrahost EIV nonsynonymous mutations present in multiple horses
| Mutation (amino acid change) | Horse (day) | Direct contact? | Horses from same chain? | Motifa |
|---|---|---|---|---|
| T26C (Leu9Prob) | 7248 (5) | No | No | |
| 5D1A (5) | ||||
| T29C (Leu10Prob) | 7D36 (5) | No | Yes | |
| 5447 (3) | ||||
| 5447 (5) | ||||
| T257C (Leu71Pro) | 5257 (3) | Yes | Yes | |
| 5D1A (6) | ||||
| T337C (Tyr98His) | 7D36 (5) | No | Yes | |
| 5D1A (6) | ||||
| T358C (Tyr105His) | 5447 (3) | Yes | Yes | |
| 7C1C (3) | ||||
| C370T (Arg109Trp) | 7C1C (3) | No | No | |
| 780C (3) | ||||
| C395G (Tyr117Arg) | 6005 (4) | No | No | |
| 7A45 (3) | ||||
| C407T (Thr121Ile) | 5447 (4) | No | No | Ag A |
| 5D1A (5) | ||||
| G409A (Ala122Thr) | 6005 (5) | No | Yes (6005 and 5D1A) | Ag A |
| 5447 (3) | ||||
| 5D1A (5) | ||||
| T460C (Cys139Arg) | 6005 (5) | No | Yes | |
| 5D1A (6) | ||||
| G478A (Asp145Asn) | 7C1C (6) | Yes | Yes | Ag A |
| 5447 (6) | ||||
| T497G (Leu151Arg) | 5257 (2) | No | No | |
| 7C1C (3) | ||||
| C545T (Thr167Ile) | 7D36 (3) | Yes | Yes | Gly |
| 7248 (3) | ||||
| A635G (Gln197Arg) | 7248 (6) | No | No | Ag B |
| 780C (3) | ||||
| T709A (Trp222Arg) | 7D36 (5) | No | Yes | |
| 5447 (6) | ||||
| A730G (Arg229Gly) | 7D36 (3) | Yes (7D36 and 7248) | Yes (all) | Receptor binding associated |
| 7248 (5) | ||||
| 2F50 (5) | ||||
| A835G (Lys264Glu) | 0443 (2) | Yes (7D36 and 0443, 7D36 and 7248) | Yes (all) | |
| 7D36 (5) | ||||
| 7248 (5) | ||||
| 2F50 (5) | ||||
| C863T (Pro273Leu) | 780C (3) | No | No | Ag C |
| 2F50 (7) | ||||
| G868A (Asp275Asn) | 0443 (2) | No | Yes? | Ag C |
| 7248 (3) | ||||
| A884G (Glu280Gly) | 0443 (2) | Yes (0443 and 7D36, 7D36 and 6005, 6005 and 5257) | Yes (all) | |
| 7D36 (5) | ||||
| 6005 (3) | ||||
| 6005 (4) | ||||
| 6005 (5) | ||||
| 6005 (6) | ||||
| 5257 (2) |
Ag, antigenic site (three antigenic sites [sites A, B, and C]); Gly, glycosylation site.
Signal peptide (absent in mature HA1).
To estimate the selection pressures affecting EIV within its natural host, we calculated the mean value of dN/dS for each sample (Table 1). Although individual values were variable, mean dN/dS for the data as a whole was 0.80, close to the expected value of ∼1 expected under an entirely neutral evolutionary process. This value (and the majority of the individual values) was higher than the 0.41 value estimated for epidemiological populations of EIV H3N8, suggesting that different selection pressures act at the individual level than at the population level, with purifying selection dominating the latter. However, since our intrahost data also contains some artifactual mutations that are invariably introduced due to the experimental procedure undertaken (see supplemental material), all conclusions on the overall extent and structure of genetic diversity need to be drawn with caution. When we compared the sites and mutations that arose in the experimental infections, most nonsynonymous mutations were observed as single changes in one sequence (singletons) in sites that were not polymorphic in the public database of equine H3N8 sequences (epidemiological-scale variation), suggesting that they may have been introduced in vivo and hence are likely to represent transient deleterious mutations or may have been introduced during the preparation of the samples. This notion is also supported by the presence of sequences containing mutations that either generated premature stop codons or frameshifts (Table 1).
Patterns of intra- and interhost viral evolution in infected horses.
We investigated viral variation during infection of animals at different stages in the transmission chain. In all cases, the majority of sequences recovered were identical to the consensus sequence. In 21 of 30 data sets from individual swabs, predominantly single-nucleotide (or occasionally double-nucleotide) changes were seen from the consensus sequence. Importantly, in nine of the sets of sequences, some of the variation was seen to give rise to small phylogenetic clusters that were distinct from the consensus sequence. Hence, during infection of individual hosts, the viral populations consist of very closely related genomes, generated by an ongoing process of mutation, some of which then acquire additional changes, producing phylogenetic structure. Such phylogenetic structure is strongly suggestive of natural, rather than artifactual, variation.
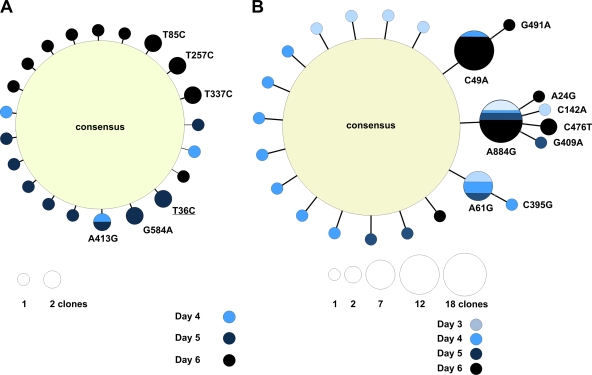
To consider the dynamic properties of the within-host influenza virus populations, we examined sequence variation from animals for which more than one virus-positive daily nasal swab was available. Again, most sequences detected were identical to the consensus sequence, and variants generated through the course of infection mostly consisted of singletons. For example, for horse 5D1A (for which we had three consecutive swabs and 214 sequences), we detected 24 clones bearing a single mutation compared to the consensus sequence and three sequences displaying two mutations. Of this entire mutational spectrum, only one mutation (A413G [Glu123Gly]) was detected on two different days, suggesting that it was either generated de novo twice, or more likely, persisted through 2 days of the infection period (Fig. 2A). Maximum likelihood phylogenetic trees for all the animals included in the transmission study are shown in Fig. S1 in the supplemental material.
FIG. 2.
Intrahost EIV populations are constituted by a mixture of closely related genomes. Minimum spanning trees of horses 5D1A (A) and 6005 (B). Each tree was inferred by compiling sequences from multiple days. With the exception of singletons, mutations are indicated on respective branches. Synonymous mutations are underlined. The size of the circle is proportional to the number of sequences that exhibit each variant.
We also repeatedly detected the same mutations in different animals. Of a total of 20 repeatedly observed nonsynonymous mutations, 17 were observed in animals linked through transmission (Table 3 and Fig. 1A). In many cases, such mutations were located at internal nodes of a phylogenetic tree linking these sequences, suggesting that they gave rise to subsequent variants. For example, nonsynonymous mutation A61G (Ser6Gly) was detected in animal 6005 on days 3, 4, and 5 after contact, with one sequence containing an additional nonsynonymous mutation (C395G [Thr117Arg]) on day 4 (Fig. 2B). Animal 5447 constitutes an interesting case, as on day 6 postcontact, two lineages that differed from the consensus sequence were observed, and the difference between both branches was a nonsynonymous mutation at Asp145 (see Fig. S1C in the supplemental material). One variant comprised six clones harboring Tyr145, while the other branch exhibited eight clones with Asn145. Interestingly, position 145 lies within antigenic site A of the HA gene (42); therefore, mutations in that position could alter the antigenic structure of the virus. More generally, the discovery that the mutations in that antigenic site were detected 6 days after contact between the infected and uninfected horses, together with their relatively high frequency, is compatible with the action of immunological selection by the nascent adaptive immune response, as a significant rise in serum antibodies is readily detected in immunologically naïve horses at day 7 postinfection (13). Mutations at this site were not detected in the donor horse (7248), but one clone bearing an Asp145Asn change was seen on day 6 postcontact in horse 7C1C, which was housed with horse 5447 and exposed at the same time, suggesting that the variant was transmitted between these two horses. This indicates that novel lineages bearing distinct antigenicity can evolve within a single individual despite the short infection period of influenza.
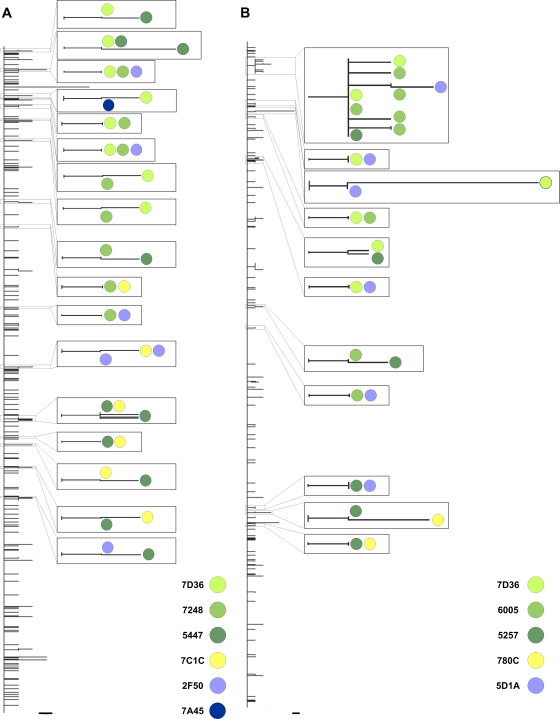
The observation of intrahost viral populations comprising more than one lineage in different horses, together with the detection of distinct variants harboring a common mutation(s) in different animals (Table 3), prompted us to determine whether distinct lineages were being transmitted between animals or if they were generated de novo within individual horses. We therefore compiled all available data sets from each branch of the transmission chain and determined their evolutionary relationships. Phylogenetic analysis revealed that some variants from different animals shared a common ancestor (Fig. 3A and B), indicating that multiple lineages were transmitted between these animals (and a pattern that is highly unlikely to occur through experimental error [see supplemental material]). This result is of great importance, as it means that multiple viral lineages are transmitted between hosts, which in turn means that transmission bottlenecks may not be especially narrow in the case of EIV.
FIG. 3.
Unrooted maximum likelihood phylogenetic trees for HA1 segment clones from samples obtained from chain 1 (A) and chain 2 (B). Branch lengths are drawn to scale, with those displaying variants detected in more than one animal magnified. Colored circles represent different animals. Horizontal branch lengths are drawn to a scale of nucleotide substitutions per site. Bars = 0.001 substitution/site.
Detection of within-host variants of equine influenza virus associated with the emergence of canine influenza virus.
The HA protein mediates attachment and entry into target cells and is a significant determinant of host range specificity (36). Canine influenza virus (CIV) emerged as a respiratory pathogen of dogs shortly after 2000 from an apparent direct interspecies transfer of an H3N8 equine influenza virus into dogs (5). Contemporary CIV and EIV share >96% sequence identity in all eight segments (5, 25), with three signature amino acid substitutions located within the stretch of 301 amino acid residues of the HA protein that we examined: Asn54Lys, Asn83Ser and Trp222Leu (5, 25). Strikingly, we found one variant bearing the Asn83Ser mutation in one horse and three other variants harboring mutations in position 222, although none of the latter exhibited leucine in that position (Trp222Gly, Trp222Arg, and Trp222stop). No mutations in the codon for Asn54 were detected. Substitution Ser92Asn, which is present in Canine/FL/04, was also detected, and a substitution of Gly7, albeit different from the one present in Canine/FL/03, was also observed.
In addition to the three HA signature substitutions that differentiate contemporary CIV and EIV isolates, there are nine further amino acid changes between the consensus sequence of the virus used in this study and other CIVs isolated in United States since 2003: Val14Ala (in the signal peptide), Thr5Ile, Thr30Ser, Ile48Met, Val58Ile, Val78Ala, Asn159Ser, Gln190Glu, and Glu193Lys. Of these, we detected substitutions Thr5Ile, Ile48Met, and Val78Ala in samples of within-host EIVs. Six clones harbored nonsynonymous mutations in the codon corresponding to Thr30, although serine was not present in any of them. Other nucleotide substitutions present in individual CIV isolates were also detected. Hence, the genetic diversity required to successfully jump the species barrier and infect a new host may sometimes be partially or completely present in viruses within individuals of the donor species.
Analysis of within-host variation in natural field cases.
To determine whether the variation observed within our experimental setting was comparable to that observed in the field, we analyzed equine H3N8 influenza virus in five nasal swabs from horses naturally infected during an outbreak that took place in England in 2003 (23). Although the within-sample variation detected was similar to that observed in the experimental infections (Table 4), important differences were observed. First, we detected two distinct consensus viral populations that differed in one amino acid position: while Arg62 was present at the consensus level in three samples, the other two samples exhibited Lys62. Further, one of the natural cases examined (horse OB151) exhibited an unusually high level of intrahost viral diversity, with nine clones harboring nine mutations, raising the possibility of a mixed infection. The mixed infection in that case was confirmed by phylogenetic analysis of sequences of 158 EIV epidemiological isolates, along with the 73 intrahost sequences derived from this sample. Two phylogenetically distinct viral lineages were observed in this isolate, and those grouped with different clades of the Florida sublineage (Fig. 4). This represents the first description of mixed infection in EIV, a process that also provides the raw material for segment reassortment. In addition, since these two viruses exhibited four nonsynonymous substitutions within HA1, it is possible that they also differ antigenically, increasing the likelihood of emergence of novel antigenic variants with outbreak potential.
TABLE 4.
Analysis of patterns of intrahost variation in EIV sequences from natural field cases
| Horse | No. of sequences | Total no. of nt mutationsa | Mean pairwise distance | Global dN/dS | No. of stop codons | No. of indelsb | No. of sites with >1 nonsyn. mutationc |
|---|---|---|---|---|---|---|---|
| OB150 | 68 | 6 | 0.0001 | 0.43 | 1 | 0 | 0 |
| OB151 | 73 | 145 | 0.002 | 0.35 | 0 | 0 | 5 |
| OB152 | 82 | 8 | 0.0002 | 0.58 | 0 | 0 | 0 |
| OB153 | 75 | 92 | 0.0005 | 0.49 | 0 | 0 | 2 |
| OB172 | 74 | 6 | 0.0001 | 0.42 | 0 | 0 | 1 |
Total number of nucleotide (nt) mutations relative to the reference sequence A/Equine/Newmarket/5/2003 (GenBank accession number FJ375213).
indels, insertions or deletions.
nonsyn., nonsynonymous.
FIG. 4.
Maximum likelihood phylogenetic tree for HA1 segment clones from a natural case of EIV (sample OB151) compared with the global phylogeny of EIV H3N8. Colored boxes represent distinct EIV phylogenetic groups according to the World Organization for Animal Health (OIE) nomenclature of H3N8 strains. Clones from sample OB151 are represented in red boxes. Bootstrap values (>70%) are shown for key nodes relating to the phylogenetic position of those sequences sampled from horse OB151. Horizontal branches are drawn to a scale of nucleotide substitutions per site, and the tree is rooted on the EIV Miami isolate from 1963. Bar = 0.001 substitution/site.
DISCUSSION
Human cases of avian H5N1 and the recently isolated H1N1 of swine origin constitute current examples of zoonotic infections with pandemic potential (2, 6). However, the determining factors that allow only a small subset of emerging viruses to become established in a new host species are still unclear. For example, although swine influenza viruses periodically spill over into humans, few have ever caused sustained outbreaks (34). Although it is clear that ecological factors can play a key role in viral emergence, it is likely that there is a genetic basis to cross-species transmission in most cases. Accordingly, it is important to understand whether the mutations required for successful host emergence can arise in the natural reservoir or if they arise to significant levels within the new host only after spillover events, particularly in long chains of transmission, should they exist (1).
Here, we applied a clonal sequencing approach to estimate the intrahost mutational spectrum of EIV in its natural host and its implications for viral emergence. The novel patterns of cross-scale viral variability revealed underlines the power of experimental transmission and beyond-consensus viral sequencing to elucidate phylodynamic patterns (12). Analysis of multiple samples taken from the same animal at different times postinfection allowed us to study the evolutionary dynamics of EIV over very short time periods. The HA protein, as the viral receptor binding protein, is a critical host range determinant and a major target of neutralizing antibodies. Even allowing for the presence of artifactual mutations introduced through error-prone reverse transcription, our results show that the HA1 segment of EIV exhibits significant variation during the course of infection, with up to 13% of the clones sequenced harboring single mutations and ∼1.4% of the total sequences exhibiting two mutations compared with the consensus population. The high frequency with which singletons fall into sites that are invariant at the population level, together with the high dN/dS values estimated for the intrahost data set and for the epidemiological-scale viruses, strongly suggest that most mutations are deleterious and will ultimately be removed by purifying selection. In contrast, some other mutations were detected on multiple days and persisted throughout infection, indicating that they were either neutral or advantageous. In particular, in one animal, we observed a high frequency of nonsynonymous mutations within an antigenic site at day 6 postcontact, suggesting that immunological selection may take place in the late stages of infection, when the evolutionary infectivity profile—the net transmission rate of immunologically selected variants, which results from the interaction between viral adaptation and immune history—will be the highest (12). Transmission studies of vaccinated or previously infected animals will be key to determine the role of immunity on within-host variation and its impact on emergence of antigenic variants.
Clearly, some of the observed mutations were likely introduced during reverse transcription and PCR amplification. However, it is highly unlikely that these experimental artifacts would result in introduction of repeated changes in the same sites in different samples, particularly in those that follow the transmission chain. On the other hand, if artifactual mutations were commonly introduced as a result of RNA secondary structure, one might expect a higher proportion of mutations at specific sites than the distribution observed here. Indeed, the finding that mutations are passed on in a manner that matches the transmission chain, together with a gradual increase in diversity along the course of infection, acts as an independent verification of the validity of this method for examining viral variation. Finally, our results from control experiments of artifactual mutations (see supplemental material) are also in agreement with those of Descloux et al. who also evaluated in vitro-introduced mutations during reverse transcription and PCR in a study of intrahost genetic diversity in dengue virus (7).
The frequency of individual mutations at any nucleotide site was only 1.8 × 10−4, which is similar to that observed by Iqbal et al. (17) where within-host variability of avian influenza viruses was assessed in vivo using a variety of avian species. However, because our sampling depth allowed consistent detection of only the more common (>5 to 10% of the population) variants, it is likely that greater variation exists at a lower frequency than is reported here. Further studies, including deeper sequencing of the entire virus genome within the host, will therefore be essential to better understand the likelihood of emergence, as all genomic segments are likely to play distinct roles in host range specificity (32, 35).
On the basis of these results, we conclude that influenza viral populations continually generate the variants that would enable them to adapt to new host species, although most are purged by purifying selection. Indeed, we detected mutations in HA1 associated with the emergence of canine influenza virus within the EIV mutational spectra. As infection progresses, the mutational spectrum gradually gains complexity, and the frequency of certain variants in the viral population increases. Hence, individual infections of EIV result in an ongoing evolutionary process. The fact that within our experimental setting, we did not detect fixed substitutions contrasts with our observations in natural cases collected from horses infected during the same outbreak, where intrahost viral populations differed in one amino acid in different animals, suggesting that replacement of the consensus population may require longer chains of transmission.
By studying genetic variation both within and among hosts, we are also able to examine the process of evolution during interhost transmission. Importantly, our observation that clones bearing common mutations are found in animals that are directly linked by transmission demonstrates that multiple viral lineages are passed among animals and hence that transmission bottlenecks are not always narrow, such as down to the level of a single virion as proposed for HIV (19). However, the size of the transmission bottlenecks may vary depending on equine management; the bottleneck may be narrower in horses with less direct contact, such as individually stabled animals or animals at pasture. The transmission of multiple influenza viral lineages will also assist in the process of viral emergence, as the probability that new host species will be exposed to mutations of adaptive value will be greater and natural selection will act more efficiently. Along the same line, loose transmission bottlenecks will allow efficient immune selection if individuals with different immunological histories are exposed to antigenic mutations. The timing of transmission may therefore also play a role in the extent of initial viral diversity, such that the later transmission takes place, the more diverse the transmitted viral population is. In addition, the occurrence of mixed infection, as we describe here for EIV in nature, provides the raw material for reassortment (3). Finally, we suggest that understanding the dynamics of within-host variation will assist in developing the theoretical tools required to predict and therefore avoid or control the emergence of new viruses in humans and animals.
Supplementary Material
Acknowledgments
This work was supported by a grant from DEFRA and HEFCE under the Veterinary Training and Research Initiative to the Cambridge Infectious Disease Consortium (CIDC) and also by a program grant from the Wellcome Trust. P.R.M. is supported by a Wellcome Trust Veterinary Postdoctoral Fellowship. J.L.N.W. is supported by the Alborada Trust. J.D. was supported by the Horserace Betting Levy Board equine influenza virus surveillance program. B.T.G. and J.L.N.W. were supported by the RAPIDD program of the Science & Technology Directorate, Department of Homeland Security, and the Fogarty International Center, National Institutes of Health. B.T.G. was also supported by grants NSF0742373 and NIH R01 GM083983-01. E.C.H. and C.R.P. were supported by grant R01 GM080533.
Footnotes
Published ahead of print on 5 May 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Antia, R., R. R. Regoes, J. C. Koella, and C. T. Bergstrom. 2003. The role of evolution in the emergence of infectious diseases. Nature 426:658-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beigel, J. H., J. Farrar, A. M. Han, F. G. Hayden, R. Hyer, M. D. de Jong, S. Lochindarat, T. K. Nguyen, T. H. Nguyen, T. H. Tran, A. Nicoll, S. Touch, and K. Y. Yuen. 2005. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 353:1374-1385. [DOI] [PubMed] [Google Scholar]
- 3.Bryant, N. A., A. S. Rash, C. A. Russell, J. Ross, A. Cooke, S. Bowman, S. Macrae, N. S. Lewis, R. Paillot, R. Zanoni, H. Meier, L. A. Griffiths, J. M. Daly, A. Tiwari, T. M. Chambers, J. R. Newton, and D. M. Elton. 2009. Antigenic and genetic variations in European and North American equine influenza virus strains (H3N8) isolated from 2006 to 2007. Vet. Microbiol. 138:41-52. [DOI] [PubMed] [Google Scholar]
- 4.Cook, R. F., R. Sinclair, and J. A. Mumford. 1988. Detection of influenza nucleoprotein antigen in nasal secretions from horses infected with A/equine influenza (H3N8) viruses. J. Virol. Methods 20:1-12. [DOI] [PubMed] [Google Scholar]
- 5.Crawford, P. C., E. J. Dubovi, W. L. Castleman, I. Stephenson, E. P. Gibbs, L. Chen, C. Smith, R. C. Hill, P. Ferro, J. Pompey, R. A. Bright, M. J. Medina, C. M. Johnson, C. W. Olsen, N. J. Cox, A. I. Klimov, J. M. Katz, and R. O. Donis. 2005. Transmission of equine influenza virus to dogs. Science 310:482-485. [DOI] [PubMed] [Google Scholar]
- 6.Dawood, F. S., S. Jain, L. Finelli, M. W. Shaw, S. Lindstrom, R. J. Garten, L. V. Gubareva, X. Xu, C. B. Bridges, and T. M. Uyeki. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605-2615. [DOI] [PubMed] [Google Scholar]
- 7.Descloux, E., V. M. Cao-Lormeau, C. Roche, and X. De Lamballerie. 2009. Dengue 1 diversity and microevolution, French Polynesia 2001-2006: connection with epidemiology and clinics. PLoS Negl. Trop. Dis. 3:e493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 9.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 10.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 11.Garten, R. J., C. T. Davis, C. A. Russell, B. Shu, S. Lindstrom, A. Balish, W. M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C. B. Smith, S. L. Emery, M. J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D. F. Burke, R. A. Fouchier, C. Pappas, C. M. Alpuche-Aranda, H. Lopez-Gatell, H. Olivera, I. Lopez, C. A. Myers, D. Faix, P. J. Blair, C. Yu, K. M. Keene, P. D. Dotson, Jr., D. Boxrud, A. R. Sambol, S. H. Abid, K. St. George, T. Bannerman, A. L. Moore, D. J. Stringer, P. Blevins, G. J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H. F. Guevara, E. A. Belongia, P. A. Clark, S. T. Beatrice, R. Donis, J. Katz, L. Finelli, C. B. Bridges, M. Shaw, D. B. Jernigan, T. M. Uyeki, D. J. Smith, A. I. Klimov, and N. J. Cox. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenfell, B. T., O. G. Pybus, J. R. Gog, J. L. Wood, J. M. Daly, J. A. Mumford, and E. C. Holmes. 2004. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303:327-332. [DOI] [PubMed] [Google Scholar]
- 13.Hannant, D., J. A. Mumford, and D. M. Jessett. 1988. Duration of circulating antibody and immunity following infection with equine influenza virus. Vet. Rec. 122:125-128. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275-2289. [DOI] [PubMed] [Google Scholar]
- 15.Holmes, E. C., E. Ghedin, N. Miller, J. Taylor, Y. Bao, K. St. George, B. T. Grenfell, S. L. Salzberg, C. M. Fraser, D. J. Lipman, and J. K. Taubenberger. 2005. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 3:e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17.Iqbal, M., H. Xiao, G. Baillie, A. Warry, S. C. Essen, B. Londt, S. M. Brookes, I. H. Brown, and J. W. McCauley. 2009. Within-host variation of avian influenza viruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:2739-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Keele, B. F., E. E. Giorgi, J. F. Salazar-Gonzalez, J. M. Decker, K. T. Pham, M. G. Salazar, C. Sun, T. Grayson, S. Wang, H. Li, X. Wei, C. Jiang, J. L. Kirchherr, F. Gao, J. A. Anderson, L. H. Ping, R. Swanstrom, G. D. Tomaras, W. A. Blattner, P. A. Goepfert, J. M. Kilby, M. S. Saag, E. L. Delwart, M. P. Busch, M. S. Cohen, D. C. Montefiori, B. F. Haynes, B. Gaschen, G. S. Athreya, H. Y. Lee, N. Wood, C. Seoighe, A. S. Perelson, T. Bhattacharya, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuiken, T., E. C. Holmes, J. McCauley, G. F. Rimmelzwaan, C. S. Williams, and B. T. Grenfell. 2006. Host species barriers to influenza virus infections. Science 312:394-397. [DOI] [PubMed] [Google Scholar]
- 21.Mumford, J. A., D. Hannant, and D. M. Jessett. 1990. Experimental infection of ponies with equine influenza (H3N8) viruses by intranasal inoculation or exposure to aerosols. Equine Vet. J. 22:93-98. [DOI] [PubMed] [Google Scholar]
- 22.Nelson, M. I., and E. C. Holmes. 2007. The evolution of epidemic influenza. Nat. Rev. Genet. 8:196-205. [DOI] [PubMed] [Google Scholar]
- 23.Newton, J. R., J. M. Daly, L. Spencer, and J. A. Mumford. 2006. Description of the outbreak of equine influenza (H3N8) in the United Kingdom in 2003, during which recently vaccinated horses in Newmarket developed respiratory disease. Vet. Rec. 158:185-192. [DOI] [PubMed] [Google Scholar]
- 24.Ning, Z., A. J. Cox, and J. C. Mullikin. 2001. SSAHA: a fast search method for large DNA databases. Genome Res. 11:1725-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Payungporn, S., P. C. Crawford, T. S. Kouo, L. M. Chen, J. Pompey, W. L. Castleman, E. J. Dubovi, J. M. Katz, and R. O. Donis. 2008. Influenza A virus (H3N8) in dogs with respiratory disease, Florida. Emerg. Infect. Dis. 14:902-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pond, S. L., and S. D. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 27.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 28.Rambaut, A., O. G. Pybus, M. I. Nelson, C. Viboud, J. K. Taubenberger, and E. C. Holmes. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roumagnac, P., F. X. Weill, C. Dolecek, S. Baker, S. Brisse, N. T. Chinh, T. A. Le, C. J. Acosta, J. Farrar, G. Dougan, and M. Achtman. 2006. Evolutionary history of Salmonella typhi. Science 314:1301-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell, C. A., T. C. Jones, I. G. Barr, N. J. Cox, R. J. Garten, V. Gregory, I. D. Gust, A. W. Hampson, A. J. Hay, A. C. Hurt, J. C. de Jong, A. Kelso, A. I. Klimov, T. Kageyama, N. Komadina, A. S. Lapedes, Y. P. Lin, A. Mosterin, M. Obuchi, T. Odagiri, A. D. Osterhaus, G. F. Rimmelzwaan, M. W. Shaw, E. Skepner, K. Stohr, M. Tashiro, R. A. Fouchier, and D. J. Smith. 2008. The global circulation of seasonal influenza A (H3N2) viruses. Science 320:340-346. [DOI] [PubMed] [Google Scholar]
- 31.Schafer, J. R., Y. Kawaoka, W. J. Bean, J. Suss, D. Senne, and R. G. Webster. 1993. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology 194:781-788. [DOI] [PubMed] [Google Scholar]
- 32.Scholtissek, C., H. Burger, O. Kistner, and K. F. Shortridge. 1985. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147:287-294. [DOI] [PubMed] [Google Scholar]
- 33.Scholtissek, C., I. Koennecke, and R. Rott. 1978. Host range recombinants of fowl plague (influenza A) virus. Virology 91:79-85. [DOI] [PubMed] [Google Scholar]
- 34.Shinde, V., C. B. Bridges, T. M. Uyeki, B. Shu, A. Balish, X. Xu, S. Lindstrom, L. V. Gubareva, V. Deyde, R. J. Garten, M. Harris, S. Gerber, S. Vagasky, F. Smith, N. Pascoe, K. Martin, D. Dufficy, K. Ritger, C. Conover, P. Quinlisk, A. Klimov, J. S. Bresee, and L. Finelli. 2009. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N. Engl. J. Med. 360:2616-2625. [DOI] [PubMed] [Google Scholar]
- 35.Subbarao, E. K., W. London, and B. R. Murphy. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki, Y., T. Ito, T. Suzuki, R. E. Holland, Jr., T. M. Chambers, M. Kiso, H. Ishida, and Y. Kawaoka. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 74:11825-11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reference deleted.
- 38.Swofford, D. L. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 39.Taubenberger, J. K., A. H. Reid, R. M. Lourens, R. Wang, G. Jin, and T. G. Fanning. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889-893. [DOI] [PubMed] [Google Scholar]
- 40.Waddell, G. H., M. B. Teigland, and M. M. Sigel. 1963. A new influenza virus associated with equine respiratory disease. J. Am. Vet. Med. Assoc. 143:587-590. [PubMed] [Google Scholar]
- 41.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiley, D. C., and J. J. Skehel. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365-394. [DOI] [PubMed] [Google Scholar]
- 43.Wood, J. M., G. C. Schild, C. Folkers, J. Mumford, and R. W. Newman. 1983. The standardization of inactivated equine influenza vaccines by single-radial immunodiffusion. J. Biol. Stand. 11:133-136. [DOI] [PubMed] [Google Scholar]
- 44.Woolhouse, M. E., D. T. Haydon, and R. Antia. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20:238-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.