Abstract
Ibalizumab is a humanized monoclonal antibody that binds human CD4, the primary receptor for human immunodeficiency virus type 1 (HIV-1). With its unique specificity for domain 2 of CD4, this antibody potently and broadly blocks HIV-1 infection in vitro by inhibiting a postbinding step required for viral entry but without interfering with major histocompatibility complex class II (MHC-II)-mediated immune function. In clinical trials, ibalizumab has demonstrated anti-HIV-1 activity in patients without causing immunosuppression. Thus, a characterization of the ibalizumab epitope was conducted in an attempt to gain insight into the underlying mechanism of its antiviral activity as well as its safety profile. By studying mouse/human chimeric CD4 molecules and site-directed point mutants of CD4, amino acids L96, P121, P122, and Q163 in domain 2 were found to be important for ibalizumab binding, with E77 and S79 in domain 1 also contributing. All these residues appear to cluster on the interface between domains 1 and 2 of human CD4 on a surface opposite the site where gp120 and the MHC-II molecule bind on domain 1. Separately, the epitope of M-T441, a weakly neutralizing mouse monoclonal antibody that competes with ibalizumab, was localized entirely within domain 2 on residues 123 to 125 and 138 to 140. The results reported herein not only provide an appreciation for why ibalizumab has not had significant adverse immunological consequences in infected patients to date but also raise possible steric hindrance mechanisms by which this antibody blocks HIV-1 entry into a CD4-positive cell.
The human immunodeficiency virus type 1 (HIV-1) epidemic continues to spread at the alarming rate of approximately 2.5 million new cases per year, despite intensive efforts from the scientific community. A safe and effective HIV-1 vaccine would be a key weapon to fight this epidemic; however, vaccine development has not yet proven successful. The extraordinary diversity of the virus, its capacity to evade adaptive immune responses, and the inability to induce broadly neutralizing antibodies against HIV-1 represent unprecedented challenges for vaccine development (3). Alternatively, the strategy of preexposure prophylaxis (PrEP) with antiretroviral drugs or even virus-specific immunoglobulins (Igs) (11) is gaining traction. Protection of rhesus macaques from challenge with simian immunodeficiency virus (SIV) has been observed after passive administration of anti-gp120 or anti-gp41 monoclonal antibodies, such as b12, 2G12, 2F5, and 4E10 (2, 20). However, the application of these antibodies as PrEP has been hindered due to their lack of potency or breadth or both. To this end, PrEP strategies could also consider antibodies to CCR5 (13) or CD4 (8, 12, 14), which have potent and broad inhibitory activities against HIV-1 without unwanted side effects.
The CD4 molecule, a cell surface glycoprotein found primarily on T lymphocytes, is the primary receptor for the HIV-1 envelope gp160 glycoprotein (7, 18). A member of the immunoglobulin superfamily (19), CD4 consists of an extracellular segment composed of four tandem immunoglobulin-like domains (D1, D2, D3, and D4), a single transmembrane span, and a short C-terminal cytoplasmic tail (15, 24). It is worth noting that both human major histocompatibility complex (MHC) class II (26) and HIV-1 gp120 (16, 24) bind to the same surface on the first domain (D1) of the CD4 molecule.
Ibalizumab (formerly known as TNX-355) is a humanized IgG4 monoclonal antibody that blocks HIV-1 entry by binding to human CD4 (8, 12, 14, 33). It was engineered from its mouse progenitor (5A8) by grafting the mouse complementary-determining region (CDR) onto a human IgG4 construct (4, 5). The IgG4 isotype was chosen to minimize the chances for CD4+ T-cell depletion by antibody- and complement-dependent cytotoxicity mediated by binding to Fc receptors. Ibalizumab or 5A8 blocks CD4-dependent virus entry and inhibits a broad spectrum of both laboratory-adapted and clinical HIV-1 isolates, including CCR5-tropic and CXCR4-tropic strains from multiple subtypes, with 50% inhibitory concentrations (IC50s) of 0.0004 to 0.152 μg/ml (4, 5). In vivo, treatment with ibalizumab prominently reduced plasma viremia in rhesus macaques infected with SIV, because this monoclonal antibody has equal affinity for rhesus CD4 (22, 23). In HIV-1 patients, single as well as multiple doses of ibalizumab resulted in substantial reductions (∼10-fold) in viral loads and increases in CD4+ T-cell counts without evidence of serious adverse effects or immunologic impairments (12, 14).
Efforts were made years ago to delineate the antibody binding site of 5A8 on human CD4 (hCD4) (5). Two stretches, amino acids (aa) 121 to 124 and aa 127 to 134, in domain 2 (D2) were found to be critical for binding. Since then, however, little work has been done to fine-map this epitope, and whether other parts of hCD4 are involved in the binding of this antibody has remained unexplored. The fact that an anti-hCD4-D2 antibody can noncompetitively, yet potently, block HIV-1 entry is intriguing, as viral gp120 binds to D1 of hCD4 (16, 24). Therefore, to gain a better understanding of the mechanism by which ibalizumab inhibits HIV-1 infection while avoiding undesired side effects, we sought to fine-map the epitope of this unique monoclonal antibody.
MATERIALS AND METHODS
Reagents.
Recombinant soluble hCD4 protein, CD4-IgG2, hCD4 gene, and TZM-bl cells were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Peptides PR14 (aa 121-PPGSSPSVQCRSPR-134) and TG26 (aa 115-TLTLESPPGSSPSVQCRSPRGKNIQG-140) were synthesized at SCI Scientific, Inc. (Maynard, MA). Peptide PR14 was also conjugated to keyhole limpet hemocyanin (KLH) at SCI Scientific. Ibalizumab was provided by TaiMed Biologics (Irvine, CA), which licensed the rights to this monoclonal antibody from Genentech (San Francisco, CA) after its acquisition of Tanox (Houston, TX). M-T441 was purchased from Ancell (Bayport, MN). HIV-1 isolates WT-1a, MDR-1a, and MDR-5a were provided by H. Mohri of the Aaron Diamond AIDS Research Center (New York, NY).
ELISA.
Soluble hCD4 or a relevant peptide at 2 μg/ml was adsorbed onto microtiter wells. The plates were then blocked with 4% dehydrated milk and 1% bovine serum albumin in phosphate-buffered saline (PBS) with Tween (blocking buffer). Ibalizumab was diluted with blocking buffer, added to the wells, and incubated. Alternatively, ibalizumab was incubated with CD4-IgG2 or PR14 in PBS and then diluted with blocking buffer before it was added to the wells. The plates were washed and incubated with alkaline phosphatase (AKP)-conjugated mouse anti-human IgG4 secondary antibodies (Alpha Diagnostic International, San Antonio, TX). Plates were then washed, developed with the AMPAK kit (Argene Inc., North Massapequa, NY), and read on an enzyme-linked immunosorbent assay (ELISA) reader at an optical density (OD) of 490 nm. The end point antibody titers were calculated as the reciprocal of the last dilution that yielded an OD at least 2-fold higher than those of mouse serum controls and an absolute absorbance of >0.1. Data are also shown as the OD at 490 nm. When mouse serum samples were used instead of ibalizumab, BALB/c mice were immunized twice at a 2-week interval with PR14 conjugated to KLH mixed first with complete Freund's adjuvant and then with incomplete Freund's adjuvant. Three weeks after the second immunization, pooled serum samples were collected, added in serial dilutions, and incubated. The plates were washed and incubated with AKP-conjugated goat anti-mouse secondary antibodies (Invitrogen, Carlsbad, CA) and then washed and developed with the AMPAK kit (Argene).
Plasmid construction.
Mouse CD4 (mCD4)/pVax was constructed by PCR amplification of pBS mCD4 (Addgene, Cambridge, MA) using primers 5′-CCCCCGAATTCCACCATGTGCCGAGCCATCTCTCTT-3′ and 5′-CCCCCCTCGAGTCAGATGAGATTATGGCTCTT-3′, which contain the EcoRI and XhoI restriction sites. The amplification product was subsequently cloned into the pVAX1 vector (Invitrogen). Mouse and human CD4 chimeric molecules were constructed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All the constructs were confirmed by DNA sequencing.
Fluorescence-activated cell sorter (FACS) analysis of ibalizumab or M-T441 binding to cell surface CD4.
Chimeric mCD4/pVax and hCD4 constructs were mixed with polyethylenimine (PEI; Polysciences, Warrington, PA) and then transiently transfected into 293T cells (human embryonic kidney 293 cells). 293T cells were harvested 2 days after transfection and stained with either ibalizumab, fluorescein isothiocyanate (FITC)-conjugated M-T441 (Ancell, Bayport, MN), or phycoerythrin (PE)-conjugated rat anti-mouse CD4 (RM4-5 and H129.19; BD Pharmingen, San Diego, CA). Ibalizumab-stained cells were then incubated with a biotin-labeled mouse anti-human IgG4 (Alpha Diagnostic) and streptavidin-conjugated PE/Cy5 (BioLegend, San Diego, CA). Flow data were acquired on a BD LSR II (BD Biosciences), and data analysis was performed using BD FACSDiva software 5.0.1 (BD Biosciences).
Virus neutralization assay using TZM-bl cells.
A neutralization assay was performed based on the methods of Wei et al. (28) with modification. TZM-bl cells were derived from a HeLa cell line (JC.53) that stably expresses CD4 and CCR5. TZM-bl cells also express luciferase and β-galactosidase under the control of the HIV-1 promoter. Briefly, 10,000 cells per well were seeded in a 96-well plate in 100 μl/well of Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (D10) and incubated overnight at 37°C. The next day, 100 μl of serial diluted ibalizumab or M-T441 was added to the cells and incubated for 1 h at 37°C. Then, 50 μl/well of 500 to 1,000 50% tissue culture infective doses (TCID50) of HIV-1 JRCSF virus were prepared in D10 containing 10 μg/ml of DEAE-dextran (Sigma, St. Louis, MO) and added to the cells. The cells were incubated for 42 h at 37°C. The β-galactosidase activity in individual wells was measured using the Galacto-Star system (Applied Biosystems, Cedarville, OH). The percentage of inhibition on viral infectivity was calculated as follows: [1 − (signal in antibody-treated wells/(signal in untreated infected wells)] × 100. The IC50 and IC80 values (the antibody concentrations that conferred 50% and 80% neutralization, respectively) were calculated by a nonlinear regression analysis. The results shown below represent those from at least three independent experiments.
FACS-based displacement assay.
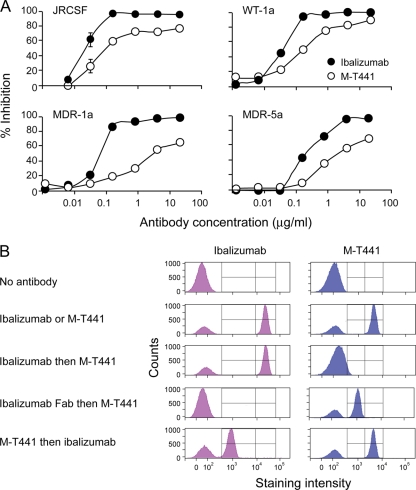
Human peripheral blood mononuclear cells (PBMCs) were first incubated with M-T441 conjugated to FITC (clone M-T441; Ancell) or ibalizumab and then conjugated to anti-human IgG4 to PE (clone HP6025; Southern Biotech, Birmingham, AL). The cells were analyzed through FACS as single-stain controls. Next, human PBMCs were incubated with ibalizumab followed by the addition of M-T441 conjugated to FITC and then stained with anti-human IgG4 conjugated to PE for the detection of ibalizumab. Alternatively, human PBMCs were incubated with ibalizumab Fab followed by the addition of M-T441 conjugated to FITC. Finally, human PBMCs were incubated with M-T441 conjugated to FITC first, followed by the addition of ibalizumab. Cells were then incubated with anti-CD45 conjugated to peridinin chorophyll protein, anti-CD3 conjugated to allophycocyanin, and a noncompeting anti-CD4 conjugated to Alexa 700 (clones 2D1, SK7, and RPA-T4, respectively; BD Biosciences). Flow data were acquired on a BD LSR II (BD Biosciences), and data analysis was performed using BD FACSDiva software 5.0.1 (BD Biosciences). Binding activities of ibalizumab and M-T441 were assessed on CD45+ CD3+ CD4+ cells.
Biacore assay.
Binding affinity analyses were performed with a Biacore 3000 optical biosensor (GE Healthcare, Piscataway, NJ). Immobilization of ibalizumab or M-T441 to CM5 sensor chips was performed following the standard amine coupling procedure. Briefly, carboxyl groups on the sensor chip surface were activated by injection of 35 μl of a solution containing 0.2 M N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide and 0.05 M N-hydroxysuccinimide at a flow rate of 5 μl/minute. Next, a protein ligand at a concentration of 2 μg/ml in 10 mM NaO-acetate buffer, pH 5.0 for ibalizumab and pH 4.5 for M-T441, respectively, was allowed to flow over the chip surface at a rate of 10 μl/minute until the desired level of response units of reacted protein was achieved. After unreacted protein was washed out, excess active ester groups on the sensor surface were capped by the injection of 35 μl of 1 M ethanolamine, pH 8.0, at a flow rate of 5 μl/minute. As background to correct instrument and buffer artifacts, a reference was generated under the same conditions with omission of the protein ligand.
Binding experiments were performed at 25°C in HBS-EP buffer (0.01 M HEPES [pH 7.4], 0.15 M NaCl, 3 mM EDTA, 0.005% vol/vol surfactant P20 (GE Healthcare). Binding kinetics were measured by passing various concentrations of analyte, human soluble CD4 (sCD4) protein, over the chip surface at a flow rate of 30 μl/minute for 2 min. Dissociation of bound analytes was monitored while the surface was washed with buffer for 15 min. Remaining analytes were removed in the surface regeneration step at a flow rate of 50 μl/minute with two 30-s injections of 10 mM glycine-HCl, pH 1.8, for the ibalizumab chip and one 30-s injection of 10 mM glycine-HCl, pH 2.0, for the M-T441 chip. For kinetics data analysis, after subtraction of the blank cell from each response value, the kinetic parameters (kon, association rate constant; koff, dissociation rate constant; KD, equilibrium dissociation constant) were determined by collectively fitting the overlaid sensograms locally by using the BIAevaluation 4.1 software to the 1:1 Langmuir binding model.
RESULTS
The epitope of ibalizumab is likely to be conformational.
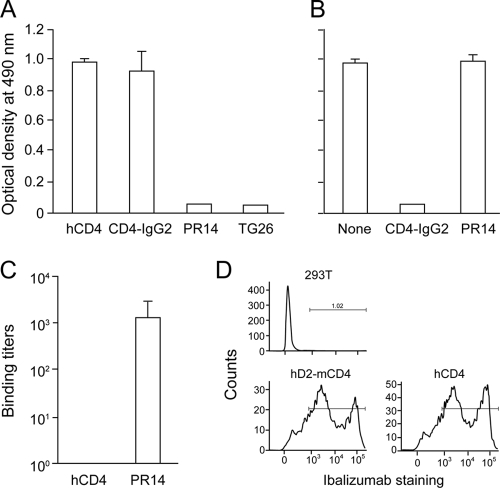
Previously, Burkly et al. (5) demonstrated that the region of aa 121 to 134 in D2 of hCD4 was essential for the binding of the ibalizumab precursor 5A8 to CD4. We began our studies by determining if a peptide named PR14, which covers this specific region (aa 121 to 134) is able to bind to ibalizumab. In a direct ELISA, ibalizumab was able to bind to both soluble hCD4 and CD4-IgG2 (a tetrameric fusion protein containing two human IgGs in which the Fv portions of both heavy and light chains have been replaced by D1 and D2 of hCD4), but not to PR14 or an even longer peptide named TG26 (aa 115 to 140) that extends PR14 into its flanking amino acids (Fig. 1A). Furthermore, peptide PR14 could not compete with soluble hCD4 in binding to ibalizumab in solution, while CD4-IgG2 could (Fig. 1B). In addition, antisera raised to peptide PR14 did not recognize soluble hCD4 protein (Fig. 1C). Together, these results demonstrate that linear peptides may not be able to present the epitope of ibalizumab. The fact that ibalizumab could bind well to CD4-IgG2 (Fig. 1A) indicates that only D1 and D2 of hCD4 are involved in the specific binding.
FIG. 1.
The epitope of ibalizumab is likely to be conformational. (A) Binding of ibalizumab to soluble hCD4, CD4-IgG2, PR14 (aa 121 to 134), or TG26 (aa 115 to 140) as measured by ELISA. (B) Competition for ibalizumab binding to sCD4 in solution by CD4-IgG2 or PR14 as measured by ELISA. (C) Binding titers of mouse anti-PR14 antiserum to hCD4 or PR14. (D) FACS analysis of cell surface staining by ibalizumab in control 293T cells or cells transfected with hD2-mCD4 or hCD4 plasmid DNA.
We next explored whether the hCD4 D2 alone is sufficient to mediate ibalizumab binding. 293T cells were transiently transfected with plasmid DNA of hCD4 or hD2-mCD4, a chimeric gene in which D2 of mCD4 was replaced with D2 of hCD4. The cells were then stained with ibalizumab, biotin-labeled mouse anti-human IgG4, and streptavidin-conjugated PE/Cy5. The observation that ibalizumab could bind to hD2-mCD4 (Fig. 1D) suggests that the epitope of ibalizumab is mainly located in D2 of hCD4. However, these findings do not exclude the possibility of D1 contribution by elements that are conserved or similar between mouse and human CD4.
A chimeric mouse/human CD4 protein, mCD4-hGACFG, can bind to ibalizumab.
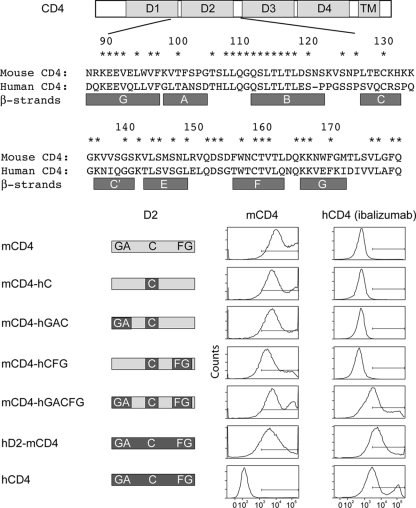
To determine if regions of hCD4 besides aa 121 to 134 are involved in ibalizumab binding, we substituted certain β-strands of mouse CD4 with corresponding human CD4 sequences and monitored their effects on the overall binding of ibalizumab using FACS analysis. As shown in Fig. 2, no ibalizumab binding was observed when either D2-C (aa 121 to 134, which includes the C strand and the turn between B and C strands) only, D2-C plus D1-G/D2-A, or D2-C plus D2-F/G β-strands were substituted with the homologous hCD4 sequences in mCD4. Only by exchanging all the previously mentioned regions simultaneously (mCD4-hGACFG) could the binding of ibalizumab be reconstituted. Anti-mCD4 staining confirmed positive protein expression from all of the chimeric genes. These results further confirmed our previous observation that the ibalizumab epitope is conformational and indicated that regions other than aa 121 to 134 in hCD4 are involved. However, these data do not suggest whether these three regions are part of the ibalizumab epitope or simply crucial in maintaining the native conformation of hCD4.
FIG. 2.
Map of the ibalizumab epitope obtained by constructing and testing mouse-human chimeric CD4 molecules in binding studies using FACS. Sequence identity between mouse and human CD4 is denoted by an asterisk above the amino acid alignment. The β-strands defined by Ryu et al. (24) are shown below the sequence alignment.
Fine-mapping the ibalizumab epitope by point mutations in chimeric mCD4-hGACFG or in hCD4.
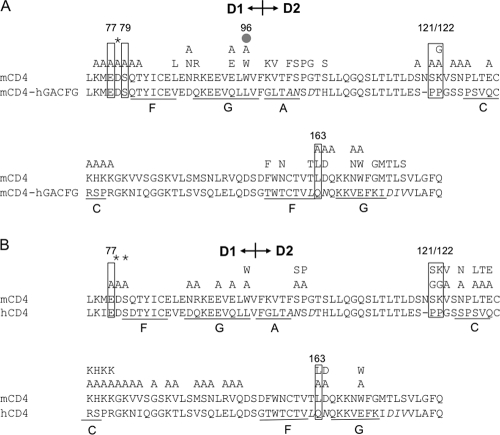
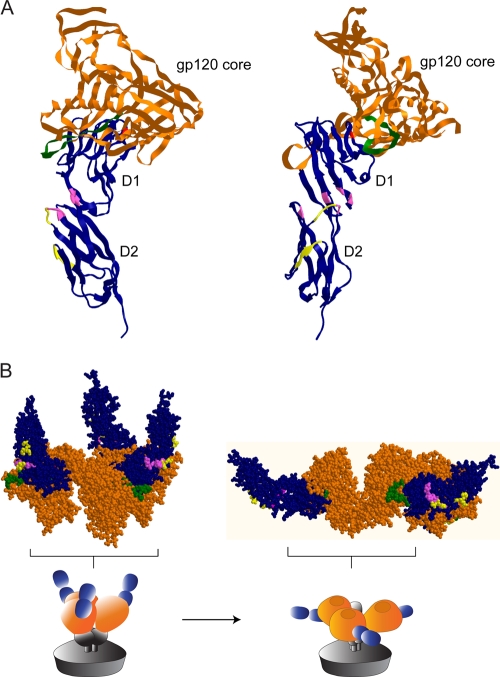
To further define the ibalizumab epitope, we introduced single amino acid changes in the chimeric mCD4-hGACFG molecule (Fig. 3A) or hCD4 protein (Fig. 3B). Their effects on CD4 expression and conformation as well as on ibalizumab binding were measured using FACS as previously described. Figure 3A and B show that E77, P121, P122, and Q163 (boxes) in CD4 are essential for ibalizumab binding. In addition, S79 and to a lesser degree L96 (gray dot) appear to be important for ibalizumab binding to mCD4-hGACFG. The substitution of many amino acids in mCD4 by alanine, glycine, or the corresponding amino acid from hCD4 did not significantly affect ibalizumab binding or CD4 expression level. However, when D78 (asterisk) was replaced by alanine, the protein expression level or overall conformation was greatly altered so that even control antibodies to other domains of CD4 could not detect the chimeric protein. These results, collectively, indicate that the ibalizumab epitope is mainly in D2 (P121, P122, and Q163), with contributions from D1 elements (E77 and possibly S79). However, it is likely that some of these amino acids are directly involved in the binding while others are crucial in maintaining the proper conformation that is essential for the binding. Taken together, these results indicate that the possible components of the ibalizumab epitope are E77/S79, L96, P121/P122, and Q163, all of which are identical between human and rhesus CD4 proteins. It is also worth noting that residues E77 and S79 are identical for hCD4 and mCD4 but not residues 96, 121, 122, and 163. Other neighboring amino acids may also contribute to the epitope either in direct binding or in maintaining the native conformation of these key sequences. Visualization of the residues forming the ibalizumab epitope in a three-dimensional structure of hCD4 (Fig. 4, pink amino acids) indicated that they cluster together, and ibalizumab most likely binds to the interface between the first and the second domains of hCD4 on a surface opposite to the site where HIV-1 gp120 and human MHC class II bind on D1 (Fig. 4A, red segments on F43 of hCD4).
FIG. 3.
Results of fine-mapping the ibalizumab epitope by point mutations in the chimeric mCD4-hGACFG (A) or hCD4 (B). Amino acids in regions of interest for ibalizumab binding were mutated to alanine, glycine, or the corresponding mouse amino acid as shown by single-letter amino acid symbols above the CD4 sequence alignment. Mutations with no impact on CD4 expression or ibalizumab binding are unmarked. Residues critical to ibalizumab binding are enclosed by boxes. A gray dot denotes the amino acid that partially affected ibalizumab binding. The asterisks indicate mutations that resulted in a substantial reduction (to <25%) in CD4 expression.
FIG. 4.
Localization of ibalizumab and M-T441 epitopes on three-dimensional structures of hCD4. The epitopes of ibalizumab (pink) and M-T441 (yellow) are highlighted on CD4 (blue) complexed to the core of gp120 (brown), with two views of the structure rotated 90° around a central vertical axis (A), or space-filling model of three CD4s (blue) bound to the trimer of gp120 core (brown), before and after a conformational rearrangement (B), as derived from the cryo-electron microscopy study of Liu et al. (17). The red dot in D1 denotes F43, a critical component that binds HIV-1 Env. The parts in green highlight the V5 loop in gp120.
Ibalizumab is more potent than M-T441 in inhibiting HIV-1 infection.
To further explore ibalizumab's inhibition of HIV-1 infection, we compared its relative virus neutralization activity with that of M-T441, a commercially available mouse monoclonal antibody purportedly directed against the second domain of hCD4. As shown in Fig. 5A, ibalizumab was able to inhibit HIV-1 JRCSF more potently than M-T441 in vitro. The IC80 for ibalizumab was 0.08 μg/ml (0.54 nM), while M-T441 could not inhibit 80% of infection even at a concentration of 20 μg/ml. In addition, the IC50 for M-T441 against HIV-1 JRCSF was 3.6 times higher than that of ibalizumab. Similar potency differences were noted for three other virus isolates (WT-1a, MDR-1a, and MDR-5a) (Fig. 5A).
FIG. 5.
Comparison of ibalizumab with M-T441 in virus neutralization and competitive binding to hCD4. (A) Neutralizing activity of ibalizumab versus M-T441 as tested against four HIV-1 isolates (JRCSF, WT-1a, MDR-1a, and MDR-5a). (B) Two-way binding competitions between ibalizumab and M-T441 based on FACS analysis.
The epitopes of ibalizumab and M-T441 are in close proximity.
To understand the mechanism behind the different neutralization activities of these two anti-hCD4 monoclonal antibodies, FACS-based competition assays were used to delineate the relative locations of their epitopes. Both antibodies were able to bind efficiently to CD4 on human PBMCs when added separately (Fig. 5B). However, when ibalizumab was added to human PBMCs followed by M-T441, the latter did not bind to hCD4. That M-T441 could not bind to the complex of hCD4 and ibalizumab suggests complete or partial overlapping of the epitopes of these two antibodies. To study whether the stereo hindrance of the whole ibalizumab antibody is responsible for this finding, the Fab portion of ibalizumab was incubated with human PBMCs before the addition of M-T441. In this case, M-T441 binding to hCD4 was partially restored. Due to the lack of reagents that can identify the Fab fragment, we were unable to detect the presence of ibalizumab Fab. However, the fact that M-T441 binding was perturbed in the presence of ibalizumab Fab strongly suggests binding of the Fab fragment to CD4. Ibalizumab binding was also affected in the presence of its Fab (data not shown). In addition, when M-T441 was incubated with human PBMCs followed by ibalizumab staining, we observed that staining of hCD4 was only partial with ibalizumab but complete with M-T441. Taken together, these results indicate that the epitopes of ibalizumab and M-T441 are overlapping and in close proximity on the hCD4 molecule.
Fine-mapping of the M-T441 epitope.
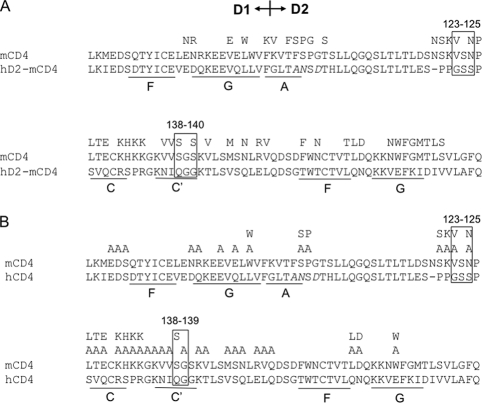
To determine the epitope of M-T441 on hCD4, we used a chimeric mCD4 molecule called hD2-mCD4 in which mouse D2 was replaced with human D2. It was observed that hD2-mCD4 could be recognized by M-T441 to a degree that was comparable to hCD4 (data not shown), supporting the notion that M-T441 is D2 specific. Next, we introduced single or multiple amino acid changes in both hD2-mCD4 (Fig. 6A) and hCD4 (Fig. 6B). Their effects on CD4 protein expression, conformation, and M-T441 binding were measured using FACS as previously described. The regions G123 to S125 and Q138 to G140 were critical for M-T441 binding to CD4, observed in both hD2-mCD4 and hCD4 (Fig. 6A and B). When the epitopes of ibalizumab and M-T441 were interpreted on the three-dimensional structure of hCD4 (Fig. 4), it was apparent that both epitopes are located in close proximity on roughly (within 45°) the same surface of hCD4, opposite to the side where HIV-1 gp120 binds in D1. These findings are consistent with the results from the competition binding assays (Fig. 5B). Thus, ibalizumab binds to the interface between the first and the second domains of hCD4 (Fig. 4, pink amino acids), whereas M-T441 binds to D2 only (Fig. 4, yellow residues). The two stretches that form the M-T441 epitope are lower, or more proximal to the membrane, on the rod-like structure of hCD4.
FIG. 6.
Results of fine-mapping of the M-T441 epitope by point mutations in the backbone of hD2-mCD4 (A) or hCD4 (B). Unmarked mutations have no impact on CD4 expression or M-T441 binding. Mutations enclosed by boxes substantially decreased M-T441 binding.
Ibalizumab has a higher affinity for hCD4 than M-T441.
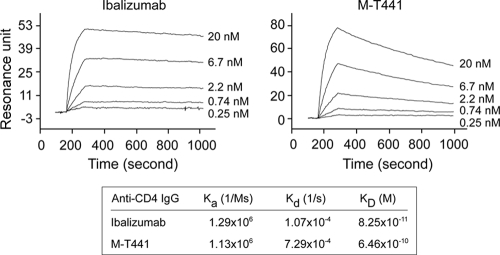
It is also possible that the differences in HIV-1 neutralization activity between ibalizumab and M-T441 are due, at least in part, to their differential binding affinities to hCD4. To explore this possibility, we conducted a Biacore study to determine the binding kinetics of ibalizumab and M-T441 to human sCD4 proteins. Each antibody was immobilized by amine coupling to the sensor chip surface to a density suitable for kinetics studies, namely, 200 resonance units. Antibody interaction with human sCD4 was measured over an sCD4 protein concentration range of 0.25 to 20 nM. As shown in Fig. 7, while ibalizumab and M-T441 bound to sCD4 at a similar on rate, M-T441 had a faster off rate (about 7-fold) than ibalizumab. Overall, ibalizumab has a KD of 82.5 pM to human sCD4, which is about 8-fold lower than that of M-T441. From these data, we can conclude that ibalizumab's higher binding affinity for CD4 may contribute, at least in part, to its greater HIV-1 neutralization potency.
FIG. 7.
Binding affinity of ibalizumab and M-T441 to hCD4 as assessed in a Biacore assay.
DISCUSSION
Ibalizumab is a humanized anti-CD4 monoclonal antibody that potently and broadly blocks in vitro infection by a large panel of HIV-1 isolates (4, 5, 8, 33). From phase 1 through phase 2b clinical trials in infected patients in need of salvage therapy, ibalizumab has demonstrated in vivo antiviral activity by consistently lowering viral load by about 1 log, without causing significant adverse side effects (8, 12, 14). Ibalizumab thus appears to be a promising agent for salvage therapy as well as for passive immunization against HIV-1 infection. Consequently, it seems important to define the epitope of this antibody in order to gain further insights into its mechanism of action as well as its safety profile.
Previous studies have shown that ibalizumab, like its murine progenitor (5A8), is able to bind to D2 of rhesus and human CD4, thereby preventing postbinding entry of the virus into CD4 T cells (5, 21). Here, extensive mutagenesis studies indicate that E77, S79, P121, P122, and Q163 are essential for ibalizumab binding to hCD4 (Fig. 3A and B). These findings do not exclude the possibility that exchanging the proline at positions 121 and 122, known to be important residues in overall protein architecture, for the mouse counterpart amino acids altered the conformation of the hCD4 protein and therefore its function. The results presented in Fig. 3 are consistent with previous findings that amino acids 121 to 124 on hCD4 are necessary for 5A8 (ibalizumab precursor) binding (5). Interestingly, amino acids 127 to 134, identified in the previous study as important for hCD4 binding (5), seem to be less crucial in our analysis. This apparent discrepancy may be due to the different methods used for epitope mapping. Furthermore, the previous study used replacement of a stretch of amino acids whereas the current study utilized single amino acid substitutions.
It is noteworthy that residues E77 and S79, both located in D1 of hCD4, as well as Q163 also seem to be essential in for ibalizumab binding. These amino acids cluster together with P121 and P122 on one surface of the three-dimensional structure of hCD4 (Fig. 4). If D1 were the head and neck and D2 were the torso, then it appears that ibalizumab would bind to the back of the “neck and upper torso,” whereas gp120 and MHC class II molecules bind to the “nose.” Thus, it is not surprising that ibalizumab does not interfere with gp120-CD4 binding (5). Perhaps more importantly, the spatial separation of the ibalizumab epitope and the MHC class II binding site helps to explain why ibalizumab is well tolerated by patients without any evidence of adverse immunological sequelae noted to date.
In this study, we also studied another monoclonal antibody, M-T441, specific for D2 on human CD4, in order to compare and contrast its properties with ibalizumab. M-T441 was found to be substantially less potent in blocking HIV-1 infection in vitro (Fig. 5A). One possible explanation for this is that its epitope is lower down on D2 at the “back of the middle torso” and ∼45° axially separated from that of ibalizumab (Fig. 4), thereby resulting in less interference of some critical step in viral entry. The lower virus neutralization potency of M-T441, however, could also be explained by its lower binding affinity as measured in a Biacore assay (Fig. 7). It is possible that both affinity and epitope differences contribute to the lower potency of M-T441 against HIV-1.
Armed with information on the epitope of ibalizumab, what could be said about the mechanism by which this antibody blocks HIV-1 infection of CD4 T cells? Specifically, which step of viral entry is inhibited? A previous study (5) showed that gp120-CD4 binding is not inhibited by ibalizumab, but it is formally possible that virion binding to CD4 is blocked because monomeric gp120 does not accurately reflect the trimeric glycoprotein spike on the virion surface. This possibility is rendered less likely because the gp120 binding site in D1 is located on a surface opposite the ibalizumab epitope (Fig. 4). Nevertheless, it would be prudent to conduct follow-up studies to examine whether ibalizumab could interfere with the binding of HIV-1 particles to CD4-positive cells.
It is known that after gp120 binds to CD4, both molecules undergo conformational changes, leading to the exposure of the so-called bridging sheet on gp120 that is important for engaging the coreceptor, either CCR5 or CXCR4 (10, 31). Thus, it is theoretically possible that ibalizumab attached to the “neck and upper torso” of CD4 could impede the formation of the bridging sheet. This possibility is, however, unlikely given that ibalizumab does not lower the affinity of CD4-gp120 binding.
Upon close inspection of the structure of gp120 core complexed with CD4 and a monoclonal antibody, 17b (which binds to the bridging sheet) (16, 25, 32), it is clear that the ibalizumab epitope is in close proximity to the V5 loop of gp120 (Fig. 4, green elements) with its one or two N-linked glycans. This spatial relationship suggests that ibalizumab could easily impinge on this part of gp120 or the glycans thereon and thereby indirectly affect the structural alterations necessary in the subsequent steps during viral entry.
Upon exposure of the bridging sheet, gp120 must then move closer to the cell surface in order to engage either CCR5 or CXCR4. One can imagine this happening by having the CD4 molecule lie down on the cell surface (flexing between D4 and the transmembrane region) or jackknife from a rod-like structure into a ball (folding at the hinge between D2 and D3). Conceivably, ibalizumab could also cross-link two CD4 molecules and thus restrain them from lying down. The latter hypothesis is supported by the original observation made by Burkly et al. (5) that Fab fragments of 5A8 do not have antiviral activity. However, our recent finding that Fab of ibalizumab is only slightly less active than the full antibody or F(ab′)2 (unpublished data) essentially rules out cross-linking of CD4 as a mode of inhibition. With ibalizumab attached to the back of the “neck and upper torso,” it is easy to envision CD4 having trouble lying down with its “face” up. But this does not explain how CD4 wouldn't be able to lie down with its “face” down. Two reports suggest that CD4 becomes a ball-like structure following binding to gp120 (1, 17), perhaps by jackknifing “backwards” with D1D2 anti-parallel to D3D4 (1). With such a scenario, it would be easy to imagine how ibalizumab could prevent such a dramatic structural rearrangement of CD4. However, it must be pointed out that CD4 may not have such a high degree of flexibility at the hinge between D2 and D3 to allow jackknifing to occur (24, 27, 30). A more subtle bending at this hinge may be possible though.
It remains possible that ibalizumab blocks an even later step in viral entry, such as coreceptor binding, unleashing of gp41, or the six-helix bundle formation associated with membrane fusion (6, 21, 29). If so, it would suggest that CD4 stays involved with the envelope glycoprotein for much longer than current models of viral entry would indicate (9).
In summary, by mapping the epitope of ibalizumab, we have gained a better appreciation for why this antibody has not had any adverse immunological consequences in infected patients to date. While the precise mechanism of its inhibition of HIV-1 entry remains elusive, it has become clear that CD4 does not merely serve as an anchor for the virus; instead, it is likely involved in a complicated dance with gp120/gp41 throughout a considerable part of the entry process. Follow-up structural and mechanistic studies are necessary to provide a better understanding of how ibalizumab blocks HIV-1 infection and how it could be further developed as a passive immunization agent to slow the spread of this pandemic.
Acknowledgments
We thank TaiMed Biologics for providing ibalizumab, D. A. Oren and B. Chen for advice on CD4 structural analysis, S. Weinheimer, S. Vasan, N. Padte, J. Verhagen, C. Pace, M. Fordyce, and Q. Fang for helpful discussions, and W. Chen for preparation of figures.
This work was supported by a grant from the Bill & Melinda Gates Foundation.
D. D. Ho is the scientific founder of TaiMed Biologics and has equity in the company.
Footnotes
Published ahead of print on 12 May 2010.
REFERENCES
- 1.Ashish, I. J. Juncadella, R. Garg, C. D. Boone, J. Anguita, and J. K. Krueger. 2008. Conformational rearrangement within the soluble domains of the CD4 receptor is ligand-specific. J. Biol. Chem. 283:2761-2772. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 3.Barouch, D. H. 2008. Challenges in the development of an HIV-1 vaccine. Nature 455:613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boon, L., B. Holland, W. Gordon, P. Liu, F. Shiau, W. Shanahan, K. A. Reimann, and M. Fung. 2002. Development of anti-CD4 MAb hu5A8 for treatment of HIV-1 infection: preclinical assessment in non-human primates. Toxicology 172:191-203. [DOI] [PubMed] [Google Scholar]
- 5.Burkly, L. C., D. Olson, R. Shapiro, G. Winkler, J. J. Rosa, D. W. Thomas, C. Williams, and P. Chisholm. 1992. Inhibition of HIV infection by a novel CD4 domain 2-specific monoclonal antibody. Dissecting the basis for its inhibitory effect on HIV-induced cell fusion. J. Immunol. 149:1779-1787. [PubMed] [Google Scholar]
- 6.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 7.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 8.Dimitrov, A. 2007. Ibalizumab, a CD4-specific mAb to inhibit HIV-1 infection. Curr. Opin. Investig. Drugs 8:653-661. [PubMed] [Google Scholar]
- 9.Doms, R. W. 2004. Unwelcome guests with master keys: how HIV enters cells and how it can be stopped. Top. HIV Med. 12:100-103. [PubMed] [Google Scholar]
- 10.Hart, T. K., R. Kirsh, H. Ellens, R. W. Sweet, D. M. Lambert, S. R. Petteway, Jr., J. Leary, and P. J. Bugelski. 1991. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc. Natl. Acad. Sci. U. S. A. 88:2189-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber, M., W. C. Olson, and A. Trkola. 2008. Antibodies for HIV treatment and prevention: window of opportunity? Curr. Top. Microbiol. Immunol. 317:39-66. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson, J. M., D. R. Kuritzkes, E. Godofsky, E. DeJesus, J. A. Larson, S. P. Weinheimer, and S. T. Lewis. 2009. Safety, pharmacokinetics, and antiretroviral activity of multiple doses of ibalizumab (formerly TNX-355), an anti-CD4 monoclonal antibody, in human immunodeficiency virus type 1-infected adults. Antimicrob. Agents Chemother. 53:450-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobson, J. M., M. S. Saag, M. A. Thompson, M. A. Fischl, R. Liporace, R. C. Reichman, R. R. Redfield, C. J. Fichtenbaum, B. S. Zingman, M. C. Patel, J. D. Murga, S. M. Pemrick, P. D'Ambrosio, M. Michael, H. Kroger, H. Ly, Y. Rotshteyn, R. Buice, S. A. Morris, J. J. Stavola, P. J. Maddon, A. B. Kremer, and W. C. Olson. 2008. Antiviral activity of single-dose PRO 140, a CCR5 monoclonal antibody, in HIV-infected adults. J. Infect. Dis. 198:1345-1352. [DOI] [PubMed] [Google Scholar]
- 14.Kuritzkes, D. R., J. Jacobson, W. G. Powderly, E. Godofsky, E. DeJesus, F. Haas, K. A. Reimann, J. L. Larson, P. O. Yarbough, V. Curt, and W. R. Shanahan, Jr. 2004. Antiretroviral activity of the anti-CD4 monoclonal antibody TNX-355 in patients infected with HIV type 1. J. Infect. Dis. 189:286-291. [DOI] [PubMed] [Google Scholar]
- 15.Kwong, P. D., S. E. Ryu, W. A. Hendrickson, R. Axel, R. M. Sweet, G. Folena-Wasserman, P. Hensley, and R. W. Sweet. 1990. Molecular characteristics of recombinant human CD4 as deduced from polymorphic crystals. Proc. Natl. Acad. Sci. U. S. A. 87:6423-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, J., A. Bartesaghi, M. J. Borgnia, G. Sapiro, and S. Subramaniam. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maddon, P. J., A. G. Dalgleish, J. S. McDougal, P. R. Clapham, R. A. Weiss, and R. Axel. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47:333-348. [DOI] [PubMed] [Google Scholar]
- 19.Maddon, P. J., D. R. Littman, M. Godfrey, D. E. Maddon, L. Chess, and R. Axel. 1985. The isolation and nucleotide sequence of a cDNA encoding the T cell surface protein T4: a new member of the immunoglobulin gene family. Cell 42:93-104. [DOI] [PubMed] [Google Scholar]
- 20.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore, J. P., Q. J. Sattentau, P. J. Klasse, and L. C. Burkly. 1992. A monoclonal antibody to CD4 domain 2 blocks soluble CD4-induced conformational changes in the envelope glycoproteins of human immunodeficiency virus type 1 (HIV-1) and HIV-1 infection of CD4+ cells. J. Virol. 66:4784-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reimann, K. A., L. C. Burkly, B. Burrus, B. C. Waite, C. I. Lord, and N. L. Letvin. 1993. In vivo administration to rhesus monkeys of a CD4-specific monoclonal antibody capable of blocking AIDS virus replication. AIDS Res. Hum. Retrovir. 9:199-207. [DOI] [PubMed] [Google Scholar]
- 23.Reimann, K. A., R. L. Cate, Y. Wu, L. Palmer, D. Olson, B. C. Waite, N. L. Letvin, and L. C. Burkly. 1995. In vivo administration of CD4-specific monoclonal antibody: effect on provirus load in rhesus monkeys chronically infected with the simian immunodeficiency virus of macaques. AIDS Res. Hum. Retrovir. 11:517-525. [DOI] [PubMed] [Google Scholar]
- 24.Ryu, S. E., P. D. Kwong, A. Truneh, T. G. Porter, J. Arthos, M. Rosenberg, X. P. Dai, N. H. Xuong, R. Axel, R. W. Sweet, et al. 1990. Crystal structure of an HIV-binding recombinant fragment of human CD4. Nature 348:419-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, J. H., R. Meijers, Y. Xiong, J. H. Liu, T. Sakihama, R. Zhang, A. Joachimiak, and E. L. Reinherz. 2001. Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule. Proc. Natl. Acad. Sci. U. S. A. 98:10799-10804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, J. H., Y. W. Yan, T. P. Garrett, J. H. Liu, D. W. Rodgers, R. L. Garlick, G. E. Tarr, Y. Husain, E. L. Reinherz, and S. C. Harrison. 1990. Atomic structure of a fragment of human CD4 containing two immunoglobulin-like domains. Nature 348:411-418. [DOI] [PubMed] [Google Scholar]
- 28.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 29.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 30.Wu, H., P. D. Kwong, and W. A. Hendrickson. 1997. Dimeric association and segmental variability in the structure of human CD4. Nature 387:527-530. [DOI] [PubMed] [Google Scholar]
- 31.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 32.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, X. Q., M. Sorensen, M. Fung, and R. T. Schooley. 2006. Synergistic in vitro antiretroviral activity of a humanized monoclonal anti-CD4 antibody (TNX-355) and enfuvirtide (T-20). Antimicrob. Agents Chemother. 50:2231-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]