Abstract
Single-genome amplification (SGA) and sequencing of HIV-1 RNA in plasma of acutely infected humans allows the identification and enumeration of transmitted/founder viruses responsible for productive systemic infection. Use of this strategy as a means for identifying transmitted viruses suggested that intrarectal simian immunodeficiency virus (SIV) inoculation of macaques recapitulates key features of human rectal infection. However, no studies have used the SGA strategy to identify vaginally transmitted virus(es) in macaques or to determine how early SIV diversification in vaginally infected animals compares with HIV-1 in humans. We used SGA to amplify 227 partial env sequences from a SIVmac251 challenge stock and from seven rhesus macaques at the earliest plasma viral RNA-positive time point after low- and high-dose intravaginal inoculation. Sequences were analyzed phylogenetically to determine the relationship of transmitted/founder viruses within and between each animal and the challenge stock. In each animal, discrete low-diversity env sequence lineages were evident, and these coalesced phylogenetically to identical or near-identical env sequences in the challenge stock, thus confirming the validity of the SGA sequencing and modeling strategy for identifying vaginally transmitted SIV. Between 1 and 10 viruses were responsible for systemic infection, similar to humans infected by sexual contact, and the set of viruses transmitted to the seven animals studied represented the full genetic constellation of the challenge stock. These findings recapitulate many of the features of sexual HIV-1 transmission in women. Furthermore, the SIV rhesus macaque model can be used to understand the factors that influence the transmission of single versus multiple SIV variants.
HIV transmission most commonly occurs at mucosal surfaces, and preventative strategies should be directed at viral variants responsible for establishing productive infection (9, 21). Until recently, identification of the individual HIV-1 variants that were transmitted and produced clinical infection was not feasible because sequencing strategies were hampered by Taq-induced nucleotide misincorporation and recombination, template resampling, and bacterial cloning bias. These errors are eliminated by single-genome amplification (SGA) and direct sequencing, which provide an accurate and proportionate view of the viral quasispecies during primary HIV infection (23, 26). In addition to single-genome amplification, recent empirical data together with mathematical models have shown that each founder HIV-1 lineage generally diversifies by a Poisson distribution of random nucleotide substitutions, leading to a star-like phylogeny with no or few shared mutations (9, 13). For each low-diversity HIV-1 lineage, the consensus of the sequences within that lineage represents the inferred ancestral sequence. Since mutations occur randomly, if sampled prior to the adaptive immune response or the onset of other selection pressures, the consensus sequence represents the actual transmitted or founder HIV-1 variant sequence (9).
Utilizing this sequencing approach and mathematical modeling, it has now been reported in eight patient cohorts representing HIV-1 subtypes A, B, C, and D that most (60 to 90%) mucosal infections originate from single-variant transmissions (1, 6, 8, 9, 26, 27). The remaining 10 to 40% of infections are initiated by a limited number of transmitted/founder HIV-1 variants. Therefore, for each individual infected, the potential viral diversity in the period of acute infection was limited to a single or a few HIV-1 lineages. This genetic bottleneck was less pronounced in individuals engaged in high-risk behaviors (anal-receptive intercourse or intravenous drug use) (9) and in patients with sexually transmitted infections (1, 6, 8, 26, 27). Importantly, acute infection with “heterogeneous” HIV populations has been linked to more rapid disease progression (4, 24, 33). The study of the number of transmitted viral variants and their overall diversity can thus have important implications for developing both prophylactic vaccines and antiviral therapy.
Studies of HIV variant transmission are challenging because the exact time of transmission can be difficult to determine, and only linked infection studies of donor and recipient pairs can define the genetic diversity in the infecting inoculum relative to the variant population that is transmitted to the recipient. The simian immunodeficiency virus (SIV) models of mucosal HIV transmission are valuable for examining the transmitted or early founder populations that establish productive infection, because the genetic composition of the virus stock used for mucosal inoculation can be readily determined and compared to the viruses transmitted to the mucosally inoculated animals. Further, because the timing of SIV transmission is defined (5, 14, 19-22), mathematical modeling is not needed to infer the timing of virus transmission in macaques. Recently Keele et al. reported the number of transmitted/founder viruses in 18 rhesus macaques infected either intrarectally (i.r.) or intravenously with either SIVmac251 or SIVsmE660 (10). Utilizing a repeated low-dose i.r. challenge system, they found that a limited number of transmitted variants (range, 1 to 5; median, 1) established productive infection after i.r. challenge. In fact, the rectal barrier provided a 2,000- to 20,000-fold decrease in the number of transmitted variants after normalizing intrarectal and intravenous virus inputs (10). In addition, since the challenge stock was characterized directly, the mathematic model of early viral diversification was corroborated and provided direct evidence that the consensus sequence obtained prior to immune selection is the transmitted/founder virus. After comparing all animals' transmitted/founder viruses, the authors concluded that no two transmitted viral lineages were identical between individuals and that each transmitted lineage was distributed throughout the phylogenetic tree of SIV stock sequences. Overall, these data suggest that the intrarectal SIV model recapitulates many of the features of HIV transmission and early diversification (10). However, three differences were noted between the viral variants in newly established SIV and HIV infections. Intrarectal SIV infection was associated with (i) an increase in low-level G-to-A mutations, (ii) a higher frequency of transmitted/founder lineages with low sequence representation, and (iii) a shorter eclipse phase leading to lower sequence diversity in early plasma samples (10).
There are obvious physiological and immunological differences between vaginal/cervical HIV exposure and rectal HIV exposure. In women and rhesus macaques, the vagina is composed of a multilayered stratified squamous epithelium that varies in thickness during the menstrual cycle (15), while the endocervix is a columnar epithelium that does not vary in thickness during the cycle (15). In contrast, the rectal mucosa is composed entirely of a columnar epithelium that is a single cell layer thick. Additionally, while the epithelium of the endocervix and rectum are both a single cell layer thick, more CD4+ T cells are present in the lamina propria of the gastrointestinal tract (7) than the vagina and cervix (15) of SIV-negative rhesus macaques.
To date, no studies have been conducted to determine if the SGA-direct sequencing modeling approach for the identification of transmitted/founder viruses is applicable to the SIV-macaque vaginal infection model. Nor have studies been performed to determine if viruses representing the entire genetic spectrum of the SIVmac251 infection stock are responsible for transmission or if actual transmitted viral env sequences can be tracked from the inoculum across the vaginal mucosa and into the circulating plasma, thus providing a formal proof for the SGA-based hypothesis (9, 13) for identifying transmitted/founder viruses. Our results answer these questions affirmatively and suggest that the vaginal SIVmac251 transmission recapitulates the key features of HIV transmission (28, 30) and may provide insight into the host and viral factors that permit HIV transmission and dissemination.
MATERIALS AND METHODS
Animals.
The rhesus macaques (Macaca mulatta) used in these studies were housed at the California National Primate Research Center in accordance with the regulations of the American Association for Accreditation of Laboratory Animal Care. These experiments were approved by the Institutional Animal Use and Care Committee of the University of California, Davis. All animals were negative for antibodies to HIV-2, SIV, type D retrovirus, and simian T-cell lymphotropic virus type 1 at the time the study was initiated. When necessary, animals were anesthetized with ketamine hydrochloride (10 mg/kg of body weight; Parke-Davis, Morris Plains, NJ) or 0.7 mg/kg tiletamine HCl and zolazepan (Telazol; Fort Dodge Animal Health, Fort Dodge, IA) injected intramuscularly. The details of the original transmission study have been described previously (17).
Vaginal SIVmac251 inoculation.
A cell-free stock of SIVmac251 (UCD-2/02) was produced by short-term expansion of a previous virus stock (SIVmac251 UCD-2/00) (18) in staphylococcal enterotoxin A (SEA)-stimulated rhesus monkey peripheral blood mononuclear cells and used for these studies. This SIVmac251 stock (UCD-2/02) contains approximately 109 viral RNA (vRNA) copies/ml and a 105 50% tissue culture infection dose (TCID50)/ml when titers are determined on CEMX174 cells. Two animals were vaginally inoculated with 1 ml of the undiluted stock (105 TCID50/ml) twice in 1 day with a 4-hour interval between the inoculations. Five animals were vaginally inoculated with 1 ml of a 100-fold dilution of the stock (103 TCID50/ml). For 13 weeks, these five animals were inoculated with the 103 TCID50 inoculum twice on a single day weekly, with a 4-hour interval between the inoculations. In all cases the virus inoculum was introduced nontraumatically into the vaginal canal by using a needleless, 1-ml tuberculin syringe.
Viral RNA isolation and cDNA synthesis.
Plasma and virus stock were thawed at room temperature and RNA was isolated using the QIAamp Ultrasens viral kit (Qiagen, Valencia, CA) according to the manufacturer's protocol and eluted in 50 μl. vRNA was reverse transcribed using SuperScript III (Invitrogen, Carlsbad, CA) from the cDNA synthesis kit. A mixture of 2 μl of 50 μM of the oligo(dT) primer with the sequence dT23VN (V, anything but T; N, any nucleotide) 2 μl of 10 mM deoxynucleoside triphosphates (dNTPs), and 22 μl of viral RNA was heated to 65°C for 5 min followed by incubation on ice for 2 min. A master mix of the following was then added: 8 μl of 5× First-Strand buffer, 2 μl of 0.1 M dithiothreitol, 2 μl of RNaseOUT recombinant RNase inhibitor (40 units/μl), and 2 μl of SuperScript III reverse transcriptase (200 units/μl). Incubation steps were 25°C for 5 min, 50°C for 60 min, and 70°C for 15 min. One microliter of Escherichia coli RNase H (5 U/μl) was added, and the mixture was incubated at 37°C for 20 min. cDNA was stored at −20°C until amplification.
Single-genome nested amplification of SIVmac251 env.
Near-full-length 2.2-kb SIVmac251 env amplicons spanning nucleotides (nt) 197 to 2429 of gp160 were obtained from cDNA of plasma vRNA using nested PCR single-genome amplification (23). A 5-fold dilution series was made from cDNA in TE buffer (10 mM Tris [pH 8.0], 0.1 mM EDTA; Integrated DNA Technologies), and nested PCR was performed. A master mix of the following was made at room temperature: 5 μl of 5× Phusion HF buffer (New England BioLabs, Ipswich, MA), 0.5 μl of 10 mM dNTPs, 0.75 μl of 10 μM 251envF1 (5′-CAG TCT TTT ATG GTG TAC CAG CTT GGA GGA ATG-3′), 0.75 μl of 10 μM 251envR1 (5′-GAG GAT CCA TCT TCC ACC TCT CCT AAG AGT C-3′), 0.2 μl of Phusion Hot Start high-fidelity DNA polymerase (New England BioLabs, Ipswich, MA), and 14.8 μl of double-distilled water (ddH2O). A 23-μl volume of master mix and 2 μl of cDNA or DNA from 5-fold serial dilutions in TE were added to 0.2-ml tubes. The following cycling conditions produced the first-round 2,519-bp PCR product: 98°C for 45 s, 35 cycles of 98°C for 15 s and 72°C for 1.5 min, with a 4°C dwell. Second-round amplification entailed a master mix of 10 μl of 5× Phusion HF buffer, 1 μl of 10 mM dNTPs, 1.5 μl of 10 μM 251envF2 (5′-GGA ACA ACT CAG TGC CTA CCA GAT AAT GGT G-3′), 1.5 μl of 10 μM 251envR2 (5′-GTA GGT CAG TTC AGT CCT GAG GAC TTC TCG-3′), 0.4 μl Phusion Hot Start high-fidelity DNA polymerase, and 33.6 μl of ddH2O. A 48-μl aliquot of this master mix and 2 μl of first-round product were added to 0.2-ml tubes and cycled under the following conditions to produce a 2,316-bp PCR product: 98°C for 45 s, 35 cycles at 98°C for 15 s and 72°C for 1.5 min, with a 4°C dwell. All PCR products were visualized on a 2.0% E-gel precast agarose gel (Invitrogen).
The end point of dilution was determined to be between the last dilution reaction mixture to show a PCR positive band on the gel and the next dilution. Replicates of 24 PCR mixtures of the last dilution to show a band and of 24 PCR mixtures one dilution below that point were performed. A positive reaction rate of 30% or lower ensured that amplicons were derived from a single template. Replicates were repeated until sufficient PCR-positive reactions were produced. PCR products were purified using a QIAquick 96 PCR purification kit (Qiagen). Amplicons were eluted in 50 μl EB, and both strands were sequenced using partially overlapping fragments by Sequetech (Mountain View, CA) using BigDye Terminator methodologies on an ABI 3730xl DNA analyzer platform. To confirm PCR amplification from a single template, chromatograms were manually examined for multiple peaks, indicating the presence of amplicons that resulted from PCR-generated recombination events, Taq polymerase errors, or multiple variant templates, and thus we could ensure proportional representation of individual env sequences circulating in vivo. Any sequences containing sites of ambiguous sequence were not included in the analysis.
Sequence analysis.
Sequences were aligned using ClustalW (29) and hand edited using Jalview (2) to improve alignment quality. All trees were constructed with Phylip (3) using the neighbor-joining method (25) with the Kimura two-parameter distance matrix (12) and bootstrapped for reliability. Sequences with large deletions were omitted from the analysis. Within-subject env diversity was analyzed in three ways, as described in detail previously (9), and fell into two distinct levels, classified as either homogeneous or heterogeneous. Briefly, diversity was determined by visually inspecting sequences by using neighbor-joining phylogenies and the Highlighter tool (www.hiv.lanl.gov). Also, distribution of pairwise Hamming distances (HD) within each sample were examined for peak modality: single peaks, indicating infection from a single viral variant, and multiple peaks, reflecting infection arising from viral variants of polyphyletic lineage. Lastly, mathematical modeling was used to test predictions of expected maximum HD against assumptions of infection variant phylogenies.
Nucleotide sequence accession numbers.
All sequences were deposited in GenBank with accession numbers GU952668 to GU952761.
RESULTS
Outcome of vaginal inoculations and SIV infection kinetics.
As previously reported seven rhesus macaques were intravaginally (i.vag.) inoculated with the cell-free virus stock of SIVmac251 (17) (Table 1). Both of the animals (30991 and 31523) that were vaginally inoculated with the undiluted stock (105 TCID50/ml) were plasma viral RNA positive (vRNA+) 1 week after inoculation. Of the five animals inoculated with the lower-titer inoculum (103 TCID50), two animals (27337 and 30991) became plasma vRNA+ at week 2 (after 2 inoculations in week 1); two animals (25479 and 25948) became plasma vRNA+ at week 5 (after 8 inoculations over 4 weeks), and one animal (29459) became plasma vRNA+ at week 7 (after 12 inoculations over 6 weeks). The number of inoculations required to establish systemic infection and the vRNA levels in the first vRNA+ plasma samples of each animal are summarized in Table 1.
TABLE 1.
Summary of animals, SIV inoculations, and results
| Animal no. | SIVmac251 dose/inoculation |
First vRNA+ plasma sample |
SGA results |
Est.b min. no. of env variants in first vRNA+ plasma | ||||
|---|---|---|---|---|---|---|---|---|
| TCID50 | No. of vRNA copies | No. of inoculations required | No. of vRNA copies/ml (log10) | No. of SGAs from first vRNA+ plasma | Number of HMa among SGAs | Range (mean) of % nt diversity among SGAs | ||
| SIV 251 stock | NAc | NA | NA | NA | 65 | NA | 0-1.1 (0.47) | NA |
| 30991 | 105 | 109 | 2 | 4.5 | 23 | 1 | 0-1.1 (0.22) | 5 |
| 31523 | 105 | 109 | 2 | 4.9 | 22 | 1 | 0-1.1 (0.22) | 7 |
| 25479 | 103 | 107 | 8 | 4.3 | 21 | 1 | 0-0.37 (0.03) | 1 |
| 29271 | 103 | 107 | 2 | 5.7 | 21 | 2 | 0-0.78 (0.24) | 6 |
| 27337 | 103 | 107 | 2 | 4.6 | 22 | 1 | 0-1.06 (0.46) | 7 |
| 25948 | 103 | 107 | 8 | 2.7 | 27 | 0 | 0-0.18 (0.03) | 1 |
| 29459 | 103 | 107 | 12 | 3.4 | 26 | 1 | 0-0.33 (0.04) | 1 |
HM, APOBEC-hypermutated viral sequences.
Note that low-diversity lineages in the animals could have arisen by recombination of transmitted/founder viruses (11), and recombination-generated lineages also could have arisen in the production of the stock. Thus, the number of unique env variants transmitted to each animal is an estimate.
NA, not applicable.
Diversity of viral env in the SIVmac251 stock.
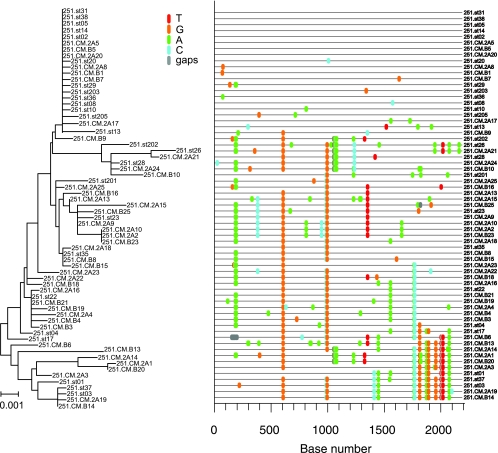
A total of 65 env nucleotide sequences were derived by SGA from the SIVmac251 stock and analyzed for sequence diversity by pairwise sequence comparison, neighbor-joining phylogenetic tree reconstruction (Phylip), and use of the Highlighter tool (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter). The Highlighter tool facilitates the tracing of common ancestry by constructing a visual representation of a nucleotide alignment where each sequence is compared to a master sequence and all polymorphisms are indicated by a colored mark. Phylogenetic relationships among the env sequences present in the SIVmac251 stock used as the inoculum in these studies are represented in a neighbor-joining tree and Highlighter plot (Fig. 1). One lineage, at the top of the tree and Highlighter plot, is represented by 20 identical or nearly identical sequences of the 65 SIV env sequences amplified from the virus stock. Because SGA provides proportional representation of viral variant frequency, approximately one-third of all the viruses in the SIVmac251 stock have identical or nearly identical env gene sequences. Overall, however, the maximum nucleotide diversity of the SIVmac251 stock used in this study was 1.1%, which is consistent with other SIVmac251 stocks (9) and with the conclusion that it is a genetically complex population of related SIV env variants (quasispecies).
FIG. 1.
Neighbor-joining tree and Highlighter plots of SGA-derived env sequences from the SIVmac251 challenge stock. Bar, 0.001 nucleotide substitutions per site. Nucleotide polymorphisms are indicated by a colored tic mark (thymine in red, guanine in orange, adenine in green, and cytosine in blue).
Viral diversity in the earliest vRNA+ plasma samples of infected animals.
A total of 162 SGA-derived 2.2-kb env sequences were obtained from plasma of seven rhesus macaques (median of 22 sequences per animal; range, 21 to 27). Each animal was sampled at the earliest plasma vRNA+ time point after vaginal inoculation with SIVmac251. The viral loads in these samples were between 103 and 105 copies/ml of plasma (Table 1). We found evidence of G-to-A mutations by using Hypermut (www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html) and Highlighter in six of the animals, representing six total hypermutated sequences. The number of mutated sequences for each animal is listed in Table 1 and indicated in brown within the phylogenetic trees. For the overall diversity calculations, individual variant hypermutated sequences and gaps (deletions) within a sequence were excluded. Maximum env diversity within the plasma samples from the animals ranged from 0.10 to 1.10%, capturing all the diversity of the inoculating stock. Maximum viral diversity of three animals (25479, 25948, and 29459) was below 0.4%, while the remaining four animals had a maximum viral diversity greater than 0.75% (Table 1).
Enumerating transmitted/founder viruses.
We sought to enumerate the total number of transmitted/founder viral lineages within each animal by using previously described criteria (26). For the enumeration of individual variants, hypermutated sequences and gaps (deletions) within a sequence were excluded. We assumed that in acute infection the virus grows exponentially with a fixed generation time with no selection pressure, no recombination, no occurrence of back mutations, and a constant mutation rate across positions and across lineages (26). Furthermore, as the kinetics of the ramp-up, peak, and set point viremia are the same in all animals, regardless of the number of times they are vaginally inoculated (16), we assumed that the inoculation 1 week before the first vRNA+ plasma sample was responsible for SIV transmission in all cases. Thus, there is sufficient time for only two nucleotide substitutions to be introduced into a transmitted sequence by the time of sampling, and based on this we assumed that if variants had unique nucleotides at more than two positions, then they arose from separately transmitted founder variants.
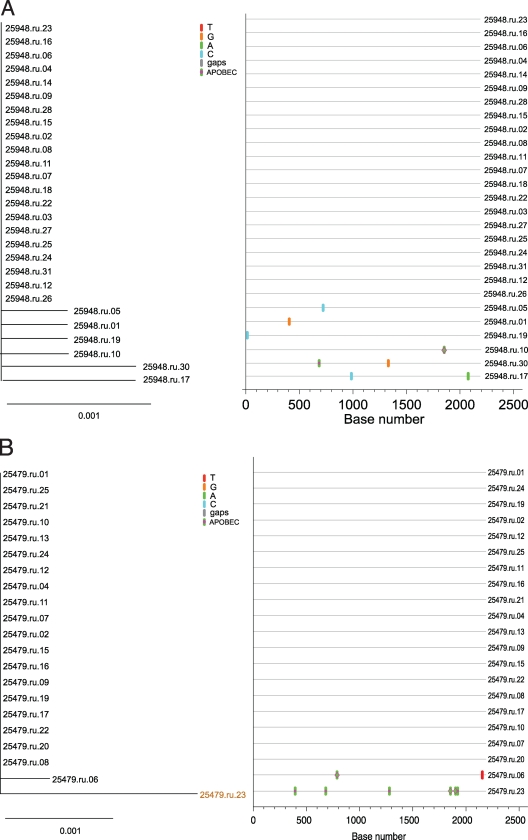
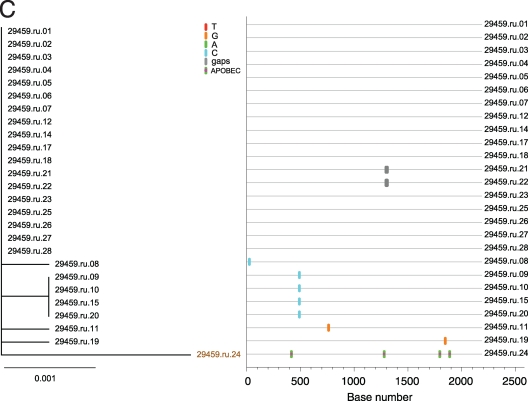
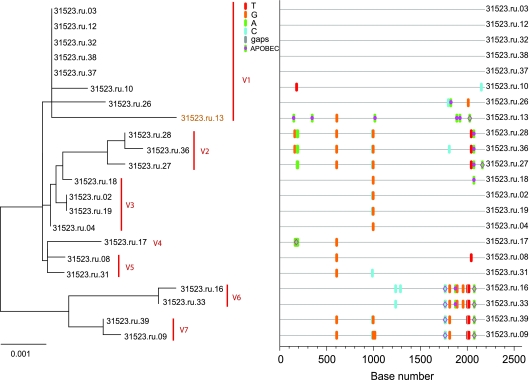
Using these criteria, the three animals with low overall viral diversity (25479, 25948, and 29459) showed phylogenetic evidence of only a single lineage at the first vRNA+ plasma sample (Fig. 2). Sequences from all three animals conformed to a star-like or near-star-like phylogeny, with most mutations randomly distributed throughout the gene. Interestingly, in animal 29459 (Fig. 2C) there are four sequences with a common polymorphism that is a mutation which is either currently being selected for or is simply a remnant of an early stochastic event predicted to have occurred within the first few rounds of replication (1). Animals with high overall viral diversity had evidence of multiple variants in the first vRNA+ plasma sample. Animals 31523 and 30991, which were inoculated with the higher-dose inoculum, were infected with at least five to seven viruses with distinct SIV env lineages representing separate transmitted/founder viruses (Fig. 3 and 4). While animals 27337 and 29271 became infected after two low-dose SIV inoculations (2 logs less virus than the high-dose group received), they had six to seven distinct, low-diversity SIV env lineages, each representing a unique transmitted/founder virus (Fig. 5 and 6). While it is formally possible that certain low-diversity lineages in these animals could have arisen as a consequence of recombination of transmitted/founder virus progeny (11), this is unlikely to have occurred within the short time period between virus transmission and sampling, and our criteria assume that recombination did not occur after transmission. Three of the animals, 25479, 25948, and 29459, were infected by a single SIV env variant, and the remaining four animals, 29271, 27337, 30991, and 31523, appear to have been infected by five to seven SIV env variants. The relative dose of the inoculum did not correlate grossly with the number of low-diversity SIV lineages in the first vRNA+ plasma sample, as three of five animals inoculated with the lower-titer inoculum became infected with multiple SIV env variants (Table 1).
FIG. 2.
Analysis of SGA-derived env sequences from macaques productively infected with a single transmitted variant. Results shown include neighbor-joining trees of SGA-derived env sequences (left panel), Highlighter plots (center panel), and phylogenetic trees of the inoculum with each animal's consensus sequence highlighted (right panel). (A) Animal 25948; (B) animal 25479; (C) animal 29459. Bar, 0.001 nucleotide substitutions per site. Nucleotide polymorphisms are indicated by a colored tic mark (thymine in red, guanine in orange, adenine in green, and cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and deletions are shown by gray tics in the Highlighter plots. ABOBEC-mediated G-to-A mutations are brown in the neighbor-joining trees.
FIG. 3.
Analysis of SGA-derived env sequences from animal 31523, which was productively infected with at least seven distinct transmitted SIV variants. The neighbor-joining tree and Highlighter plot indicate the presence of multiple low-level diversity lineages, and two lineages are represented by a single viral sequence. Bar, 0.001 nucleotide substitutions per site. Nucleotide polymorphisms are indicated by a colored tic mark (thymine in red, guanine in orange, adenine in green, and cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and deletions are shown by gray tics in the Highlighter plot. ABOBEC-mediated G-to-A mutations are brown in the neighbor-joining tree.
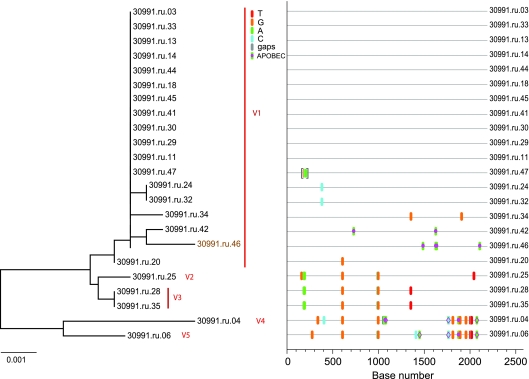
FIG. 4.
Analysis of SGA-derived env sequences from animal 30991, which was productively infected with at least five distinct transmitted variants. The neighbor-joining tree and Highlighter plot indicate the presence of at least five low-level diversity lineages, and two lineages are represented by a single viral sequence. Bar, 0.001 nucleotide substitutions per site. Nucleotide polymorphisms are indicated by a colored tic mark (thymine in red, guanine in orange, adenine in green, and cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and deletions are shown by gray tics in the Highlighter plot. ABOBEC-mediated G-to-A mutations are brown in the neighbor-joining tree.
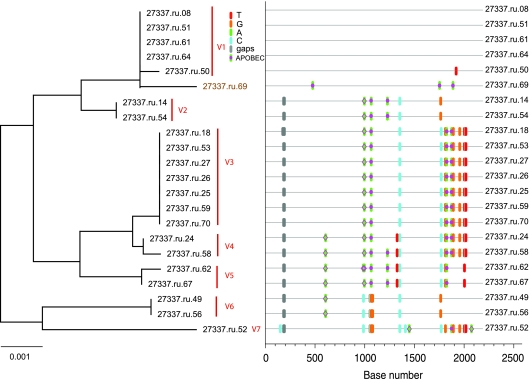
FIG. 5.
Analysis of SGA-derived env sequences from animal 27337, which was productively infected with at least seven distinct transmitted SIV variants. The neighbor-joining tree and Highlighter plot indicate the presence of multiple low-level diversity lineages. Bar, 0.001 nucleotide substitutions per site. Nucleotide polymorphisms are indicated by a colored tic mark (thymine in red, guanine in orange, adenine in green, and cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and deletions are shown by gray tics in the Highlighter plot. ABOBEC-mediated G-to-A mutations are brown in the neighbor-joining tree.
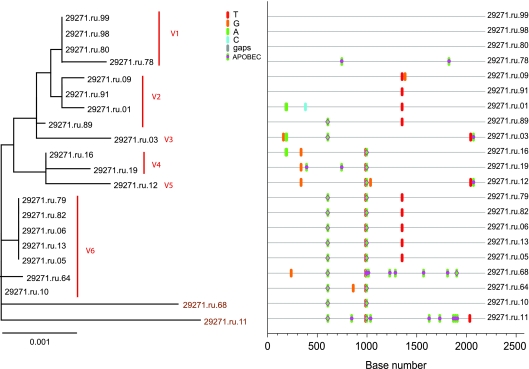
FIG. 6.
Analysis of SGA-derived env sequences from animal 29271, which was productively infected with at least six distinct transmitted variants. The neighbor-joining tree and Highlighter plot indicate the presence of at least six low-level diversity lineages, and five lineages are represented by a single viral sequence. Bar, 0.001 nucleotide substitutions per site. Nucleotide polymorphisms are indicated by a colored tic mark (thymine in red, guanine in orange, adenine in green, and cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and deletions are shown by gray tics in the Highlighter plot. ABOBEC-mediated G-to-A mutations are brown in the neighbor-joining tree.
Transmitted/founder viruses compared to the SIVmac251 stock.
The model of early HIV-1 diversification (9) allows predictions that can be tested directly in the SIV/macaque model. One of these predictions is that the consensus of SGA-derived HIV-1 sequences represents the transmitted or founder virus. In three animals there were six transmitted SIV env lineages that were identical to SIV env variants found in the SIVmac251 stock inoculum (Fig. 7 and 8). This represents the first direct evidence confirming the predictions of the mathematical model of early HIV diversification after intravaginal inoculation of SIV. Among three other animals infected by single viruses (25479, 25948, and 29459), two were infected with SIV env variants that are closely related to sequences in the SIVmac251 stock. Interestingly, the transmitted virus from monkey 29459 did not closely cluster with any virus sequenced from the inoculating stock (Fig. 7). In fact, the nearest stock env sequence is nearly 1% removed from this transmitted SIV env variant. We generated a large number of sequences from the challenge stock without detecting this particular SIV env lineage, so it was most likely present in the stock at low frequency (less than 2%), although the possibility that it was generated by recombination of two transmitted viruses cannot be ruled out.
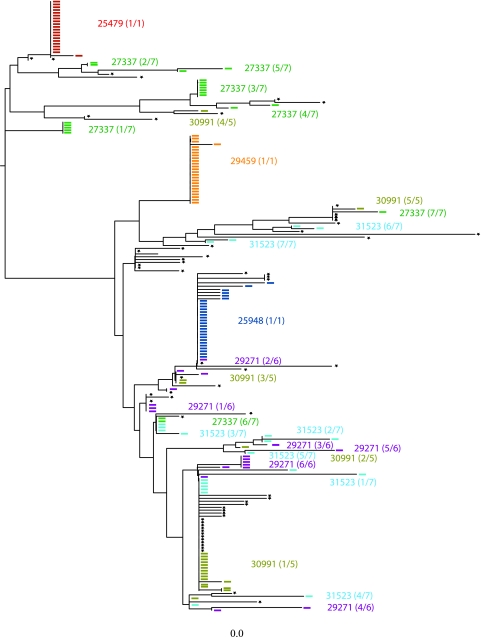
FIG. 7.
Composite neighbor-joining analysis of env diversity in SIVmac251 stock and the earliest plasma vRNA+ samples from infected animals. Variants from individual animals are represented as colored ticks on the phenogram; the colors of the ticks correspond to the animal that was the source of the variant. The number of the transmitted variants over the number of transmitted lineages observed in each animal is noted. Black stars indicate the variants that were amplified from the SIVmac251 stock. Bar, 0.001 nucleotide substitutions per site.
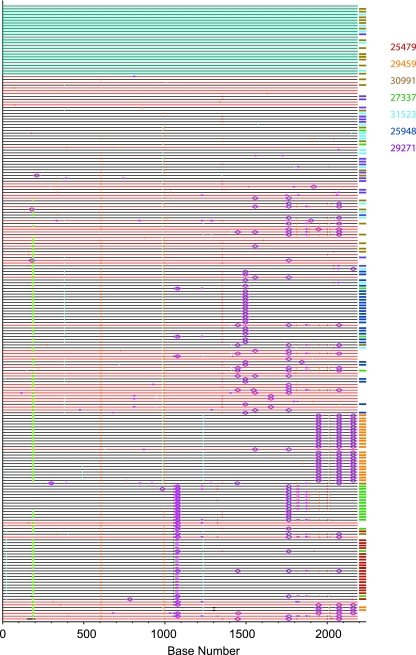
FIG. 8.
Composite alignment of the SIVmac251 stock env sequences and plasma SIV env sequences. Results shown are the Highlighter plot of SIVmac251 stock (red lines) and earliest plasma vRNA+ sample from infected animals (black lines). Variants isolated from the individual animals are indicated by the different-colored tic marks to the right of each line. Individual nucleotide polymorphisms are indicated by green (adenine), red (thymine), yellow (guanine), or blue (cytosine) tics and are in comparison to master sequence 251st01. Gaps are indicated in gray. To simplify the figure, APOBEC signatures and G-to-A conversions are not specifically indicated. The blue box denotes the group of identical sequences found in the SIVmac251 stock inoculum and the plasma samples from three animals.
Interanimal and SIVmac251 stock env diversity.
All 65 sequences from the SIVmac251 stock and 162 sequences from plasma samples from seven rhesus macaques amplified from the first vRNA+ plasma sample were aligned and analyzed by neighbor-joining (Fig. 7) and Highlighter (Fig. 8) analyses. Low-diversity SIV env lineages represent transmitted SIV env variants with only a few randomly accumulated mutations. SIVmac251 stock sequences are interspersed among the animal plasma sequences, and there appears to be only one lineage that was transmitted to more than one animal. However, this SIV env lineage is overrepresented in the challenge stock and therefore may be consistently transmitted simply due to its prevalence in the inoculum.
DISCUSSION
The nature of the virus transmitted from an HIV-infected person to an uninfected partner has been intensely investigated, because this virus must be targeted by microbicides and/or vaccine-induced immunity to prevent viral infection from becoming established in the naïve host. Based on studies of limited numbers of people, it has been known for some time that newly infected individuals have relatively few HIV genetic variants compared to the large number of variants forming the HIV quasispecies in individuals infected for longer periods (31-34). Of note, the direction of heterosexual transmission does not seem to affect this genetic bottleneck, as it has been reported in naïve women partnered with infected men and in naïve men partnered with infected females (31-34). Recent studies of viral variants in relatively large numbers of newly infected people, using the SGA technique to avoid the artifacts inherent in bacterial cloning of viral sequences, have confirmed and extended these initial results to the point that it is now clear that the majority of humans are infected with a few HIV-1 env variants. The consistent observation that only a limited number of HIV-1 variant viruses are commonly transmitted can be explained by four possible scenarios that are not mutually exclusive: (i) the infecting partner could shed a limited number of infectious viruses; (ii) there could be selection for a specific infecting variant at the mucosal barrier; (iii) stochastic exclusion of most variants in a complex inoculum could occur at the mucosal barrier; (iv) there could be competition for a limited number of target cells in the mucosa among transmitted HIV-1 variants with a single variant outcompeting the others (31-34). Mucosal inoculation of nonhuman primates with SIV offers an excellent opportunity to verify and extend the conclusions of the HIV transmission studies, because the SIV env variants in the virus stock used for inoculation can be determined, the route of virus exposure can be strictly controlled, and the time of inoculation is known. In fact, the first nonhuman primate study to explore these issues using the SGA technology demonstrated that rhesus macaques inoculated i.r. with either SIVmac251 or SIVsmE660 became infected with one or a few viruses that diversified in a manner similar to HIV-1 in humans (10).
In a previous study using much less robust heteroduplex mobility assay technology, we concluded that after intravenous SIVmac251 inoculation rhesus monkeys became infected with highly diverse populations of SIV env genetic variants (5), and while three of five i.vag.-inoculated monkeys became infected with a homogeneous population of SIV env variants, two of five monkeys were infected with complex populations of env variants that were similar to the intravenously inoculated animals (5). In the present study, we found that after i.vag. SIVmac251 inoculation, three of seven rhesus macaques became infected with 1 viral variant, while four of seven animals became infected with ≥5 variants. As in our previous study (5), here we found that some i.vag.-inoculated animals became infected with the most common variant in the stock inoculum. Thus, two studies using very different technologies to define either the complexity of transmitted viral populations or the exact viral variants after i.vag. inoculation with a genetically diverse SIVmac251 stock have yielded very similar results: approximately 50% of animals became infected with one to two env variants, and 50% became infected with complex viral env populations. Consistent with the results of HIV transmission studies (1, 6, 8, 9, 26, 27), the number of variants infecting rhesus macaques after i.vag SIVmac251 inoculation does not form a normal distribution around some mean number of transmitted variants, with a disproportionate number of animals infected with a relatively large number of variants. Further, the relative number of SIV variants transmitted did not correlate with the dose of virus in the inoculum, as two animals inoculated with the relatively low, but not limiting, dose of SIVmac251 became infected with at least six env variants. Finally, in both the current study and our previous study (5) of SIV variant transmission after vaginal SIVmac251 inoculation, a specific viral variant in the SIVmac251 stock was not consistently transmitted by i.vag. inoculation.
Indeed, in Fig. 7 it is apparent that the transmitted SIV env variants are randomly distributed along the neighbor-joining tree and that there are no distinctive phylogenetic relationships present among the transmitted variants.
In the present study, there was an inverse association between the number of SIV inoculations that produced infection and the number of viral variants transmitted. Thus, among the five animals inoculated with 103 TCID50 of SIVmac251, the two animals that were infected with multiple SIV env variants became infected after only two intravaginal inoculations, while the three animals that were infected with one SIV env variant required 8 to 12 virus inoculations to become infected. Although the number of animals studied is small, this relationship suggests that after vaginal inoculation, some animals are readily susceptible to infection while others are relatively resistant. Although infection is generally initiated by a single HIV-1 variant that is derived from the quasispecies of the transmitting partner, infection with multiple HIV-1 variants does occur (6, 9), and an association between infection by multiple genetic variants and inflammatory genital infections in the newly infected individual has been reported (6). Although this association could provide an explanation for the findings in the present study, direct experimental evidence to support the hypothesis that genital tract inflammation increases the number of variants transmitted is lacking. Vaginal SIV transmission can provide a ready model to test the putative association between genital tract inflammation and infection with a relatively large number of viral variants.
The results of this study on vaginal SIV transmission and of a previous study of rectal SIV transmission (10) demonstrate that one to two env variants establish productive infection in rhesus macaques after mucosal challenge with an inoculum containing numerous SIV env variants. Thus, it is possible to conclude that exposure to a limited number of viral variants does not explain the extremely limited number of HIV viruses in people after HIV transmission. However, the results of the present study and similar studies of SIV and HIV variant transmission have not distinguished between exclusion of viral variants at the mucosal surface and the competitive selection among transmitted variants as they disseminate from the mucosa to the systemic compartment. Finally, although assessing the viral variants in plasma is convenient and may be the only practical option in humans, we have shown that after vaginal SIV inoculation the first virus detected in the plasma can be a recombinant variant of two viruses inoculated onto the mucosa rather than a strain present in the inoculum (11). Thus, additional studies are needed to determine if variant selection at the mucosal surface or in the mucosa and draining lymph nodes during the earliest stages of infection accounts for the limited number of viruses in infected individuals after mucosal HIV and SIV transmission. Taken together, the results reported here demonstrate that the SIVmac251 vaginal transmission model can recapitulate the low number of HIV-1 variants transmitted mucosally and that the model can be used to assess the effects of host and viral factors on HIV-1 variant transmission.
Acknowledgments
We thank the Primate Services Unit at the CNPRC and Katy Lantz, Ding Lu, Lili Guo, Tracy Rourke, Linda Fritts, and Veronique de Silva for excellent technical assistance.
This work was supported by NIH grants RR00169, AI066314, AI44480, and AI087383, a grant from the Bill & Melinda Gates Foundation (number 37874), National Cancer Institute contract no. HHSN266200400088C, and a gift from the James B. Pendleton Charitable Trust.
Footnotes
Published ahead of print on 12 May 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Abrahams, M. R., J. A. Anderson, E. E. Giorgi, C. Seoighe, K. Mlisana, L. H. Ping, G. S. Athreya, F. K. Treurnicht, B. F. Keele, N. Wood, J. F. Salazar-Gonzalez, T. Bhattacharya, H. Chu, I. Hoffman, S. Galvin, C. Mapanje, P. Kazembe, R. Thebus, S. Fiscus, W. Hide, M. S. Cohen, S. A. Karim, B. F. Haynes, G. M. Shaw, B. H. Hahn, B. T. Korber, R. Swanstrom, and C. Williamson. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 83:3556-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 3.Felsenstein, J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22:521-565. [DOI] [PubMed] [Google Scholar]
- 4.Ganeshan, S., R. E. Dickover, B. T. Korber, Y. J. Bryson, and S. M. Wolinsky. 1997. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J. Virol. 71:663-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenier, J. L., C. J. Miller, D. Lu, P. J. Dailey, F. X. Lu, K. J. Kunstman, S. M. Wolinsky, and M. L. Marthas. 2001. Route of simian immunodeficiency virus inoculation determines the complexity but not the identity of viral variant populations that infect rhesus macaques. J. Virol. 75:3753-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haaland, R. E., P. A. Hawkins, J. Salazar-Gonzalez, A. Johnson, A. Tichacek, E. Karita, O. Manigart, J. Mulenga, B. F. Keele, G. M. Shaw, B. H. Hahn, S. A. Allen, C. A. Derdeyn, and E. Hunter. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 5:e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heise, C., C. J. Miller, A. Lackner, and S. Dandekar. 1994. Primary acute simian immunodeficiency virus infection of intestinal lymphoid tissue is associated with gastrointestinal dysfunction. J. Infect. Dis. 169:1116-1120. [DOI] [PubMed] [Google Scholar]
- 8.Kearney, M., F. Maldarelli, W. Shao, J. B. Margolick, E. S. Daar, J. W. Mellors, V. Rao, J. M. Coffin, and S. Palmer. 2009. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J. Virol. 83:2715-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keele, B. F., E. E. Giorgi, J. F. Salazar-Gonzalez, J. M. Decker, K. T. Pham, M. G. Salazar, C. Sun, T. Grayson, S. Wang, H. Li, X. Wei, C. Jiang, J. L. Kirchherr, F. Gao, J. A. Anderson, L. H. Ping, R. Swanstrom, G. D. Tomaras, W. A. Blattner, P. A. Goepfert, J. M. Kilby, M. S. Saag, E. L. Delwart, M. P. Busch, M. S. Cohen, D. C. Montefiori, B. F. Haynes, B. Gaschen, G. S. Athreya, H. Y. Lee, N. Wood, C. Seoighe, A. S. Perelson, T. Bhattacharya, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keele, B. F., H. Li, G. H. Learn, P. Hraber, E. E. Giorgi, T. Grayson, C. Sun, Y. Chen, W. W. Yeh, N. L. Letvin, J. R. Mascola, G. J. Nabel, B. F. Haynes, T. Bhattacharya, A. S. Perelson, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, E. Y., M. Busch, K. Abel, L. Fritts, P. Bustamante, J. Stanton, D. Lu, S. Wu, J. Glowczwskie, T. Rourke, D. Bogdan, M. Piatak, Jr., J. D. Lifson, R. C. Desrosiers, S. Wolinsky, and C. J. Miller. 2005. Retroviral recombination in vivo: viral replication patterns and genetic structure of simian immunodeficiency virus (SIV) populations in rhesus macaques after simultaneous or sequential intravaginal inoculation with SIVmac239Δvpx/Δvpr and SIVmac239Δnef. J. Virol. 79:4886-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 13.Lee, H. Y., E. E. Giorgi, B. F. Keele, B. Gaschen, G. S. Athreya, J. F. Salazar-Gonzalez, K. T. Pham, P. A. Goepfert, J. M. Kilby, M. S. Saag, E. L. Delwart, M. P. Busch, B. H. Hahn, G. M. Shaw, B. T. Korber, T. Bhattacharya, and A. S. Perelson. 2009. Modeling sequence evolution in acute HIV-1 infection. J. Theor. Biol. 261:341-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lifson, J. D., M. A. Nowak, S. Goldstein, J. L. Rossio, A. Kinter, G. Vasquez, T. A. Wiltrout, C. Brown, D. Schneider, L. Wahl, A. L. Lloyd, J. Williams, W. R. Elkins, A. S. Fauci, and V. M. Hirsch. 1997. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J. Virol. 71:9508-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma, Z., F. X. Lu, M. Torten, and C. J. Miller. 2001. The number and distribution of immune cells in the cervico-vaginal mucosa remain constant throughout the menstrual cycle of rhesus macaques. Clin. Immunol. 100:240-249. [DOI] [PubMed] [Google Scholar]
- 16.Ma, Z. M., K. Abel, T. Rourke, Y. Wang, and C. J. Miller. 2004. A period of transient viremia and occult infection precedes persistent viremia and antiviral immune responses during multiple low-dose intravaginal simian immunodeficiency virus inoculations. J. Virol. 78:14048-14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma, Z. M., M. Stone, M. Piatak, Jr., B. Schweighardt, N. L. Haigwood, D. Montefiori, J. D. Lifson, M. P. Busch, and C. J. Miller. 2009. High specific infectivity of plasma virus from the pre-ramp-up and ramp-up stages of acute simian immunodeficiency virus infection. J. Virol. 83:3288-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marthas, M. L., D. Lu, M. C. Penedo, A. G. Hendrickx, and C. J. Miller. 2001. Titration of an SIVmac251 stock by vaginal inoculation of Indian and Chinese origin rhesus macaques: transmission efficiency, viral loads, and antibody responses. AIDS Res. Hum. Retrovir. 17:1455-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, C. J., N. J. Alexander, S. Sutjipto, A. A. Lackner, A. G. Hendrickx, A. Gettie, L. J. Lowenstine, M. Jennings, and P. A. Marx. 1989. Genital mucosal transmission of simian immunodeficiency virus: animal model for heterosexual transmission of human immunodeficiency virus. J. Virol. 63:4277-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, C. J., Q. Li, K. Abel, E. Y. Kim, Z. M. Ma, S. Wietgrefe, L. La Franco-Scheuch, L. Compton, L. Duan, M. D. Shore, M. Zupancic, M. Busch, J. Carlis, S. Wolinsky, and A. T. Haase. 2005. Propagation and dissemination of infection after vaginal transmission of simian immunodeficiency virus. J. Virol. 79:9217-9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, C. J., M. Marthas, J. Torten, N. J. Alexander, J. P. Moore, G. F. Doncel, and A. G. Hendrickx. 1994. Intravaginal inoculation of rhesus macaques with cell-free simian immunodeficiency virus results in persistent or transient viremia. J. Virol. 68:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowak, M. A., A. L. Lloyd, G. M. Vasquez, T. A. Wiltrout, L. M. Wahl, N. Bischofberger, J. Williams, A. Kinter, A. S. Fauci, V. M. Hirsch, and J. D. Lifson. 1997. Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J. Virol. 71:7518-7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer, S., M. Kearney, F. Maldarelli, E. K. Halvas, C. J. Bixby, H. Bazmi, D. Rock, J. Falloon, R. T. Davey, Jr., R. L. Dewar, J. A. Metcalf, S. Hammer, J. W. Mellors, and J. M. Coffin. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sagar, M., L. Lavreys, J. M. Baeten, B. A. Richardson, K. Mandaliya, B. H. Chohan, J. K. Kreiss, and J. Overbaugh. 2003. Infection with multiple human immunodeficiency virus type 1 variants is associated with faster disease progression. J. Virol. 77:12921-12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 26.Salazar-Gonzalez, J. F., E. Bailes, K. T. Pham, M. G. Salazar, M. B. Guffey, B. F. Keele, C. A. Derdeyn, P. Farmer, E. Hunter, S. Allen, O. Manigart, J. Mulenga, J. A. Anderson, R. Swanstrom, B. F. Haynes, G. S. Athreya, B. T. Korber, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salazar-Gonzalez, J. F., M. G. Salazar, B. F. Keele, G. H. Learn, E. E. Giorgi, H. Li, J. M. Decker, S. Wang, J. Baalwa, M. H. Kraus, N. F. Parrish, K. S. Shaw, M. B. Guffey, K. J. Bar, K. L. Davis, C. Ochsenbauer-Jambor, J. C. Kappes, M. S. Saag, M. S. Cohen, J. Mulenga, C. A. Derdeyn, S. Allen, E. Hunter, M. Markowitz, P. Hraber, A. S. Perelson, T. Bhattacharya, B. F. Haynes, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shattock, R. J., and J. P. Moore. 2003. Inhibiting sexual transmission of HIV-1 infection. Nat. Rev. Microbiol. 1:25-34. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wawer, M. J., R. H. Gray, N. K. Sewankambo, D. Serwadda, X. Li, O. Laeyendecker, N. Kiwanuka, G. Kigozi, M. Kiddugavu, T. Lutalo, F. Nalugoda, F. Wabwire, M. P. Meehan, and T. C. Quinn. 2005. Rates of HIV-1 transmission per coital act, by stage of HIV, HIV-1 infection, in Rakai, Uganda. J. Infect. Dis. 191:1403-1409. [DOI] [PubMed] [Google Scholar]
- 31.Wolinsky, S. M., C. M. Wike, B. T. Korber, C. Hutto, W. P. Parks, L. L. Rosenblum, K. J. Kunstman, M. R. Furtado, and J. L. Munoz. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134-1137. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, L. Q., P. MacKenzie, A. Cleland, E. C. Holmes, A. J. Leigh Brown, and P. Simmonds. 1993. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J. Virol. 67:3345-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, T., N. Wang, A. Carr, D. S. Nam, R. Moor-Jankowski, D. A. Cooper, and D. D. Ho. 1996. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for virus compartmentalization and selection during sexual transmission. J. Virol. 70:3098-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]