Abstract
Nonprimate animal models of HIV-1 infection are prevented by missing cellular cofactors and by antiviral actions of species-specific host defense factors. These blocks are profound in rodents but may be less abundant in certain Carnivora. Here, we enabled productive, spreading replication and passage of HIV-1 in feline cells. Feline fibroblasts, T-cell lines, and primary peripheral blood mononuclear cells supported early and late HIV-1 life cycle phases in a manner equivalent to that of human cells, except that produced virions had low infectivity. Stable expression of feline immunodeficiency virus (FIV) Vif-green fluorescent protein (GFP) in HIV-1 entry receptor-complemented feline (CrFK) cells enabled robust spreading HIV-1 replication. FIV Vif colocalized with feline APOBEC3 (fA3) proteins, targeted them for degradation, and prevented G→A hypermutation of the HIV-1 cDNA by fA3CH and fA3H. HIV-1 Vif was inactive against fA3s as expected and even paradoxically augmented restriction in some assays. In an interesting contrast, simian immunodeficiency virus SIVmac Vif had substantial anti-fA3 activities, which were complete against fA3CH and partial against fA3H. Moreover, both primate lentiviral Vifs colocalized with fA3s and could be pulled down from cell lysates by fA3CH. HIV-1 molecular clones that encode FIV Vif or SIVmac Vif (HIV-1VF and HIV-1VS) were then constructed. These viruses replicated productively in HIV-1 receptor-expressing CrFK cells and could be passaged serially to uninfected cells. Thus, with the exception of entry receptors, the cat genome can supply the dependency factors needed by HIV-1, and a main restriction can be countered by vif chimerism. The results raise the possibility that the domestic cat could yield an animal model of HIV-1 infection.
To improve the relevance of macaque models to human immunodeficiency virus type 1 (HIV-1)/AIDS research, simian immunodeficiency viruses (SIVs) that contain various portions of HIV-1 have been developed, beginning with simian/human immunodeficiency viruses (SHIVs) that incorporated HIV-1 tat, rev, vpu, and env into SIVmac (83). More recently, HIV-1 clones in which only the vif gene or vif and capsid sequences from the SIVmac/SIVsm/HIV-2 lineage were introduced, which allowed the viruses to evade macaque intrinsic immunity defenses, were developed (23, 24, 30, 33). In a promising recent iteration, peak HIV-1 viremia in the range of 105 to 106 RNA copies/ml followed by gradually declining replication for approximately 6 months was achieved in pig-tailed macaques with a vif-only chimera (23). Their vital roles notwithstanding, macaque models are not without limitations, such as relative scarcity and expense, a breeding time of 5 to 6 months, risk of a casually transmissible lethal zoonosis (B virus) to handlers (27), and so far, inability of modified HIV-1 to produce two hallmarks of human infection: disease and chronic, sustained viral replication (1). Whether or not macaques become fully realized as HIV-1/AIDS disease models, a nonprimate animal model of HIV-1 replication in a highly available, small, and readily bred species, e.g., transgenic mice, could be extremely useful for vaccine, pathogenesis, and therapeutics research. However, progress in this direction has been distinctly limited. HIV-1, like all lentiviruses, displays a narrow host tropism that reflects viral requirements for specific host cell factors and antiviral activities of species-specific restriction factors. For example, introduction of human CD4 and a chemokine coreceptor into rodent fibroblasts overcomes the entry block to HIV-1 (17), and expression of human cyclin T1 restores efficient Tat-dependent proviral transcription (91). However, there is marked attenuation of virus production, a defect attributed mainly to a lack of factors needed for proper viral assembly (4, 51, 89). Murine T cells also display a postentry block to HIV-1 (3).
Even if all necessary human dependency factors can be identified and made to function in a nonprimate animal with transgenic methods, HIV-1 replication is inhibited in the cells of many primate (29, 74, 87) and nonprimate (32, 50, 61, 76, 84) species by the antiviral activity of species-specific restriction factors, such as TRIM5α, TRIMCyp, and APOBEC3 (A3) proteins. The APOBEC3 proteins are DNA and RNA cytidine deaminases, some of which have antiviral activity. In human cells, Vif-deficient HIV-1 is inhibited by APOBEC3G (81), which is encapsidated into budding virions through interactions with Gag and viral and/or small cellular RNAs (36, 45, 75, 88, 93, 95; see also reference 47 for a recent review). During subsequent target cell reverse transcription, APOBEC3G deaminates minus-strand cytidines to uridine, resulting in the accumulation of deleterious G→A mutations on the plus strand (6, 21, 48, 50, 96). APOBEC3G has also been reported to interfere with reverse transcription at various steps (5, 20, 31, 41, 53). The degree to which the observed DNA synthesis impairments are separable from the effects of deamination on nascent genomes is not yet clear. In particular, APOBEC3G and -F have been reported to exert deaminase-independent antiviral activities, although the contribution of these to the overall antiviral activity is currently controversial (5, 7, 11, 26, 59, 62, 63, 79). Inhibition of HIV-1 reverse transcript elongation has been proposed as a dominant mechanism (7).
With the exception of equine infectious anemia virus (EIAV), all primate, ungulate, and feline lentiviruses encode a Vif protein. The primate lentiviral Vif proteins function primarily to deplete cellular APOBEC3G/F by recruiting an E3 ubiquitin ligase complex comprised of cullin5 (cul5), elongin B, elongin C, and an unidentified E2-conjugating enzyme, thereby inducing APOBEC3G polyubiquitination and proteasomal degradation (13, 52, 57, 82, 86, 94). In the absence of Vif, or in a species for which a particular lentiviral Vif protein is ineffective, APOBEC3G potently restricts viral replication.
The domestic cat genome encodes a truncated TRIM5α protein that lacks the capsid-engaging B30.2 domain and does not restrict HIV-1, SIVmac, or N-tropic murine leukemia virus (N-MLV) (54). Consistent with this, feline cells lack significant postentry restricting activity toward primate lentiviruses and have frequently been used as null backgrounds for testing antiviral properties of primate TRIM5 proteins or for HIV entry receptor studies (34, 55). The domestic cat genome encodes five APOBEC3 proteins (60). Three closely related feline A3C (fA3C) proteins (fA3Ca, fA3Cb, and fA3Cc) are active against feline foamy virus but not vif-deficient FIV (60). In contrast, the two other proteins (fA3H and fA3CH) have antilentiviral effects, as they restrict Δvif FIV and HIV-1 with vif intact (60, 61). fA3CH, the only two-domain feline A3 protein, is an unusual hybrid encoded by exons 1 to 3 of fA3Ca, exon 4 of fA3Cb, and exons 2 to 5 of fA3H (60). fA3H and fA3CH mediate hypermutation of wild-type HIV-1 (61). Whether fA3 proteins act through other mechanisms as well, whether any Vif protein of any lentivirus triggers fA3 degradation, or whether any Vif can protect HIV-1 against them has not been determined.
In the present study, we analyzed the limits to HIV-1 propagation in a variety of feline cells. We characterized biochemical and virological properties of FIV, HIV-1, and SIVmac Vif proteins with respect to fA3Ca, fA3H, and fA3CH. We established that FIV Vif acts similarly to primate Vifs, by reducing A3 levels and preventing hypermutation. We demonstrated that productive, spreading replication of fully wild-type HIV-1 can be enabled in a feline cell line (CrFK) by stable in trans expression of FIV Vif, identifying fA3 proteins as the principal restriction to HIV-1 replication in these cells. We show further that SIVmac Vif can also interact with, degrade, and block hypermutation by fA3 proteins and that chimeric HIV-1 molecular clones that express either FIV Vif or SIVmac Vif can replicate and be continuously passaged in the HIV-1 receptor-complemented feline cells. The data establish that the feline genome can provide all dependency factors needed for HIV-1 replication once viral entry is enabled by expression of cell surface receptors.
MATERIALS AND METHODS
fA3 nomenclature.
In the present work, we use the initial C/H/CH nomenclature for fA3 proteins (61) because of its verbal and lexical simplicity as well as to facilitate comparison with prior publications and fA3 sequence database information. “fA3Ca,” “fA3H,” and “fA3CH” correspond to suggested “fA3Z2b,” “fA3Z3,” and “fA3Z2b-Z3” names in a recent proposal (39) that Z domain composition-based names of the type recently assigned by LaRue and colleagues to artiodactyl A3s (40) be henceforth used for all nonprimate A3s. We concur with LaRue et al. (39) that specific orthologous relationships should not be inferred between proteins designated by same A-to-H letter (e.g., the respective feline and human C and H proteins). In regard to Z domain composition (Z1, Z2, or Z3), the initial feline A3 nomenclature captures the fact that human and feline H proteins are both unidomain Z3 proteins and the only Z3 proteins in their respective repertoires. Similarly, the human C protein and the feline C proteins are each unidomain Z2 proteins and are also the only such proteins in their respective repertoires. However, amino acid homology is limited. A3 repertoires are outcomes of complex evolutionary processes in each species. Accordingly, there is no correlation between Z domain composition and antiretroviral properties.
Construction of feline APOBEC3 and lentiviral Vif expression plasmids.
To generate C-terminally FLAG epitope-tagged feline APOEBC3 cDNAs, total RNA was isolated from CrFK cells (RNeasy; Qiagen), reverse transcribed using random hexamers (SuperScript III; Invitrogen), and PCR amplified (Phusion; Finnzymes) with the following primer sets: S-fA3Ca (5′-ATATCTCGAGACCATGGAGCCCTGGCGCCCCAGC-3′) and AS-fA3Ca (5′-ATATGAATTCTCATTTGTCGTCATCGTCTTTGTAGTCCCTAAGGATTTCTTGAAGCTC-3′), S-fA3H (5′-ATATCTCGAGACCTGGAGGCAGCCTGGGAGGTG-3′) and AS-fA3H/CH (5′-ATATGAATTCTCATTTGTCGTCATCGTCTTTGTAGTCTTCAAGTTTCAAATTTCTGAAGTCATTCC-3′), and S-fA3CH (5′-ATATACCCTCGAGACCAAGGCTGGAGAGAGGAATGG-3′) and AS-fA3H/CH. PCR products were cloned into pCDNA3.1(−) (Invitrogen) with the use of XhoI and EcoRI to create pcDNA3.1-fA3Ca(FLAG), pcDNA3.1-fA3H(FLAG), and pcDNA3.1-fA3CH(FLAG), which were confirmed by sequencing. The fA3 cDNA nucleotide sequences we obtained matched precisely the GenBank entries corresponding to fA3Ca (EU109281.1; protein ID ABW83272.1), fA3H (EU109281; protein ID ABW83274.1), and fA3CH (EF173021; protein ID ABO82577.1). A codon-optimized FIV C36 allele (termed vifCO) was synthesized (GenScript Corporation) to optimize expression in human cells, remove potential RNA splice donors and acceptors, and include a C-terminal hemagglutinin (HA) epitope. C-terminally HA epitope-tagged HIV-1 NL4-3, SIVmac, and FIV C36 Vif expression plasmids were made by PCR-based cloning into the polylinker of a human cytomegalovirus (CMV) promoter expression plasmid p1012 (22). Constructed plasmids were confirmed by Sanger sequencing.
Cells.
Adherent cell lines were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and T-cell lines in RPMI medium supplemented with 10% FCS, with penicillin/streptomycin and l-glutamine. FeT-J cells were a gift of J. Yamamoto, and KE-R cells (61) were a gift of Carsten Münk. Primary blood mononuclear cells of normal cat and human donors were Ficoll purified from whole blood and maintained in RPMI medium supplemented with 10% FCS, 2 mM glutamine, 1 mM sodium pyruvate, essential and nonessential amino acids, 10 mM HEPES, 0.05 mM β-mercaptoethanol, phytohemagglutinin E (PHA-E; 2 μg/ml), and human IL-2 (50 U/ml, used in experiment 1 shown in Fig. 1B). An alternative source of IL-2 (used in experiments 2 and 3 shown in Fig. 1B) was conditioned medium from murine L2.23 feeder cells (92), which were a kind gift of T. Miyazawa. PHA-E was discontinued 48 h after isolation. For macrophages, peripheral blood mononuclear cells (PBMCs) were obtained by Ficoll centrifugation of feline PBMCs obtained by blood drawing or human PBMCs eluted from Mayo Clinic Blood Bank apheresis machine leukoreduction system chambers (10), which were cultured in RPMI medium with 10% heat-inactivated filtered human AB serum (Irvine Scientific) or fetal bovine serum (Gibco). Cells were plated at a density of 5 × 106 cells per ml and allowed to differentiate to macrophages by plastic adherence. At day 7 postisolation, cells formed a confluent adherent monolayer with characteristic fried-egg morphology. A 1:1 trypsin-Versene solution was used to replate the macrophages into 24-well plates at 2 × 105 cells per well. Transduction with challenge vector HIV-1luc was done with six serial 1:3 dilutions. At day 3 posttransduction, cells were removed from wells with trypsin-Versene solution, counted, and lysed for luciferase activity measurements by using SteadyGlo or BrightGlo (Promega). Luciferase activities were normalized for total protein (measured with the Bio-Rad agent) or for cell number counted at the time of lysate harvest.
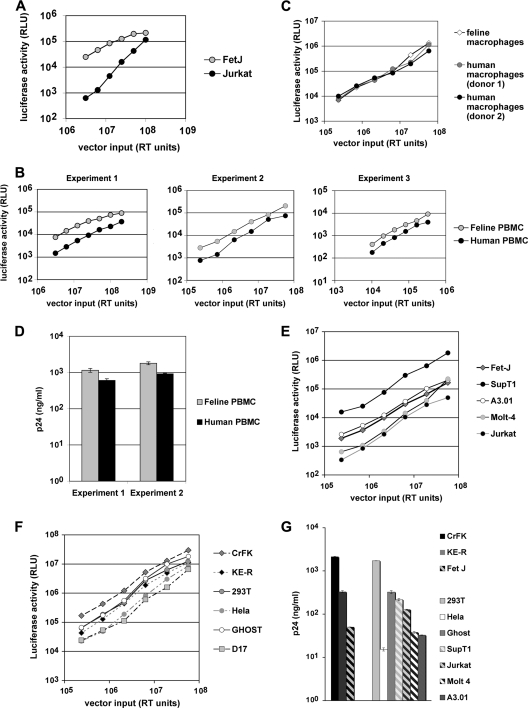
FIG. 1.
Early and late HIV-1 viral gene expression in primary PBMCs, primary macrophages, CD4+ T-cell lines, and fibroepithelial cell lines. For panels A to C, the indicated cells were infected with increasing doses of VSV-G-pseudotyped HIV-1luc reporter virus. RT, reverse transcriptase; RLU, relative light units. Cell lysates from equal numbers of cells were assayed for luciferase activity 72 h later. (A) Jurkat and FeT-J cells. (B) Ficoll-purified, PHA-E- and IL-2-activated PBMCs from healthy feline and human donors. The three PBMC experiments demonstrated strong robustness of the main experimental variable (feline PBMC susceptibility versus human PBMC susceptibility) to particular conditions. Each was carried out at a different time using in each case entirely different human donors, feline donors, HIV-1luc(VSV-G) stocks, and RT assay components. Accordingly, absolute luciferase activities/RT units vary somewhat between the three experiments. Input ranges also varied between experiments (x axes), as did sources of IL-2 (see Materials and Methods). However, the slopes of the curves are very similar, and in each experiment, feline PBMCs exhibited greater luciferase production than the human PBMCs over a wide range of inputs, without evident saturation. (C) HIV-1luc susceptibilities of human and feline monocyte-derived macrophages. (D) Late HIV-1 gene product (p24 antigen) in feline and human PBMC supernatants. Totals of 4 × 104 feline and human PBMCs were infected for 16 h with the same amounts of HIV-1luc(VSV-G) and then washed five times to remove input virus. Five days later, filtered supernatants were assayed for capsid by p24 antigen enzyme-linked immunosorbent assay (ELISA). (E to G) Experiments were repeated as described above in a wider variety of human and feline cell lines. Equal numbers of cells were infected for any comparison, and for panel G, cells were also counted on the day of supernatant sampling (day 4 after infection) and p24 values are per 0.9 × 106 cells of each line.
Generation of stable cell lines with retroviral vectors.
CrFK.X4.CD4 and CrFK.X4.CD4.Vif-GFP cell lines were derived from CrFK cells by using FIV-based lentiviral vectors (44) with the following gene arrangements: CD4-internal ribosome entry site (IRES)-neomycin phosphotransferase gene (neo), CXCR4-IRES-puromycin N-acetyltransferase gene (pac), or the above-described VifCO-GFP fusion protein gene. FIV vectors used are derivatives of pGiNWF, with particles produced as described previously (67, 69). For stable expression of human CD4, a pT4-iNWF FIV transfer vector was constructed by cleaving a human CD4 cDNA from pZ-CD4 (66) with BglII and ApaI and inserting this fragment between BamHI and ApaI of pGiNWF (44). To transduce human CXCR4, we first replaced the EcoRI-EcoRI IRES-neo segment of pGiNWF with the Klenow fragment-blunted EcoRI-HpaI IRES-pac fragment from pEFIRESP, creating pGiPWF. This was followed by ligation of the BglII-ApaI human CXCR4 cDNA fragment of pZ.CXCR4 (66) between BamHI and ApaI of pGiPWF, creating pX4-iPWF. The transfer vector expressing VifCO-GFP was created by PCR amplification of the synthetic vifCO cDNA by using S.Fus.Uni (5′-ATATACCGGTGGATCCATGGGTGGCGCGGCCGC-3′) and AS.Fus.Uni (5′-ATATACCGGTATAGCGTAGTCTGGGACGTCGTATGGGTA-3′). The resulting PCR product was cleaved with AgeI and inserted into the AgeI site of pGiPWF (thus, in frame with GFP). Stable cell lines were established by transduction and appropriate drug selection (800 μg/ml G418 and/or 0.5 μg/ml puromycin). VifCO-GFP-expressing cells were selected for GFP expression five days after transduction and confirmed by immunoblotting with anti-GFP antibody.
Viruses.
Reverse transcriptase (RT) activity was determined using a 32P-based RT assay as described previously (70). Either this assay or HIV-1 p24 antigen capture assays were used for normalization. The HIV-1 luciferase reporter virus HIV-1luc (pNL4-3R−E−Δ426luc) has been described previously (43). Δvif HIV-1luc was derived from HIV-1luc by blunt-end closure of the NdeI and PflMI restriction sites within vif. Vesicular stomatitis virus G protein (VSV-G)-pseudotyped viruses were generated by cotransfecting 293T cells with pMD.G by using calcium phosphate coprecipitation. Replication-competent NL4-3 viruses expressing each of three vif allele products, HIV-1 Vif (HIV-1VH), codon-optimized FIV C36 Vif (HIV-1VF), and SIVmac239 Vif (HIV-1VS) were derived from HIV-1 NLSX, a generous gift of Akio Adachi, University of Tokushima, Japan. NLSX was originally constructed to separate the 5′ integrase gene and 3′ vpr overlapping reading frames from vif and to introduce unique SmaI and XbaI restriction sites (72). We modified this clone to restore Vpr expression by replacing the PflMI-EcoRI fragment with that from HIV-1 NL4-3, generating HIV-1VH. HA-tagged SIVmac Vif was PCR amplified from SIVmac239 by using primers S-SIV.Vif.Xma (5′-ATATCCCGGGATGGAGGAGGAAAAGAGGTGG-3′) and AS-SIV.Vif.HA (5′ ATATCCATTCTATGGTTAAGCGTAGTCTGGGACGTCGTATGGGTATGCCAGTATTCCCAAGACCTTTGCC-3′) and cloned into HIV-1VH by using XmaI and PflMI to create HIV-1VS. A similar strategy was employed to create viruses which expressed native FIV C36 Vif or codon-optimized FIV C36 Vif from the natural HIV-1 genomic location, but both viruses were rendered noninfectious because of aberrant splicing. Alternatively, we created HIV-1VF with a codon-optimized FIV vif gene (vifCO) in the native nef genomic location. This virus contains a frameshift mutation and a deletion of the initiation methionine deletion in HIV-1 vif. To permit cloning into the nef gene, a BamHI-XhoI fragment from NL4-3R−E−Δ426 was cloned into HIV-1 ΔVif to create HIV-1 ΔVif luciferase. Codon optimized FIV vif was then cleaved from the p1012-coFIV.Vif(HA) expression plasmid (NotI-XhoI) to replace the luciferase gene. All proviral plasmids were confirmed by Sanger sequencing of modified segments. Replication-competent viruses were produced by transfection (with 293T cells, 10 μg plasmid DNA, and a 75-cm2 flask). Medium was changed 12 to 16 h after transfection, and virus-containing supernatants were filtered (pore size, 0.45 μm) after an additional 36 h. VSV-G pseudotypes were created using the same conditions plus 3 μg pMD.G.
Infectivity per ng of p24 was determined by titration on GHOST cells according to the NIH AIDS Research and Reference Reagent Program protocol. For infections with p24-normalized viruses, 3 × 105 CrFK.X4.CD4 cells were infected with 1 ng of p24 of each virus in six-well plates. Twenty-four to thirty-six hours later, cells were washed five times with DMEM to remove input virus, and a time zero p24 sample was collected. Cultures were maintained by splitting 1:10 as needed, and supernatants were collected every 2 to 3 days for p24 measurements. After approximately 75% or more of the cellular monolayer was occupied by syncytia, supernatants were collected and filtered (pore size, 0.45 μm) before passage to uninfected CrFK.X4.CD4 cells.
Infectivity and protein degradation assays.
Δvif HIV-1luc virus stocks were created by calcium phosphate cotransfection of 293T cells in six-well plates by using 1 μg HIV-1 proviral DNA, 0.5 μg pMD.G, 1 μg pcDNA3.1(−) control or FLAG-tagged APOBEC3 expression plasmid, and 0.5 μg p1012 control or lentiviral HA-tagged Vif expression plasmid. Medium was changed 12 h after transfection, and viruses were collected 24 h later. 293T or SupT1 cells were immediately infected with filtered (pore size, 0.45 μm) supernatants for 24 h before cells were assayed for relative luciferase activity (SteadyGlo; Promega). To assay for degradation of feline APOBEC3 proteins, cells were transfected as described above without proviral or envelope expression plasmids. Twelve hours after transfection, the medium was changed, and 8 to 12 h later, cells were washed with phosphate-buffered saline (PBS) and lysed with radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 1% NP-40, 150 mM Tris-HCl, pH 8.0) plus protease inhibitors (complete-Mini; Boehringer). Protein concentrations were determined using Coomassie blue reagent (Bio-Rad), and equal quantities were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) before being transferred to Immobilon P membranes (Millipore). Blocked membranes were probed with mouse anti-FLAG (Sigma), or mouse antitubulin (Sigma) primary antibodies followed with an anti-mouse antibody-peroxidase conjugate.
Hypermutation analysis.
A total of 4.5 × 105 293T cells/well were plated in six-well plates the day before infection with VSV-G-pseudotyped Δvif HIV-1luc reporter virus as described for the infectivity assays. RT- or p24-normalized (ZeptoMetrix) input virus was treated with Ambion Turbo DNase (1 μl/ml at 37°C for 30 min) to remove residual plasmid DNA and used to inoculate the 293T cells for 12 h. Cells were washed and suspended in PBS, and DNA was harvested with the DNeasy kit (Qiagen). DpnI digestion was then used to eradicate remaining plasmid DNA carryover. A 600-nucleotide (nt) 5′ long terminal repeat (LTR)-Gag fragment was amplified using Phusion Hot Start DNA polymerase, with initial denaturation of 98°C for 30 s and then cycling with 98°C for 10 s, annealing at 55°C for 1 s, and extension at 72°C for 90 s for 35 cycles, followed by a final extension at 72°C for 10 min. The primers were MH531 (5′-TGTGTGCCCGTCTGTTGTGT-3′) and CM100 (5′-TGGAGGTTCTGCACTATAGGG-3′) as described by Münk et al. (61). PCR products were gel purified (Qiagen) and then blunt-end cloned with the StrataClone Ultra Blunt PCR cloning kit (Stratagene). At least five independent clones were sequenced for each infection and compared to the original Δvif HIV-1luc for hypermutation analysis.
Immunoblotting and co-IP.
For immunoblotting, cell lysates were prepared by washing cells in phosphate-buffered saline and lysing in RIPA buffer (150 mM NaCl, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate, 1% NP-40, 150 mM Tris-HCl [pH 8.0]) with protease inhibitors (complete-Mini; Boehringer). Protein concentrations were determined using Coomassie blue reagent (Bio-Rad), and equal quantities were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis before being transferred to Immobilon P membranes (Millipore). For coimmunoprecipitations (co-IPs), 3 × 106 293T cells were transfected (CaPO4) with 5 μg each APOBEC3-CH(FLAG) or p1012-Vif(HA) expression plasmid where indicated. Medium was changed 14 h later, and cells were treated with 10 μM MG132 (InSolution; Calbiochem) for 8 h, washed with PBS, and then lysed in RIPA buffer plus protease inhibitor. Protein concentrations were determined as described above, and 200 μg protein was incubated with 25 μg anti-FLAG monoclonal antibody (MAb; Sigma) and 30 μl blocked magnetic beads (Dynabeads with sheep anti-mouse IgG; Invitrogen) in 1 ml PBS plus 10% milk for 1 h at 4°C. Beads were washed five times with ice-cold RIPA buffer before boiling with Laemmli sample buffer (Bio-Rad) plus β-mercaptoethanol for 5 min. Total cell lysates were also prepared with 10 μg of total cellular extract. Proteins were separated by SDS-PAGE and transferred to Immobilon P membranes. Blocked membranes were probed with mouse anti-FLAG (Sigma), rat anti-HA (Roche), or mouse antitubulin (Sigma) primary antibodies followed by an anti-mouse or anti-rat horseradish peroxidase (HRP)-conjugated secondary antibody.
Immunofluorescence and confocal microscopy.
COS7 cells were transfected with 1 μg each feline FLAG-tagged APOBEC3 and/or 1 μg lentiviral HA-tagged Vif expression plasmid (FuGENE6; Roche) in Labtek II two-chamber slides (Nalge Nunc International) for 12 h, and then 10 μM MG132 was added for 8 h. Cells were fixed in 4% paraformaldehyde and permeabilized in ice-cold methanol followed by blocking in 10% FCS, 20 mM ammonium chloride, and PBS for 30 min. Primary antibody staining was done with mouse anti-FLAG (Sigma), rabbit anti-HA (Santa Cruz Biotechnology), and rabbit anti-DDX6 (Bethyl) followed by mouse and rabbit Alexa 594 or 488 secondary antibodies (Invitrogen). Prolong Gold mounting solution plus DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen) was added to visualize nuclei, and slides were imaged on an LSM 510 microscope.
RESULTS
Feline peripheral blood mononuclear cells and immortalized T cells support early and late HIV-1 life cycle events.
In agreement with published studies, (e.g., reference 34), we have found that feline fibroblast cell lines do not exhibit detectable postentry restriction of HIV-1, FIV, EIAV, or N-tropic MLV (71; data not shown). We wished to pursue the issue of feline and human cell tropisms further by directly comparing HIV-1 in human and feline T-cell lines as well as in primary peripheral blood mononuclear cells (PBMCs) and macrophages of both species. Cells were infected with HIV-1luc, a single-cycle HIV-1 reporter virus that was pseudotyped to bypass receptor requirements. As the only alterations to the HIV-1 genome are a 426-nt deletion in env and replacement of nef with a firefly luciferase (luc) transgene, HIV-1luc provides a quantitative marker for successful entry, reverse transcription, integration, and early, Rev-independent viral gene expression (43). At equivalent HIV-1luc inputs, the feline CD4+ T-cell line FeT-J supported robust luciferase expression that exceeded that of Jurkat cells (Fig. 1 A). In contrast to these results, Münk et al. recently reported 5- to 12-fold less early HIV-1 transgene expression in the feline T-cell lines Mya-1 and Fet1C when these were compared to human A3.01 cells (61). The use of different immortalized cell lines most likely accounts for this discrepancy, so we repeated the comparison with PBMCs from both species, using three separate sets of donors. In agreement with our T-cell line findings, the primary feline PBMCs supported high-level luciferase expression after HIV-1luc challenge (Fig. 1B). Moreover, primary human and feline macrophages displayed equivalent luciferase activities (Fig. 1C). To investigate late life cycle events, including particle egress, HIV-1 p24 production was assayed. In contrast to the marked attenuation of virus production that occurs in rodent cells (4, 51, 89), feline PBMCs produced HIV-1 p24 equivalent to human PBMCs (Fig. 1D). Repetition of these experiments while extending them to a broader panel of human and feline suspension and adherent cell lines was corroborative. Infected lymphoid (Fig. 1E) and fibroblast (Fig. 1F) feline cell lines supported abundant HIV-1 early gene expression and virion production (Fig. 1G) at a level comparable to that from human cells.
Stable FIV Vif expression enables HIV-1 replication in feline cells.
The results described above indicated that diverse primary and immortalized feline cells do not possess major postentry restrictions to HIV-1 early life cycle events or postintegration blocks to virion production. However, in agreement with Münk and colleagues (61), we found that engineering expression of human CD4 and a suitable chemokine coreceptor enabled efficient single-cycle infection with wild-type HIV-1 NL4-3 but did not enable spreading infection in any feline cells and that virus produced in feline cells had low infectivity (data not shown). These results were consistent with the known feline APOBEC3 protein restriction (61). fA3H and fA3CH restrict Δvif FIV, the infectivity of which can be increased by FIV Vif expression (60). These two fA3 proteins also restrict HIV-1, although the ability of any lentiviral Vif proteins to prevent this inhibition has not been examined (61).
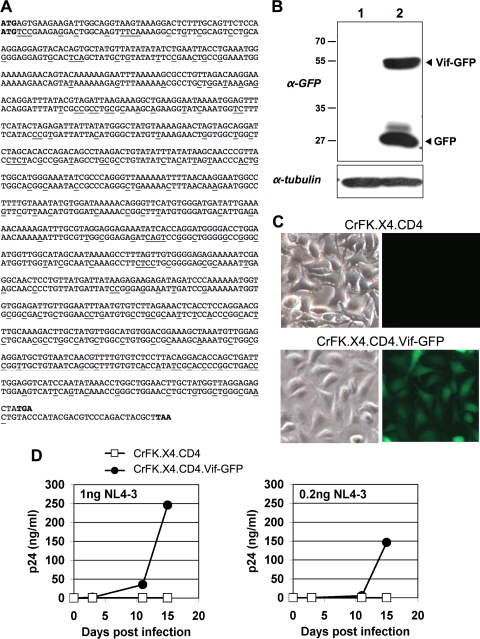
We therefore hypothesized that productive HIV-1 replication could be enabled by expressing FIV Vif protein stably in feline cells that we had previously complemented for human CXCR4 and CD4 (CrFK.X4.CD4 cells). As FIV vif is Rev dependent and proved to be poorly expressed from standard expression plasmids (data not shown), we synthesized a mammalian codon usage-optimized vif cDNA (Fig. 2 A). We chose the vif allele of FIV C36 (15), an infectious molecular clone of a clade C FIV isolate (16). Prospective confidence in robust biological activity of this Vif protein was supported by evidence that FIV C36 and its parental isolate generate persistent infection with rapid progression to AIDS in 75% and 60% of domestic cats, respectively (15, 16). The codon-optimized gene (vifCO) was fused in frame with gfp, and the resulting FIV Vif-GFP fusion protein was expressed in CrFK.X4.CD4 cells. Because subsequent HIV-1 challenge was planned, this was done with an FIV-based lentiviral vector (67). After stable selection for vector-coconferred puromycin resistance, uniformly GFP-positive cells were enriched by flow cytometry, and immunoblotting and microscopy were used to confirm expression of the fusion protein (Fig. 2B and C). Some cleavage of Vif-GFP to yield free GFP (and presumably free Vif) was also evident (Fig. 2B). HIV-1 infection of CrFK.X4.CD4.Vif-GFP cells and CrFK.X4.CD4 cells at multiplicities of infection (MOIs) of 0.05 and 0.01 showed that FIV Vif-GFP expression in trans enabled productive, spreading HIV-1 replication (Fig. 2D). HIV-1 could be readily passaged and serially amplified in these cells (data not shown). Taken together, the results of Fig. 1 and 2 suggest that feline APOBEC3 proteins are the principal inhibitors of spreading HIV-1 replication in feline cells.
FIG. 2.
Stable FIV Vif expression in feline cells rescues HIV-1 replication. (A) FIV vif codon optimization. The vif gene of FIV C36 (GenBank accession number AY600517.1), shown on the upper lines, was altered to optimize expression, remove potential RNA splice donors and acceptors, and include a C-terminal HA epitope. Nucleotide changes are underlined. (B) Immunoblotting of stable CrFK cell lines. Lane 1, CrFK.X4.CD4 cells; lane 2, CrFK.X4.CD4.Vif-GFP cells. The upper panel corresponds to antibody to GFP, and the lower panel corresponds to antibody to tubulin. α, anti. Molecular weights (in thousands) are indicated to the left. (C) CrFK.X4.CD4.Vif-GFP cells visualized by epifluorescence microscopy, with Nomarski images to the left. (D) A total of 3 × 105 CrFK.X4.CD4 or CrFK.X4.CD4.Vif-GFP cells were infected at MOIs of 0.05 and 0.01 with HIV-1 NL4-3 (1.0- or 0.2-ng p24 input) for 16 h before being washed five times with medium. p24 antigen was assayed by ELISA in filtered supernatants at the time points indicated.
FIV and SIVmac Vif, but not HIV-1 Vif, antagonize feline APOBEC3 mediated restriction.
Of the five feline APOBEC3 proteins, two (fA3H and fA3CH) inhibit the infectivities of HIV-1 and Δvif FIV (60, 61). In contrast, the three fA3Cs do not restrict Δvif FIV (60) and the one fA3C tested (fA3Ca) does not restrict HIV-1 (61). The activity of fA3H and fA3CH against HIV-1 presumably reflects a failure of HIV-1 Vif to counteract them. To test whether the specificity of replication may be attributed to the ability of various lentiviral Vif proteins to counteract the feline APOBEC3 proteins, we constructed six plasmids that express HA-tagged versions of the Vif proteins of HIV-1 NL4-3, FIV C36, and SIVmac and FLAG-tagged versions of fA3C (using fA3Ca), fA3H, and fA3CH. We confirmed that as previously reported (60), fA3CH and fA3H strongly restricted the Δvif FIV vector, while fA3Ca had a minimal effect. In side-by-side comparisons, fold decreases in RT-normalized Δvif FIV vector infectivity were 1.2, 46, and 55 for fA3Ca, fA3H, and fA3CH, respectively.
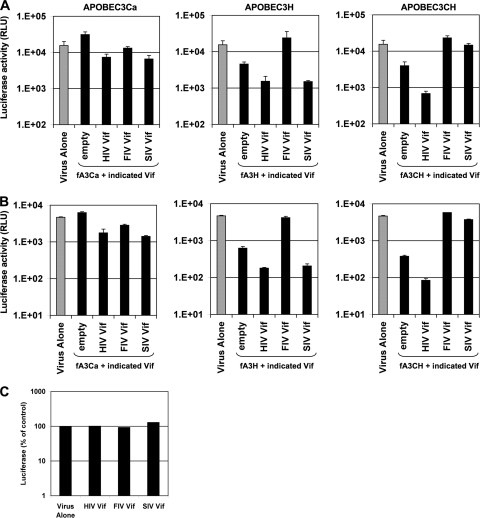
Combinations of these six plasmids were then cotransfected with Δvif HIV-1luc and a VSV-G expression plasmid. Supernatants were collected at 36 h and used to infect SupT1 (Fig. 3 A) or 293T (Fig. 3B) target cells, which were then assayed 3 days later for luciferase activity. In the absence of fA3 proteins, the Vif proteins did not have significant intrinsic inhibitory effects on HIV-1 infectivity (Fig. 3C). In the absence of Vif, fA3H and fA3CH reduced infectivity 8-fold and 12-fold, respectively, while fA3Ca had no detectable restriction activity (Fig. 3A and B). Interestingly, we found that expression of HIV-1 Vif paradoxically augmented the antiviral activities of fA3H and fA3CH (26-fold and 55-fold inhibitions, respectively) and also induced the otherwise inactive fA3Ca to display minor (3-fold) restriction. In contrast, FIV Vif counteracted fA3H and fA3CH well, restoring infectivity completely. Moreover, although SIVmac Vif shares almost no amino acid identity with FIV Vif (and no primate Vif has previously been shown to counter a nonprimate A3), SIVmac Vif was also able to counteract fA3CH. In addition, note that SIVmac Vif acted like HIV-1 Vif in producing an apparent paradoxical increase in the restriction activity of fA3H but not fA3CH in these assays (Fig. 3). Similar to HIV-1 Vif, a small 3-fold induction of fA3Ca activity was also seen. In summary, we found that HIV-1 Vif offered no protection against feline APOBEC3 proteins while FIV Vif was completely protective and SIVmac Vif displayed an intermediate phenotype.
FIG. 3.
FIV and SIVmac Vif, but not HIV-1 Vif, antagonize fA3 restriction of HIV-1. Δvif HIV-1luc(VSV-G) reporter virus stocks were produced in 293T cells by cotransfection of 0.5 μg Vif and 1.0 μg A3 expression plasmids. Thirty-six hours after transfection, supernatants were collected and filtered. p24-normalized inputs were used to infect SupT1 (A) or 293T (B) target cells. Gray bars, virus alone (no fA3 or Vif coexpressed); black bars, fA3 plus indicated Vif or empty vector. Twenty-four hours after infection, equal numbers of target cells were lysed and assayed for luciferase expression. (C) Lentiviral Vif proteins were assayed with the same conditions, with A3 plasmids replaced with empty vector.
FIV and SIVmac Vif-induced degradation of feline APOBEC3 proteins augments HIV-1 infectivity.
HIV-1 Vif acts as an adaptor between human A3G and the cullin5-elongin B/C-Rbx ubiquitin ligase, thus mediating polyubiquitination and proteasomal degradation of the host restriction factor (13, 52, 57, 82, 86, 94). Two highly conserved C-terminal motifs within HIV-1 Vif are critical for the ubiquitin ligase interaction: the SLQ(Y/F)LA (BC box), which mediates the elongin C interaction, and an upstream zinc-coordinating HCCH motif required for cullin5 interaction. Although primate lentiviral Vifs have limited sequence identity otherwise, the HCCH motif and BC box of HIV-1 Vif are well conserved in SIV and HIV-2 Vif proteins and are required for function (46, 58).
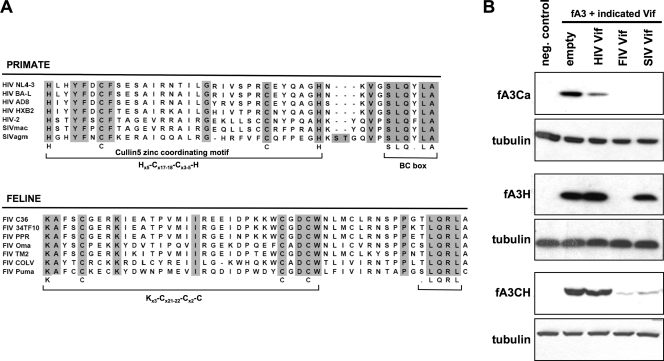
Whether FIV Vif (or the Vif of any lentivirus) triggers degradation of feline APOBEC proteins has not been examined. Aligning Vif protein sequences from diverse FIV serogroups (Fig. 4 A), we could identify C-terminal peptide motifs of interest, including a putative HCCH-resembling motif (KCCC) and an adjacent potential BC box [(T/S)LQRL], suggesting that the fA3-antagonizing mechanism of FIV Vif may resemble that of primate lentivirus Vifs. Outside of this region, we identified no significant homologies between primate and feline lentiviral Vif proteins, e.g., in N-terminal segments known to mediate A3G interaction for HIV-1 Vif. To evaluate the capacities of HIV-1, FIV, and SIVmac Vifs to target fA3 proteins for degradation, combinations of Vif and fA3 proteins were coexpressed. In the absence of Vif, each fA3 protein was robustly expressed (Fig. 4B). With the exception of a modest reduction of fA3Ca, HIV-1 Vif had no impact on intracellular fA3 levels. In clear contrast, FIV Vif depleted each fA3, resulting in undetectable fA3Ca and fA3H and drastic reduction of fA3CH. These results suggest that FIV Vif functions analogously to HIV-1 Vif to reduce the intracellular pool of restrictive fA3 proteins. Interestingly and fully consistent with the infectivity results (Fig. 3), SIVmac Vif reduced intracellular levels of fA3CH but fA3H was considerably less sensitive (Fig. 4B). Thus, the ability of a lentiviral Vif to rescue infectivity depends upon its ability to target restrictive fA3 proteins for degradation. Inexplicably, although SIVmac Vif consistently and paradoxically unmasked a modest fA3Ca antiviral effect (Fig. 3), it also effectively targeted fA3Ca for degradation. In contrast, the greater paradoxical infectivity reduction produced by SIVmac Vif in the presence of fA3H correlated with its minimal degradative activity toward this protein.
FIG. 4.
Degradation of feline APOBEC3 proteins is triggered by FIV Vif and SIVmac Vif. (A) The top panel shows alignment of the C-terminal portion of primate lentiviral Vif proteins, with highly conserved cullin5 and the BC box annotated. The bottom panel shows alignment of diverse FIV Vif proteins in the same region with the putative cul5 and BC box. The BC box of primate lentiviral Vif proteins is one of two conserved components of the SOCS (suppressor of cytokine signaling) box in these proteins; a proline-leucine (P/L)-rich region dispensable for elongin C binding by HIV-1 Vif (56) is not present in FIV Vif proteins (data not shown). (B) Lentiviral Vif induced degradation of feline APOBEC3 proteins. 293T cells were cotransfected with 1 μg pcDNA3.1 control plasmid or indicated FLAG epitope-tagged fA3 expression plasmid and 0.5 μg control plasmid or indicated lentiviral HA-tagged Vif expression plasmid. Sixteen hours after transfection, cells were lysed and immunoblotting was performed with anti-FLAG or antitubulin monoclonal antibody. The experiments were repeated three times with the same results, and those of a representative experiment are shown. neg., negative.
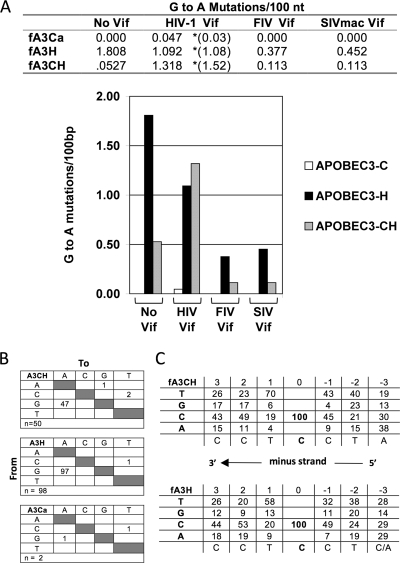
Feline APOBEC3-induced G-to-A hypermutation is counteracted by FIV and SIVmac Vifs.
A main mechanism of A3 protein restriction of primate lentiviruses is deamination of cytidines to uridines in the viral minus-strand DNA prior to plus-strand synthesis (6, 21, 48, 50, 61, 96). To directly assay the ability of each feline APOBEC3 protein to induce G-to-A hypermutation of HIV-1 and the capacity of each of the three lentiviral Vifs to prevent it, we produced stocks of Δvif HIV-1luc(VSV-G) by using combinations of the lentiviral Vif and feline APOBEC3 expression plasmids. p24-normalized preparations were then used to infect target cells, and the resulting proviral DNAs were analyzed by PCR and sequencing. In agreement with our protein degradation results, feline APOBEC3 protein-induced G-to-A hypermutation was readily observed and correlated strongly and inversely with infectivity (Fig. 5 A). Whether Vif was present or not, fA3Ca had no detectable cytidine deamination activity. In contrast, in the absence of Vif, expression of fA3H and fA3CH during viral production induced 1.808 and 0.527 G-to-A mutations per 100 bp, respectively. Cytidine deamination by fA3H and fA3CH was also substantial in the presence of HIV-1 Vif, producing 1.092 and 1.318 G-to-A mutations per 100 bp, respectively (Fig. 5A). These values determined with HIV-1 Vif are in excellent quantitative agreement with mutation rates determined by Münk et al. (61) when the feline A3s were coexpressed with HIV-1 with Vif intact (values in parentheses in Fig. 5). Nucleotide substitution preferences and local sequence contexts with respect to the dC residue deaminated on the minus strand are shown in Fig. 5B and C. The paradoxically augmented hypermutation induced by fA3CH in the presence of HIV-1 Vif (Fig. 5A) correlates with the augmented restriction seen in Fig. 3A and B. In addition, we found that FIV Vif was uniformly protective, reducing mutations generated by fA3H and fA3CH approximately 5-fold. SIVmac Vif was also protective, reducing fA3H and fA3CH mutations 4.0-fold and 4.7-fold, respectively. In general, these findings demonstrate that protection from cytidine deamination rescues infectivity. Again, SIVmac Vif, which did not induce efficient fA3H degradation (Fig. 4B) or rescue viral infectivity (Fig. 3), yielded unexpected results by reducing fA3H-induced mutations (Fig. 5). These results suggest that fA3H may have an editing-independent mechanism of restriction that is dominant over its SIVmac Vif-antagonized editing capacity. It is additionally noteworthy that in this instance, SIVmac Vif can block editing while triggering minimal degradation in the cotransfection assay.
FIG. 5.
Lentiviral Vif antagonism of feline APOBEC3-mediated hypermutation. (A) VSV-G-pseudotyped DNase-treated ΔVif HIV-1luc virus stocks created by cotransfection with indicated APOBEC3 and Vif expression plasmid combinations in 293T cells were used to infect 293T target cells. Twelve hours postinfection, target cell DNA was isolated and digested with DpnI and a 600-nt LTR-gag fragment was PCR amplified, isolated, and cloned into a vector for sequencing. At least five independent clones per APOBEC3-Vif combination were analyzed for G-to-A mutations, which are displayed as G→A mutations per 100 bp in table and graphical formats. *, values in parentheses for HIV-1 Vif are the G→A hypermutation rates determined by Münk et al. (61), in whose study HIV-1 Vif was expressed from the HIV-1 NL4-3 provirus. (B) Nucleotide substitution preferences for fA3Ca, fA3H, and fA3CH. (C) Preferred local sequence contexts with respect to the dC residue deaminated on the minus strand. Values are the frequencies (as percentages) at which each of the four bases was found at adjacent positions. 5′-TCCTCC-3′ (deaminated C underlined) was the consensus for both fA3H and fA3CH, with both having a preference for T at +1 (i.e., AG→AA on the plus strand).
HIV-1 Vif and feline APOBEC3 proteins interact and colocalize in cells, but this is insufficient to rescue infectivity.
This editing analysis, the fA3 degradative effects, and the apparently paradoxical effects of primate lentivirus Vif proteins prompted us to analyze Vif-fA3 protein-protein interactions. Polyubiquitination and proteasome-mediated degradation of human A3G (hA3G) require direct interaction between particular N-terminal residues of HIV-1 Vif and hA3G (8, 25, 28, 49, 68, 77, 78, 93). Therefore, we hypothesized that defective binding of HIV-1 Vif to feline APOBEC3 proteins could account for the inability to rescue infectivity. Such a defect would be consistent with the lack of sequence identity between FIV and HIV-1 Vif in the relevant N-terminal region. For example, the DRMR and YRHHY motifs at HIV-1 Vif residues 14 to 17 and 40 to 44, respectively, are not conserved in FIV Vif, although the critical D128 of human A3G, along with seven flanking residues (RLYYFWDP), is found in fA3Ca and fA3CH but not fA3H.
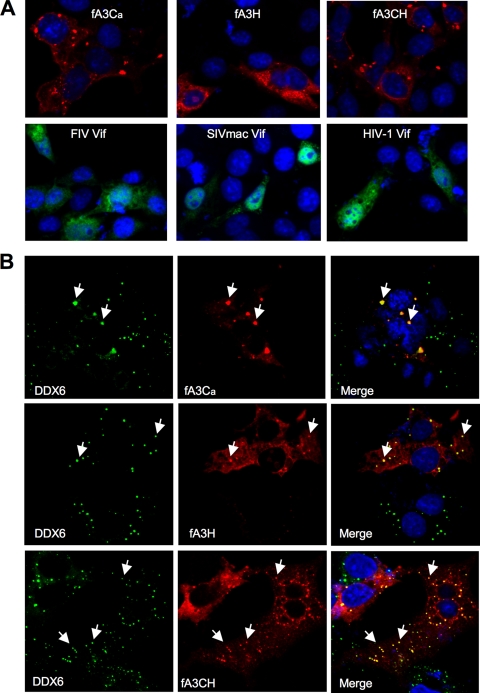
To examine this question, a series of confocal imaging and coimmunoprecipitation studies were performed. FIV Vif has been reported to be either cytoplasmic or nuclear depending upon fixation and staining methods (10), and fA3 locations in cells have not been previously described. Therefore, we first imaged fA3s and Vif proteins in the absence of one another. Using 4% paraformaldehyde fixation and labeling with either mouse monoclonal or rabbit anti-HA antibodies, we found that each lentiviral Vif was present in both the nucleus and the cytoplasm (Fig. 6 A). fA3Ca, fA3H, and fA3CH were found exclusively in the cytoplasm, where they were distributed both diffusely and in small puncta (Fig. 6A). These patterns are similar to that reported for human A3G (18); also like hA3G, the fA3s are found in P bodies (Fig. 6B).
FIG. 6.
Intracellular distributions of feline A3 proteins and three lentiviral Vif proteins. COS7 cells expressing fA3s or one of three lentiviral Vif proteins (FIV, SIVmac, or HIV-1) were analyzed for protein localization. (A) Confocal immunofluorescence microscopy with mouse anti-FLAG for fA3s and rabbit anti-HA for Vifs. The same results were obtained in 293T cells (data not shown). (B) fA3 proteins colocalize with a P-body marker as assessed by coimaging of endogenous DDX6 (rck/p54), a dead-box helicase that facilitates P-body formation and acts as a translational repressor (12). Confocal immunofluorescence was performed as described for panel A for fA3s but with rabbit anti-DDX6 to identify P bodies.
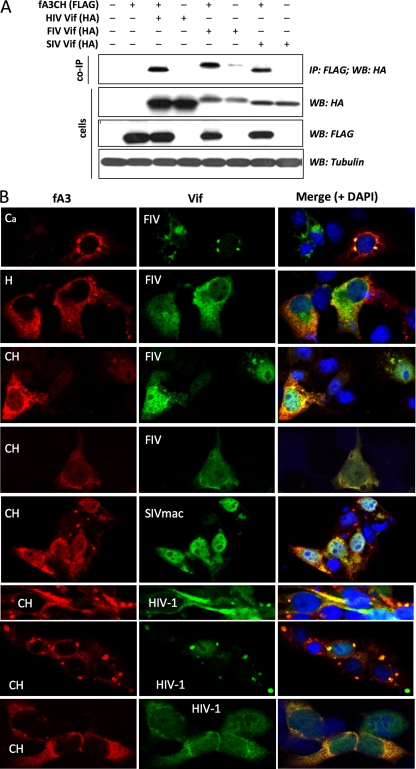
We next determined whether the feline and primate lentivirus Vif proteins and fA3s can interact in cells and whether they colocalize. Immunoprecipitation experiments were performed with fA3CH and each Vif in the presence of the proteasome inhibitor MG132. FIV Vif and SIVmac Vif were each pulled down by fA3CH from cell lysates (Fig. 7 A). This is consistent with the rescue of HIV-1 infectivity these two Vif proteins cause in the presence of fA3CH (Fig. 3), their strong promotion of fA3CH degradation (Fig. 4B), and their prevention of fA3CH-induced hypermutation (Fig. 5). Interestingly and contrary to our initial hypothesis, HIV-1 Vif was pulled down by fA3CH as well.
FIG. 7.
Analysis of fA3 protein interaction and colocalization with feline and primate lentivirus Vif proteins. WB, Western blot. (A) Interactions of fA3CH with lentiviral Vif proteins were assessed by immunoprecipitation. 293T cells coexpressing fA3CH and Vifs were treated with MG132 for 8 h before being lysed and immunoprecipitated for fA3CH, using mouse anti-FLAG. The immunoprecipitates were analyzed by immunoblotting for Vifs, using rat anti-HA. Cell lysates were simultaneously probed to verify inputs. Total transfected DNA was kept constant, with fA3-negative and Vif-negative transfections containing empty plasmid vector. (B) Feline A3 proteins and lentiviral Vif proteins colocalize in cytoplasmic compartments. COS7 cells were treated with the proteasome inhibitor MG132 starting 16 h after transfection. Eight hours later, the cells were analyzed by confocal immunofluorescence microscopy using mouse anti-FLAG and rabbit anti-HA. fA3Ca (Ca) and fA3H (H) produced the same colocalizations as fA3CH (CH) with the primate Vifs (data not shown). Immunofluorescence controls were negative (not shown); note also the untransfected (DAPI-positive, label-negative) cells as well as single-protein-expressing cells (e.g., bottom row) that serve as internal labeling controls.
Confocal imaging also showed that each of the lentiviral Vifs colocalizes to a significant extent with each fA3 protein, in discrete puncta as well as in more diffuse patterns (Fig. 7B). Note that while the location of the Vif proteins could be variable, in any one cell, the A3 protein sequesters with it to a substantial extent (for an example, compare the bottom three rows of Fig. 7B for the HIV-1 Vif-fA3CH combination). A second consistent feature was that Vifs were also more focally localized in the presence of a coexpressed fA3 (for an example, compare the A3Ca-positive and -negative cells in the top row of Fig. 7B). These effects are not diagnostic of direct or indirect protein-protein interaction, but they are consistent with the interaction observed in the immunoprecipitation experiment.
These results demonstrate that the capacity of a lentiviral Vif protein to interact with or to colocalize in cells with fA3 proteins does not correlate directly with rescue of infectivity and raised the possibility that HIV-1 Vif can interact with fA3 proteins but is unable to recruit or correctly orchestrate the E3 ubiquitin ligase machinery necessary for polyubiquitination and degradation. The paradoxical augmentation of restriction seen with coexpressed HIV-1 Vif (Fig. 3) could fit with such a model.
Productive spreading infection of HIV-1 with vif-chimeric in feline cells.
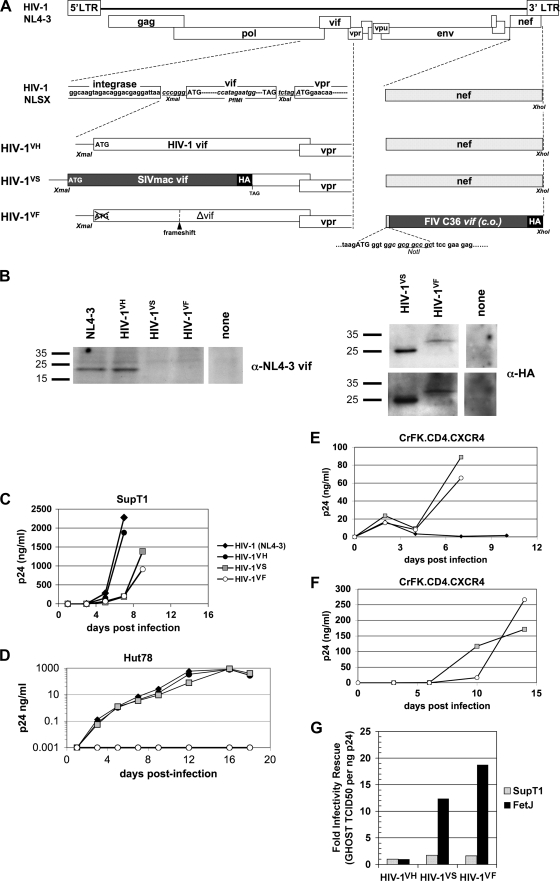
Taken together, the results presented in Fig. 1 through 7 suggested that fA3 proteins could represent the only substantial block (other than entry receptors) to productive replication and spread of HIV-1 in feline cells. To further test this hypothesis, we constructed a set of HIV-1 NL4-3 molecular clones that express HIV-1, FIV, or SIVmac Vif. Ideally, non-HIV-1 Vifs in such chimeras would be expressed from the natural vif open reading frame to achieve tight fidelity to the normal temporal and quantitative expression profiles of this accessory gene. For FIV Vif, this proved difficult despite extensive construction and testing of numerous candidate chimeras bearing both native and codon-optimized FIV vif alleles (data not shown). The complete lack of nucleotide sequence homology between FIV and HIV-1 vif genes, compounded by the critical viral splice sites in primate lentivirus vif genes, resulted in aberrant splicing and noninfectious viruses (data not shown). Provision of HIV-1 Rev in trans boosted the Gag/Pol expression of these defective clones substantially (data not shown). Attempts to construct hybrid Vifs (various portions of HIV-1 and FIV Vifs) were unsuccessful. For an alternative, we found that replacement of the nef gene with the codon-optimized, HA-tagged FIV C36 vif allele was tolerated and enabled infectious virus production. HIV-1 Vif expression was abolished in this clone, HIV-1VF, by a frameshift mutation and deletion of the initiation methionine (Fig. 8A).
FIG. 8.
HIV-1 chimeric viruses expressing FIV or SIVmac Vif replicate in feline cells. (A) Structures of HIV-1 expressing chimeras of NL4-3 (GenBank accession number M19921) Vif, SIVmac239 (M33262) Vif, or the codon-optimized version of the Vif protein of FIV C36 (AY600517.1). See Materials and Methods for details of construction. (B) Expression of Vif proteins by infectious HIV-1 clones. Proviral DNAs were transfected in parallel in 293T cells, which were lysed for immunoblotting 48 h later. The viruses yielded roughly equivalent supernatant p24 levels (HIV-1 NL4-3, HIV-1VH, HIV-1VS, and HIV-1VF produced 315, 268, 532, and 178 ng of p24/ml, respectively, also measured at 48 h), and the immunoblot loading of cell lysates was normalized using these p24 levels. Molecular weights (in thousands) are indicated to the left. (C to E) Replication of parental and chimeric viruses in CrFK.X4.CD4 cells and in human cells that are either HIV-1 Vif permissive (SupT1) or not Vif permissive (HUT78). A total of 3 × 105 cells were infected with 1-ng p24 equivalents of wild-type HIV-1 (NL4-3), HIV-1VH, HIV-1VF, or HIV-1VS for 24 h and then washed five times to remove input. Cell-free supernatants were collected at the time points indicated and assayed for replication by p24 antigen-capture ELISA. Syncytium formation and cell death also correlated directly with virus replication (data not shown). Note that CrFK cells are not permissive for Vif-negative FIV (80; data not shown) and express substantial levels of mRNA for the three fA3s (60; data not shown). (F) Chimeric virus replication after filtering and passage in feline cells. Initial cultures were started as described for panel B and passaged by filtering supernatants (pore size, 0.45 μm) and infecting fresh feline CrFK.X4.CD4 cells when CPE involved ≥75% of the monolayer. HIV-1VS (0.2 ng; passage 8) and HIV-1VF (2.5 ng; passage 4) were used to infect CrFK.X4.CD4 cells. Replication was monitored as described for panel C. (G) FIV and SIVmac Vif expression in chimeric viruses rescues HIV-1 infectivity in feline T cells. FeT-J and SupT1 T cells were infected with VSV-G-pseudotyped viruses at an MOI of 5.0 for 16 h before being washed five times to remove input virus. Five days after infection, supernatants were collected and filtered, and their titers were determined on GHOST indicator cells, which were scored for titer calculation at three days. TCID50, 50% tissue culture infective doses.
In contrast to the complications we encountered with FIV vif, and consistent with the work of others (23, 24, 30, 33), HIV-1 NL4-3 was found to be amenable to local exchange of its vif gene with SIVmac vif. For this purpose, we modified the HIV-1 NLSX molecular clone of Sakurai et al., which separates the normally overlapping integrase and Vif reading frames and flanks HIV-1 Vif with unique restriction sites but abrogates Vpr expression (72). We first restored vpr by reestablishing the HIV-1 Vif/Vpr overlap region, creating HIV-1VH (Fig. 8A). We then derived HIV-1VS by placing the SIVmac239 vif allele in the natural genome location while preserving a noncoding 3′ portion of the HIV-1 vif sequence so as to avoid interfering with Vpr expression (Fig. 8A).
We verified that HIV-1VH, HIV-1VS, and HIV-1VF expressed the respective encoded Vif proteins (Fig. 8B). Note that HIV-1VH produced as much Vif as wild-type NL4-3, showing that the rearranged molecular context of the clones with chimeric vif (Fig. 8A) allows equivalent Vif expression levels. These viruses were then analyzed functionally, first in informative human T-cell lines that do or do not express A3G (HUT78 or SupT1 cells, respectively). Each was replication competent in SupT1 cells (Fig. 8C) and in GHOST cells (data not shown). However, while HIV-1VH and HIV-1VS replicated in non-Vif-permissive HUT78 cells as well as wild-type HIV-1 NL4-3, HIV-1VF was restricted (Fig. 8D). This result shows that FIV Vif cannot ablate human A3G/F restriction and also that the two primate Vifs were expressed at functionally adequate levels by these infectious molecular clones, thus corroborating the immunoblots of Fig. 8B.
To assay the infectivity of these chimeric viruses in feline cells, 3 × 105 CrFK.X4.CD4 or GHOST cells were infected with HIV-1 NL4-3, HIV-1VF, or HIV-1VS (Fig. 8E). Thirty-six hours after infection, cells were washed five times to remove input virus and cultures were maintained for 10 days or until extensive syncytium formation was evident. During the first few days of infection, scant and equivalent cytopathic effects (CPEs) and supernatant p24 levels were observed in CrFK.X4.CD4 cells regardless of which of the three viruses was inoculated (Fig. 8E). By day 4, both CPE and p24 were lost in HIV-1 NL4-3 cultures (or HIV-1VH cultures [data not shown]), signifying an inability to propagate through secondary rounds of infection. In contrast, HIV-1VF replicated robustly. To further document spreading replication, small amounts of filtered HIV-1VF supernatant were repeatedly passaged to fresh CrFK.X4.CD4 cells. Each time, exponential amplification of p24, in tandem with extensive CPE, overwhelmed the culture, whereas HIVVH never amplified or passaged. An example is shown in Fig. 8F, for which 3 × 105 CrFK.X4.CD4 cells were infected with passage four HIV-1VF. We also carried out PCR amplification and sequencing of the HIV-1VF capsid open reading frame and the genome segment spanning vif to vpu after nine passages and identified no consistent or predominant changes. These experiments indicate that fA3 proteins represent the only significant restriction to productive HIV-1 replication and continuous passage in these cells and that incorporating FIV Vif into HIV-1 rescues the ability of the virus to propagate.
Interestingly, HIV-1VS also replicated well in feline cells (Fig. 8E and F). This result is intriguing because we found that although expression of SIVmac Vif in trans prevented fA3H-induced hypermutation, it neither appreciably rescued HIV-1 infectivity from cotransfected fA3H nor triggered efficient degradation of fA3H (Fig. 3 and 4B). We (data not shown) and others (60) readily detected fA3H mRNA in CrFK cells, indicating that a lack of fA3H expression is unlikely to explain this result. However, the spreading replication of HIV-1VS provides functional corroboration of the evidence for infection rescue (Fig. 3), degradation (Fig. 4), hypermutation prevention (Fig. 5), interaction (Fig. 7A), and colocalization (Fig. 7B) of this primate Vif protein with fA3CH.
We were able to derive feline T-cell lines that were stably positive for human CD4, CXCR4, or CCR5 by using retroviral vectors, but our attempts to derive stable double-positive lines were unsuccessful (data not shown), precluding direct assessment of HIV-1 spreading infection. To circumvent this problem, we pseudotyped replication-competent HIV-1VH, HIV-1VF, and HIV-1VS with VSV-G and infected T-cell lines of each species (SupT1 and FeT-J) at GHOST cell MOI equivalents of 5.0. Sixteen hours after infection, cells were washed five times to remove all pseudotyped virion input, and 4 days later, culture supernatants were filtered and p24 normalized, and their titers were determined on GHOST indicator cells. All three viruses produced in SupT1 cells were equally infectious and incurred no advantage or disadvantage from expressing FIV Vif or SIVmac Vif. In contrast, HIV-1VF and HIV-1VS produced in feline FeT-J T cells were 19- and 12-fold more infectious than HIV-1VH produced in these cells (Fig. 8G).
DISCUSSION
Exogenous lentiviruses have been identified in four orders of Mammalia: primates (HIV-1/2 and SIVs), Artiodactyla (visna virus, caprine arthritis encephalitis virus [CAEV], and bovine immunodeficiency virus [BIV]), Perissodactyla (EIAV), and Carnivora (FIV). Within each of these orders, cross-species infection and disease causation are highly circumscribed. These barriers are best characterized in the primate lentivirus group, particularly in the case of HIV-1, which does not undergo sustained replication or cause disease in any species other than Homo sapiens. In natural settings, overt lentiviral disease appears to reflect comparatively recent acquisition, e.g., the rise of the HIV-1 main group by transmission of SIVcpz from chimpanzees in West Africa approximately a century ago (19). Among nonprimate lentiviruses, an analogous example is FIV, which has likely infected felids in the Panthera lineage since the early Pleistocene (2). FIV is highly pathogenic only in the domestic cat, with the preponderance of evidence indicating comparatively recent acquisition (65).
While there is no evidence for transmission of any extant or recently ancestral lentiviruses between mammalian orders, it has been known for some years that cells of certain carnivoran species, e.g., the domestic cat and dog, display little if any impediment to HIV-1 postentry life cycle events up to and including integration. McEwan et al. recently determined one basis, namely, that felidae lack a functional TRIM5α protein due to a truncated B30.2 domain (54). Dogs also have this locus disrupted (73). Münk and colleagues showed that domestic cat cells produce HIV-1 with low infectivity and that this is at least partially accounted for by minus-strand editing by two of the five fA3 proteins; the greatest activity was attributable to the two-domain fA3CH, with fA3H making a lesser yet significant contribution (61). fA3Ca, at the time the only fA3C cloned, was inactive against HIV-1 (61). A second study (60) showed that Δvif FIV was also inhibited by fA3CH and fA3H but not any of the fA3Cs (a, b, or c type). We confirmed here that fA3H and fA3CH, but not fA3Ca, possess the main HIV-1- and Δvif FIV-restricting activity. We went on to address several questions not previously examined, including whether FIV Vif acts analogously to primate lentiviral Vifs by reducing fA3 levels and/or preventing hypermutation, whether Vifs interact with or colocalize with fA3s, and if this correlates with degradation or infectivity rescue. We also asked whether primate lentivirus Vifs might counteract fA3 restriction. We show that FIV Vif triggers degradation of all three fA3s and prevents HIV-1 genome hypermutation, while HIV-1 Vif does not. We also show for the first time that a primate lentiviral Vif (that of SIVmac) can counter the APOBEC3 proteins of a nonprimate species. Finally, we examined feline cells that are not permissive for Δvif FIV and express the restricting fA3s at substantial levels (CrFK). We demonstrated that productive replication of HIV-1 can be enabled in these cells by vif chimerism or by FIV Vif expression in trans. Therefore, HIV-1 can replicate productively with the dependency factors supplied by the domestic cat proteome if A3 restriction is countered.
Primary feline cells (PBMCs and macrophages) and a variety of fibroblast and lymphoid feline cell lines robustly supported early and late viral gene expression at levels equal to or greater than those of equivalent human cells (Fig. 1). These results contrast in part with those of Münk et al., who reported a relative postentry block to internally promoted minimal genome HIV-1-based GFP vectors in some feline T-cell lines compared to human cell lines but did not examine PBMCs (61). We also did not observe restricted HIV-1 particle release from KE-R and feline lymphoid cell lines (61). However, in full agreement with Münk et al., we found that HIV-1 receptor-complemented feline cells are susceptible to an initial round of HIV-1 infection, but spreading infection does not occur.
Stable expression of FIV Vif-GFP in receptor-complemented CrFK cells rendered them permissive to productive, spreading HIV-1 replication (Fig. 2). Confirming the sufficiency of FIV Vif, HIV-1VF could replicate productively in these cells (Fig. 8). A second Vif protein, from SIVmac, was also sufficient. In contrast, only the primate lentivirus Vif-encoding HIV-1VH and HIV-1VS were able to replicate in non-Vif-permissive human cells (Fig. 8D, HUT78 cells), showing that FIV Vif cannot counter human A3s. The replication competence of HIV-1VS in HUT78 cells is consistent with the known activity of SIVmac Vif against human A3G (50) and the prior demonstration that an SIVmac Vif-encoding HIV-1 replicated productively in non-Vif-permissive human CEMx174 cells (24).
Table 1 summarizes the actions of the three Vif proteins against HIV-1. These results with HIV-1VF and HIV-1VS are the first demonstration of productive spreading replication and passage of HIV-1 in feline cells. To the best of our knowledge, they are only the third reported example in any nonprimate cells. The two precedents we can identify also involved carnivoran cells, in the Caniformia lineage rather than the Feliformia lineage. Sodroski and colleagues reported HIV-1 replication and passage in receptor-complemented canine Cf2Th cells (38, 64). We also acknowledge the precedent of Koito et al., who reported that an HIV-1 entry receptor-complemented mink cell line could support spreading HIV-1 replication (37). However, as we interpret this last experiment, it cannot be determined clearly if spreading replication occurred, as the authors monitored the culture to an endpoint of eight days after inoculation of a 15-ng p24 equivalent of human cell-produced HIV-1. They documented accumulation of more than 100 ng/ml of p24 over days 1 to 8 but did not demonstrate serial passage or exponential amplification of virus emerging from the mink cells, which leaves open the alternative possibility that only a single round of provirus establishment by the initial human cell-produced inoculum occurred (37). Nevertheless, taken together, these studies in cells of species that now encompass both suborders of the Carnivora suggest that further examination of carnivoran cells may be of interest. Generalization between members of the felidae or other carnivoran groups is unwarranted, however. For example, primate lentiviruses encounter a strong postentry block in lion cells (W. McEwan and B. Willett, personal communication).
TABLE 1.
Summary of HIV-1, SIVmac, and FIV Vif effects against HIV-1
| Vif | Effect of Vifa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Eradicated intracellular fA3 |
Rescued HIV-1luc infectivity from fA3 |
Blocked HIV-1 G→A hypermutation by fA3 |
Enabled HIV-1 replication in CrFK.4.CD4 cells |
||||||
| fA3Ca | fA3H | fA3CH | fA3H | fA3CH | fA3H | fA3CH | In cis | In trans | |
| HIV-1 | − | − | − | − | − | − | − | − | ND |
| SIVmac | + | − | + | − | + | + | + | + | ND |
| FIV | + | + | + | + | + | + | + | + | + |
ND, not done.
Taking the replication properties of HIV-1VF and HIV-1VS into account with the remainder of our results, we emphasize five points. First, it is clear from this work that with the exception of the specific entry receptors, the domestic cat genome encodes the essential complement of dependency factors needed for HIV-1 replication. In this respect, this nonprimate species differs markedly from mice (4, 14, 51, 89). Second, the productive replication of HIV-1VF and HIV-1VS in CrFK.X4.CD4 cells (and wild-type HIV-1 in CrFK.X4.CD4.Vif-GFP cells) demonstrates that expression of an active Vif alone alleviates the restrictions to HIV-1 replication in this cell line. Third, despite our additional data pinpointing the restricting activity in feline T cells to an A3-characteristic mechanism, i.e., producer cell-specific attenuation of virion infectivity, further blocks may well be identified in other particular domestic cat cell lines and primary cell types. We have so far been unsuccessful in achieving stable complementation of both human CD4 and a coreceptor in feline T-cell lines to conclusively determine the answer to this question, but we were able to use pseudotyped viruses to show that the FIV and SIVmac Vif chimeras produced in such cells were 19- and 12-fold more infectious, respectively, than the virus expressing HIV-1 Vif (Fig. 8G). This correlates with FIV and SIVmac Vif activities in other assays (Fig. 3, 4, and 5). A block to particle egress, e.g., BST-2/tetherin restriction, may exist in some feline cells. This deserves additional investigation, but we observed equivalent vpu-positive HIV-1 p24 production in primary feline and human PBMCs (Fig. 1D), and feline cell lines also produced comparatively large amounts of virus (Fig. 1G). Fourth, HIV-1 Vif interacts intracellularly with at least one restricting fA3, as judged by the fA3CH coimmunoprecipitation, but this triggers no degradation, hypermutation rescue, or infectivity rescue, suggesting defective assembly of a competent fA3-Vif-ubiquitin ligase complex.
Fifth, the finding that a primate lentivirus Vif (of SIVmac) can interact with, trigger the degradation of, and counter the antiviral activities of a nonprimate APOBEC3 protein is both novel and unexpected. Vif action against A3 proteins is in general highly species specific (8, 49, 77, 85, 93), with very limited and so far only interprimate cross-activity reported (24, 50, 90, 97). This issue was most thoroughly explored in our study for fA3CH and involves six lines of evidence that are consistent with each other. SIVmac Vif (i) rescues HIV-1 infectivity from fA3CH in transient transfection (Fig. 3), (ii) protects against G→A hypermutation by fA3CH (Fig. 5), (iii) triggers effective fA3CH degradation (Fig. 4), (iv) pulls down fA3CH from cells (Fig. 7A), (v) occupies the same cellular compartments as fA3CH (Fig. 7B), and providing the clearest functional proof, (vi) enables productive HIV-1 replication in a non-Vif-permissive feline cell line that expresses fA3CH as well as fA3H and fA3Ca (Fig. 8). The ineffective interaction of HIV-1 Vif with fA3CH is also intriguing and is discussed further below.
fA3CH and fA3H, but not fA3Ca, induced cytidine deamination of the HIV-1 genome (Fig. 5) at rates that agree very well with the previously published data for these three proteins with HIV-1 with intact Vif (61). We found that FIV and SIVmac Vif significantly reduce this hypermutation for both fA3CH and fA3H. Our hypermutation data are therefore in excellent agreement with our viral replication data. fA3Ca did not produce HIV-1 hypermutation (Fig. 5) and had little restricting activity, although it did interact with each of the Vif proteins in cells. SIVmac Vif has a distinctive, complex phenotype relative to the feline A3s. SIVmac Vif did not counteract fA3H restriction effectively in the cotransfection experiments and in fact augmented it (Fig. 3). This Vif protein also had lower degradative activity toward fA3H (Fig. 4B). However, it strongly antagonized fA3H hypermutation (Fig. 5). fA3H may therefore also have an editing-independent mechanism of restriction. The ability of the SIVmac Vif chimera to replicate in CrFK cells raises the possibility that endogenous fA3H does not significantly contribute to HIV-1 restriction in these feline cells or that antagonism of hypermutation is sufficient. Other experiments could be informative in the future if technically feasible. We attempted to knock down the fA3s in CrFK cells by using small hairpin RNAs (shRNAs) encoded by lentiviral vectors but found that this triggered confounding toxicity (data not shown). This could be due to off-target effects of the shRNAs we used, and/or it may mean that loss of fA3 expression is deleterious. One observation consistent with the latter possibility is that the puromycin-stable CrFK.X4.CD4.Vif-GFP cells showed steadily diminishing Vif-GFP expression with time in culture (data not shown). Nevertheless, the relief of the restriction to productive replication by Vif proteins is strong evidence that the fA3s are the key blockers.
For several reasons, it will be of interest to continue efforts to construct HIV-1 strains that express FIV Vif in the usual genomic position, without co-opting the nef open reading frame. SIVmac Vif is not an optimal anti-fA3, by definition and by the evidence presented here. FIV Vif rather than SIVmac Vif produced the most consistent and strongest fA3-countering effects, extending in all assays to fA3H as well as fA3CH. The activity of SIVmac Vif against fA3Cb and fA3Cc (the alternative names are fA3Z2c and fA3Z2a, respectively) has also not been specifically tested. fA3Cb and fA3Cc have not been tested for restricting activity against HIV-1, but they do not inhibit Δvif FIV (60).
Our primate lentiviral Vif data indicate that interaction with A3 proteins can occur without being sufficient for A3 degradation or relief of restriction. Whatever cellular factors FIV Vif interacts with to target fA3H or fA3CH for degradation (e.g., feline equivalents of Cul5 and elongin B/C), it is likely that HIV-1 Vif is unable to correctly recruit them or establish a functional complex despite its ability to connect with the fA3s. In this regard, the paradoxical augmentation of fA3 antiviral activity by coexpressed primate Vif proteins (Fig. 3) is intriguing, although determining its significance will require further investigation. Some reports suggest that Vif is incorporated into viral particles (9, 35, 42). We speculate that HIV-1 Vif complexing with fA3 proteins without targeting them for degradation could increase the efficiency of their particle incorporation or otherwise accentuate antiviral activity.
Primate models that utilize viruses that are chimeric for vif genes and capsid segments have considerable promise and a natural advantage of higher phylogenetic relatedness of these species to Homo sapiens (1). Nevertheless, development of a nonprimate small animal model would aid studies of HIV-1 transmission, pathogenesis, and vaccination. Considerable efforts have been devoted to developing such models, but to date, all attempts have failed due either to the lack of dependency factors required for replication or to complex antiviral restrictions. Domestic cats have been characterized in the FIV model for properties of relevance to AIDS pathology. This animal is also readily available, breeds prolifically, and can be kept specific-pathogen-free, and its maintenance is less resource-intensive than maintenance of nonhuman primates. Our study demonstrates that feline cells can supply all necessary cellular factors, with the exception of entry receptors, needed by HIV-1. If more subtle or cell type-specific restrictions do not turn out to be limiting and if workable transgenic technology for entry receptors can be devised, cats could have potential as a small, accessible animal model of HIV-1 infection.
Acknowledgments
We thank Takayasu Miyazawa, Carsten Münk, Brian Willet, and Janet Yamamoto for providing cell lines, Brian Willet for permission to cite unpublished data, John Elder for FIV C36, Akio Adachi for HIV-1 NLSX, Andrew Harrison for a helpful reading of the manuscript, and the NIH AIDS Research and Reference Reagent Program and its contributing investigators for antibodies, cell lines, and various plasmids.
Footnotes
Published ahead of print on 12 May 2010.
REFERENCES
- 1.Ambrose, Z., V. N. KewalRamani, P. D. Bieniasz, and T. Hatziioannou. 2007. HIV/AIDS: in search of an animal model. Trends Biotechnol. 25:333-337. [DOI] [PubMed] [Google Scholar]
- 2.Antunes, A., J. L. Troyer, M. E. Roelke, J. Pecon-Slattery, C. Packer, C. Winterbach, H. Winterbach, G. Hemson, L. Frank, P. Stander, L. Siefert, M. Driciru, P. J. Funston, K. A. Alexander, K. C. Prager, G. Mills, D. Wildt, M. Bush, S. J. O'Brien, and W. E. Johnson. 2008. The evolutionary dynamics of the lion Panthera leo revealed by host and viral population genomics. PLoS Genet. 4:e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann, J. G., D. Unutmaz, M. D. Miller, S. K. Breun, S. M. Grill, J. Mirro, D. R. Littman, A. Rein, and V. N. KewalRamani. 2004. Murine T cells potently restrict human immunodeficiency virus infection. J. Virol. 78:12537-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D., and B. R. Cullen. 2000. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 74:9868-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 7.Bishop, K. N., M. Verma, E. Y. Kim, S. M. Wolinsky, and M. H. Malim. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4:e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. U. S. A. 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camaur, D., and D. Trono. 1996. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J. Virol. 70:6106-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterji, U., C. K. Grant, and J. H. Elder. 2000. Feline immunodeficiency virus Vif localizes to the nucleus. J. Virol. 74:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 12.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 14.Coskun, A. K., M. van Maanen, V. Nguyen, and R. E. Sutton. 2006. Human chromosome 2 carries a gene required for production of infectious human immunodeficiency virus type 1. J. Virol. 80:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Rozières, S., C. K. Mathiason, M. R. Rolston, U. Chatterji, E. A. Hoover, and J. H. Elder. 2004. Characterization of a highly pathogenic molecular clone of feline immunodeficiency virus clade C. J. Virol. 78:8971-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diehl, L. J., C. K. Mathiason-Dubard, L. L. O'Neil, L. A. Obert, and E. A. Hoover. 1995. Induction of accelerated feline immunodeficiency virus disease by acute-phase virus passage. J. Virol. 69:6149-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 18.Gallois-Montbrun, S., B. Kramer, C. M. Swanson, H. Byers, S. Lynham, M. Ward, and M. H. Malim. 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81:2165-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 20.Guo, F., S. Cen, M. Niu, Y. Yang, R. J. Gorelick, and L. Kleiman. 2007. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J. Virol. 81:11322-11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 22.Hartikka, J., M. Sawdey, F. Cornefert-Jensen, M. Margalith, K. Barnhart, M. Nolasco, H. L. Vahlsing, J. Meek, M. Marquet, P. Hobart, J. Norman, and M. Manthorpe. 1996. An improved plasmid DNA expression vector for direct injection into skeletal muscle. Hum. Gene Ther. 7:1205-1217. [DOI] [PubMed] [Google Scholar]
- 23.Hatziioannou, T., Z. Ambrose, N. P. Chung, M. Piatak, Jr., F. Yuan, C. M. Trubey, V. Coalter, R. Kiser, D. Schneider, J. Smedley, R. Pung, M. Gathuka, J. D. Estes, R. S. Veazey, V. N. KewalRamani, J. D. Lifson, and P. D. Bieniasz. 2009. A macaque model of HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 106:4425-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatziioannou, T., M. Princiotta, M. Piatak, Jr., F. Yuan, F. Zhang, J. D. Lifson, and P. D. Bieniasz. 2006. Generation of simian-tropic HIV-1 by restriction factor evasion. Science 314:95. [DOI] [PubMed] [Google Scholar]
- 25.He, Z., W. Zhang, G. Chen, R. Xu, and X. F. Yu. 2008. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 381:1000-1011. [DOI] [PubMed] [Google Scholar]
- 26.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 282:2587-2595. [DOI] [PubMed] [Google Scholar]
- 27.Huff, J. L., and P. A. Barry. 2003. B-virus (Cercopithecine herpesvirus 1) infection in humans and macaques: potential for zoonotic disease. Emerg. Infect. Dis. 9:246-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huthoff, H., and M. H. Malim. 2007. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and virion encapsidation. J. Virol. 81:3807-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huthoff, H., and G. J. Towers. 2008. Restriction of retroviral replication by APOBEC3G/F and TRIM5alpha. Trends Microbiol. 16:612-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Igarashi, T., R. Iyengar, R. A. Byrum, A. Buckler-White, R. L. Dewar, C. E. Buckler, H. C. Lane, K. Kamada, A. Adachi, and M. A. Martin. 2007. Human immunodeficiency virus type 1 derivative with 7% simian immunodeficiency virus genetic content is able to establish infections in pig-tailed macaques. J. Virol. 81:11549-11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwatani, Y., D. S. Chan, F. Wang, K. S. Maynard, W. Sugiura, A. M. Gronenborn, I. Rouzina, M. C. Williams, K. Musier-Forsyth, and J. G. Levin. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 35:7096-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jónsson, S. R., G. Hache, M. D. Stenglein, S. C. Fahrenkrug, V. Andresdottir, and R. S. Harris. 2006. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 34:5683-5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamada, K., T. Igarashi, M. A. Martin, B. Khamsri, K. Hatcho, T. Yamashita, M. Fujita, T. Uchiyama, and A. Adachi. 2006. Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. U. S. A. 103:16959-16964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. U. S. A. 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan, M. A., C. Aberham, S. Kao, H. Akari, R. Gorelick, S. Bour, and K. Strebel. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 75:7252-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan, M. A., S. Kao, E. Miyagi, H. Takeuchi, R. Goila-Gaur, S. Opi, C. L. Gipson, T. G. Parslow, H. Ly, and K. Strebel. 2005. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 79:5870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koito, A., Y. Kameyama, C. Cheng-Mayer, and S. Matsushita. 2003. Susceptibility of mink (Mustera vision)-derived cells to replication by human immunodeficiency virus type 1. J. Virol. 77:5109-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolchinsky, P., T. Mirzabekov, M. Farzan, E. Kiprilov, M. Cayabyab, L. J. Mooney, H. Choe, and J. Sodroski. 1999. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J. Virol. 73:8120-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaRue, R. S., V. Andresdottir, Y. Blanchard, S. G. Conticello, D. Derse, M. Emerman, W. C. Greene, S. R. Jonsson, N. R. Landau, M. Lochelt, H. S. Malik, M. H. Malim, C. Munk, S. J. O'Brien, V. K. Pathak, K. Strebel, S. Wain-Hobson, X. F. Yu, N. Yuhki, and R. S. Harris. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 83:494-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaRue, R. S., S. R. Jonsson, K. A. Silverstein, M. Lajoie, D. Bertrand, N. El-Mabrouk, I. Hotzel, V. Andresdottir, T. P. Smith, and R. S. Harris. 2008. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 9:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, X. Y., F. Guo, L. Zhang, L. Kleiman, and S. Cen. 2007. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 282:32065-32074. [DOI] [PubMed] [Google Scholar]
- 42.Liu, H., X. Wu, M. Newman, G. M. Shaw, B. H. Hahn, and J. C. Kappes. 1995. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J. Virol. 69:7630-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Llano, M., D. T. Saenz, A. Meehan, P. Wongthida, M. Peretz, W. H. Walker, W. Teo, and E. M. Poeschla. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314:461-464. [DOI] [PubMed] [Google Scholar]
- 44.Loewen, N., R. Barraza, T. Whitwam, D. T. Saenz, I. Kemler, and E. Poeschla. 2003. FIV vectors. Methods Mol. Biol. 229:251-271. [DOI] [PubMed] [Google Scholar]
- 45.Luo, K., B. Liu, Z. Xiao, Y. Yu, X. Yu, R. Gorelick, and X. F. Yu. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 78:11841-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo, K., Z. Xiao, E. Ehrlich, Y. Yu, B. Liu, S. Zheng, and X. F. Yu. 2005. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. U. S. A. 102:11444-11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malim, M. H. 2009. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:675-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 49.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 50.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 51.Mariani, R., B. A. Rasala, G. Rutter, K. Wiegers, S. M. Brandt, H. G. Krausslich, and N. R. Landau. 2001. Mouse-human heterokaryons support efficient human immunodeficiency virus type 1 assembly. J. Virol. 75:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 53.Mbisa, J. L., R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McEwan, W. A., T. Schaller, L. M. Ylinen, M. J. Hosie, G. J. Towers, and B. J. Willett. 2009. Truncation of TRIM5 in the Feliformia explains the absence of retroviral restriction in cells of the domestic cat. J. Virol. 16:8270-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKnight, A., P. R. Clapham, and R. A. Weiss. 1994. HIV-2 and SIV infection of nonprimate cell lines expressing human CD4: restrictions to replication at distinct stages. Virology 201:8-18. [DOI] [PubMed] [Google Scholar]
- 56.Mehle, A., J. Goncalves, M. Santa-Marta, M. McPike, and D. Gabuzda. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18:2861-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 58.Mehle, A., E. R. Thomas, K. S. Rajendran, and D. Gabuzda. 2006. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 281:17259-17265. [DOI] [PubMed] [Google Scholar]
- 59.Miyagi, E., S. Opi, H. Takeuchi, M. Khan, R. Goila-Gaur, S. Kao, and K. Strebel. 2007. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 81:13346-13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Münk, C., T. Beck, J. Zielonka, A. Hotz-Wagenblatt, S. Chareza, M. Battenberg, J. Thielebein, K. Cichutek, I. G. Bravo, S. J. O'Brien, M. Lochelt, and N. Yuhki. 2008. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 9:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Münk, C., J. Zielonka, H. Constabel, B. P. Kloke, B. Rengstl, M. Battenberg, F. Bonci, M. Pistello, M. Lochelt, and K. Cichutek. 2007. Multiple restrictions of human immunodeficiency virus type 1 in feline cells. J. Virol. 81:7048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Navarro, F., B. Bollman, H. Chen, R. Konig, Q. Yu, K. Chiles, and N. R. Landau. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374-386. [DOI] [PubMed] [Google Scholar]
- 63.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 64.Pacheco, B., S. Basmaciogullari, J. A. Labonte, S. H. Xiang, and J. Sodroski. 2008. Adaptation of the human immunodeficiency virus type 1 envelope glycoproteins to new world monkey receptors. J. Virol. 82:346-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pecon-Slattery, J., J. L. Troyer, W. E. Johnson, and S. J. O'Brien. 2008. Evolution of feline immunodeficiency virus in Felidae: implications for human health and wildlife ecology. Vet. Immunol. Immunopathol. 123:32-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poeschla, E., and D. Looney. 1998. CXCR4 is required by a non-primate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent and feline cells. J. Virol. 72:6858-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poeschla, E., F. Wong-Staal, and D. Looney. 1998. Efficient transduction of nondividing cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 4:354-357. [DOI] [PubMed] [Google Scholar]
- 68.Russell, R. A., and V. K. Pathak. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81:8201-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saenz, D. T., R. Barraza, N. Loewen, W. Teo, and E. Poeschla. 2006. Production and use of feline immunodeficiency virus (FIV)-based lentiviral vectors, p. 57-74. In J. Rossi and T. Friedman (ed.), Gene transfer: a Cold Spring Harbor laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 70.Saenz, D. T., N. Loewen, M. Peretz, T. Whitwam, R. Barraza, K. Howell, J. H. Holmes, M. Good, and E. M. Poeschla. 2004. Unintegrated lentiviral DNA persistence and accessibility to expression in nondividing cells: analysis with class I integrase mutants. J. Virol. 78:2906-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saenz, D. T., W. Teo, J. C. Olsen, and E. Poeschla. 2005. Restriction of feline immunodeficiency virus by Ref1, LV1 and primate TRIM5a proteins. J. Virol. 79:15175-15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakurai, A., A. Jere, A. Yoshida, T. Yamada, A. Iwamoto, A. Adachi, and M. Fujita. 2004. Functional analysis of HIV-1 vif genes derived from Japanese long-term nonprogressors and progressors for AIDS. Microbes Infect. 6:799-805. [DOI] [PubMed] [Google Scholar]
- 73.Sawyer, S. L., M. Emerman, and H. S. Malik. 2007. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 3:e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 75.Schäfer, A., H. P. Bogerd, and B. R. Cullen. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328:163-168. [DOI] [PubMed] [Google Scholar]
- 76.Schaller, T., S. Hue, and G. J. Towers. 2007. An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J. Virol. 81:11713-11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schröfelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. U. S. A. 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schröfelbauer, B., T. Senger, G. Manning, and N. R. Landau. 2006. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 80:5984-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schumacher, A. J., G. Hache, D. A. Macduff, W. L. Brown, and R. S. Harris. 2008. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 82:2652-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shacklett, B. L., and P. A. Luciw. 1994. Analysis of the vif gene of feline immunodeficiency virus. Virology 204:860-867. [DOI] [PubMed] [Google Scholar]
- 81.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 82.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 83.Shibata, R., M. Kawamura, H. Sakai, M. Hayami, A. Ishimoto, and A. Adachi. 1991. Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J. Virol. 65:3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Si, Z., N. Vandegraaff, C. O'Huigin, B. Song, W. Yuan, C. Xu, M. Perron, X. Li, W. A. Marasco, A. Engelman, M. Dean, and J. Sodroski. 2006. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc. Natl. Acad. Sci. U. S. A. 103:7454-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simon, J. H., D. L. Miller, R. A. Fouchier, M. A. Soares, K. W. Peden, and M. H. Malim. 1998. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 17:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 87.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 88.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W. S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822-35828. [DOI] [PubMed] [Google Scholar]
- 89.Swanson, C. M., B. A. Puffer, K. M. Ahmad, R. W. Doms, and M. H. Malim. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 23:2632-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takeuchi, H., S. Kao, E. Miyagi, M. A. Khan, A. Buckler-White, R. Plishka, and K. Strebel. 2005. Production of infectious SIVagm from human cells requires functional inactivation but not viral exclusion of human APOBEC3G. J. Biol. Chem. 280:375-382. [DOI] [PubMed] [Google Scholar]
- 91.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 92.Willett, B. J., E. L. McMonagle, S. Ridha, and M. J. Hosie. 2006. Differential utilization of CD134 as a functional receptor by diverse strains of feline immunodeficiency virus. J. Virol. 80:3386-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. U. S. A. 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 95.Zennou, V., D. Perez-Caballero, H. Gottlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78:12058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang, W., M. Huang, T. Wang, L. Tan, C. Tian, X. Yu, W. Kong, and X. F. Yu. 2008. Conserved and non-conserved features of HIV-1 and SIVagm Vif mediated suppression of APOBEC3 cytidine deaminases. Cell. Microbiol. 10:1662-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]