Abstract
To get more insight into the role of APOBEC3 (A3) cytidine deaminases in the species-specific restriction of feline immunodeficiency virus (FIV) of the domestic cat, we tested the A3 proteins present in big cats (puma, lion, tiger, and lynx). These A3 proteins were analyzed for expression and sensitivity to the Vif protein of FIV. While A3Z3s and A3Z2-Z3s inhibited Δvif FIV, felid A3Z2s did not show any antiviral activity against Δvif FIV or wild-type (wt) FIV. All felid A3Z3s and A3Z2-Z3s were sensitive to Vif of the domestic cat FIV. Vif also induced depletion of felid A3Z2s. Tiger A3s showed a moderate degree of resistance against the Vif-mediated counter defense. These findings may imply that the A3 restriction system does not play a major role to prevent domestic cat FIV transmission to other Felidae. In contrast to the sensitive felid A3s, many nonfelid A3s actively restricted wt FIV replication. To test whether VifFIV can protect also the distantly related human immunodeficiency virus type 1 (HIV-1), a chimeric HIV-1.VifFIV was constructed. This HIV-1.VifFIV was replication competent in nonpermissive feline cells expressing human CD4/CCR5 that did not support the replication of wt HIV-1. We conclude that the replication of HIV-1 in some feline cells is inhibited only by feline A3 restriction factors and the absence of the appropriate receptor or coreceptor.
In the family of Retroviridae, the vif gene is present in most members of the genus Lentivirus. Its absence in Equine infectious anemia virus and in endogenous lentiviruses of the order Lagomorpha might indicate that vif appeared later in lentivirus evolution and strongly emphasizes that lentivirus replication is not strictly dependent on Vif (20, 58). Nevertheless, in the well-studied primate and feline lentiviruses, viruses with a vif deletion barely replicate in vivo and are not pathogenic (10, 46). Functional studies revealed that the Vif protein of the human immunodeficiency virus type 1 (HIV-1) binds in the virus-producing cells the cellular cytidine deaminases APOBEC3F (A3F) and APOBEC3G (A3G) and induces their polyubiquitination and subsequent degradation by the proteasome (27, 45, 54, 55). In addition, it is reported that Vif of HIV-1 inhibits APOBEC3 (A3) proteins by other, nondegrading mechanisms (for a review, see reference 6).
During viral infections when either no Vif protein is made or when HIV-1 faces A3 proteins normally absent in T cells or macrophages, A3s can be incorporated in the viral particles and often inhibit the next round of infection. Other than the cytidine deaminase activity on single-stranded viral DNA during reverse transcription, human A3F and A3G inhibit Δvif HIV-1 by other means (2, 15, 16, 18, 28, 34). The finding that HIV-1 does not infect species besides Homo sapiens and Pan troglodytes is based on the species-specific adaptation of Vif to human A3s and other host-virus restriction mechanisms. In support of this model, HIV-1 is inhibited by A3s of rhesus macaques, African green monkeys (AGM), pigs, mouse, cats, and horses (4, 19, 25, 33). In the case of A3G from AGM, a single amino acid at position 128 determines whether Vif of HIV-1 or simian immunodeficiency virus (SIV) of AGM can or cannot counteract its antiviral activity (3, 24, 43, 53).
Similar to the situation of SIVs in primates, specific types of the feline immunodeficiency virus (FIV) can be isolated from many Felidae (49). Little is known about whether the diverse FIV isolates have a restricted host range as do HIV-1 and the SIVs (32). The phylogenetic data indicate that most of these FIV types isolated from different felids are monophyletic (summarized in references 39 and 48). However, recent FIV cross-species transmissions have been observed in pumas infected by the FIVs of domestic cats and bobcats, in leopards and tigers infected with the FIVs of lions, and in free-ranging leopard cats infected with FIV strains of domestic cats, indicating that repeated and/or multiple historic FIV cross-species felid-to-felid transmission events can occur in the wild or in captivity (5, 13, 35, 39, 48, 51). With the exception of repeated FIV transmissions from bobcats to pumas, these reports mostly describe singular events supporting that FIV is not frequently cross-species transmitted.
Recently, we characterized the feline Felis catus A3 (FcaA3) locus showing that it is less complex than that of primates but significantly more evolved than that of rodents (31). Based on an evolution-based nomenclature that reflects the identity of the zinc (Z) coordinating domain(s) (21), cats encode a triplicate Z2 and a single Z3 A3 gene. In addition, a complex process of readthrough transcription and alternative splicing results in two-domain feline A3Z2-Z3 molecules (31; see also below). These different feline A3 restriction factors display a specific pattern of restriction toward wild-type (wt) feline retroviruses (FIV, Feline leukemia virus [FeLV], and Feline foamy virus [FFV]) and variants devoid of their counterdefense proteins: while wt FIV and FFV are almost resistant against any of the feline A3s, FeLV is moderately inhibited by FcaA3Z2-Z3s. In contrast, Vif-deficient FIV is restricted by the two-domain FcaA3Z2-Z3 and slightly by the Z3 form, while Bet-deficient FFV is restricted to variable degrees by the three FcaA3Z2 proteins (22, 31).
In the present study, we sought to determine whether the Vif protein of the domestic cat FIV is counteractive against A3s of four lineages within Felidae. Using single-round replicating FIV-reporter viruses, we detected a very broad activity of VifFIV inhibiting almost all tested A3s of Felidae similar to their own species A3s. We further found that vifFIV cloned into HIV to replace the expression of authentic vif enables such chimeric HIV-1 to overcome the A3 restriction in feline cells, allowing productive replication in cat cells engineered to express the human receptors for HIV-1.
MATERIALS AND METHODS
Cells and transfections.
The human cell lines 293T, HeLa, HOS (ATCC CRL-1543), HOS.CD4.CCR5 (NIBSC ARP078), and the modified feline cell line CrFK.CD4.CCR5 (33) were maintained in Dulbecco high-glucose modified Eagle medium (Biochrom, Berlin, Germany) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). Ficoll-enriched human peripheral blood mononuclear cells (PBMC) were stimulated with 5 μg of phytohemagglutinin (Murex Diagnostics, Ltd., Dartfield, United Kingdom)/ml and incubated with 30 IU of human IL-2 (Proleukin; Chiron, Emeryville, CA)/ml in RPMI 1640 containing 10% FBS and 2 mM Glutamax I (Invitrogen, Karlsruhe, Germany) for 48 h before infection. Plasmid transfections into 293T cells were performed with Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions. Transfections were done in 12-well plates seeded with 2.5 × 105 cells/well the day before transfection and 2.2 μg of plasmid DNA. The standard transfection experiment contained 0.5 μg of FIV packaging construct, 0.5 μg of FIV-luciferase vector, 0.5 μg of A3 expression plasmid, 0.5 μg of Vif expression plasmid, and 0.2 μg of VSV-G expression plasmid; in some experiments pcDNA3.1(+) (Invitrogen) was used instead of Vif and/or A3 plasmids. For HIV-luciferase transfections, 1 μg of HIV-Luc plasmid was used instead of both FIV plasmids. At 48 h posttransfection, cells and supernatants were collected.
A3 and Vif plasmids.
All A3s are expressed as carboxy-terminal hemagglutinin (HA)-tagged proteins, except human (Homo sapiens) APOBEC3DE (HsaA3DE), a gift from Y.-H. Zheng, that carries a carboxy-terminal V5-tag (9) and human A3H splice variants (SV), gifts from Viviana Simon, that carry amino-terminal Flag tags (14). The human A3G expression construct was provided by N. R. Landau (25). FcaA3Z2s and -Z3, big cat (Panthera tigris corbetti [Pti], Lynx lynx [Lly], Panthera leo bleyenberghi [Ple], Puma concolor [Pco], Panthera pardus japonensis [Ppa])-A3Z2s and -Z3s, and Equus caballus (Eca)-, Mus musculus (Mmus)-, and Sus scrofa (Ssc)-A3 expression plasmids and human A3B, -C, and -F were previously described (22, 30, 31, 58). FcaA3Z2b-Z3 (SV-B), FcaA3Z2c-Z3 and big cat A3Z2-Z3 cDNAs were identified by reverse transcriptase PCR (RT-PCR) with the forward (fw) primer fAPO-30 (5′-TGCATCGGTACCACCAAGGCTGGAGAGAGGAATGG-3′) and the reverse (rv) primer fAPO-28 (5′-AGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGATATTCAAGTTTCAAATTTCTGAAG-3′) using cDNA of total RNA from feline PBMC and Pfu polymerase (Fermentas, St. Leon-Rot, Germany) as described previously (31). Each 30 cycles were run at 94°C for 30 s, 58°C for 1 min, and 72°C for 2 min, and PCR products were cloned into the KpnI and XhoI sites of pcDNA3.1(+) and sequence verified. A3Z2c-Z3 variants including polymorphic sequences of exon 4 of four different Felis catus breeding lines were generated by exchanging the following amino acids through overlapping extension PCR: for Birman (BIR), G179A, F182S, and K186E; for Japanese Bobtail (BOB), K186E; for British Shorthair (SHO), R172Q, G179D, F182L, and K186E; for Turkish Van (VAN), K157E, H158Y, D165Y, H166N, and K186E (see Fig. 1B, top panel). The resulting full-length constructs were cloned into pcDNA3.1(+) using the KpnI and XhoI restriction sites. Human and feline A3Z2-Z3 chimera were made by exchanging the N-terminal or C-terminal or both zinc (Z)-coordinating domains of the FcaA3Z2c-Z3 template through overlapping extension PCR. The resulting cloned PCR products were sequence verified. The 5′ and 3′ fragments were amplified separately by using primer pairs fApo3F-18 x fe3C/hu3H.rv (5′-GTTAACAGAGCCATTGTGGGTCTGGGCAA-3′) and fe3C/hu3H.fw (5′-TTGCCCAGACCCACAATGGCTCTGTTAAC-3′) x huA3H-HA.rv (5′-TTCAGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGATAGGACTGCTTTATCCTGTCAAG-3′) for FcaA3Z2c-HsaA3Z3 (FcaA3Z2c-HsaA3H); CEM15-CEM13 (5′-TAAGCGGAATTC TATCTAAGAGGCTGAACATG-3′) x hu3C/fe3H.rv (5′-CTTTGTTGGCCGGGATGGAGACTCTCCCGT-3′) and hu3C/fe3H.fw (5′-ACGGGAGAGTCTCCATCCCGGCCAACAAAG-3′) x fAPO-28 for HsaA3Z2b-FcaA3Z3 (HsaA3C-FcaA3Z3) and the plasmids human A3C or A3H (HapII RDD-SV-183 [FJ376614] [14]) and FcaA3Z2c-Z3 as a template. To test the relevance of the interdomain linker of FcaA3Z2c-Z3, both feline domains were replaced by human sequences: HsaA3Z2b-HsaA3Z3 (HsaA3C-HsaA3H) was constructed by overlapping PCR using the primer pairs CEM15-CEM13 x fe3C/hu3H.rv and fe3C/hu3H.fw x huA3H-HA.rv and the plasmids human A3H (HapII RDD-SV-183) and HsaA3Z2b-FcaA3Z3 as a template. The 5′ and 3′ fragments were then mixed and amplified with the two external primers. The resulting full-length mutants were cloned into pcDNA3.1(+) using KpnI and XhoI restriction sites. V5-tagged pVifHIV-1 of HIV-1NL4-3 was generated by amplifying the vif gene from pNL-LucR−E− (7) using fw primer HIV1_5′_HH (5′-TGCAGGTACCATGGAAAACAGATGGCAGGTGATGAT-3′) and rv primer HIV1_3′_HH (5′-AATGGCGGCCGCTCACGTAGAATCCAGTCCCAAGAGCGGGTTTGGGATAGGCTTGCCGTGTCCATTCATTGTATGGCTC-3′). The amplicon was cloned into pcDNA3.1(+) carrying a Woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) (11) via KpnI and NotI restriction sites. pVifSIVagm was provided by N. R. Landau and the V5 tag containing Vif of FIV (pVifFIV) was described previously (31). pVIFFIV expresses the codon-optimized vif of FIV-34TF10, and it was generated by cloning the codon-optimized vif gene containing a V5 tag sequence at the 3′ end, into pcDNA3.1(+) using the KpnI and NotI restriction sites; a 3′-WPRE element was included in the NotI and ApaI sites. Vif of FIV-34TF10 and Vif of FIV-PPR (expressed in the packaging construct pCPRΔEnv) share 90.4% identical amino acids. All sequences of plasmid DNA were obtained by dye terminator sequencing on an ABI 3730xl (Applied Biosystems, Darmstadt, Germany).
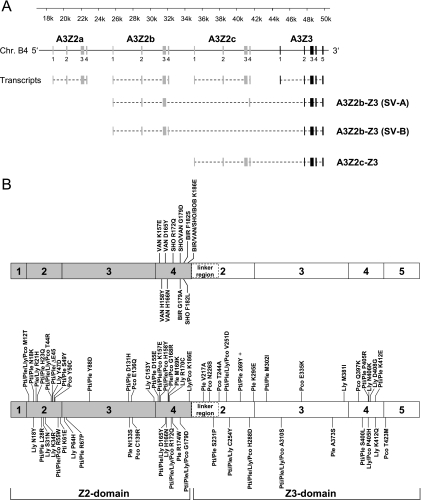
FIG. 1.
Representation of APOBEC3 (A3) coding regions in the genome of Felis catus. (A) Transcripts with translated exons (Z2, gray rectangles; Z3, black rectangles) and spliced-out introns (dashed lines) are indicated. Three readthrough transcripts were found: the already described two-domain FcaA3Z2c-Z3 (31) and the two splice variants (SV) A3Z2b-Z3 (SV-A) and A3Z2b-Z3 (SV-B) that include the exon 4-derived sequence of either A3Z2b or -Z2c in an additionally spliced version (for details, see reference 31). (B) Schematic representation of the exon structure of the A3Z2-Z3 readthrough transcripts. The locations of amino acid replacements in FcaA3Z2c-Z3 are indicated with respect to exon 4 variability of major cat breeds (upper figure) and A3Z2-Z3 genes of big cats (lower figure). A plus sign indicates the insertion of an amino acid. Numbers in shaded boxes indicate exons from Z2c, and those in open boxes indicate exons from Z3. The region of Z2-Z3 proteins designated “linker region” protein is encoded by exon 2 of A3Z3, a sequence that is untranslated in single-domain A3Z3. BIR, Birman; BOB, Japanese Bobtail; SHO, British Shorthair; VAN, Turkish Van; Pti, Panthera tigris corbetti; Ppa, Panthera pardus japonensis; Ple, Panthera leo bleyenberghi; Lly, Lynx lynx; Pco, Puma concolor.
Viruses and infections.
FIV single-cycle luciferase vectors (FIV-Luc) were produced by cotransfecting 293T cells with the following: the replication-deficient packaging construct pFP93 (derived from clone FIV-34TF10 [GenBank accession number M25381], a gift from Eric M. Poeschla [23]), which expresses gag, pol, and rev but does not express env or vif (referred as FIVΔvif in Results); the replication-deficient packaging construct pCPRΔEnv (derived from clone FIV-PPR [GenBank accession number M36968], a gift from Garry P. Nolan [8]), which expresses all FIV genes, including Vif, but not env (referred as wt FIV in Results); the FIV luciferase vector pLiNSin (31); a VSV-G expression plasmid pMD.G; an A3 expression plasmid; or empty vector pcDNA3.1(+). Vector pLiNSin was derived from pGiNSin, a self-inactivating (Sin) vector variant of pGiNWF (23), which is a minimal bicistronic FIV transfer vector plasmid coding for enhanced green fluorescent protein (EGFP) and neomycin phosphotransferase containing the posttranscriptional regulatory element WPRE and the FIV central DNA flap. In addition, the luciferase reporter vectors FIVΔvif plus Vif were produced by cotransfecting 293T cells with pFP93, pLiNSin, a Vif expression plasmid (pMD.G), and an A3 expression plasmid or empty vector pcDNA3.1(+). Gag-Pol of strains FIV-34TF10 and FIV-PPR show ∼96% identical amino acids. HIV-Luc Δvif was produced by cotransfecting 293T cells with pNL-LucR−E−Δvif (25) and pMD.G, together with defined A3 expression plasmids or pcDNA3.1(+) and a Vif expression plasmid or empty vector pcDNA3.1(+). The RT activity of viruses was quantified by using the Cavidi HS Lenti RT kit (Cavidi Tech, Uppsala, Sweden). For reporter virus infections, HOS cells were seeded at 2.0 × 103 cells/well 1 day before transduction in 96-well plates and then infected with reporter virus stocks normalized for RT activity. Firefly luciferase activity was measured 3 days posttransduction with the Steadylite HTS reporter gene assay system (Perkin-Elmer, Cologne, Germany) according to the manufacturer's instructions on a MicroLumat Plus luminometer (Berthold Detection Systems, Pforzheim, Germany). Transductions were performed in triplicates; the means and standard deviations of each triplicate are shown. Replication-competent NL-BaL (NL4.3 with the env BaL) (26) virus stocks were prepared by harvesting the supernatant of transfected 293T cells. The cloning used to generate pNL-BaL.vifFIV and pNL-LucR−E−vifFIV results in an in-frame insertion of FIV vif in the HIV-1 vif coding region. The constructs were designed to express VifFIV by internal initiation. Three fragments were amplified separately using pVIFFIV and pNL-LucR−E− as template DNA and the primer pairs Vpu_mut_5′out_HH (5′-GAACCGGTACATGGAGTGTATTAT-3′) and HIV1_vif_FIV1.rv (5′-CTCTTCGCTCATGGCCTATGTATGCAGACC-3′); HIV1_vif_FIV2.fw (5′-GGTCTGCATACATAGGCCATGAGCGAAGAG-3′) and HIV1_vif_FIV2a.rv (5′-CTCCATTCTATGGTCAGGTGCTGTCC-3′); and HIV1_vif_FIV3.fw (5′-GGACAGCACCTGACCATAGAATGGAG-3′) and HIV1_vif_FIV3.rv (5′-TGCAGAATTCTTATTATGGCTTCCA-3′). The forward primer HIV1_vif_FIV2.fw was used to insert a stop codon upstream of the FIV Vif-V5 start codon. The fragments were fused through overlapping extension PCR, and the final fragment was amplified by using the forward Vpu_mut_5′out_HH and reverse HIV1_vif_FIV3.rv primers. The amplicon was cloned into restriction sites AgeI and EcoRI of the HIV-1 plasmids. The sequence identity of the final HIV-derived plasmids was confirmed: vifFIV was inserted without disturbing the reading frames of the overlapping pol and vpr genes. The final vif region encodes for the first 76 amino acids of VifHIV, followed by the vif-V5 of FIV (see Fig. 5A for details). 3′ of the stop codon of FIV vif-V5, the natural reading frame of the terminal 107 amino acids of vifHIV were maintained but are assumed to be nonfunctional. The kinetics of viral spreading replication was determined with feline CrFK.CD4.CCR5 cells, human HOS.CD4.CCR5 cells, and human PBMC by infection with 50-pg equivalents of RT from NL-BaL and NL-BaL.vifFIV. Spreading virus replication was quantified over 15 days using the Zeptometrix Retro-Tek HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) (Zeptometrix, New York, NY) according to the instructions of the manufacturer. For all transfections, transductions and infections, representative data are shown, and all experiments were repeated independently at least three times.
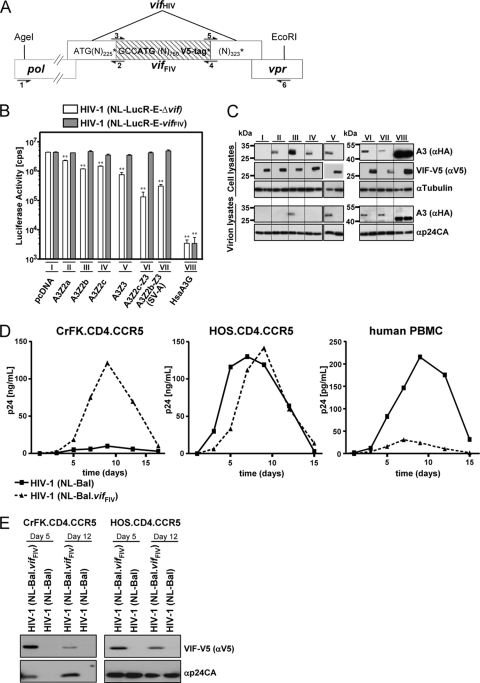
FIG. 5.
HIV-1 is protected by VifFIV in feline cells. (A) Schematic representation of the insertion of the vifFIV (shaded) in the vif gene of the HIV-derived constructs (NL-LucR−E−vifFIV and NL-Bal.vifFIV) by fusion PCR. Overlapping pol (schematically depicted) and vpr genes are shown. Restriction enzyme recognition sites AgeI and EcoRI were used to clone the fusion PCR product. Position of PCR primers used are shown as arrows: primer 1, vpu_mut_5′out_HH; primer 2, HIV1_vif_FIV1.rv; primer 3, HIV1_vif_FIV2.fw; primer 4, HIV1_vif_FIV2a.rv; primer 5, HIV1_vif_FIV3.fw; and primer 6, HIV1_vif_FIV3.rv. “N” represents adenine (A), cytosine (C), guanine (G), or thymidine (T). (N)number represents the number of Ns. *, Stop codon. (B and C) The effect of VifFIV in cis for the infectivity of the chimeric HIV-1 constructs was tested by using single-cycle luciferase reporter viruses. VSV-G pseudotyped virus was produced by cotransfection of pNL-LucR−E−Δvif or pNL-LucR−E−vifFIV in the presence or absence (pcDNA) of the indicated expression plasmids for feline A3 and human A3G. Infectivity of the viruses was determined by quantification of luciferase in HOS cells infected with normalized amounts of viruses (B). Asterisks represent statistically significant differences (*, P < 0.05; **, P < 0.01 [Dunnett t test]) relative to the pcDNA control. Corresponding immunoblots of cell lysates and virion lysates were probed with anti-HA antibody for A3 expression (C). The expression of VifFIV was confirmed by probing the blot with an anti-V5 antibody. Cell lysates were also analyzed for equal amounts of total proteins by using an anti-tubulin antibody. Virus lysates were analyzed for equal amounts of viral proteins by using anti-p24CA (HIV-1 capsid) antibody. Roman numbers indicate a set of transfections with the same A3 plasmid. (D and E) Replication of wild-type HIV-1(NL-Bal) and HIV-1(NL-Bal.vifFIV). Human HOS.CD4.CCR5 cells, feline CrFK.CD4.CCR5 cells, and PHA/IL-2 activated human PBMC were infected with NL-BaL and NL-BaL.vifFIV at a multiplicity of infection of 0.05. (D) Culture supernatants were quantified every 2 or 3 days by p24CA ELISA. (E) Immunoblot analysis of virus lysates from infected CrFK.CD4.CCR5 and HOS.CD4.CCR5 cells on days 5 and 12 postinfection. Expression of VifFIV was confirmed by probing the filter with an anti-V5 antibody, the same blot was also stained with anti-p24CA (capsid) antibody to detect HIV-1. α, anti.
Immunoblot analysis.
Transfected cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], protease inhibitor cocktail set III [Calbiochem, Darmstadt, Germany]). Virions (filtered, 0.45-μm pore size) culture supernatant) were pelleted by centrifugation through a 20% sucrose cushion at 35,000 rpm in a SW40 Ti rotor for 1.5 h and resuspended in RIPA buffer. The protein concentration of the lysates was quantified by using Bradford reagent (Applichem, Darmstadt, Germany). Then, 20 μg of each sample was separated by SDS-PAGE and transferred to polyvinylidene difluoride filters. Filters were probed with mouse anti-hemagglutinin (anti-HA) antibody (1:10,000 dilution, MMS-101P; Covance, Münster, Germany), mouse anti-Flag M2 antibody (1:1,000, clone M2; Sigma-Aldrich, Taufkirchen, Germany), mouse anti-V5 antibody (1:5,000 dilution, MCA1360; ABDserotec, Düsseldorf, Germany), mouse anti-α-tubulin antibody (1:4,000 dilution, clone B5-1-2; Sigma-Aldrich), mouse anti-capsid p24 hybridoma supernatant (1:50 dilution, α-p24 183-H12-5C; provided by Egbert Flory), or mouse anti-VSV-G antibody (1:10,000 dilution; clone P5D4; Sigma-Aldrich), followed by horseradish peroxidase-conjugated rabbit anti-mouse antibody (α-mouse-IgG-HRP; GE Healthcare, Munich, Germany), and developed with ECL chemiluminescence reagents (GE Healthcare).
Statistical analysis.
The data were analyzed by using a Dunnett t test using GraphPad InStat 3 software (GraphPad Software, San Diego, CA). This is a multiple-comparison method that uses a pooled standard deviation and allows a comparison of multiple values to a single control. P values of <0.05 were considered significant.
Data deposition.
The novel sequences reported here have been deposited in the GenBank database (accession number): FcaA3Z2b-Z3 SV-B (HM100128), FcaA3Z2c-Z3 (GU097660), PtiA3Z2-Z3 (GU097663), PleA3Z2-Z3 (GU097662), LlyA3Z2-Z3 GU097661), and PcoA3Z2-Z3 (GU097659).
RESULTS
Complex splicing and readthrough transcription of feline A3s.
Recently, we described five different Felis catus A3 cDNAs (FcaA3Z2a, -Z2b, -Z2c, -Z3, and the readthrough splice variant [SV]-Z2b-Z3 [now designated FcaA3Z2b-Z3 SV-A]) representing the genes of the sequenced Abyssinian cat genome (22, 31, 33). During the present study, we identified the alternative splice variant B (SV-B) of A3Z2b-Z3 with exons 1 to 4 of A3Z2b, whereas the previously described A3Z2b-Z3 splice variant A (SV-A) differs in exon 4 that is derived from the A3Z2c gene (Fig. 1 A). Via RT-PCR we identified another two-domain A3 readthrough transcript in feline PBMC, A3Z2c-Z3. The A3Z2c-Z3 cDNA is composed of the fused open reading frames of A3Z2c and A3Z3 (Fig. 1A). All readthrough A3s differ moderately from each other in three or six amino acids. In these two-domain feline A3 proteins, the Z2 and Z3 domains are naturally separated by a linker region that is derived from the 156-nucleotide (nt) long unique 5′ region of the A3Z3 Exon 2 (Fig. 1). In addition, we cloned expression plasmids of A3Z2c-Z3s containing the polymorphic sequences detected in exon 4 of A3Z2c of the four domestic cat breeds Japanese Bobtail (BOB), Birman (BIR), British Shorthair (SHO), and Turkish Van (VAN) (31). The exon 4-derived variability of the cat breeds with respect to the reference Abyssinian sequence affected up to five amino acids (Fig. 1B, top panel; for details, see Materials and Methods).
FIV Vif broadly counteracts Felidae A3s.
To test these A3s against FIV, we analyzed the infectivity of wt, Δvif FIV, and Δvif FIV plus VifFIV produced in the presence of the described feline A3s and human A3G using single-round luciferase reporter vectors. In the wt FIV vector system (based on FIV-PPR), the Vif protein is expressed in its natural genomic context, whereas the Δvif FIV vector system (based on FIV-34TF10) lacks the vif gene. To complement Vif in trans, plasmid pVifFIV expressing a codon-optimized Vif protein of FIV-34TF10 with a C-terminal V5 tag was used. The Vifs of FIV-34TF10 and of FIV-PPR share 90.4% identical amino acids and Gag-Pol proteins of both FIV strains are ∼96% identical.
293T cells were cotransfected either with wt FIV or Δvif FIV or Δvif FIV plus the VifFIV expression plasmid and one of the A3 expression plasmids. At 2 days posttransfection, virus-containing cell supernatants were collected, normalized for RT activity, and used to transduce HOS cells. The intracellular luciferase activity in transduced HOS cells was analyzed 3 days postinfection (dpi), a time when provirus formation had finished. Similar to recent findings (31), expression of the three A3Z2 isoforms did not inhibit wt or Δvif FIV (compare the first bar with the second and third bars in sets II to IV, Fig. 2 A; variations are not statistically significant). A3Z3 reduced the infectivity of Vif-deficient FIV ∼5-fold (statistically significant, P < 0.01), but not if Vif was expressed (compare the first bar with the second and third bars in set V, Fig. 2A). Both the Vif of the wt FIV packaging construct and of the VifFIV expression plasmid induced a depletion of A3Z2s and of A3Z3 proteins in the producer cells, indicating that the V5 tag in Vif did not compromise the Vif function (compare the first lane with the second and third lanes in sets II to IV of cell lysate, Fig. 2B). Interestingly, not only A3Z3 also A3Z2 proteins were easily detectable in particles of Δvif FIV (see first lane of set II to V, virion lysates, Fig. 2B). In analyzing the double-domain A3s, we found that both A3Z2-Z3s were active against Δvif FIV (6- to 12-fold inhibition; P < 0.01) but were completely or mostly inactive when Vif was coexpressed (compare the first bar with the second and third bars in sets VI and VII, Fig. 2A). A3Z2b-Z3 splice variants A and B showed indistinguishable expression levels and activities (data not shown); therefore, all figures show only experiments performed with the A3Z2b-Z3 (SV-A).
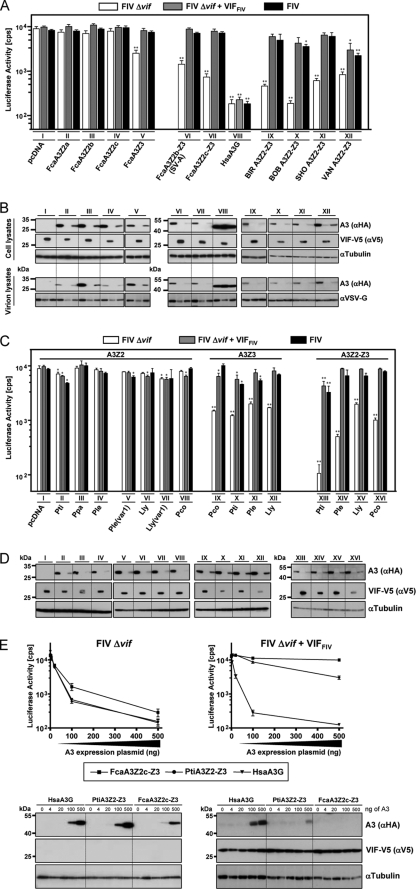
FIG. 2.
FIV Vif of domestic cats overcomes the antiviral activity of Felidae A3s. VSV-G pseudotyped wt FIV (expressing Vif), Δvif, and Δvif plus VifFIV luciferase reporter vectors were produced in 293T cells cotransfected with HA-tagged A3 expression plasmids of the domestic cat and chimeric A3s with major amino acid exchanges of exon 4 of A3Z2c of four cat breeding lines (A and B) and big cats (C and D). Roman numbers indicate a set of transfections with the same A3 plasmid. Plasmid pVifFIV expresses a V5-tagged Vif protein. pcDNA3.1(+) (pcDNA) was included as a control. Reporter vector infectivity was determined by quantification of luciferase activity in HOS cells transduced with vector particles normalized for RT. Asterisks represent statistically significant differences (*, P < 0.05; **, P < 0.01 [Dunnett t test]) relative to the pcDNA control. A3 and Vif (Vif-V5) expression in the transfected 293T producer cells were detected by immunoblotting with anti-HA antibody for A3 or anti-V5 for Vif (B and D). Cell lysates were also analyzed for equal amounts of total proteins by using anti-tubulin antibody. For panel B, Felis catus A3s proteins were also analyzed for encapsidation into released FIV particles. Encapsidated A3 proteins were detected on immunoblots probed with anti-HA antibody. Vector particle lysates were analyzed for equal amounts of viral proteins by using anti-VSV-G antibody. (E) Increasing amounts of A3 (0, 4, 20, 100, and 500 ng of plasmid) were tested against Δvif FIV and Δvif FIV plus Vif (500 ng of pVifFIV plasmid) and alternatively Δvif FIV in the presence of 500 ng of A3 plasmid with increasing amounts of VifFIV (0, 4, 20, 100, and 500 ng of plasmid) was analyzed (F). Corresponding immunoblots of lysates of the vector producer cells are shown. A3 and Vif (Vif-V5) expression in transfected 293T producer cells were detected by immunoblotting with anti-HA antibody for A3 or anti-V5 for Vif. Cell lysates were also analyzed for equal amounts of total proteins by using anti-tubulin antibody. α, anti; cps, counts per second; BIR, Birman; BOB, Japanese Bobtail; SHO, British Shorthair; VAN, Turkish Van; Fca, Felis catus; Pti, Panthera tigris corbetti; Ppa, Panthera pardus japonensis; Ple, Panthera leo bleyenberghi; Lly, Lynx lynx; Pco, Puma concolor; Hsa, Homo sapiens; (var1), A3Z2 variant transcript detected in the indicated species.
The high genetic variability in exon 4 of A3Z2c in some cat breeds (31) made us wonder whether these amino acids are the result of a positive selection in response to FIV. We found that the A3Z2c-Z3 constructs called BIR, BOB, SHO, and VAN (see above) were at least as active as the parental A3Z2c-Z3 (Abyssinian cat) under the experimental conditions. The A3 constructs containing the exon 4 sequences from BIR, SHO, and VAN significantly inhibited up to 15-fold, and the most active variant with sequences of BOB reduced the infectivity of Δvif FIV by 38-fold (first bar in sets IX, XI, and XII, Fig. 2A). Coexpression of VifFIV fully counteracted the A3Z2c-Z3s from the Abyssinian cat and constructs with sequences of BIR and SHO (second and third bars in sets IX and XI, Fig. 2A) and mostly rescued FIV also from A3Z2-Z3s with the exon 4 variability of BOB, with a difference of no statistical significance for Δvif FIV plus VifFIV and significance for wt FIV (P < 0.05) (second and third bars, set X, Fig. 2A). Vif could not completely restore the infectivity of Δvif FIV+VifFIV or wt FIV made in the presence of VAN (statistically significant, P < 0.05 or P < 0.01, respectively), and it remained reduced by ∼5-fold (second and third bars, set XII, Fig. 2A). Immunoblots of the virus producer cells demonstrated equal expression of the A3 proteins and a Vif-dependent depletion (compare the first lane with the second and third lanes in sets II to VII and IX to XII, Fig. 2B). As expected, the antiviral activity of HsaA3G was not counteracted by VifFIV. Taken together, these findings suggest that the genetic variability in exon 4 of A3Z2c-Z3 has a minor effect on the infectivity of domestic cat FIV. However, amino acids derived from exon 4 of Turkish Van do have some impact on the counteraction of VifFIV. Because our studies, described below, implicate that VifFIV interacts with both Z domains of A3Z2c-Z3, the mechanism of the moderate Vif resistance of VAN cannot be explained and requires further investigation.
Our finding that Vif of FIV potently inhibited mostly all tested A3s of the domestic cat prompted us to evaluate the ability of VifFIV to counteract A3 proteins of felid species different from the domestic cat lineage. For this purpose, A3Z2, A3Z3, and A3Z2-Z3 expression plasmids of Panthera tigris corbetti, Lynx lynx, Panthera leo bleyenberghi, Puma concolor, and A3Z2 of Panthera pardus japonensis, representing three lineages within Felidae, were analyzed for their anti-FIV activity. Because only the F. catus chromosomal A3 locus is characterized (31), the felid A3 cDNAs of big cats cannot be related to a specific Z2 gene or to a specific Z2-Z3 readthrough transcript. Comparing the protein sequence of FcaA3Z2c-Z3 to those of big cats, many amino acids changes become obvious, especially in exons 2 and 4 of A3Z2 and exon 4 of A3Z3 (Fig. 1B, bottom panel). Testing the big cat A3s in the FIV luciferase vector system, we surprisingly detected no strong inhibitory activity against the wt domestic cat FIV (see the second and third bars in sets II to XVI, Fig. 2C). The felids A3Z2s did not much affect the infectivity of wt or Δvif FIV (sets II to VIII, Fig. 2C) similar to the inert antiviral activities of the domestic cat counterparts (sets II to IV, Fig. 2A). However, we found for some felid A3Z2s a mild, sometimes significant, inhibition of ∼1.5- to 2-fold of FIV (see sets II to VIII, Fig. 2C). Vif-deficient FIV was significantly (P < 0.01) inhibited by A3Z3s and A3Z2-Z3s, but in all cases the antiviral activity was efficiently suppressed by Vif (compare the first bar to the second and third bars, set IX to XVI, Fig. 2C). Compared to the infectivity data (Fig. 2C), Vif induced a robust depletion of all big cat A3s in the corresponding producer cells (compare the first lane to the second and third lanes in sets II to XVI, Fig. 2D). The tiger A3s were slightly, but significantly, more active than those of the other big cat A3s (see sets II, X, and XIII, Fig. 2C), such as PtiA3Z2-Z3, inhibited in this experiment the infectivity of Δvif FIV by 85-fold and inhibited FIV ca. 3- to 5-fold despite the coexpression of Vif (P < 0.01, compare the first bar to the second and third bars, set XIII, Fig. 2C).
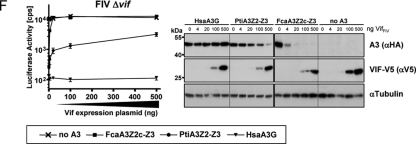
To demonstrate that the moderate inhibition of FIV by tiger A3Z2-Z3 is relevant, a titration of cat Vif and tiger A3 plasmids in the context of Δvif FIV was performed (Fig. 2E and F). In contrast to the standard assays, where each 500 ng of pVifFIV and A3 expression plasmids were transfected, 4, 20, 100, or 500 ng each of FcaA3Z2c-Z3, PtiA3Z2-Z3, and VifFIV was used. Human A3G was included as a control for a VifFIV-insensitive A3. The titration curves of human A3G and tiger A3 in the absence of Vif were very similar to each other and showed that in the range from 100 to 500 ng of plasmid tiger A3Z2-Z3 and A3G were ca. 2- to 3-fold more inhibitory than FcaA3Z2c-Z3 (Fig. 2E, left panel). We redid these experiment in the presence of 500 ng of Vif expression plasmid (Fig. 2E, right panel) and saw that only the highest amount of the tiger A3Z2-Z3 plasmid (500 ng) inhibited FIV by ∼5-fold.
To further study whether a reduced Vif expression would render FIV replication more vulnerable to A3 restriction, a fixed amount of 500 ng of A3 expression plasmid and varied concentrations of the Vif plasmid were used (Fig. 2F). We found that FcaA3Z2c-Z3 was very sensitive to Vif and that 20 ng of pVifFIV could fully restore FIV infectivity. In stark contrast, the heterologous tiger A3Z2-Z3 inhibited FIV ∼10-fold in the presence of low amounts of Vif (20 and 100 ng of Vif plasmid). Only with the highest amount of pVifFIV (500 ng) was the antiviral activity of tiger A3Z2-Z3 reduced to an ∼4-fold inhibition (Fig. 2F). The corresponding immunoblots of the vector producer cells (Fig. 2F) showed that the degree of Vif-induced depletion of the tiger and cat A3Z2-Z3s correlated with the differences of these A3 to inhibit FIV. Whereas 20 ng of pVifFIV was sufficient to deplete all detectable levels of FcaA3Z2c-Z3, small amounts of tiger A3Z2-Z3 were detectable even in the presence of 25-fold more Vif plasmid (Fig. 2F). Together, these results indicate that, depending on the expression levels of A3 and of Vif, even a moderate Vif-insensitive A3 (such as the tiger A3Z2-Z3) weakly inhibits FIV replication. However, considering the results of the experiments with the wt FIV vector system that expresses in cis natural amounts of Vif (set XIII in Fig. 2C), it is likely that FIV under in vivo conditions could counteract most of the inhibitory effect of tiger A3Z2-Z3. Thus, we conclude that, in contrast to the reported species-specific interaction of VifHIV-1 to human but not to nonhuman primate A3Gs (25, 52), VifFIV showed barely any species specificity for felid A3s (Fig. 2C). More studies are required to determine whether Vif expression levels vary during natural infections, e.g., due to the presence or absence of cellular host factors for splicing or posttranslational protein modifications, and whether these variations are relevant to preventing cross-species transmissions among felids.
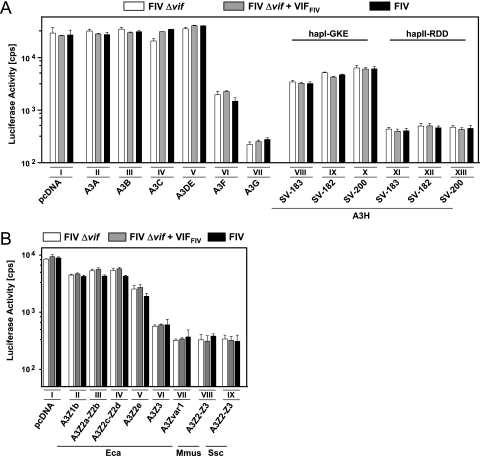
Nonfelid A3s are resistant to Vif of FIV.
To further characterize the specificity of VifFIV, we tested A3s from human and other nonfelid species. These experiments were also done to identify the nonfelid A3s that likely restrict cross-species transmission of FIV in vivo. We generated FIV particles as described above together with one of the human, equine, porcine, or murine A3 expression plasmids. Again, HOS cells were transduced with the FIV vector normalized for RT, and at 3 dpi the intracellular luciferase activities were quantified. As shown in Fig. 3, human A3F, A3G, and haplotype (hap) and splice variants (SV) of A3H, equine A3Z3, porcine A3Z2-Z3, and murine A3Z2-Z3 proteins were found to exert a significant inhibitory effect on the infectivities of wt and vif-deficient FIV. In contrast to the activity of Vif against the antiviral effect of A3s from Felidae (Fig. 2A and C), all nonfelid A3s showed resistance to VifFIV (Fig. 3). Although human A3A, A3B, A3C, and A3DE did not restrict FIV, we found that human A3H alleles encoding a single nucleotide polymorphism cluster (G105R, K121D, E178D, hapII-RDD) restricted FIV more efficiently than “wild-type” A3H (hapI-GKE), which is very similar to the described restriction pattern against HIV-1 (14). The activity of A3H against FIV was independent of the tested splice isoforms (SV-182, SV-183, and SV-200 [14]). Under these experimental conditions, the human A3G reduced the infectivity of FIV by 130-fold, human A3F and A3H (hapI-GKE) by ∼10-fold, and pig and mouse A3Z2-Z3 by 25-fold, and the hapII-RDD variant of the human A3H reduced FIV infectivity by 60-fold. The equine A3Z1b, A3Z2-Z2, and A3Z2e proteins moderately inhibited FIV only 2- to 3-fold. Together, the results demonstrate that humans, horses, mice, and pigs have at least one A3 gene that likely forms part of the barrier against a cross-species transmission of FIV.
FIG. 3.
Inhibition of FIV by nonfeline A3s. VSV-G pseudotyped FIV luciferase reporter vectors (wt FIV, expressing Vif; Δvif; and Δvif plus VifFIV) were produced in 293T cells in the presence or absence (pcDNA) of human A3s (A) or nonprimate equine, murine, and porcine A3s (B). The infectivity of the vector particles was determined by quantification of luciferase activity in HOS cells transduced with equal amounts of particles. pcDNA, pcDNA3.1(+); SV, splice variant; hapI, haplotype I; hapII, haplotype II; cps, counts per second; Eca, Equus caballus; Mmus, Mus musculus; Ssc, Sus scrofa. Roman numerals indicate a set of transfections with the same A3 plasmid.
The double-domain FcaA3Z2c-Z3 protein contains two Vif interaction sites.
In order to induce proteasomal degradation, VifHIV-1 binds either the C-terminal Z2f domain of human A3F (HsaA3Z2e-Z2f) or the N-terminal Z2g domain of human A3G (HsaA3Z2g-Z1c) (17, 42, 56, 57). Protection of FIV replication by VifFIV is always associated with depletion of the antiviral feline A3 proteins, and VifFIV can even induce depletion of the feline A3 proteins that do not restrict FIV (Fig. 2B, compare A3 detection in cell lysates in the first, second, and third lanes in sets II, III, and IV). These findings suggest that VifFIV can interact with feline Z3 and Z2 domains.
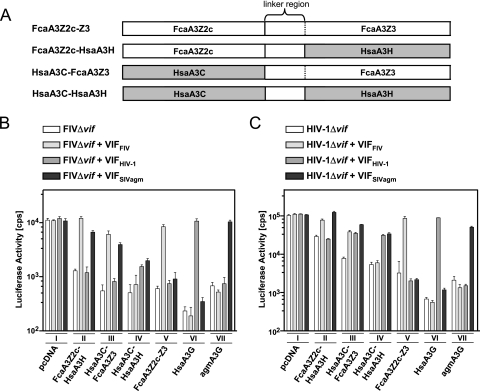
To test this hypothesis, Vif interaction with FcaA3Z2c-Z3 (Abyssinian breed) was assayed using feline and human A3Z2- Z3 chimeras (Fig. 4 A). For chimera FcaA3Z2c-HsaA3H (FcaA3Z2c-HsaA3Z3), the feline A3Z3 was substituted by HsaA3H (HsaA3Z3, hapII-RDD SV-183). Conversely, in HsaA3C-FcaA3Z3 (HsaA3Z2b-FcaA3Z3), the N-terminal feline A3Z2 was exchanged by human A3C (HsaA3Z2b). To test the relevance of the interdomain linker of the parental feline protein, both feline domains were replaced by human sequences, termed HsaA3C-HsaA3H (HsaA3Z2b-HsaA3Z3) (Fig. 4A). These A3s, together with A3Gs of human and AGM origin, were tested by using the single-cycle Δvif FIV- and Δvif HIV-1-luciferase reporter viruses to determine their antiviral activities and their susceptibilities to FIV, HIV-1, and SIVagm Vif proteins. Viral vector stocks were produced in 293T cells, and the infectivities of normalized particles were measured on HOS cells by quantification of the intracellular luciferase activity.
FIG. 4.
Analyses of human/feline A3 chimeras reveal Vif interactions with both Z-domains of FcaA3Z2c-Z3. (A) Schematic representation of human/feline A3Z2-Z3 chimera. Open bars denote feline sequences; filled bars denote human sequences. In FcaZ2-Z3, the Z2 and Z3 domains are naturally separated by a linker region (demarcated by dotted lines) that is encoded by exon 2 of Z3, which is untranslated in FcaA3Z3 (see Fig. 1A). FIVΔvif (B) and HIVΔvif (C) luciferase reporter particles were produced in 293T cells in the presence or absence (pcDNA) of feline and chimeric human/feline A3Z2-Z3s expression plasmids and expression plasmids for VifFIV, VifHIV-1, or VifSIVagm. To control the specificity of primate Vifs, human and AGM A3G expression plasmids were included. The infectivities of the vector particles were determined by quantification of luciferase in HOS cells transduced with equal amounts of vector particles. pcDNA, pcDNA3.1(+); cps, counts per second. Roman numerals indicate a set of transfections with the same A3 plasmid.
All A3 chimera inhibited both viruses in the absence of Vif proteins (Fig. 4B and C, compare the first bars in sets I, II, III, and IV). VifFIV counteracted FcaA3Z2c-HsaA3H and HsaA3C-FcaA3Z3, but not HsaA3C-HsaA3H, in both viral systems (see the second bars in sets II, III, and IV of Fig. 4B and C). The Vif of HIV-1 failed to inhibit feline-human chimera FcaA3Z2c-HsaA3H (compare the first and third bars in set II of Fig. 4B and C), the wt feline A3Z2c-Z3, and the agmA3G (see the third bars in sets V and VII, Fig. 4B and C). These results were expected because human A3H, feline A3s, and agmA3G are resistant to Vif of HIV-1 (14, 25, 33). The human-feline (HsaA3C-FcaA3Z3) and the human-human (HsaA3C-HsaA3H) chimeras were sensitive to VifHIV-1 in the HIV vector system (see the third bars in sets III and IV, Fig. 4C). However, in the FIV vector system VifHIV-1 insufficiently counteracted the human-feline A3 (HsaA3C-FcaA3Z3) and, like VifSIVagm, only weakly inhibited the human-human (HsaA3C-HsaA3H) A3 chimera (compare third bar in set III and IV of Fig. 4B and 4C). These unexpected results implicate that Vif of HIV-1 can interact with the A3C domain if it is part of an engineered double-domain A3 and that this interaction can be modulated by viral proteins of the packaging constructs. It is thus possible that conformational changes in the human-feline A3 (HsaA3C-FcaA3Z3) required for VifHIV-1 interaction are induced or stabilized by binding of this A3 protein to HIV-1 Gag but not by binding to FIV Gag. The SIVagm Vif acted as an active inhibitor of agmA3G, human-feline A3 (HsaA3C-FcaA3Z3), feline-human A3 (FcaA3Z2c-HsaA3H) in both viral vector systems, but was less efficient against the human-human (HsaA3C-HsaA3H) chimera (compare the fourth bars in Fig. 3B and C). Although the results support that VifFIV can use both Z domains to inhibit feline single- and double-domain A3s, conformational problems of the human and feline chimeras might have influenced the sensitivity to Vifs of HIV-1 and SIVagm. Taken together, we conclude that both Z domains of FcaA3Z2c-Z3, but not the linker region, interact with VifFIV.
FIV Vif protects HIV-1 in feline cells.
To corroborate the observation that VifFIV can protect Δvif HIV-1 against feline A3s (see the second bar in set V, Fig. 4C), we sought to determine whether VifFIV expressed in cis in the proviral context would also rescue HIV-1 from feline A3s. Therefore, two HIV-1-derived constructs, termed NL-LucR−E−vifFIV (HIV-Luc vifFIV) and NL-BaL.vifFIV, were generated, in which the HIV-1 vif gene was replaced by the codon-optimized FIV-34TF10 vif gene expressed by internal initiation (for details of the construction, see Materials and Methods and Fig. 5A).
To quantify protection by VifFIV in cis, we compared the infectivities of the HIV-1 luciferase reporter viruses NL-LucR−E−Δvif and NL-LucR−E−.vifFIV. The data in Fig. 5B show that in human cells the six feline A3 proteins inhibited Δvif HIV-1-mediated gene transfer to different degrees, depending on the type of A3 protein, similar to previous results (33). FcaA3Z2a isoforms reduced the infectivity of Δvif HIV-Luc by 3- to 4-fold; A3Z3 inhibited the infectivity by 6-fold, and the double-domain A3Z2-Z3 proteins inhibited the infectivity by 14- to 33-fold. Importantly, VifFIV expressed by the chimeric HIV-Luc fully protected HIV-1 against the antiviral activity of all feline A3s under these experimental conditions (see the second bars in sets II to VII, Fig. 5B). In these cases, the expression of VifFIV induced the depletion of feline A3s in the producer cells and likely thereby rescued vector infectivity (compare A3 detection in cell lysates in the first and second lanes of sets II to VII in Fig. 5C). A3-mediated restriction of marker gene expression was found to correlate with the presence of A3Z3 and A3Z2-Z3 proteins in released particles (see A3 detection in virion lysates in the first lanes of sets V, VI, and VII in Fig. 5C). The moderately active A3Z2s were only weakly detectable in lysates of the virions (see A3 detection in virion lysates, first lanes of sets II, III and IV, Fig. 5C). Human A3G was encapsidated in both particles, Δvif HIV-Luc and HIV-Luc vifFIV (see A3 detection in virion lysates in set VIII in Fig. 5C). Also, both reporter viruses were equally inhibited by A3G (see both bars in set VIII of Fig. 5B), thereby strongly supporting that HIV-Luc vifFIV does not express any residual VifHIV-1 activity.
Next, we evaluated the ability of VifFIV to support spreading replication of HIV-1 in feline cells. Wild-type HIV-1 (NL-BaL) and NL-BaL.vifFIV were used to infect feline CrFK.CD4.CCR5 and human HOS.CD4.CCR5 cells that are both genetically modified to express human CD4/CCR5. In addition, we infected PHA/IL-2-activated human PBMC with both viruses. From previous experiments, we knew that in contrast to human PBMC, HOS.CD4.CCR5 cells do support replication of Δvif HIV-1 (data not shown). Feline CrFK or CrFK.CD4.CCR5 cells support replication of neither Δvif FIV nor wt HIV-1 (33, 38, 44). In the supernatants of infected cells, the amount of released CA.p24 was measured over a period of 15 days. The results shown in Fig. 5D demonstrated that wt HIV-1 replicated efficiently in HOS.CD4.CCR5 cells and in human PBMC but not in the CrFK.CD4.CCR5 cells (Fig. 5D). In contrast, the VifFIV-encoding virus (NL-BaL.vifFIV) was able to replicate to similar levels in the permissive human HOS.CD4.CCR5 and the nonpermissive feline CrFK.CD4.CCR5 cells (Fig. 5D). Human PBMC did not support efficient spreading of NL-BaL. vifFIV (Fig. 5D). The PBMC cultures showed highest permissivity toward wt HIV-1 but yielded in very low p24 levels in the supernatant after inoculation with NL-BaL.vifFIV (Fig. 5D), which may be derived from the initial first round of infection. These data confirm (see above) that the VifFIV-chimeric viruses carry a HIV vif gene that is genetically and functionally fully inactivated. Expression of VifFIV was confirmed in virion lysates of the infected HOS.CD4.CCR5 and CrFK.CD4.CCR5 cultures in which NL-BaL.vifFIV showed spreading replication (Fig. 5E). At 12 dpi, the signal for the Vif-V5 was weaker than the V5 signal of samples of day 5 in both cultures, which may be due to a partial loss of the nonessential V5 tag of the VifFIV protein. We conclude that, at least in some feline cell types, A3 proteins constitute the major restriction mechanism acting inhibitory against HIV-1.
DISCUSSION
Examples of FIV cross-species transmissions have been reported and are currently studied to gain increased understanding regarding the permissiveness of selected and sometimes endangered feline species to diverse, molecularly characterized isolates of pathogenic FIV. These studies also serve as a valid and experimentally easier-to-handle model for the evolution of HIV from SIV. From these investigations of felids and their immunodeficiency viruses, we know that lentiviruses of lion (FIV-Ple) and puma (FIV-Pco) differentially replicate in activated PBMC of cats: FIV-Ple shows spreading replication, but FIV-Pco did not replicate in cat PBMC (50). Subsequently, the FIV-Pco isolate from experimentally infected cats contained extensive G-to-A mutations reminiscent of A3 activity (41). It was also described that pumas support the replication of FIV from bobcats that are frequently transmitted between these two species (12). In the cases where FIVs do replicate in heterologous species or PBMC, one can assume that the Vif protein can counteract the A3s of the novel, heterologous hosts. Alternatively, but less likely, it is possible that certain A3s are not active against some of the heterologous FIVs.
Based on our experiments, it appears that most of the diverse felid A3s of the domestic cat and big cats (lion, tiger, puma, and lynx) are probably not major determinants to prevent cross-species transmission of FIV of domestic cats to these closely related animal species. The A3Z2c-Z3 protein of tiger and the A3Z2-Z3 proteins containing amino acids of the polymorphic exon 4 of some cat breeds were found to be moderately resistance to the Vif of domestic cat FIV. For the tiger A3 we showed that low levels of cat Vif insufficiently counteracted its antiviral activity and regular Vif levels mostly inhibited this A3. Further experiments analyzing spreading FIV replications will be necessary to learn more about the inhibitory impact of these moderately resistant A3s during a longer observation period. We predict that FIV would quickly adapt its Vif protein to a complete A3 counteraction after a few rounds of replication in cells expressing, e.g., tiger A3Z2-Z3 protein. These assumptions are in fact supported by the ease with which FIV of the domestic cats infects other Felidae. A contributing factor may be that restriction of FIV replication and interspecies transmission by TRIM5 proteins is lacking in Felidae due to an in-frame stop mutation in the TRIM5 gene (29). However, other feline retroviruses and nonretroviruses might be more sensitive to the tested felid A3s in a species-specific manner, possibly explaining the positive selection on the felid A3 genes and the high prevalence of nonsilent mutations in the A3Z2 genes in different cat breeding lines (31). Polymorphic A3 genes are found also in humans, horses, and mice and show differential antiviral activities (1, 14, 36, 37, 58). Experiments with wt and Δvif FIVs of other felids will be required to ultimately demonstrate whether the FIV of the domestic cat is unique or whether many FIVs have the capacity to counteract heterologous felid A3s. The monophyly of the FIV strains of the diverse Felidae is a strong argument against frequent cross-species transmissions and, beside A3, other cellular and noncellular factors such as contact rates between different species likely determine or even limit interspecies FIV spread (51).
However, independent of whether feline A3s restrict FIV or not, the VifFIV protein recognizes diverse feline A3 proteins and induces their depletion (FcaA3Z3 versus FcaA3Z2 proteins). Due to this broad reactivity, VifFIV has the capacity to counteract also engineered two-domain A3Z2-Z3s that carried only one domain of feline origin (Fig. 4B and C). It is currently unclear whether feline A3Z2 and -Z3 proteins share a Vif interaction domain, whether Vif interacts independently with the Z2 and Z3 domains or whether the A3-Vif interaction in a single molecule of FcaA3Z2-Z3 happens with both Z-domains simultaneously or preferentially only with one domain. For the virus, it is an obvious advantage that Vif interacts with both Z domains. As a disadvantage for the feline host, the feline A3Z2-Z3 proteins need to evolve at two regions at the same time to escape the Vif-induced degradation. In contrast to FIV, the HIV-1 Vif protein binds only a single Z domain in the human A3F (HsaA3Z2e-Z2f) and A3G (HsaA3Z2g-Z1c): Z2f or the Z2g, respectively (17, 42, 56, 57). It is hard to guess whether the interaction of VifHIV-1 with only a single Z domain is advantageous for either the virus or the host, and we do not know why Vif of FIV appears to interact with both Z domains. The interaction of VifFIV with A3Z2s may be a relict of an FIV ancestor virus that was refractory also to these A3 types and had to evolve a Vif protein that targets also Z2 proteins. In this model, modern FIVs preserved the “ancient” function to interact with A3Z2. Alternatively, the limited reagents used in the present study might not properly reflect the FIV-cat interaction. Here, we tested reporter vectors based on the domestic cat strains FIV-PPR (40) and FIV-34TF10 (47). We cannot exclude that wt or Δvif variants of other FIV strains, e.g., from other feline hosts, are more restricted by feline A3Z2 proteins than these two FIV isolates. Moreover, cats might carry thus far unidentified A3Z2 alleles that show a stronger inhibitory activity than the described Z2 proteins.
Because cats are resistant to HIV-1, we wanted to understand how many genetic changes in HIV-1 or cats would be required to circumvent this species block. We recently found that the feline CD4 receptor does not support HIV-1 infection, and we described the feline A3 proteins as restriction factors for HIV-1 (33). In order to prevent HIV-1 restriction by feline A3s, we replaced the vif gene of HIV-1 by vif of FIV. This HIV-1vifFIV replicated in nonpermissive, feline A3-expressing CrFK.CD4.CCR5 cells to levels of wt HIV-1 replication in the permissive human HOS.CD4.CCR5 cells. Since the Felidae, including the domestic cat, do not encode a functional copy of the TRIM5 gene (29), cats likely will not exhibit a TRIM5-like restriction against HIV-1. Other limitations and restrictions for HIV-1 in cats might exist but, together, these results are encouraging to strengthen research on an animal model for HIV-1 based on the domestic cat.
Acknowledgments
We thank Wioletta Hörschken and Melanie Krämer for expert technical assistance and Egbert Flory, Nathaniel R. Landau, Garry P. Nolan, Erik M. Poeschla, Viviana Simon, and Jens Thielebein for the gift of reagents. We thank Dieter Häussinger for support.
This project was funded by the DFG grant MU 1608/4-1 to C.M. C.M. is supported by the Heinz-Ansmann Foundation for AIDS Research.
Footnotes
Published ahead of print on 5 May 2010.
REFERENCES
- 1.An, P., G. Bleiber, P. Duggal, G. Nelson, M. May, B. Mangeat, I. Alobwede, D. Trono, D. Vlahov, S. Donfield, J. J. Goedert, J. Phair, S. Buchbinder, S. J. O'Brien, A. Telenti, and C. A. Winkler. 2004. APOBEC3G genetic variants and their influence on the progression to AIDS. J. Virol. 78:11070-11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. U. S. A. 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., R. L. Tallmadge, J. L. Oaks, S. Carpenter, and B. R. Cullen. 2008. Equine infectious anemia virus resists the antiretroviral activity of equine APOBEC3 proteins through a packaging-independent mechanism. J. Virol. 82:11889-11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter, M. A., E. W. Brown, M. Culver, W. E. Johnson, J. Pecon-Slattery, D. Brousset, and S. J. O'Brien. 1996. Genetic and phylogenetic divergence of feline immunodeficiency virus in the puma (Puma concolor). J. Virol. 70:6682-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu, Y. L., and W. C. Greene. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317-353. [DOI] [PubMed] [Google Scholar]
- 7.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type 1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 8.Curran, M. A., S. M. Kaiser, P. L. Achacoso, and G. P. Nolan. 2000. Efficient transduction of nondividing cells by optimized feline immunodeficiency virus vectors. Mol. Ther. 1:31-38. [DOI] [PubMed] [Google Scholar]
- 9.Dang, Y., X. Wang, W. J. Esselman, and Y. H. Zheng. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80:10522-10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel, M. D., F. Kirchhoff, S. C. Czajak, P. K. Sehgal, and R. C. Desrosiers. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938-1941. [DOI] [PubMed] [Google Scholar]
- 11.Donello, J. E., J. E. Loeb, and T. J. Hope. 1998. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J. Virol. 72:5085-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin, S. P., J. L. Troyer, J. A. Terwee, L. M. Lyren, W. M. Boyce, S. P. Riley, M. E. Roelke, K. R. Crooks, and S. Vandewoude. 2007. Frequent transmission of immunodeficiency viruses among bobcats and pumas. J. Virol. 81:10961-10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin, S. P., J. L. Troyer, J. A. Terwee, L. M. Lyren, R. W. Kays, S. P. Riley, W. M. Boyce, K. R. Crooks, and S. Vandewoude. 2007. Variability in assays used for detection of lentiviral infection in bobcats (Lynx rufus), pumas (Puma concolor), and ocelots (Leopardus pardalis). J. Wildl. Dis. 43:700-710. [DOI] [PubMed] [Google Scholar]
- 14.Harari, A., M. Ooms, L. C. Mulder, and V. Simon. 2009. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 83:295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation: comparisons with APOBEC3G. J. Biol. Chem. 282:2587-2595. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, R. K., M. H. Malim, and K. N. Bishop. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32:118-128. [DOI] [PubMed] [Google Scholar]
- 17.Huthoff, H., and M. H. Malim. 2007. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J. Virol. 81:3807-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwatani, Y., D. S. Chan, F. Wang, K. S. Maynard, W. Sugiura, A. M. Gronenborn, I. Rouzina, M. C. Williams, K. Musier-Forsyth, and J. G. Levin. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 35:7096-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonsson, S. R., G. Hache, M. D. Stenglein, S. C. Fahrenkrug, V. Andresdottir, and R. S. Harris. 2006. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 34:5683-5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katzourakis, A., M. Tristem, O. G. Pybus, and R. J. Gifford. 2007. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. U. S. A. 104:6261-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larue, R. S., V. Andresdottir, Y. Blanchard, S. G. Conticello, D. Derse, M. Emerman, W. C. Greene, S. R. Jonsson, N. R. Landau, M. Löchelt, H. S. Malik, M. H. Malim, C. Münk, S. J. O'Brien, V. K. Pathak, K. Strebel, S. Wain-Hobson, X. F. Yu, N. Yuhki, and R. S. Harris. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 83:494-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Löchelt, M., F. Romen, P. Bastone, H. Muckenfuss, N. Kirchner, Y. B. Kim, U. Truyen, U. Rosler, M. Battenberg, A. Saib, E. Flory, K. Cichutek, and C. Münk. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 102:7982-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loewen, N., R. Barraza, T. Whitwam, D. T. Saenz, I. Kemler, and E. M. Poeschla. 2003. FIV vectors. Methods Mol. Biol. 229:251-271. [DOI] [PubMed] [Google Scholar]
- 24.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 25.Mariani, R., D. Chen, B. Schröfelbauer, F. Navarro, R. König, B. Bollman, C. Münk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 26.Mariani, R., B. A. Rasala, G. Rutter, K. Wiegers, S. M. Brandt, H. G. Krausslich, and N. R. Landau. 2001. Mouse-human heterokaryons support efficient human immunodeficiency virus type 1 assembly. J. Virol. 75:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 28.Mbisa, J. L., R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McEwan, W. A., T. Schaller, L. M. Ylinen, M. J. Hosie, G. J. Towers, and B. J. Willett. 2009. Truncation of TRIM5 in the Feliformia explains the absence of retroviral restriction in cells of the domestic cat. J. Virol. 83:8270-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Löwer, K. Cichutek, E. Flory, G. G. Schumann, and C. Münk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161-22172. [DOI] [PubMed] [Google Scholar]
- 31.Münk, C., T. Beck, J. Zielonka, A. Hotz-Wagenblatt, S. Chareza, M. Battenberg, J. Thielebein, K. Cichutek, I. G. Bravo, S. J. O'Brien, M. Löchelt, and N. Yuhki. 2008. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 9:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Münk, C., T. Hechler, S. Chareza, and M. Löchelt. 2010. Restriction of feline retroviruses: lessons from cat APOBEC3 cytidine deaminases and TRIM5alpha proteins. Vet. Immunol. Immunopathol. 134:14-24. [DOI] [PubMed] [Google Scholar]
- 33.Münk, C., J. Zielonka, H. Constabel, B. P. Kloke, B. Rengstl, M. Battenberg, F. Bonci, M. Pistello, M. Löchelt, and K. Cichutek. 2007. Multiple restrictions of human immunodeficiency virus type 1 in feline cells. J. Virol. 81:7048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 35.Nishimura, Y., Y. Goto, K. Yoneda, Y. Endo, T. Mizuno, M. Hamachi, H. Maruyama, H. Kinoshita, S. Koga, M. Komori, S. Fushuku, K. Ushinohama, M. Akuzawa, T. Watari, A. Hasegawa, and H. Tsujimoto. 1999. Interspecies transmission of feline immunodeficiency virus from the domestic cat to the Tsushima cat (Felis bengalensis euptilura) in the wild. J. Virol. 73:7916-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.OhAinle, M., J. A. Kerns, M. M. Li, H. S. Malik, and M. Emerman. 2008. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4:249-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okeoma, C. M., A. Low, W. Bailis, H. Y. Fan, B. M. Peterlin, and S. R. Ross. 2009. Induction of APOBEC3 in vivo causes increased restriction of retrovirus infection. J. Virol. 83:3486-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul, T. A., J. W. Casey, R. J. Avery, and C. A. Sutton. 2007. Expression of feline immunodeficiency virus Vif is associated with reduced viral mutation rates without restoration of replication of vif mutant viruses. Virology 361:112-122. [DOI] [PubMed] [Google Scholar]
- 39.Pecon-Slattery, J., J. L. Troyer, W. E. Johnson, and S. J. O'Brien. 2008. Evolution of feline immunodeficiency virus in Felidae: implications for human health and wildlife ecology. Vet. Immunol. Immunopathol. 123:32-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips, T. R., R. L. Talbott, C. Lamont, S. Muir, K. Lovelace, and J. H. Elder. 1990. Comparison of two host cell range variants of feline immunodeficiency virus. J. Virol. 64:4605-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poss, M., H. A. Ross, S. L. Painter, D. C. Holley, J. A. Terwee, S. Vandewoude, and A. Rodrigo. 2006. Feline lentivirus evolution in cross-species infection reveals extensive G-to-A mutation and selection on key residues in the viral polymerase. J. Virol. 80:2728-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell, R. A., J. Smith, R. Barr, D. Bhattacharyya, and V. K. Pathak. 2009. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J. Virol. 83:1992-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schröfelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. U. S. A. 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shacklett, B. L., and P. A. Luciw. 1994. Analysis of the vif gene of feline immunodeficiency virus. Virology 204:860-867. [DOI] [PubMed] [Google Scholar]
- 45.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 46.Shen, X., C. M. Leutenegger, C. K. Stefano, N. C. Pedersen, and E. E. Sparger. 2007. A feline immunodeficiency virus vif-deletion mutant remains attenuated upon infection of newborn kittens. J. Gen. Virol. 88:2793-2799. [DOI] [PubMed] [Google Scholar]
- 47.Talbott, R. L., E. E. Sparger, K. M. Lovelace, W. M. Fitch, N. C. Pedersen, P. A. Luciw, and J. H. Elder. 1989. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc. Natl. Acad. Sci. U. S. A. 86:5743-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Troyer, J. L., J. Pecon-Slattery, M. E. Roelke, W. Johnson, S. Vandewoude, N. Vazquez-Salat, M. Brown, L. Frank, R. Woodroffe, C. Winterbach, H. Winterbach, G. Hemson, M. Bush, K. A. Alexander, E. Revilla, and S. J. O'Brien. 2005. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J. Virol. 79:8282-8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vandewoude, S., and C. Apetrei. 2006. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin. Microbiol. Rev. 19:728-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandewoude, S., S. J. O'Brien, K. Langelier, W. D. Hardy, J. P. Slattery, E. E. Zuckerman, and E. A. Hoover. 1997. Growth of lion and puma lentiviruses in domestic cat cells and comparisons with FIV. Virology 233:185-192. [DOI] [PubMed] [Google Scholar]
- 51.Vandewoude, S., J. Troyer, and M. Poss. 2009. Restrictions to cross-species transmission of lentiviral infection gleaned from studies of FIV. Vet. Immunol. Immunopathol. 134:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Virgen, C. A., and T. Hatziioannou. 2007. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J. Virol. 81:13932-13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. U. S. A. 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 55.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, L., J. Saadatmand, X. Li, F. Guo, M. Niu, J. Jiang, L. Kleiman, and S. Cen. 2008. Function analysis of sequences in human APOBEC3G involved in Vif-mediated degradation. Virology 370:113-121. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, W., G. Chen, A. M. Niewiadomska, R. Xu, and X. F. Yu. 2008. Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host antiviral proteins. PLoS One 3:e3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zielonka, J., I. G. Bravo, D. Marino, E. Conrad, M. Perkovic, M. Battenberg, K. Cichutek, and C. Münk. 2009. Restriction of equine infectious anemia virus by equine APOBEC3 cytidine deaminases. J. Virol. 83:7547-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]