Abstract
Penicillium chrysogenum virus (PcV), a member of the Chrysoviridae family, is a double-stranded RNA (dsRNA) fungal virus with a multipartite genome, with each RNA molecule encapsidated in a separate particle. Chrysoviruses lack an extracellular route and are transmitted during sporogenesis and cell fusion. The PcV capsid, based on a T=1 lattice containing 60 subunits of the 982-amino-acid capsid protein, remains structurally undisturbed throughout the viral cycle, participates in genome metabolism, and isolates the virus genome from host defense mechanisms. Using three-dimensional cryoelectron microscopy, we determined the structure of the PcV virion at 8.0 Å resolution. The capsid protein has a high content of rod-like densities characteristic of α-helices, forming a repeated α-helical core indicative of gene duplication. Whereas the PcV capsid protein has two motifs with the same fold, most dsRNA virus capsid subunits consist of dimers of a single protein with similar folds. The spatial arrangement of the α-helical core resembles that found in the capsid protein of the L-A virus, a fungal totivirus with an undivided genome, suggesting a conserved basic fold. The encapsidated genome is organized in concentric shells; whereas the inner dsRNA shells are well defined, the outermost layer is dense due to numerous interactions with the inner capsid surface, specifically, six interacting areas per monomer. The outermost genome layer is arranged in an icosahedral cage, sufficiently well ordered to allow for modeling of an A-form dsRNA. The genome ordering might constitute a framework for dsRNA transcription at the capsid interior and/or have a structural role for capsid stability.
Viruses with double-stranded RNA (dsRNA) genomes are found in a broad range of organisms, ranging from bacteria to simple (fungi and protozoa) and complex (animals and plants) eukaryotes. The genome of most dsRNA viruses is contained within a specialized icosahedrally symmetric nanocompartment with a triangulation number equal to 1. This T=1 lattice has a protein dimer as the asymmetric unit and is also referred to as a “T=2” layer (e.g., see reference 11). T=1 inner cores of dsRNA viruses are generally known to be critical for genome replication (minus-strand synthesis) and transcription (plus-strand synthesis), since the viral-RNA-dependent RNA polymerase(s) (RdRp) is frequently packaged as an integral component of the capsid. Nanocompartments built entirely of protein subunits with enzymatic activity are found in all domains of life, from the bacterial carboxysome (65) to the eukaryotic vault particle (64). dsRNA, single-stranded RNA (ssRNA) with secondary structure, and dsRNA-containing replication intermediates synthesized during viral infections in plants, animals, and fungi are potent inducers of host cell responses (27, 38). As their structural integrity is undisturbed throughout the viral cycle (24), dsRNA virus T=1 capsids prevent triggering of host defense mechanisms and, therefore, constitute a dsRNA-sequestering mechanism.
dsRNA virus T=1 layers consist of 60 asymmetric dimers of a single protein, and the stringent requirements of dsRNA metabolism might explain the similarities observed in capsid architecture among a broad spectrum of dsRNA viruses. These capsids are an extended feature of dsRNA viruses (Fig. 1); they are described (i) in members of the family Reoviridae (37, 44, 69, 71), which infect mostly higher eukaryotic systems, (ii) in viruses of the family Picobirnaviridae (18), a recently established taxonomic family whose members infect humans and other vertebrates, (iii) in bacteriophages of the family Cystoviridae, which infect the prokaryote Pseudomonas syringae (26, 28), (iv) and in viruses of the families Totiviridae (12, 13, 67) and Partitiviridae (48), which mostly infect unicellular and simple eukaryotes (fungi and protozoa). A notable exception is the Penicillium chrysogenum virus (PcV) of the family Chrysoviridae, which has an authentic T=1 capsid formed by just 60 copies of one polypeptide (11). In the study presented here, we found that the PcV capsid protein (CP) has two similar domains that can be considered vestiges of gene duplication. Our results are consistent with the accepted idea that a 120-subunit T=1 capsid is a conserved architecture for managing dsRNA metabolism.
FIG. 1.
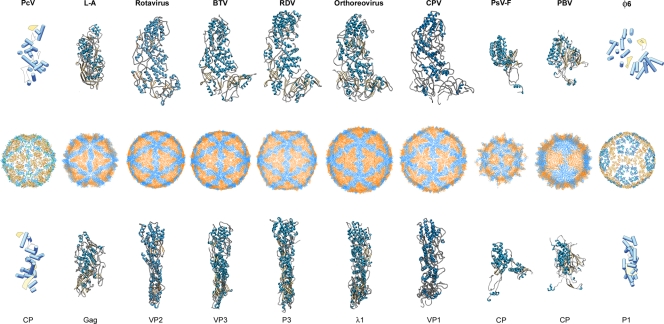
T=1 capsid protein structures, based on X-ray and cryo-EM data. T=1 capsids of Penicillium chrysogenum virus (PcV) (this study), L-A virus, rotavirus, bluetongue virus (BTV), rice dwarf virus (RDV), orthoreovirus, cytoplasmic polyhedrosis virus (CPV), Penicillium stoloniferum virus F (PsV-F), picobirnavirus (PBV), and φ6 phage are viewed along a 2-fold axis of icosahedral symmetry (center row). PcV CP half-protein A (this study), Gag (PDB accession no. 1m1c; 680 residues), VP2 (PDB accession no. 3kz4; 880 residues), VP3 (PDB accession no. 2btv; 901 residues), P3 (PDB accession no. 1uf2; 1,019 residues), λ1 (PDB accession no. 1ej6; 1,275 residues), VP1 (EMDB accession no. 3cnf; 1,333 residues), PsV-F CP (PDB accession no. 3es5; 420 residues), PBV CP (PDB accession no. 2vf1; 590 residues), and P1 (PDB accession no. emd-1206; 769 residues) are shown from a top view (top row). Side views of the same structures are shown (bottom row; the T=1 shell exteriors are to the right). α-Helices are represented by blue cylinders and β-sheets by yellow planks for cryo-EM-based structures. SSE of the φ6 CP were determined as described in Materials and Methods, based on the published 7.5-Å-resolution nucleocapsid cryo-EM structure (PDB accession no. emd-1206).
At present, eight 120-subunit T=1 capsid layers have been resolved at an atomic resolution (Fig. 1): the VP3 cores of the orbivirus bluetongue virus (BTV) (23), λ1 cores of the reovirus in the genus Orthoreovirus (54), P3 of the rice dwarf virus (RDV) (46), the capsid shell protein (CPS) of the cytoplasmic polyhedrosis virus (CPV) (69), VP2 inner capsid of rotavirus (44) (all five Reoviridae), the CP of Penicillium stoloniferum virus F (PsV-F, of the family Partitiviridae) (49), the CP of picobirnavirus (PBV) (18), and Gag of the yeast L-A virus (45) (of the family Totiviridae). Sequence similarities among these viruses are negligible, and comparative analyses are inconclusive. All eight proteins are nevertheless predominantly α-helical; VP3, λ1, P3, CPS, and VP2 show a similar fold (53), PsV-F and PBV CP are also based on a similar fold, and Gag has a distinct tertiary structure bearing only a faint resemblance to the reovirus proteins (below).
Members of the families Birnaviridae are exceptions to the rule that all dsRNA viruses have a T=1 core made of 60 dimers of the CP; birnaviruses have a single T=13 shell (16, 59) that surrounds a polyploid dsRNA genome (41) organized as a ribonucleoprotein complex (42).
PcV belongs to a family of symptomless mycoviruses with a multipartite genome consisting of four monocistronic dsRNA segments (with genome sizes ranging from 2.4 to 3.6 kbp). Each segment is separately encapsidated in a similar particle (29). dsRNA-1 (3.6 kbp) encodes the RdRp (1,117 amino acid residues with a molecular mass of 128,548 Da; there are 1 or 2 copies per virion); dsRNA-2 (3.2 kbp) encodes the CP (982 amino acid residues; 108,806 Da); and dsRNA-3 and -4 (3 and 2.9 kbp, respectively) code for virion-associated proteins of unknown function. Chrysoviruses accumulate in the cytoplasm and are transmitted intracellularly during cell division, formation of asexual spores, and cytoplasmic fusion following hyphal anastomosis. Based on multiple alignment of the RdRp conserved motifs, chrysoviruses appear to be more closely related to the totiviviruses (with a single genome segment) than to the partitiviruses (with two segments) (21).
We report the three-dimensional (3D) structure of the PcV capsid at 8 Å resolution. The identification of secondary-structure elements (SSE) shows that the PcV CP has two structurally duplicated domains. This basic repeated domain could provide insight into the structural evolutionary relationships between dsRNA viruses. Due to its numerous interactions with the inner surface of the capsid, the encapsidated genome conforms to the icosahedral symmetry and is partially visualized as tubular densities corresponding to an A-form duplex.
MATERIALS AND METHODS
Virion preparation.
PcV virions were purified from Penicillium chrysogenum strain ATCC 9480 by two cycles of differential centrifugation and a rate zonal centrifugation in a sucrose density gradient, as described elsewhere (11).
Cryo-EM.
For cryoelectron microscopy (Cryo-EM), samples (5 μl) were applied to one side of a holey carbon grid, blotted, and plunged into liquid ethane, as described previously (43, 59). Full and empty PcV particles were imaged together under low-dose conditions (∼10 e−/Å2) in a Tecnai G2 electron microscope operating at 200 kV and equipped with a field emission gun, using a nominal magnification of ×50,000.
Image processing.
General image-processing operations were performed using Bsoft (http://lsbr.niams.nih.gov/bsoft/ [25]), Xmipp (http://xmipp.cnb.csic.es/ [60]), and Spider (http://www.wadsworth.org/spider_doc/spider/docs/ [63]), as described previously (12, 43). Graphic representations were produced by using the Chimera visualization software package (http://www.cgl.ucsf.edu/chimera/ [50]).
A Zeiss TD scanner was used to digitize micrographs at a 7-μm step size to yield a 1.4-Å pixel size in the specimen. A total of 56 micrographs with a 0.9- to 3.3-μm defocus range, determined using Bshow (http://lsbr.niams.nih.gov/bsoft/bshow/bshow.html [25]) were selected for processing. X3d (15) was used to extract and normalize 6,322 and 4,530 individual particle images for PcV empty and full particles, respectively. Initial estimates of origin and orientation parameters for each image were determined using polar Fourier transform (pft) procedures (6) with the previously published PcV capsid structure (11), low pass filtered to 30 Å, as a starting model. These parameters were improved using consecutive cycles of PFT refinement of origin and orientation and subsequent computation of a 3D reconstruction and assessment of resolution. At the end of each refinement cycle, two independent reconstructions were computed using the Em3dr2 Fourier-Bessel program (http://people.chem.byu.edu/belnap/pft3dr/pft2_em3dr2.html [10]), and the new resolution limit was assessed. The subsequent refinement cycle included all Fourier data to this resolution limit. Refinement progressed until no further improvement in resolution was achieved. At the end of refinement, 5,692 and 4,156 particle images were included in the empty and full PcV capsid reconstructions, respectively.
Amplitude decay was calculated using the spatial frequency components from the cryo-EM maps (with an effective resolution determined by the Fourier shell correlation [FSC] criterion) and the X-ray L-A capsid map (PDB accession number 1m1c). The decay profile of the cryo-EM maps was then adjusted to match the profile of the X-ray maps, and the fitted function was applied to the cryo-EM maps in the frequency range from 245 Å to the maximum resolution achieved, and a soft low-pass filter was applied. In addition, amplitude decay was calculated and corrected with the Embfactor program (19), with similar results. These sharpening methods, used for identification and interpretation of high-resolution structural features, are analogous to adjusting the temperature factor (or B factor) of the two structures. Resolution was assessed by FSC between independent half-data-set maps, applying a correlation limit of 0.5.
Structural analysis.
The structural asymmetric unit boundaries were established by contouring the map at different σ levels, based on its compactness and contacts with neighboring densities. To segment the asymmetric subunit and the half subunits, a dummy atom model was manually generated in Chimera and iteratively refined to avoid overlapping between subunits or loose density. To test that the selected density corresponded to a single asymmetric unit, the complete capsid map was restored in each cycle after applying icosahedral symmetry to the selected density. Using a similar approach, we deduced that the asymmetric unit is formed by two elongated ellipsoid-like structures.
The secondary structure features in the PcV capsid subunit were identified and modeled using the AIRS programs (http://ncmi.bcm.tmc.edu/software/AIRS [5]), which provides a graphic interface to the SSE Hunter and SSE Builder in the Chimera visualization software package. Secondary structural elements (SSE) were displayed as virtual reality modeling language (VRML) cylinders and planar faces (α-helices and β-sheets, respectively), and prototypical ribbon-style helices were constructed as well.
PcV CP elements A and B were initially aligned manually between both VRML SSE models in Chimera, using the ∼37-Å-long α-helices as a reference. These results were verified twice by computational structure comparison analysis between prototypical helix models generated for each subunit using LGA (local-global alignment) (http://predictioncenter.org/local/lga/lga.html [70]) with a root mean square (RMS) deviation of 1.53 Å, a level of structural similarity (LGA_S) of 14.33%, and an alignment quality score (LGA_Q) of 2.15 and by exhaustive six-dimensional search between the segmented map halves using the Foldhunter program (30) (correlation coefficient [CC] = 0.43). These approaches consistently yielded essentially the same results.
The L-A capsid protein Gag (20) X-ray structure was manually aligned to the matched VRML SSE models of PcV CP elements A and B, treated as a rigid body. The PcV 37-Å long α-helices were used as an initial anchor position for the L-A virus monomer, whose orientation was manually refined to optimize the number of overlapping SSE between both structures. The spatial arrangement for both capsid proteins in the shell was maintained. These alignments were verified using LGA, with RMS deviations of 1.67 and 1.72 Å for half-proteins A and B, respectively.
Segmentation and analysis of dsRNA densities were carried out graphically with Chimera. A dsRNA model was built using a dsRNA 34-bp fragment from BTV (PDB accession number 1h1k) (17), adjusted manually and optimized in Chimera.
Secondary-structure predictions.
Secondary structure predictions were generated with the following programs (web addresses are indicated): UniProt, http://www.uniprot.org/; PsiPred, http://bioinf.cs.ucl.ac.uk/psipred/; Jnet, http://www.compbio.dundee.ac.uk/www-jpred/; Porter, http://distill.ucd.ie/porter; Sable, http://sable.cchmc.org; Gor, http://gor.bb.iastate.edu; Yaspin, http://www.ibi.vu.nl/programs/yaspinwww/; Profsec, http://www.predictprotein.org/; and GeneSilico, http://genesilico.pl/meta2/.
Protein structure accession numbers.
3D reconstructions are deposited in EMBL-EBI (accession numbers emd-1610 and emd-1611 [full and empty particles, respectively]).
RESULTS
PcV capsid three-dimensional structure at 8-Å resolution.
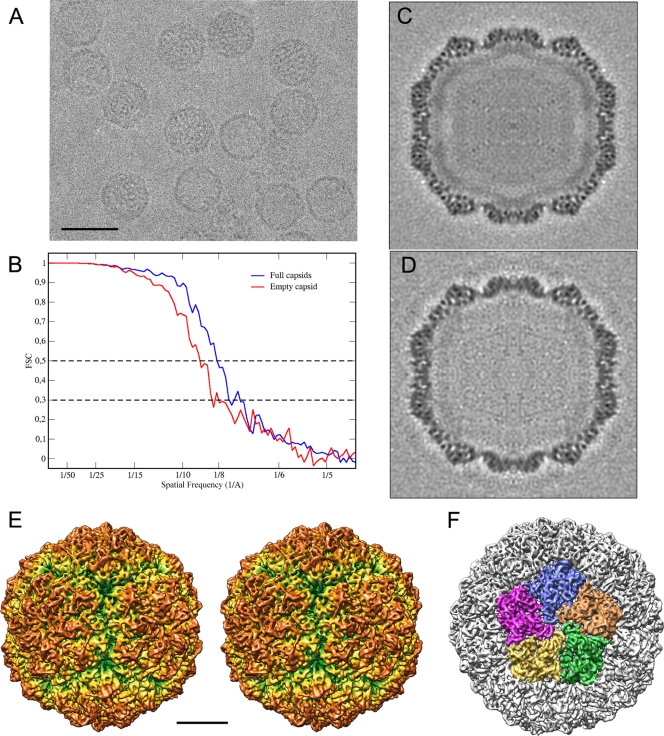
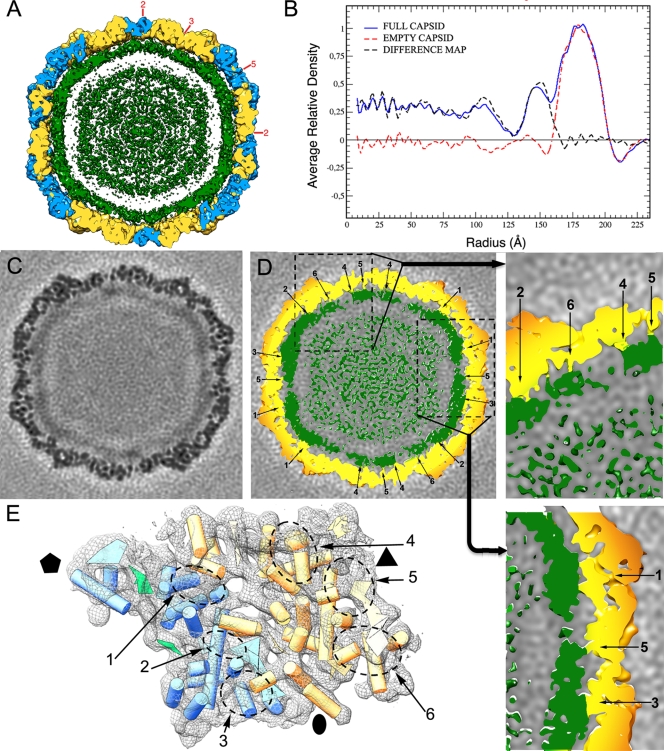
PcV particles were purified and analyzed by cryo-EM as described previously (11). Full and empty particles were imaged together (Fig. 2A), and their 3D reconstructions were calculated separately. The final resolution of the reconstruction for full PcV virions was estimated to be 8.0 Å based on a Fourier-shell correlation (FSC) threshold of 0.5 (and 8.9-Å resolution for empty particles) (Fig. 2B). The central sections of the 3D maps showed no marked structural differences between full and empty capsids in the protein shell (Fig. 2C and D). This interpretation was further supported when full and empty particles were subjected to joint refinement cycles; the estimated resolution of the combined 3D map was 7.5 Å, indicating that both maps are virtually identical (not shown). The quality of the information provided by the resolution profile is better for the full-capsid 3D reconstruction; the map images shown are taken from this reconstruction.
FIG. 2.
Three-dimensional cryo-EM of PcV virions. (A) Representative cryoelectron micrograph of native PcV virions (2-μm underfocus; first zero of the contrast transfer function at 22 Å). Bar = 50 nm. (B) Assessment of the resolution of full and empty PcV reconstructions. FSC resolution curves were calculated for the full (blue) and empty (red) capsids. Each set of particle images was subdivided randomly into two subsets, and independent reconstructions were computed from these data. The resolutions at which the correlations dropped below 0.5 and 0.3 are indicated. For the 0.5 threshold, the values for full and empty PcV were 8.0 and 8.9 Å, respectively; values for the 0.3 threshold were 7.15 and 8.0 Å, respectively. 1/A, 1/Å. (C, D) Central sections from the 3D reconstructions of full (C) and empty (D) capsids, viewed along a 2-fold axis. Protein and/or RNA are dark. The two protein shells are virtually identical, and the RNA density of the full capsid is quite low compared to that of the protein shell. (E) Stereo view of the radially color-coded outer surface of the full capsid, viewed along a 2-fold axis of icosahedral symmetry. The most prominent features are 12 outward-protruding pentamers. The map is contoured at 2 σ above the mean density. Bar = 100 Å. (F) Surface view of PcV virion, viewed along an icosahedral 5-fold axis and showing the five CP structural subunits in a pentamer (green, orange, blue, pink, and yellow).
The particle molecular architecture is essentially as previously described at lower resolution (11). The capsid's diameter is 400 Å, and the protein shell is 48 Å thick. The most prominent features of the T=1 capsid were 12 outwardly protruding pentons that correspond to the pentameric positions of the icosahedral lattice (Fig. 2E). The enantiomorph maps shown in this study were chosen based on preliminary X-ray analysis that, in addition, matched well with our docking analysis (below). The structural asymmetric unit boundaries were established by contouring the map at different levels, based on its compactness and contacts with neighboring densities (Fig. 2F). To confirm that the selected density corresponded to a single asymmetric unit, the complete capsid map was fully restored after applying icosahedral symmetry to the selected density.
At 8 Å resolution, numerous rod- and sheet-like densities were found on the relatively uneven outer surface, similar to L-A virus. This is in contrast with the smooth outer surface of the reovirus particles, in which the CP has a plate-like structure.
Structural duplication in the PcV capsid protein.
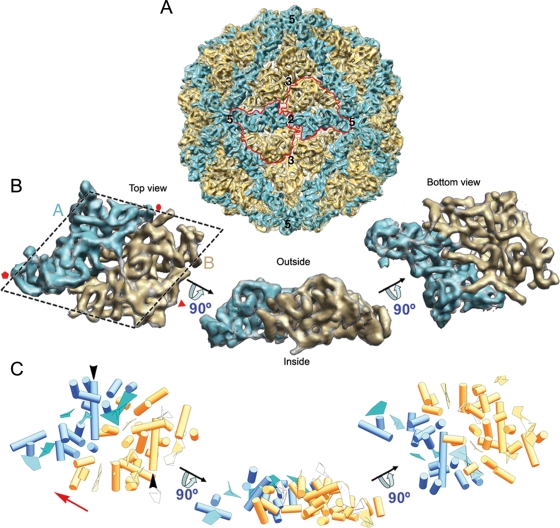
The asymmetric unit is formed by two ellipsoid-like structures with roughly similar morphology, although they differ somewhat in size (Fig. 3 A, blue and yellow). The asymmetric unit is a rhomboidal prism, ∼46 Å high and ∼130 Å long (Fig. 3B). The smaller part makes up 43% of the total unit volume (blue; half-protein A) and the larger one represents 57% (yellow; half-protein B). These elements are arranged in two sets of five; five half-protein A structures directly surround the icosahedral 5-fold axis, and five half-protein B structures are intercalated between them, forming a pseudodecamer (Fig. 3A). This quaternary organization is similar to the 120-subunit T=1 shell in reoviruses and totiviruses (Fig. 1).
FIG. 3.
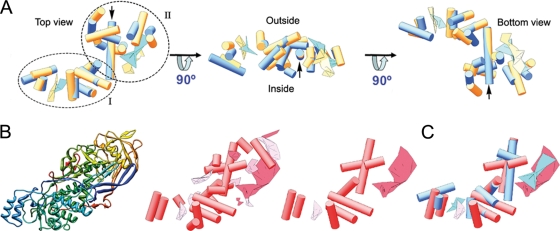
Structure of the PcV T=1 capsid and model of the capsid protein fold. (A) Surface-shaded virion capsid viewed along an icosahedral 2-fold axis with capsid protein (CP) elements in cyan and yellow; boundaries for two CPs are outlined in red. Icosahedral symmetry axes are numbered. (B) Segmented asymmetric unit (PcV CP monomer). The dashed line highlights the rhomboidal shape. Protein halves A (cyan) and B (yellow) are indicated. Red symbols indicate icosahedral symmetry axes. (C) PcV CP secondary structural elements (SSE), using the same color scheme and orientations as in panel B. Cylinders, α-helices; planks, β-sheets. The red arrow indicates translation direction to superimpose half-protein B on A. Black arrows indicate the ∼37-Å-long α-helices of both PcV CP elements.
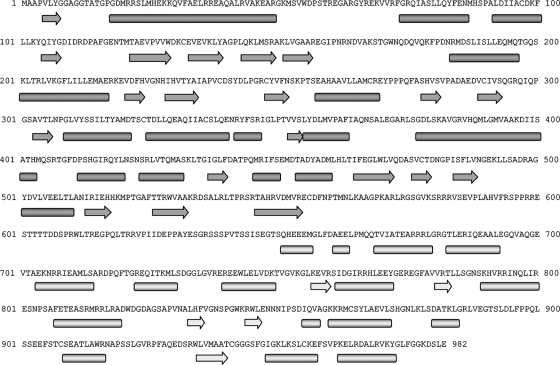
The SSE Hunter program showed a high α-helical content for the PcV CP; we identified 16 α-helices and 5 β-sheets in the half-protein A structure and 24 α-helices and 7 β-sheets in the larger half-protein B structure (Fig. 3C). We used various sequence-based SSE prediction methods (PsiPred [32], Jnet [14], Porter [51], Sable [2], Gor [62], Yaspin [39], and Profsec [58]) on the PcV CP sequence (982 amino acids) to test the correlation with our structural subunit model (Fig. 4). SSE predictions from the PcV CP sequence indicate a high α-helical content, as directly visualized in our cryo-EM map and identified with the SSE Hunter program. In addition to a consensus SSE prediction compatible with our fold model, we found an unstructured central region that divides the PcV CP into two parts, reflecting a structurally disordered region (amino acids ∼550 to ∼650). The SSE gap in the middle of the protein sequence was confirmed by the consensus prediction of protein order with the GeneSilico metaserver (35). The unstructured region might represent the linker sequence between the two unequal halves of the PcV CP.
FIG. 4.
Secondary structure consensus prediction for the PcV CP. The PcV CP sequence was obtained from the UniProt database (982 amino acids; accession no. Q8JVC1). Several SSE prediction methods (PsiPred, Jnet, Porter, Sable, Gor, Yaspin, and Profsec) were used to correlate our model of the structural subunit. A consensus SSE prediction was obtained by simple majority at each sequence position. The results were very similar to the consensus SSE prediction obtained with the GeneSilico fold prediction metaserver. The secondary structure gap in the middle of the protein sequence (segment Glu555-Ser650) reflects a structurally disordered region, as confirmed by the consensus prediction of protein order with the GeneSilico metaserver, and divides the PcV coat protein into two parts. Arrow, β-chain; bar, α-helix.
Both PcV CP elements appeared to have a similar ∼37-Å-long α-helix tangential to the capsid surface (Fig. 3C, black arrowheads). Careful inspection showed that both protein halves, though not identical, have substantial resemblance. When one of the halves (treated as a rigid body) is superimposed on the other by an ∼45-Å translation (Fig. 3C; the red arrow indicates translation direction), the relative spatial locations of 13 α-helices and two planar regions are close and require only minor local adjustment (Fig. 5A; also see Movie S1 in the supplemental material). Structurally preserved SSE between both PcV CP halves consist of two domains, domain I with a characteristic bundle of four α-helices, located toward the 5-fold axis, and domain II, which contains the longest α-helix, as well as other SSE. Whereas the conserved core establishes lateral contacts with the neighboring units, the interface between halves A and B is less similar, with a larger number of variations. This analysis indicates that despite a lack of sequence similarity, the two PcV CP elements have a comparable structural signature, suggesting ancestral gene duplication.
FIG. 5.
Structural matching of the PcV CP elements. Comparison of PcV and L-A capsid proteins. (A) Superimposed views of conserved SSE in PcV protein halves A (cyan) and B (yellow); the relative spatial locations of 13 α-helices and two planar regions are very close. PcV protein elements are subdivided into domains I and II. (B) L-A virus Gag structure. Shown are the polypeptide chain path (top view), as a gradient from blue (N terminus) to red (C terminus) (left); a cylinder-and-plank representation of Gag, as for PcV CP, with 17 α-helices and 10 β-sheets (center); and selected Gag SSE (9 α-helices and 2 β-sheets) (right). (C) Superimposed views of helical and planar regions of selected Gag SSE (red) and PcV half-protein A (blue). This structural matching preserves the spatial orientation of both units.
Structural comparison of PcV and L-A capsid proteins.
With the exception of the ∼430-amino-acid CP of the partitivirus PsV-F, the PcV CP structural halves are the smallest polypeptides of the dsRNA virus T=1 proteins (Gag is almost twice as large and VP3, P3, CPS, and λ1 are almost four times as large as the PcV CP C-terminal element). They might represent a fold that evolved from the T=1 CP ancestral fold to the highly varied structures observed today. If this hypothesis is correct, “structural remnants” of the conserved PcV motif might be found in the modern T=1 CP.
PcV CP was compared to L-A virus Gag CP for several structural and functional reasons: (i) PcV and LA viruses both infect lower eukaryotes and lack an extracellular phase in their life cycles; (ii) they share not only an uneven outer surface, in contrast to the smooth outer surface of reovirus particles, but also a roughly similar side view (Fig. 1); and (iii) PcV and L-A CP have no outer shell, whereas Reoviridae T=1 cores serve as a template to prime the assembly of the T=13 surrounding capsid.
Overlaying L-A Gag (Fig. 5B) on either of the PcV elements, while maintaining the same spatial arrangement in the shell, showed many SSE with similar spatial distributions, including nine α-helices (of a total of 13) and two β-sheets (Fig. 5C; also see Movie S2 in the supplemental material). These structural similarities are located in the Gag N terminus (residues 1 to 414); the entire C-terminal region (residues 415 to 680) would be a terminal insertion to this conserved fold. This structural matching, in which CP orientations are maintained (i.e., the outer and inner surfaces are coincident), indicates a shared motif for these two fungal virus T=1 CPs.
dsRNA packaged within the capsid.
We studied the organization of the packaged RNA at relatively low resolution using data from our 3D cryo-EM studies. Analysis of PcV full capsids showed that the inner surface of the protein shell is in close contact with the underlying density shell that is ascribed to encapsidated dsRNA (Fig. 6A). Spherically averaged radial density plots computed from the 3D reconstructions of full and empty PcV particles indicated that, as a result of these extensive interactions, there is almost no space between the two layers (the average distance of the empty space is 9 Å). The innermost RNA shells are more diffuse, as the RNA density is quite low compared to that of the protein shell (Fig. 6B). There is clear correspondence in the peaks centered at a radius of ∼180 Å, suggesting little if any variance in the overall architecture of the protein shell.
FIG. 6.
Genomic dsRNA within the PcV virion particle. (A) A PcV 50-Å-thick slab. Capsid shell coloring is the same as in Fig. 3, contoured at a lower contour threshold (1.0 σ above the mean density) to highlight the locations of the dsRNA densities. dsRNA (green) is seen as approximately four concentric layers contoured at 1.0 σ. There are numerous contacts between the inner surface of the capsid and the outer surface of the nearest dsRNA layer. (B) Radial density profiles from 3D maps of full and empty PcV particles. Both profiles are practically superimposable at the protein shell (radius ∼162 to 200 Å). A difference map (full subtracted from empty capsid) was calculated by arithmetic subtraction of the density values. (C) A transverse section, 1.4 Å thick, taken from the 3D map of a full PcV capsid, parallel to but displaced ∼7 Å from the central section viewed along a 2-fold axis (darker, denser). (D) Same section as in panel C, but contoured at 1.0 σ above the mean (protein in yellow and RNA in green), which shows the six types of CP-dsRNA interactions (1 to 6). The two adjacent boxes show magnified views of the interactions between the inner surface of the capsid and the dsRNA outermost layer. (E) PcV capsid asymmetric unit (shown as wire frames, contoured at 2.8 σ, viewed from the inside) with the SSE as shown in Fig. 2C (cyan and yellow for half-proteins A and B, respectively). dsRNA interacts with six defined areas of the capsid inner surface (dashed ovals, 1 to 6). Observe that the cross-section size of the contacting areas is larger than that corresponding to an α-helix. Black symbols indicate icosahedral symmetry axes.
The existence of RNA-protein contacts was reported previously (11); here we define the sites (SSE) on the icosahedral protein shell involved in these interactions. The interactions of each CP half with the RNA show overall similarities (for example, three interacting areas along each one). This is consistent with the suggested structural duplication, although there are differences that reflect structural changes in the protein halves. There exist at least 360 interacting areas in total, as found after imposing icosahedral symmetry (Fig. 6C and D). These observations agree with the overall basic character of PcV CP (theoretical pI, 8.24). Specific RNA-protein interactions are evident in six defined areas of the PcV monomer inner surface (Fig. 6E, labeled 1 to 6) that will be ascribed to specific sequences when high-resolution data are available. These areas are located within the conserved SSE, whereas the interface between the CP elements (with many variations) is free of interactions.
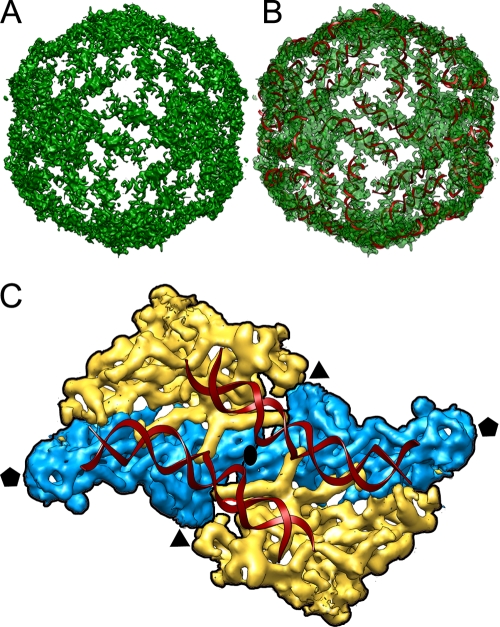
Due to the numerous icosahedral positions that interact with the genome, the outermost RNA layer has strong features of icosahedral symmetry conforming to an icosahedral cage (Fig. 7A). A difference map calculated by subtracting the full from the empty capsid shows this layer as a conspicuous peak beneath the shell protein peak. The bulk of the genome density appears diffuse toward the center of the core, in two or three concentric layers; this density weakening suggests greater RNA disorder at greater distances from the capsid shell (Fig. 6B). RNA-protein contacts appear to form tracks on the inside of the CP shell, resembling those described in the X-ray structure of BTV (see Fig. 4 in reference 22). This layer appears to be made of tube-like densities with dimensions of dsRNA helices. A-form helices of dsRNA fitted remarkably well into the cryo-EM density map (Fig. 7B). Whereas tubular densities are concentric around the 5-fold axis in BTV, in PcV they converge into the 5-fold axes and connect with neighboring strands from adjacent 5-fold axes, describing a T=1 genome shell (Fig. 7A). These dsRNA strands reinforce the connectivity between two monomers that contributes to adjacent pentamers (Fig. 7C) and, therefore, increase capsid stability.
FIG. 7.
Outer layer of dsRNA. (A) Genome density ordered into an icosahedral cage in close contact with the capsid inner surface. The capsid protein has been stripped away, showing the top half of the first layer of ordered dsRNA density. (B) A-form helices of dsRNA docked into the tubular densities corresponding to the dsRNA density. (C) Close-up view down a 2-fold axis from inside, showing two adjacent structural subunits colored as in Fig. 3. The phosphate backbone is traced as a red ribbon for the two dsRNA A-form strands. Icosahedral symmetry axes are indicated.
DISCUSSION
The 120-subunit capsid with a T=1 lattice is a peculiar but usual architecture for the layer surrounding the genome of many (or most) dsRNA viruses. The PcV capsid is atypical and based on an authentic T=1 layer composed of 60 monomers rather than of 60 asymmetric dimers. Our analyses indicate that PcV CP is formed by two elongated structures that can be envisaged as the result of domain duplication. Many SSE in each domain have a similar conformation because they are easily superposed by a translation. This organization is clearly reminiscent of the T=1 lattice in reovirus and totivirus capsids, in which CP are arranged as approximately parallel dimers (54). Our results reconcile the structural details observed in the PcV capsid with the concept that a T=1 layer with a dimer as the asymmetric unit is a conserved arrangement for managing dsRNA metabolism.
Partitiviruses and picobirnaviruses have a different quaternary organization, as the CP dimer has almost-perfect local 2-fold symmetry (18, 49). These two well-established tendencies among dsRNA virus CPs, based on their 3D atomic structures, have major consequences for the virus assembly pathway. Viruses of the first group (reo-, toti-, and chrysoviruses) are thought to be nucleated by pentamers of dimers, whereas partiti- and picobirnavirus capsids are probably assembled from dimers of CP dimers as intermediates (49). Strikingly, the capsid protein P1 of the dsRNA bacteriophage φ8, a cystovirus, is found as a soluble tetramer (33) in an in vitro assembly system.
Joined folds were first observed in the adenovirus trimeric capsomer (56). Each monomer consists of two successive jelly roll motifs, producing a pseudohexameric structure. Many other large dsDNA viruses have similar trimeric capsomers, including Paramecium bursaria chlorella virus 1 (PBCV1) (47) and bacteriophage PRD1 (1). The large subunit of comoviruses, ssRNA viruses that infect plants, is also made by the fusion of two β-barrel domains (40). Similar to the PcV CP elements, there is a lack of sequence similarity between these joined motifs.
The capsid stoichiometry is probably related to the numerous activities related to RNA synthesis (36), as an organizer of the genome and replicative complex(es) within it and as molecular sieves to isolate dsRNA molecules or replicative intermediates. The reovirus particle also serves as a template for the assembly of a T=13 outer protein shell, whereas dsRNA viruses of unicellular and simple eukaryotes, such as PcV and L-A virus, which are transmitted by cytoplasmic interchange and never leave the host, have no outer shell and lack a strategy for entry into a host cell. These different roles are directly reflected in the distinct features of the particle surfaces. PcV and L-A CP outer surfaces are highly corrugated, and Gag has a decapping activity (66). It is plausible that PcV CP, in addition to its structural role, might also have enzymatic activity, like L-A Gag. PcV is thought to accumulate in the cytoplasm in large aggregates enclosed in vesicles (9). The protruding domains might have a role in virus particle aggregation and in binding to the enclosing membranes. It could be important for virus survival to remain membrane bound, since dsRNA released from free virions is a potent inducer of host defense responses.
Previous sequence analysis of the PcV CP vaguely suggested that it has two roughly similar halves, with conserved segments in both halves of the protein after the introduction of numerous gaps, although the importance of this finding was not conclusive (11). Lack of sequence similarity is common among viral structural proteins, and there is a growing accumulation of data supporting the existence of basic tertiary structures that reflect evolutionary relationships much better than plane sequence similarity analyses. For example, the picornavirus capsid is built of three different CPs with negligible sequence similarity; nevertheless, like many other plant virus T=3 CPs, they have the same canonical jelly roll β-barrel fold (57). Portal complexes through which DNA enters the capsid are another well-known structure in which a signature fold is shared, although there is a great sequence diversity (3).
Structural comparison of CPs is used to establish relatedness when sequence conservation is limited (4, 7, 57) and has led to the detection of relationships among viruses that infect organisms widely separated in evolution (7, 8, 34, 55). Icosahedral viruses are currently grouped in four separate lineages, the PRD1-, HK97-, and BTV-like viruses and the picornavirus-like family (7). Although many viruses are not included in these four lineages, the number of folds that satisfies the assembly constraints for a viable viral shell is nonetheless thought to be limited. Whereas the Reoviridae T=1 CP has a recognizably similar fold, supporting a possible common ancestor, the L-A virus Gag has a rather different structure, and matching of any domain using traditional methods appears unlikely. Nevertheless, the fold of L-A virus is grouped within the BTV-like lineage, although at the furthest distance within the structure-based phylogenetic tree. Using a structure-based comparison with the SHP program (7, 8, 34, 55), 219 residues were matched between L-A virus and BTV, allowing L-A virus to be sensibly placed in the structure-based phylogeny of the BTV-like lineage. The conserved fold of PcV CP in L-A virus might provide insights into the ancestral fold and reveal structural evolutionary relationships of the dsRNA virus lineage. A succession of divergent evolutionary events in the ancestral PcV fold, by means of insertions in preferential sites of the preserved α-helical core, might have led to the more complex structures observed today. Partiti- and picobirnavirus CPs show an overall structural resemblance between themselves but not with other dsRNA viruses and would constitute a model of convergent evolution to a similar T=1 capsid with the same functional requirements but distinct quaternary structure.
The jelly roll β-barrel, an eight-stranded antiparallel β-barrel (with two opposing BIDG and CHEF β-sheets), is the most extended fold among capsid proteins of spherical viruses (57). Due in part to this built-in flexibility, the jelly roll motif-based CP can assemble into icosahedral capsids with highly variable size (by increasing the T number), whereas CPs with a PcV-based fold are restricted to T=1 icosahedral particles and the capsid size is strictly determined by the CP size. The PcV-like α-helical CP motif, observed only for dsRNA virus T=1 CPs, was trapped during evolution in a suboptimal stage (“local minimum”) for capsid assembly, whereas the jelly roll domain, prevalent among many spherical viruses, appears to be closer to the best-suited conformation (“final evolutionary minimum”).
Many dsRNA viruses might share an ancestor, based on a PcV fold that has been conserved throughout evolution, perhaps as a consequence of intracellular transmission in simple eukaryotes, which evades the high mutation rates of extracellularly transmitted viruses in higher eukaryotes. New data at atomic resolution for the PcV CP will indicate whether the PcV duplicated domain is merely a structural building block or whether it also comprises a conserved functional element. Whereas there are numerous functional and structural studies of the family Reoviridae, it is clear that deeper characterization of many fungal and protozoal dsRNA viruses is needed. Recent studies of fungal viruses of the family Partitiviridae identified the first T=1 capsid formed by quasisymmetric CP dimers (48, 49); an equivalent structure in viruses in higher eukaryotes, the picobirnaviruses, was also recently described (18). Another notable example is infectious myonecrosis virus (IMNV), a shrimp pathogen with a totivirus-like 120-subunit capsid. The IMNV capsid surface has fibers at the 5-fold axis, which are probably involved in cell entry, as this virus is transmitted extracellularly (67).
Some positive-sense ssRNA viruses, such as invertebrate nodaviruses and other plant viruses (61, 68), have genome regions that interact with CP subunits at symmetrically equivalent positions. The average structure of these ordered regions has been visualized at high resolution as a duplex RNA. The size of the visualized genome is variable, and the ordered ssRNA is located at the icosahedral 3-fold or 2-fold axis of the virion. In Pariacoto virus (PaV), the ssRNA forms a dodecahedral cage in close contact with the protein shell that constitutes ∼35% of the encapsidated RNA (31, 68). Cryo-EM studies with rotaviruses also showed dsRNA organized into a dodecahedral shell beneath the protein capsid (37, 52). A similar arrangement for the outer layer of RNA was described in BTV cores by X-ray crystallography (22). This high organizational level is probably related to the independent, simultaneous transcription/replication reactions of the genome segments inside the T=1 core.
Members of the family Reoviridae have a genome of 10, 11, or 12 dsRNA segments, packed at the same ratio. Individual genome segments must be transported through the active sites of the RdRp complexes at the 5-fold axes, and template motion is a limiting factor. L-A virus is a simplified version of these viruses, with a single-segment genome; there is one (or two) copy of the RdRp complex, covalently bound to the inner capsid surface as it is expressed as a fusion protein with the CP. No ordered RNA segments were observed in the structure, however, probably due to the more spacious L-A virus capsid, in which dsRNA is loosely packed in comparison to the dsRNA density in reovirus replicative cores. Previous calculations showed that the PcV capsid has a dsRNA density similar to that of L-A virus (considering an average dsRNA molecule/particle of 3.2 kbp). The PcV average genome (2.2 MDa) would occupy a volume of 2,270 nm3, assuming a dsRNA density of 1.61 g cm−3 (11). When the mass density of the full 3D map is represented at ∼1 σ from the mean (Fig. 6), the genome volume is 2,300 nm3, ∼50% of which accounts for the ordered RNA layer. These calculations indicate that the disordered dsRNA is packed at a slightly lower density (∼17 bp/100 nm3) in the remaining available volume than if there were no ordered dsRNA layer (19 bp/100 nm3). Unlike in L-A virus, PcV RdRp is not expressed as a fusion protein with CP; the lower density at the central region and the associated slight increase in dsRNA mobility might be necessary for maximum RNA polymerase activity.
Additional studies are needed, first, to solve the atomic structure of the PcV CP and, second, to test whether this fold signature is a characteristic structural feature preserved in other chrysoviruses. Meeting these objectives will allow us to evaluate whether the PcV conserved core is a hallmark fold preserved in dsRNA viruses.
Supplementary Material
Acknowledgments
We thank A. Zlotnick and J. E. Johnson for critical reading of the manuscript and C. Mark for editorial assistance.
This work was supported by grants from the Spanish Ministry of Science and Innovation (BFU 2008-02328/BMC and S-0505-Mat-0238 to J.L.C. and BIO2008-02361 to J.R.C.) and the NIH Intramural Research Program with support from the Center for Information Technology.
Footnotes
Published ahead of print on 12 May 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abrescia, N. G., J. J. Cockburn, J. M. Grimes, G. C. Sutton, J. M. Diprose, S. J. Butcher, S. D. Fuller, C. San Martin, R. M. Burnett, D. I. Stuart, D. H. Bamford, and J. K. Bamford. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68-74. [DOI] [PubMed] [Google Scholar]
- 2.Adamczak, R., A. Porollo, and J. Meller. 2005. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins 59:467-475. [DOI] [PubMed] [Google Scholar]
- 3.Agirrezabala, X., J. Martin-Benito, M. Valle, J. M. Gonzalez, A. Valencia, J. M. Valpuesta, and J. L. Carrascosa. 2005. Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. J. Mol. Biol. 347:895-902. [DOI] [PubMed] [Google Scholar]
- 4.Baker, M. L., W. Jiang, F. J. Rixon, and W. Chiu. 2005. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 79:14967-14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, M. L., T. Ju, and W. Chiu. 2007. Identification of secondary structure elements in intermediate-resolution density maps. Structure 15:7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 7.Bamford, D. H., J. M. Grimes, and D. I. Stuart. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15:655-663. [DOI] [PubMed] [Google Scholar]
- 8.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673-685. [DOI] [PubMed] [Google Scholar]
- 9.Border, D. J., K. W. Buck, E. B. Chain, G. F. Kempson-Jones, P. Lhoas, and G. Ratti. 1972. Viruses of Penicillium and Aspergillus species. Biochem. J. 127:4P-6P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bubeck, D., D. J. Filman, N. Cheng, A. C. Steven, J. M. Hogle, and D. M. Belnap. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79:7745-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castón, J. R., S. A. Ghabrial, D. Jiang, G. Rivas, C. Alfonso, R. Roca, D. Luque, and J. L. Carrascosa. 2003. Three-dimensional structure of Penicillium chrysogenum virus: a double-stranded RNA virus with a genuine T=1 capsid. J. Mol. Biol. 331:417-431. [DOI] [PubMed] [Google Scholar]
- 12.Castón, J. R., D. Luque, B. L. Trus, G. Rivas, C. Alfonso, J. M. Gonzalez, J. L. Carrascosa, P. Annamalai, and S. A. Ghabrial. 2006. Three-dimensional structure and stoichiometry of Helmintosporium victoriae 190S totivirus. Virology 347:323-332. [DOI] [PubMed] [Google Scholar]
- 13.Castón, J. R., B. L. Trus, F. P. Booy, R. B. Wickner, J. S. Wall, and A. C. Steven. 1997. Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J. Cell Biol. 138:975-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole, C., J. D. Barber, and G. J. Barton. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197-W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conway, J. F., B. L. Trus, F. P. Booy, W. W. Newcomb, J. C. Brown, and A. C. Steven. 1993. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J. Struct. Biol. 111:222-233. [DOI] [PubMed] [Google Scholar]
- 16.Coulibaly, F., C. Chevalier, I. Gutsche, J. Pous, J. Navaza, S. Bressanelli, B. Delmas, and F. A. Rey. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761-772. [DOI] [PubMed] [Google Scholar]
- 17.Diprose, J. M., J. M. Grimes, G. C. Sutton, J. N. Burroughs, A. Meyer, S. Maan, P. P. Mertens, and D. I. Stuart. 2002. The core of bluetongue virus binds double-stranded RNA. J. Virol. 76:9533-9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duquerroy, S., B. Da Costa, C. Henry, A. Vigouroux, S. Libersou, J. Lepault, J. Navaza, B. Delmas, and F. A. Rey. 2009. The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J. 28:1655-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez, J. J., D. Luque, J. R. Caston, and J. L. Carrascosa. 2008. Sharpening high resolution information in single particle electron cryomicroscopy. J. Struct. Biol. 164:170-175. [DOI] [PubMed] [Google Scholar]
- 20.Fujimura, T., J. C. Ribas, A. M. Makhov, and R. B. Wickner. 1992. Pol of gag-pol fusion protein required for encapsidation of viral RNA of yeast L-A virus. Nature 359:746-749. [DOI] [PubMed] [Google Scholar]
- 21.Ghabrial, S. A., D. Jiang, and J. R. Caston. 2005. Family Partitiviridae, p. 591-595. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. VIIIth Report of the International Committee on taxonomy of viruses. Academic Press, San Diego, CA.
- 22.Gouet, P., J. M. Diprose, J. M. Grimes, R. Malby, J. N. Burroughs, S. Zientara, D. I. Stuart, and P. P. Mertens. 1999. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell 97:481-490. [DOI] [PubMed] [Google Scholar]
- 23.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. Mertens, and D. I. Stuart. 1998. The atomic structure of the Bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 24.Harrison, S. C. 2007. Principles of virus structure, p. 59-98. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Strauss (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 25.Heymann, J. B., and D. M. Belnap. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157:3-18. [DOI] [PubMed] [Google Scholar]
- 26.Huiskonen, J. T., F. de Haas, D. Bubeck, D. H. Bamford, S. D. Fuller, and S. J. Butcher. 2006. Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure 14:1039-1048. [DOI] [PubMed] [Google Scholar]
- 27.Hutvagner, G., and M. J. Simard. 2008. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9:22-32. [DOI] [PubMed] [Google Scholar]
- 28.Jaalinoja, H. T., J. T. Huiskonen, and S. J. Butcher. 2007. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages phi6 and phi8. Structure 15:157-167. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, D., and S. A. Ghabrial. 2004. Molecular characterization of Penicillium chrysogenum virus: reconsideration of the taxonomy of the genus Chrysovirus. J. Gen. Virol. 85:2111-2121. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, W., M. L. Baker, S. J. Ludtke, and W. Chiu. 2001. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol. 308:1033-1044. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, K. N., L. Tang, J. E. Johnson, and L. A. Ball. 2004. Heterologous RNA encapsidated in Pariacoto virus-like particles forms a dodecahedral cage similar to genomic RNA in wild-type virions. J. Virol. 78:11371-11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 33.Kainov, D. E., S. J. Butcher, D. H. Bamford, and R. Tuma. 2003. Conserved intermediates on the assembly pathway of double-stranded RNA bacteriophages. J. Mol. Biol. 328:791-804. [DOI] [PubMed] [Google Scholar]
- 34.Krupovic, M., and D. H. Bamford. 2008. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat. Rev. Microbiol. 6:941-948. [DOI] [PubMed] [Google Scholar]
- 35.Kurowski, M. A., and J. M. Bujnicki. 2003. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 31:3305-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawton, J. A., M. K. Estes, and B. V. Prasad. 2000. Mechanism of genome transcription in segmented dsRNA viruses. Adv. Virus Res. 55:185-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawton, J. A., M. K. Estes, and B. V. Prasad. 1997. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat. Struct. Biol. 4:118-121. [DOI] [PubMed] [Google Scholar]
- 38.Lemaire, P. A., E. Anderson, J. Lary, and J. L. Cole. 2008. Mechanism of PKR activation by dsRNA. J. Mol. Biol. 381:351-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, K., V. A. Simossis, W. R. Taylor, and J. Heringa. 2005. A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics 21:152-159. [DOI] [PubMed] [Google Scholar]
- 40.Lomonossoff, G. P., and J. E. Johnson. 1991. The synthesis and structure of comovirus capsids. Prog. Biophys. Mol. Biol. 55:107-137. [DOI] [PubMed] [Google Scholar]
- 41.Luque, D., G. Rivas, C. Alfonso, J. L. Carrascosa, J. F. Rodriguez, and J. R. Caston. 2009. Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc. Natl. Acad. Sci. U. S. A. 106:2148-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luque, D., I. Saugar, M. T. Rejas, J. L. Carrascosa, J. F. Rodriguez, and J. R. Caston. 2009. Infectious bursal disease virus: ribonucleoprotein complexes of a double-stranded RNA virus. J. Mol. Biol. 386:891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luque, D., I. Saugar, J. F. Rodriguez, N. Verdaguer, D. Garriga, C. S. Martin, J. A. Velazquez-Muriel, B. L. Trus, J. L. Carrascosa, and J. R. Caston. 2007. Infectious bursal disease virus capsid assembly and maturation by structural rearrangements of a transient molecular switch. J. Virol. 81:6869-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClain, B., E. Settembre, B. R. Temple, A. R. Bellamy, and S. C. Harrison. 2010. X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution. J. Mol. Biol. 397:587-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naitow, H., J. Tang, M. Canady, R. B. Wickner, and J. E. Johnson. 2002. L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism. Nat. Struct. Biol. 9:725-728. [DOI] [PubMed] [Google Scholar]
- 46.Nakagawa, A., N. Miyazaki, J. Taka, H. Naitow, A. Ogawa, Z. Fujimoto, H. Mizuno, T. Higashi, Y. Watanabe, T. Omura, R. H. Cheng, and T. Tsukihara. 2003. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure 11:1227-1238. [DOI] [PubMed] [Google Scholar]
- 47.Nandhagopal, N., A. A. Simpson, J. R. Gurnon, X. Yan, T. S. Baker, M. V. Graves, J. L. Van Etten, and M. G. Rossmann. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. U. S. A. 99:14758-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ochoa, W. F., W. M. Havens, R. S. Sinkovits, M. L. Nibert, S. A. Ghabrial, and T. S. Baker. 2008. Partitivirus structure reveals a 120-subunit, helix-rich capsid with distinctive surface arches formed by quasisymmetric coat-protein dimers. Structure 16:776-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan, J., L. Dong, L. Lin, W. F. Ochoa, R. S. Sinkovits, W. M. Havens, M. L. Nibert, T. S. Baker, S. A. Ghabrial, and Y. J. Tao. 2009. Atomic structure reveals the unique capsid organization of a dsRNA virus. Proc. Natl. Acad. Sci. U. S. A. 106:4225-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 51.Pollastri, G., and A. McLysaght. 2005. Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics 21:1719-1720. [DOI] [PubMed] [Google Scholar]
- 52.Prasad, B. V., R. Rothnagel, C. Q. Zeng, J. Jakana, J. A. Lawton, W. Chiu, and M. K. Estes. 1996. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature 382:471-473. [DOI] [PubMed] [Google Scholar]
- 53.Reinisch, K. M. 2002. The dsRNA Viridae and their catalytic capsids. Nat. Struct. Biol. 9:714-716. [DOI] [PubMed] [Google Scholar]
- 54.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the Reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 55.Riffel, N., K. Harlos, O. Iourin, Z. Rao, A. Kingsman, D. Stuart, and E. Fry. 2002. Atomic resolution structure of Moloney murine leukemia virus matrix protein and its relationship to other retroviral matrix proteins. Structure 10:1627-1636. [DOI] [PubMed] [Google Scholar]
- 56.Roberts, M. M., J. L. White, M. G. Grutter, and R. M. Burnett. 1986. Three-dimensional structure of the adenovirus major coat protein hexon. Science 232:1148-1151. [DOI] [PubMed] [Google Scholar]
- 57.Rossmann, M., and J. Johnson. 1989. Icosahedral RNA virus structure. Annu. Rev. Biochem. 58:533-573. [DOI] [PubMed] [Google Scholar]
- 58.Rost, B., G. Yachdav, and J. Liu. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321-W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saugar, I., D. Luque, A. Ona, J. F. Rodriguez, J. L. Carrascosa, B. L. Trus, and J. R. Caston. 2005. Structural polymorphism of the major capsid protein of a double-stranded RNA virus: an amphipathic alpha helix as a molecular switch. Structure 13:1007-1017. [DOI] [PubMed] [Google Scholar]
- 60.Scheres, S. H., R. Nunez-Ramirez, C. O. Sorzano, J. M. Carazo, and R. Marabini. 2008. Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 3:977-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneemann, A. 2006. The structural and functional role of RNA in icosahedral virus assembly. Annu. Rev. Microbiol. 60:51-67. [DOI] [PubMed] [Google Scholar]
- 62.Sen, T. Z., R. L. Jernigan, J. Garnier, and A. Kloczkowski. 2005. GOR V server for protein secondary structure prediction. Bioinformatics 21:2787-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shaikh, T. R., H. Gao, W. T. Baxter, F. J. Asturias, N. Boisset, A. Leith, and J. Frank. 2008. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3:1941-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka, H., K. Kato, E. Yamashita, T. Sumizawa, Y. Zhou, M. Yao, K. Iwasaki, M. Yoshimura, and T. Tsukihara. 2009. The structure of rat liver vault at 3.5 angstrom resolution. Science 323:384-388. [DOI] [PubMed] [Google Scholar]
- 65.Tanaka, S., C. A. Kerfeld, M. R. Sawaya, F. Cai, S. Heinhorst, G. C. Cannon, and T. O. Yeates. 2008. Atomic-level models of the bacterial carboxysome shell. Science 319:1083-1086. [DOI] [PubMed] [Google Scholar]
- 66.Tang, J., H. Naitow, N. A. Gardner, A. Kolesar, L. Tang, R. B. Wickner, and J. E. Johnson. 2005. The structural basis of recognition and removal of cellular mRNA 7-methyl G “caps” by a viral capsid protein: a unique viral response to host defense. J. Mol. Recognit. 18:158-168. [DOI] [PubMed] [Google Scholar]
- 67.Tang, J., W. F. Ochoa, R. S. Sinkovits, B. T. Poulos, S. A. Ghabrial, D. V. Lightner, T. S. Baker, and M. L. Nibert. 2008. Infectious myonecrosis virus has a totivirus-like, 120-subunit capsid, but with fiber complexes at the fivefold axes. Proc. Natl. Acad. Sci. U. S. A. 105:17526-17531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang, L., K. N. Johnson, L. A. Ball, T. Lin, M. Yeager, and J. E. Johnson. 2001. The structure of Pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol. 8:77-83. [DOI] [PubMed] [Google Scholar]
- 69.Yu, X., L. Jin, and Z. H. Zhou. 2008. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature 453:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zemla, A. 2003. LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res. 31:3370-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, X., S. B. Walker, P. R. Chipman, M. L. Nibert, and T. S. Baker. 2003. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 A. Nat. Struct. Biol. 10:1011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.