Abstract
In lung transplant patients undergoing immunosuppression, more than one human cytomegalovirus (HCMV) genotype may emerge during follow-up, and this could be critical for the outcome of HCMV infection. Up to now, many cases of infection with multiple HCMV genotypes were probably overlooked due to the limitations of the current genotyping approaches. We have now analyzed mixed-genotype infections in 17 clinical samples from 9 lung transplant patients using the highly sensitive ultradeep-pyrosequencing (UDPS) technology. UDPS genotyping was performed at three variable HCMV genes, coding for glycoprotein N (gN), glycoprotein O (gO), and UL139. Simultaneous analysis of a mean of 10,430 sequence reads per amplicon allowed the relative amounts of distinct genotypes in the samples to be determined down to 0.1% to 1% abundance. Complex mixtures of up to six different HCMV genotypes per sample were observed. In all samples, no more than two major genotypes accounted for at least 88% of the HCMV DNA load, and these were often accompanied by up to four low-abundance genotypes at frequencies of 0.1% to 8.6%. No evidence for the emergence of new genotypes or sequence changes over time was observed. However, analysis of different samples withdrawn from the same patients at different time points revealed that the relative levels of replication of the individual HCMV genotypes changed within a mixed-genotype population upon reemergence of the virus. Our data show for the first time that, similar to what has been hypothesized for the murine model, HCMV reactivation in humans seems to occur stochastically.
Human cytomegalovirus (HCMV) is the most important viral pathogen affecting patients after solid organ transplantation (12, 18, 29). Lung transplant recipients have an especially high risk of acquiring severe and sometimes fatal HCMV infections, which are also associated directly or indirectly with graft rejection and bronchiolitis obliterans syndrome (6, 53).
Human cytomegalovirus is a double-stranded DNA virus with a genome size of about 236 kb that is predicted to contain 165 protein-coding genes (9). Although most of the HCMV genome is highly conserved among the various HCMV strains, a subset of genes exhibits a high degree of variability, as has been shown by genome-wide sequence analysis as well as by examination of individual genes. A high level of genetic heterogeneity usually occurs in a limited number of distinct genotypes (9, 32, 33). Among the most variable genes are the viral envelope glycoprotein N (gN) and gO genes and the gene encoding the predicted membrane glycoprotein UL139. Sequence analysis of highly polymorphic regions within these three genes allows the discrimination of seven distinct gN genotypes (30, 31) and eight distinct gO and UL139 genotypes (3, 24, 35, 45). The existence of such a large number of distinct genotypes provides a useful tool for investigating HCMV population diversity within a host.
Reinfection with different HCMV strains is possible in the human host, and several strains can accumulate during a person's lifetime (26). In transplant patients, not only the recipient's own pretransplant latent HCMV strains but also donor-acquired HCMV strains can accumulate and may replicate when the patient is undergoing immunosuppressive therapy. There is mounting evidence that the occurrence of mixed HCMV infections in lung transplant recipients is disadvantageous for these patients (17, 23, 34). So far, however, the extent to which patients are typically infected with multiple HCMV strains is not known. Various methods have been applied for discriminating genetically distinct HCMV strains in clinical material during HCMV infection by taking advantage of sequence polymorphisms within variable genes. However, the most frequently used HCMV genotyping methods, such as traditional Sanger DNA sequencing (3, 30, 45), restriction fragment length polymorphism of PCR products (27, 31, 35), and genotype-specific PCR assays (24), lack sensitivity and are not quantitative. Even cloning of PCR products and subsequent Sanger sequencing do not allow a sensitive assessment of the genotypes present unless an extremely large number of individual clones is sequenced. Improved genotype-specific real-time-PCR-based assays have been established recently for gB and gH genotyping, and these allow simultaneous detection and quantitation of distinct genotypes in mixed infections down to a level of 5% or even less (14, 28).
In the present study, we determined the relative amounts of different HCMV genotypes in clinical samples from lung transplant patients by using the highly sensitive ultradeep-pyrosequencing (UDPS) technology. UDPS genotyping of three of the most variable genes within the HCMV genome, the gN, gO, and UL139 genes, revealed that there is a substantially higher diversity of HCMV genotype populations in patients with mixed HCMV infection than has been recognized previously and that the HCMV strains show a distinct pattern in their quantitative distribution relative to each other. The present findings support the hypothesis that HCMV reactivation in humans occurs stochastically, as has previously only been shown using a mouse model.
MATERIALS AND METHODS
Patients and samples.
UDPS analysis was performed on clinical samples from nine patients receiving lung transplants at the Medical University of Vienna between 2001 and 2008. The patients received immunosuppressive treatment including prednisolone, mycophenolate, and either cyclosporine or tacrolimus as well as antiviral prophylaxis with ganciclovir and valganciclovir for 3 months posttransplantation. The patients' donor/recipient HCMV serostatus is shown in Table 1. All patients were routinely screened for HCMV DNA load in plasma and bronchoalveolar lavage (BAL) samples as mentioned in the next paragraph and for HCMV infectivity by shell-vial culture of the BAL specimens (13) in the posttransplant follow-up. In all of the nine patients included in the study active HCMV infection was observed within the first year after transplantation, after cessation of antiviral prophylaxis (Table 1), as defined by detection of HCMV DNA load in plasma and/or BAL samples accompanied by a positive viral culture result of the BAL specimen. We have investigated plasma samples from three of the patients, BAL samples from another three patients, and paired plasma and BAL samples from the remaining three patients. In all cases the investigations were performed during episodes of active HCMV infection. HCMV DNA-positive follow-up samples were obtained from four of the patients in later episodes of active HCMV infection and included four BAL samples and one plasma sample. Between the first and the follow-up samples investigated in the present study, all patients showed periods of undetectable plasma HCMV DNA. Additional BAL samples between the follow-up time points were available from patients B2 and B7, and these were negative for HCMV DNA and viral culture. Thus, the HCMV DNA-positive follow-up samples were considered to reflect independent replication events which were caused by reactivation of HCMV from latency or from low persistence (below the detection limit). Additional criteria for inclusion of the samples for UDPS analysis were (i) an HCMV DNA load of at least 5 × 104 copies/ml to ensure that low-abundance genotypes at levels of 0.1 to 1% could be detected and (ii) the presence of a mixed gB and/or gH genotype population in the samples, which was determined by prior testing of the samples with a genotype-specific PCR assay.
TABLE 1.
Clinical and virological features of the nine lung transplant patients
| Patient | Clinical sample | Donor/recipient HCMV serostatus | HCMV DNA load (copies/ml) | Time posttransplantation (mo) |
|---|---|---|---|---|
| 1 | BAL (B1) | D+/R+ | 2.4E+05 | 6.5 |
| 2 | BAL (B2) | D+/R+ | 2.0E+05 | 12.0 |
| 3 | BAL (B3) | D+/R− | 5.2E+04 | 6.0 |
| 4 | Plasma (P4) | D+/R− | 1.3E+05 | 4.0 |
| 5 | Plasma (P5) | D+/R+ | 5.2E+04 | 4.0 |
| 6 | Plasma (P6) | D−/R+ | 5.0E+04 | 11.0 |
| 7 | BAL (B7) | D+/R+ | 5.5E+06 | 5.0 |
| Plasma (P7) | 1.1E+06 | 5.5 | ||
| 8 | BAL (B8) | D+/R− | 2.6E+05 | 5.0 |
| Plasma (P8) | 8.6E+06 | 4.0 | ||
| 9 | BAL (B9) | D+/R+ | 1.8E+05 | 6.0 |
| Plasma (P9) | 5.0E+04 | 5.5 |
HCMV DNA isolation and gB and gH genotyping.
The HCMV DNA load in plasma and BAL samples from the patients was determined using a Cobas Amplicor CMV monitor test kit with a COBAS Amplicor analzyer (both from Roche Molecular Systems, Branchburg, NJ). For this test, HCMV DNA was isolated directly from patient samples using a QIAamp RNA isolation kit (Qiagen, Hilden, Germany). Samples with a viral DNA load of at least 5 × 104 copies/ml were then tested using gB and gH genotype-specific quantitative PCR assays, which have been described previously (14), to preselect clinical samples with mixed HCMV infections.
PCR amplification for library preparation and UDPS.
UDPS was used to sequence highly variable regions within the gN, gO, and UL139 genes after PCR amplification. To prepare PCR-derived amplicons, HCMV DNA was extracted directly from 140 μl of stored plasma and BAL fluid using a QIAamp RNA isolation kit as described above and eluted with 70 μl water. For PCR, 10 μl of the eluted HCMV DNA was used in a total of 50 μl per reaction well. Samples with viral DNA loads of 5 × 104 copies/ml provided enough viral copies per reaction for reliable detection of genotype sequences occurring at a frequency of at least 1%, and samples with viral DNA loads of ≥5 × 105 copies/ml allowed the detection of low-abundance genotype sequences down to a frequency of 0.1% or lower. Primers were chosen to sequence a region within each gene of 217 to 240 bases, which allowed the discrimination of seven gN genotypes (30, 31), eight gO genotypes (24, 35, 45), and eight UL139 genotypes (3) using specific polymorphic informative sites. Target-specific gN, gO, and UL139 primers were designed based on sequences from the GenBank database to anneal to sequences conserved among all genotypes. All of the primers used included 5′ extensions, which provided binding sites for the 454A and -B primers and 6-bp nucleotide bar code sequences, allowing multiple samples to be tested in a single sequencing experiment. Primer information is presented in detail in Table 2. gN, gO, and UL139 amplicons were generated from isolated HCMV DNA using Taq-Gold polymerase (Roche Diagnostics, Basel, Switzerland). For the gO amplicon, two separate PCRs were performed using the primer pairs gOforudps/gOMERrevudps and gOforudps/gOADrevudps. The resulting PCR products were pooled prior to purification. The gN and UL139 amplicons were each produced by a single PCR. The conditions used for PCR were 1 cycle of 95°C for 10 min followed by 40 cycles of denaturation for 1 min at 95°C, annealing of primers for 1 min at the temperatures listed in Table 2, and extension for 1 min, with a final 5-min extension at 72°C. The PCR amplicons were then purified using Microcon 500 columns (Millipore, Bedford, MA), quality controlled using an 2100 Bioanalyzer (Agilent Life Sciences and Chemical Analysis, Santa Clara, CA), and quantified using a Quant-iT PicoGreen assay kit (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. PCR amplicons were pooled according to bar code and amplicon compatibility and subjected to UDPS, which was carried out on a Genome Sequencer FLX (Roche 454 Life Sciences, Branford, CT) using GS-FLX-emPCR Kit II or III (GS-FLX-emPCR method manual) according to the manufacturer's GS-FLX sequencing method manual. A mean of 10,430 clonal reads (ranging from 4,738 to 26,216) were obtained per sample after quality control.
TABLE 2.
Primers used for ultradeep-pyrosequencing library preparation and for traditional Sanger sequencing
| Gene | Primer | Sequence (5′-3′)a | Annealing temp (°C) | Genome location |
|---|---|---|---|---|
| gN | gNforudps | gcctccctcgcgccatcagNNNNNNTGGCGGTGGTGTGATGGA | 107038-107055b | |
| gN | gNrevudps | gccttgccagcccgctcagAGTTCTGGAARCAGCAATGTCG | 61 | 107429-107408b |
| gO | gOforudps | gcctccctcgcgccatcagNNNNNNTACCTGGTTAACGCCATGAG | 108158-108139b | |
| gO | gOMERrevudps | gccttgccagcccgctcagGGCCATAATGGATTTCATAAAGTT | 60 | 107862-107885b |
| gO | gOADrevudps | gccttgccagcccgctcagGTGGCCATAATGGACTTCATA | 57 | 106555-106575c |
| UL139 | UL139forudps | gcctccctcgcgccatcagACCCCAGAATGAAAGAGTAT | 187052-187033b | |
| UL139 | UL139revudps | gccttgccagcccgctcagNNNNNNAAGGAGTCCAGATAATGTCC | 55 | 186666-186685b |
Lowercase letters, 5′ extensions for 454A and -B primers; N, 6-bp bar code; capital letters, target-specific sequences and traditional sequencing primer.
With reference to HCMV strain Merlin (GenBank accession no. AY446894.2).
With reference to HCMV strain AD169 (GenBank accession no. X17403.1).
Sanger sequencing.
The sequencing reactions were performed using a BigDye Terminator version 1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) with 5 μl of each purified amplicon and 8 pmol of each of the corresponding primers (Table 2) and analyzed on an ABI Prism 3100 genetic analyzer (Applied Biosystems). The resulting DNA sequences were aligned to reference sequences, proofread, and edited using SeqScape software 1.1 (Applied Biosystems).
Analysis of sequence data.
GS Amplicon Variant Analyzer (AVA) version 2.0.01 software from Roche was used to analyze the ultradeep-sequencing data based on multiple alignments to the reference sequences, performed with the total number of amplicon sequence reads. The AVA software was used to automatically scan for and quantify the well-defined genotype sequences. Therefore, nucleotide sequence reads from each individual amplicon were aligned to reference genotype sequences obtained from the GenBank database. Additionally, already known variations, including known genotypes, were entered to automatically determine the overall frequency of each genotype. The highly polymorphic regions within the gN, gO, and UL139 genes that were chosen for UDPS are characterized by insertions, deletions, and homopolymeric runs. Thus, the global alignment accuracy was limited by multiple indels and the high pyrosequencing error rate in the homopolymeric regions (average pyrosequencing error rate, 4.4 × 10−3) (51). To overcome these limitations, the following strategy for discovery of potential de novo variants/genotypes was used. To reduce the typical sequencing error, estimated as approximately 4 to 5 errors per kb (19), an error correction step was applied. All reads that had ambiguous bases or whose lengths lay outside the main distribution, as well as inexact matches to the primer or bar-coding sequence, were discarded (19). About 15% of the total number of amplicon sequence reads were randomly chosen using a custom-programmed Perl script and realigned to reference genotype sequences using ClustalX 1.83 (50). Identical sequence reads were combined into single sequences and edited manually using BioEdit 7.0.1 to correct obvious alignment errors and remove sections of homopolymeric runs. The unique sequence reads identified for each sample and amplicon were then combined into single gN, gO, and UL139 gene alignments, from which neighbor-joining trees were inferred using the Kimura 2-parameter method in MEGA 4.0 (47). All gN, gO, and UL139 sequences clustered with published prototype sequences, and the percent identity was calculated using MEGA 4.0. Genotype sequences were judged to be authentic if at least 10 identical sequence reads were obtained. To exclude potential variants created by amplification and pyrosequencing errors, sequences that varied from the consensus genotype sequences were considered true variants only when identical genotype sequences were identified in two or more independent samples or when the variant sequences differed from known sequences at more than 2 nucleotides. To validate the PCR amplification and deep-sequencing assay for genotype quantitation of each of the three genes, two genotypically distinct standard HCMV DNAs were mixed at a ratio of 99:1 and tested. The measured values obtained with these standards ranged from 0.5% and 99.5% to 1.6% and 98.4%, indicating that the input ratio was well preserved.
Reference genotype designation and nucleotide sequence accession numbers.
The designation of gN, gO, and UL139 genotypes was done as reported previously (3, 24, 30, 31, 35, 45). GenBank accession numbers of published reference genotype sequences used in this study were as follows: gO, GT1a_AD169 (X17403.1), GT1a_SW4 (AF531347.1), GT1b_SW990 (AF531354.1), GT1c_TB40E (EF999921.1), GT2a_FUK19U (EU348354.1), GT2b_SW1102 (AF531339.1), GT3_1960 (AF531318.1), GT3_SW5 (AF531350.1), GT4_Towne (AY315197.2), and GT5_Merlin (AY446894.2); gN, GT1_AD169 (X17403.1), GT1_ST (AF390789.1), GT1_HR (AF309974.1), GT2_N21b (EU686422.1), GT2_DL (AF309975.1), GT3a_BD (AF309980.1), GT3b_HAN38 (GQ396662.1), GT3b_A8-27F (AF390802.1), GT4a_MS (AF396731.1), GT4a_K57 (EU686453.1), GT4b_TB40E (EF999921.1), and GT4c_Merlin (AY446894.2); UL139, GT1_Merlin (AY446894.2), GT1_U11 (GU179290.1), GT2_JP (GQ221975.1), GT2_76 (AY805300.1), GT3_175 (AY805267.1), GT3_88 (AY805303.1), GT4_TB40E (EF999921.1), GT5_44 (AY805296.1), GT6_283 (AY805288.1), GT7_W (AY446865.1), and GT8_HAN13 (GQ221973.1).
RESULTS
UDPS analysis of gN, gO, and UL139 genotype populations in clinical samples.
Ultradeep pyrosequencing (UDPS) was applied to characterize the diversity of HCMV gN, gO, and UL139 genotype populations in clinical samples obtained from nine lung transplant recipients. The patient and sample characteristics are shown in Table 1. From the HCMV DNA isolated directly from plasma and bronchoalveolar lavage (BAL) samples, highly polymorphic regions within the gN, gO, and UL139 genes were analyzed to identify sequences of well-defined genotypes (Fig. 1 A). The sequence data generated by UDPS resulted in about 10,000 sequence reads, with an average length of 226 bases per amplicon and sample. Based on global alignments of the distinct amplicon sequence reads using AVA software, the numbers and the percentages of the individual gN, gO, and UL139 genotypes present in the samples were evaluated by searching for signature sequence patterns that allow the distinct gN, gO, and UL139 prototype genotype sequences to be discriminated.
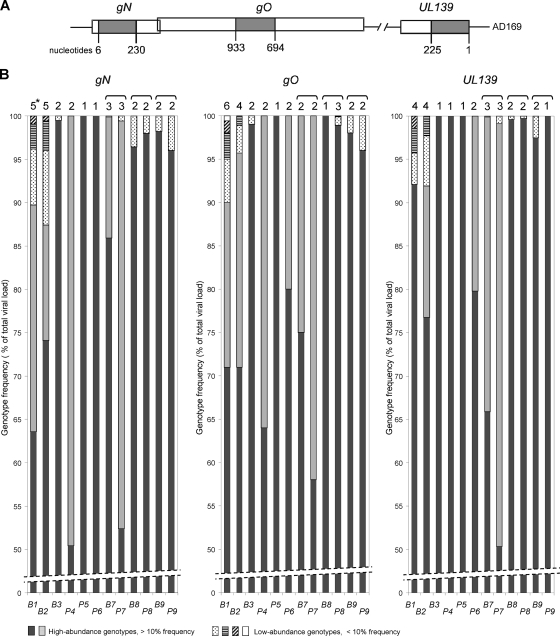
FIG. 1.
Relative abundances of gN, gO, and UL139 genotype populations determined by UDPS. (A) Schematic representation of the highly polymorphic regions analyzed. Nucleotide numbers are based on the open reading frames (ORFs) of the AD169 gene sequence (GenBank accession number X17403.1). (B) Illustration of the number and quantitative relationship of the individual gN, gO, and UL139 genotypes present in the 12 samples indicated on the x axis. Patient samples are numbered as described in Table 1, with “B” and “P” indicating BAL samples and plasma, respectively. The genotype frequencies are percentages of the total viral DNA load. *, number of genotypes detected; brackets, paired BAL and plasma samples.
All gN, gO, and UL139 amplicon sequence reads could be grouped and quantitated according to their genotype sequence patterns. The genotype population frequencies for each sample are shown in Fig. 1B. Complex mixtures of more than 3 distinct genotypes were detected only in patients with a donor- and recipient-positive (D+/R+) HCMV serostatus (Table 1 and Fig. 1B). The number of genotypes detected for each sample and genetic region ranged from 1 to 6. Quantitation of the individual gN, gO, and UL139 genotypes present in the 12 patient samples revealed that in all cases only one or two major genotypes accounted for at least 88% of the total viral DNA load. In 11 out of 12 samples, up to 4 additional low-abundance genotypes were identified at frequencies of 0.1% to 8.6%.
All amplicons were also subjected to standard Sanger sequencing. By this method, only the major genotypes were detected and none of the genotypes that were detected by UDPS at a frequency of <24% were detected.
Screening for variants of individual gN, gO, and UL139 genotype sequences.
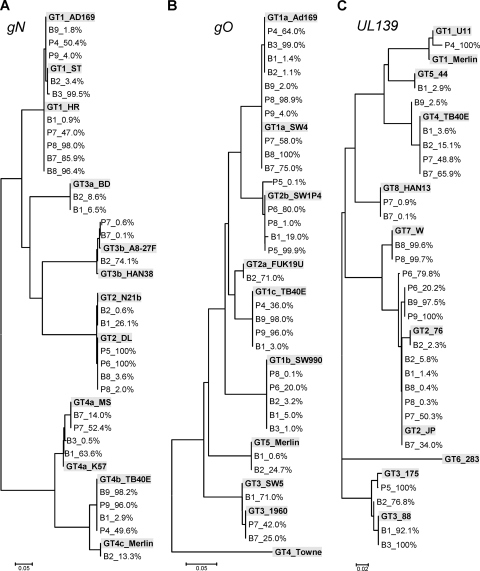
To identify variations in the sample-specific genotype sequences with respect to published prototype genotype sequences, all of the sequence data were reanalyzed in detail. For this, about 15% of the total number of sequence reads per amplicon and sample were randomly chosen, aligned, and manually edited. The unique sequence reads were combined into single gN, gO, and UL139 alignments, from which neighbor-joining trees were inferred. As presented in Fig. 2, these phylogenetic analyses revealed that all sample-specific genotype sequences clustered with prototype genotype sequences and showed 98.6% to 100% identity at the nucleotide level.
FIG. 2.
Phylogenetic analysis of gN, gO, and UL139 genotypes. Shown are neighbor-joining trees constructed using all unique genotype sequences for each genomic region found in the 12 patient samples. The trees illustrate both the frequency of the genotypes as a percentage of the total viral DNA load and their clustering with known prototype genotype sequences, as shown by boldface and gray shading. Genotype designations are used as reported previously (3, 24, 30, 31, 35, 45). Patient samples are numbered as described in Table 1.
In patient samples B2 and P6, two different variants of the same UL139 genotype 2 were present in the same sample (Fig. 2C). Identical sequences of these genotype 2 variants have been published previously or have been detected in unrelated patients. Thus, it is likely that the patients were coinfected with the different variants. Analysis of gO sequences revealed that a gO genotype 2b variant was present in sample P5 at a frequency of 0.1%, simultaneously with the well-defined gO genotype 2b sequence, which was present at 99.9% (Fig. 2B). The low-abundance gO2b variant differed at four nucleotide positions from the major gO2b genotype, and so far, no sequence identical to that of this variant has been published. Thus, it is possible that the gO2b variant might be a novel form that evolved from the major gO2b genotype in this patient.
Distribution of genotypes in blood and lung.
In lung transplant recipients, distinct HCMV gN, gO, and/or UL139 genotypes may be unequally distributed in the lungs and in blood. To assess whether HCMV genotypes in these two compartments differ within a patient, paired plasma and bronchoalveolar lavage samples from the three patients of our study cohort for which these were available (B7/P7, B8/P8, and B9/P9) (Table 1) were analyzed. The distribution and the relative frequencies of the gN, gO, and UL139 genotypes detected in the lung and in the blood compartment are shown in Fig. 1B (sample pairs are indicated by brackets above the bar charts) and Fig. 2. These data show that the relative distribution of the major gN, gO, and UL139 genotypes in these three patients was not dependent on the compartment in which they were found, and only minor differences were observed for the low-abundance genotypes (frequencies of less than 2.5%). Also, the quantitative relations between the strains were overall similar between the compartments; only in patient 7 did the relationships between the two major genotypes differ between blood and lung. Thus, it appears that nearly identical HCMV genotype populations circulate in both compartments.
Changes in gO genotype populations over time.
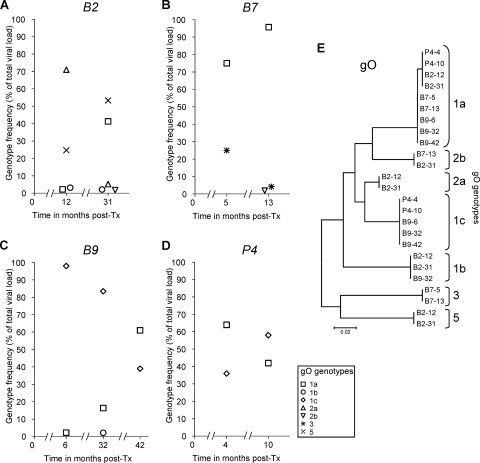
Finally, we investigated whether changes in the number, frequency, and sequences of gO genotypes, which are the most polymorphic of the ones investigated, occur during recurrent HCMV replication in individual patients over time. Additional HCMV-positive samples were collected from four of the lung transplant patients in the follow-up after transplantation at a second, and in one case also at a third, time point, at which HCMV DNA replication was routinely diagnosed. In these samples, the total HCMV DNA load reached >5 × 104 copies/ml, which enabled the detection of genotypes down to 1% or less (Fig. 3). UDPS HCMV genotyping was performed from these samples, and the gO sequences obtained by UDPS were analyzed as described above. The sequences and frequencies of the individual gO genotypes detected at the different time points are presented in Fig. 3A to D. In all cases, the relative frequencies of the resident gO genotypes had changed substantially over time (in a time interval of between 6 and 26 months), additional genotypes had emerged, and previously detected ones had become undetectable. In contrast to the high variability of the observed gO genotype frequencies, the nucleotide sequences of the individual gO genotypes were found to be completely stable over time (Fig. 3E).
FIG. 3.
Quantitative distribution and nucleotide sequence stability of gO genotype populations at different times posttransplantation. (A to D) The relative frequency of each gO genotype as a percentage of the total viral DNA load for each of the four lung transplant patients at each time point tested is displayed. (E) Neighbor-joining tree inferred from an alignment of unique gO genotype sequences obtained for each patient at different time points. Patient samples, numbered as described in Table 1, are followed by the times of sampling in months posttransplantation (post-Tx).
DISCUSSION
To the best of our knowledge, this is the first study in which the new and highly sensitive pyrophosphate-based ultradeep-sequencing technology has been applied to achieve detailed insight into the complexity of HCMV infection with mixed-genotype populations in clinical samples from transplant patients. By investigating three highly polymorphic regions of the HCMV genome, the gN, gO, and UL139 regions, quantitative deep sequencing down to genotype frequencies of 0.1% gave a clearly more complex picture than ever seen before, due to its substantial advantages over current genotyping and sequencing techniques. The ultradeep-pyrosequencing data presented in this study revealed that multiple HCMV gN, gO, and UL139 genotypes may cocirculate in plasma and bronchoalveolar lavage samples from lung transplant recipients during active HCMV infection posttransplantation. In particular, they revealed that, in addition to the major HCMV genotypes usually diagnosed, there are a number of low-abundance genotypes present at frequencies of 0.1% to 10%. Genotype populations at these low frequencies have certainly always escaped direct sequencing by the Sanger method and often also by other genotyping methods such as cloning and sequencing, restriction fragment length polymorphism analysis of PCR fragments, and genotype-specific PCR assays.
Complex mixtures of up to 6 different HCMV genotypes were observed. The highest genotype population diversity was seen in D+/R+ lung transplant recipients, most likely due to a mixture of donor-derived and recipient-derived genotypes. But as the donor and recipient HCMV genotype populations were undetectable at the time of transplantation, reinfection with another HCMV genotype posttransplantation cannot be completely excluded. Also, antiviral prophylaxis could have some impact on the development of the genotype composition, and thus the observed diversity may not exactly reflect the patients' genotype compositions at transplantation. In spite of the large number of possible genotype combinations detected in these patient samples, the individual genotype sequences were strikingly stable. Identical gN, gO, and UL139 genotype sequences were observed in different patients, and this was independent of their abundance in the sample and the body compartment from which they were obtained. The gO sequences remained stable even in sequential isolates obtained over a long-term follow-up period of several years. These findings are in agreement with data from other studies showing that hypervariable genes are stable (3, 16, 22, 45), as well as with the perception that herpesvirus genomes evolve slowly (25).
In previous publications dealing with infections with HCMV of different genotypes, there have been attempts to identify significant relationships between virus genotypes and the clinical presentation of patients (32, 33). In the light of the present data, it seems likely that in such studies the recognition of a possible relationship between certain HCMV genotypes and the outcome of HCMV infection and disease might have been hampered by the lack of ability to identify all of the genotype populations present in the samples.
Earlier studies have addressed the question of whether there are different overall distributions of genotypes in different body compartments (14, 34, 48). Our present data confirm now at a very sensitive level that there seems to be no compartmentalization between blood and lungs: overall, the genotype populations are present at similar frequencies in the lungs and blood of lung transplant patients during an episode of virus replication.
The detection of a variety of minor, low-abundance HCMV genotypes in clinical samples is also important with regard to previous data showing that infection with more than one HCMV genotype can be a negative determinant of the outcome of HCMV infection, especially in transplant recipients (17, 23, 34). It is not yet clear whether minor genotypes present at such low abundance in a background of major HCMV strains are of substantial direct clinical importance during the observed episode of HCMV replication or whether they contribute to indirect effects leading to chronic transplant rejection (11, 46, 53), and this needs to be further elucidated. In any case, however, it is conceivable that all distinct genotypes within a mixed infection can interfere with each other. Although the underlying mechanism of virus strain interaction has not been determined for HCMV, it has been demonstrated in a mouse model that distinct murine CMV variants can interact with each other by functional trans-complementation and that this may result in altered virus fitness (7).
An especially interesting outcome of this study was that the distinct genotypes present within a sample exhibited a constant pattern of frequency distribution during mixed infection at a given time point. In each patient sample tested, no more than two major genotypes accounted for about 90% of the total viral DNA load, while in the background, a number of distinct genotypes coexisted at low abundance, and this finding was independent of the genomic region tested. Intriguingly, our data further showed that, when clinical samples from the same patient were investigated during a later HCMV replication event, the relative frequencies of the individual genotypes had mostly changed: while the overall pattern of a maximum of two major genotypes and various other minor ones was maintained, the order of prevalence of these strains was different.
There are several possible explanations for these findings. First, it is possible that the observed quantitative differences among the virus populations could be associated with different replication efficiencies of specific genotypes. Our data, however, show that almost all well-defined gN, gO, and UL139 genotypes were present among the nine lung transplant patients and that none of them was found to be present exclusively as a major or minor genotype per se. Furthermore, the relative frequencies of the individual genotypes within a patient did not remain stable over time, a finding which has also been described for Epstein-Barr virus strains in mixed infections (44) and which makes it even more unlikely that a specific polymorphism at a specific viral genomic region might have influenced the level of replication of a particular genotype. As has already been demonstrated for the four major gN and two gH genotypes (2, 4, 20), host factors, such as differences in genotype-specific immune responses, could also have a certain influence on the rate of replication of individual strains, but from our data it does not seem that this effect would be strong enough to provide a constant selection of major genotypes over time. This holds true of course only for the HCMV genes tested in the present study. We cannot exclude the possibility that the genotype replication behavior is influenced in a more pronounced way by sequence variations in regions of the HCMV genome that are important for cell tropism, such as the UL128/UL130/UL131A region or RL11 family member regions (10, 15, 52). These variations may occur in genes with known or proposed roles in immune modulation, such as the UL111A gene (5); a mutant of this gene from a clinical sample, whose potential effects on pathogenesis have not been revealed so far, has recently been described (8).
Another explanation for our findings may be a quantitative difference in the latent virus genotype load at the onset of virus replication. It is likely that, in patients in which different HCMV strains are present, as in our study cohort, the frequencies of cells that are latently infected by each genotype differ substantially. Hence, the extent of replication might depend on the preexisting latent virus load levels for each genotype in the individual patient. However, the finding that the ratios of major and minor virus strains change over time in the same patient also argues against this hypothesis. Therefore, the latent virus level in the patient does not seem to play a major role.
A third hypothesis for our findings is that HCMV reactivation from latency occurs stochastically, largely independent of the number of cells that are latently infected with a specific genotype. In the current model of HCMV reactivation from latency, it is proposed that chromatin remodeling of the latent HCMV genome plays a crucial role in reactivation and that this is intimately linked to the differentiation of latently infected myeloid progenitor cells (38-43). Changes in histone modifications around the HCMV major immediate early promoter are a critical step in the initiation of the productive infection cycle (39-41), but subsequent steps may also play an important role in full reactivation to productive infection. In experimental HCMV latency models, it has been shown that the efficiency of reactivation of infectious virus is relatively low (40) and that reactivation may often end up being abortive (49). Hence, it is feasible that the emergence of individual genotypes at a given time point in active infection, as observed in our lung transplant patients, is less of a reflection of differences in latent genotype loads than it is a product of sporadic and stochastic events. What clearly supports this hypothesis is the murine model of CMV latency and reactivation postulated by Reddehase and his collegues (36, 37). This model proposes that mouse CMV reactivation may occur independently for each latently infected cell and may be putatively defined by a random pattern of silenced and desilenced genes (1, 21, 42). Consequently, reactivation from latency can be regarded as a stochastic process, and the number of latent genomes cannot be estimated by analysis at a single time point. Our data now provide the first evidence that in vivo reactivation of multiple human cytomegalovirus strains within a human host may behave in a way that is similar to that postulated for the murine model of latency and reactivation.
The highly sensitive UDPS technology has been proven to be a very useful tool to improve our understanding of the composition and replication behavior of multiple HCMV genotypes within a complex HCMV strain population. The observed frequent presence of minor subpopulations, which clearly have been underestimated so far, raises questions about their role in infection and disease and awaits further studies. A more comprehensive knowledge of HCMV infection in vivo and reactivation of mixed infections with different HCMV genotypes is an important prerequisite for developing further prophylactic strategies and therapeutic concepts to prevent HCMV disease.
Acknowledgments
We thank Barbara Dalmatiner and Daniela Schmidt for excellent technical assistance with gB-gH genotyping and UDPS amplicon library preparation, and we are grateful to Theresa Maierhofer and Gabriele Michelitsch for performing Ultra Deep 454 sequencing.
Footnotes
Published ahead of print on 12 May 2010.
REFERENCES
- 1.Böhm, V., C. K. Seckert, C. O. Simon, D. Thomas, A. Renzaho, D. Gendig, R. Holtappels, and M. J. Reddehase. 2009. Immune evasion proteins enhance cytomegalovirus latency in the lungs. J. Virol. 83:10293-10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boppana, S. B., L. B. Rivera, K. B. Fowler, M. Mach, and W. J. Britt. 2001. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 344:1366-1371. [DOI] [PubMed] [Google Scholar]
- 3.Bradley, A. J., I. J. Kovacs, D. Gatherer, D. J. Dargan, K. R. Alkharsah, P. K. Chan, W. F. Carman, M. Dedicoat, V. C. Emery, C. C. Geddes, G. Gerna, B. Ben-Ismaeil, S. Kaye, A. McGregor, P. A. Moss, R. Pusztai, W. D. Rawlinson, G. M. Scott, G. W. Wilkinson, T. F. Schulz, and A. J. Davison. 2008. Genotypic analysis of two hypervariable human cytomegalovirus genes. J. Med. Virol. 80:1615-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkhardt, C., S. Himmelein, W. Britt, T. Winkler, and M. Mach. 2009. Glycoprotein N subtypes of human cytomegalovirus induce a strain-specific antibody response during natural infection. J. Gen. Virol. 90:1951-1961. [DOI] [PubMed] [Google Scholar]
- 5.Chang, W. L., P. A. Barry, R. Szubin, D. Wang, and N. Baumgarth. 2009. Human cytomegalovirus suppresses type I interferon secretion by plasmacytoid dendritic cells through its interleukin 10 homolog. Virology 390:330-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chmiel, C., R. Speich, M. Hofer, D. Michel, T. Mertens, W. Weder, and A. Boehler. 2008. Ganciclovir/valganciclovir prophylaxis decreases cytomegalovirus-related events and bronchiolitis obliterans syndrome after lung transplantation. Clin. Infect. Dis. 46:831-839. [DOI] [PubMed] [Google Scholar]
- 7.Cicin-Sain, L., J. Podlech, M. Messerle, M. J. Reddehase, and U. H. Koszinowski. 2005. Frequent coinfection of cells explains functional in vivo complementation between cytomegalovirus variants in the multiply infected host. J. Virol. 79:9492-9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham, C., D. Gatherer, B. Hilfrich, K. Baluchova, D. J. Dargan, M. Thomson, P. D. Griffiths, G. W. Wilkinson, T. F. Schulz, and A. J. Davison. 2010. Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J. Gen. Virol. 91:605-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolan, A., C. Cunningham, R. D. Hector, A. F. Hassan-Walker, L. Lee, C. Addison, D. J. Dargan, D. J. McGeoch, D. Gatherer, V. C. Emery, P. D. Griffiths, C. Sinzger, B. P. McSharry, G. W. Wilkinson, and A. J. Davison. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301-1312. [DOI] [PubMed] [Google Scholar]
- 10.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeman, R. B., Jr. 2009. The ‘indirect’ effects of cytomegalovirus infection. Am. J. Transplant. 9:2453-2458. [DOI] [PubMed] [Google Scholar]
- 12.Gandhi, M. K., and R. Khanna. 2004. Human cytomegalovirus: clinical aspects, immune regulation, and emerging treatments. Lancet Infect. Dis. 4:725-738. [DOI] [PubMed] [Google Scholar]
- 13.Gleaves, C. A., and J. D. Meyers. 1989. Rapid detection of cytomegalovirus in bronchoalveolar lavage specimens from marrow transplant patients: evaluation of a direct fluorescein-conjugated monoclonal antibody reagent. J. Virol. Methods 26:345-349. [DOI] [PubMed] [Google Scholar]
- 14.Görzer, I., H. Kerschner, P. Jaksch, C. Bauer, G. Seebacher, W. Klepetko, and E. Puchhammer-Stöckl. 2008. Virus load dynamics of individual CMV-genotypes in lung transplant recipients with mixed-genotype infections. J. Med. Virol. 80:1405-1414. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, G., M. G. Revello, M. Patrone, E. Percivalle, G. Campanini, A. Sarasini, M. Wagner, A. Gallina, G. Milanesi, U. Koszinowski, F. Baldanti, and G. Gerna. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heo, J., S. Petheram, G. Demmler, J. R. Murph, S. P. Adler, J. Bale, and T. E. Sparer. 2008. Polymorphisms within human cytomegalovirus chemokine (UL146/UL147) and cytokine receptor genes (UL144) are not predictive of sequelae in congenitally infected children. Virology 378:86-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humar, A., D. Kumar, C. Gilbert, and G. Boivin. 2003. Cytomegalovirus (CMV) glycoprotein B genotypes and response to antiviral therapy, in solid-organ-transplant recipients with CMV disease. J. Infect. Dis. 188:581-584. [DOI] [PubMed] [Google Scholar]
- 18.Husain, S., C. E. Pietrangeli, and A. Zeevi. 2009. Delayed onset CMV disease in solid organ transplant recipients. Transpl. Immunol. 21:1-9. [DOI] [PubMed] [Google Scholar]
- 19.Huse, S. M., J. A. Huber, H. G. Morrison, M. L. Sogin, and D. M. Welch. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishibashi, K., T. Tokumoto, H. Shirakawa, K. Hashimoto, N. Kushida, T. Yanagida, K. Shishido, K. Aikawa, O. Yamaguchi, H. Toma, K. Tanabe, and T. Suzutani. 2008. Strain-specific seroepidemiology and reinfection of cytomegalovirus. Microbes Infect. 10:1363-1369. [DOI] [PubMed] [Google Scholar]
- 21.Kurz, S. K., and M. J. Reddehase. 1999. Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J. Virol. 73:8612-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lurain, N. S., A. M. Fox, H. M. Lichy, S. M. Bhorade, C. F. Ware, D. D. Huang, S. P. Kwan, E. R. Garrity, and S. Chou. 2006. Analysis of the human cytomegalovirus genomic region from UL146 through UL147A reveals sequence hypervariability, genotypic stability, and overlapping transcripts. Virol. J. 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manuel, O., A. Asberg, X. Pang, H. Rollag, V. C. Emery, J. K. Preiksaitis, D. Kumar, M. D. Pescovitz, A. A. Bignamini, A. Hartmann, A. G. Jardine, and A. Humar. 2009. Impact of genetic polymorphisms in cytomegalovirus glycoprotein B on outcomes in solid-organ transplant recipients with cytomegalovirus disease. Clin. Infect. Dis. 49:1160-1166. [DOI] [PubMed] [Google Scholar]
- 24.Mattick, C., D. Dewin, S. Polley, E. Sevilla-Reyes, S. Pignatelli, W. Rawlinson, G. Wilkinson, P. Dal Monte, and U. A. Gompels. 2004. Linkage of human cytomegalovirus glycoprotein gO variant groups identified from worldwide clinical isolates with gN genotypes, implications for disease associations and evidence for N-terminal sites of positive selection. Virology 318:582-597. [DOI] [PubMed] [Google Scholar]
- 25.McGeoch, D. J., F. J. Rixon, and A. J. Davison. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90-104. [DOI] [PubMed] [Google Scholar]
- 26.Meyer-Konig, U., K. Ebert, B. Schrage, S. Pollak, and F. T. Hufert. 1998. Simultaneous infection of healthy people with multiple human cytomegalovirus strains. Lancet 352:1280-1281. [DOI] [PubMed] [Google Scholar]
- 27.Novak, Z., S. A. Ross, R. K. Patro, S. K. Pati, R. A. Kumbla, S. Brice, and S. B. Boppana. 2008. Cytomegalovirus strain diversity in seropositive women. J. Clin. Microbiol. 46:882-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang, X., A. Humar, and J. K. Preiksaitis. 2008. Concurrent genotyping and quantitation of cytomegalovirus gB genotypes in solid-organ-transplant recipients by use of a real-time PCR assay. J. Clin. Microbiol. 46:4004-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel, R., and C. V. Paya. 1997. Infections in solid-organ transplant recipients. Clin. Microbiol. Rev. 10:86-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pignatelli, S., P. Dal Monte, and M. P. Landini. 2001. gpUL73 (gN) genomic variants of human cytomegalovirus isolates are clustered into four distinct genotypes. J. Gen. Virol. 82:2777-2784. [DOI] [PubMed] [Google Scholar]
- 31.Pignatelli, S., P. Dal Monte, G. Rossini, S. Chou, T. Gojobori, K. Hanada, J. J. Guo, W. Rawlinson, W. Britt, M. Mach, and M. P. Landini. 2003. Human cytomegalovirus glycoprotein N (gpUL73-gN) genomic variants: identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J. Gen. Virol. 84:647-655. [DOI] [PubMed] [Google Scholar]
- 32.Pignatelli, S., P. Dal Monte, G. Rossini, and M. P. Landini. 2004. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev. Med. Virol. 14:383-410. [DOI] [PubMed] [Google Scholar]
- 33.Puchhammer-Stöckl, E., and I. Görzer. 2006. Cytomegalovirus and Epstein-Barr virus subtypes—the search for clinical significance. J. Clin. Virol. 36:239-248. [DOI] [PubMed] [Google Scholar]
- 34.Puchhammer-Stöckl, E., I. Görzer, A. Zoufaly, P. Jaksch, C. C. Bauer, W. Klepetko, and T. Popow-Kraupp. 2006. Emergence of multiple cytomegalovirus strains in blood and lung of lung transplant recipients. Transplantation 81:187-194. [DOI] [PubMed] [Google Scholar]
- 35.Rasmussen, L., A. Geissler, C. Cowan, A. Chase, and M. Winters. 2002. The genes encoding the gCIII complex of human cytomegalovirus exist in highly diverse combinations in clinical isolates. J. Virol. 76:10841-10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddehase, M. J., M. Balthesen, M. Rapp, S. Jonjic, I. Pavic, and U. H. Koszinowski. 1994. The conditions of primary infection define the load of latent viral genome in organs and the risk of recurrent cytomegalovirus disease. J. Exp. Med. 179:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddehase, M. J., C. O. Simon, C. K. Seckert, N. Lemmermann, and N. K. Grzimek. 2008. Murine model of cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 325:315-331. [DOI] [PubMed] [Google Scholar]
- 38.Reeves, M., and J. Sinclair. 2008. Aspects of human cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 325:297-313. [DOI] [PubMed] [Google Scholar]
- 39.Reeves, M. B., P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J. Gen. Virol. 86:2949-2954. [DOI] [PubMed] [Google Scholar]
- 40.Reeves, M. B., P. A. MacAry, P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. U. S. A. 102:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeves, M. B., and J. H. Sinclair. 2010. Analysis of latent viral gene expression in natural and experimental latency models of human cytomegalovirus and its correlation with histone modifications at a latent promoter. J. Gen. Virol. 91:599-604. [DOI] [PubMed] [Google Scholar]
- 42.Simon, C. O., C. K. Seckert, D. Dreis, M. J. Reddehase, and N. K. Grzimek. 2005. Role for tumor necrosis factor alpha in murine cytomegalovirus transcriptional reactivation in latently infected lungs. J. Virol. 79:326-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinclair, J. 2010. Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection. Biochim. Biophys. Acta 1799:286-295. [DOI] [PubMed] [Google Scholar]
- 44.Sitki-Green, D., M. Covington, and N. Raab-Traub. 2003. Compartmentalization and transmission of multiple Epstein-Barr virus strains in asymptomatic carriers. J. Virol. 77:1840-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanton, R., D. Westmoreland, J. D. Fox, A. J. Davison, and G. W. Wilkinson. 2005. Stability of human cytomegalovirus genotypes in persistently infected renal transplant recipients. J. Med. Virol. 75:42-46. [DOI] [PubMed] [Google Scholar]
- 46.Streblow, D. N., S. L. Orloff, and J. A. Nelson. 2007. Acceleration of allograft failure by cytomegalovirus. Curr. Opin. Immunol. 19:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 48.Tarrago, D., C. Quereda, and A. Tenorio. 2003. Different cytomegalovirus glycoprotein B genotype distribution in serum and cerebrospinal fluid specimens determined by a novel multiplex nested PCR. J. Clin. Microbiol. 41:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor-Wiedeman, J., P. Sissons, and J. Sinclair. 1994. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J. Virol. 68:1597-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, C., Y. Mitsuya, B. Gharizadeh, M. Ronaghi, and R. W. Shafer. 2007. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 17:1195-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, D., and T. Shenk. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 79:10330-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zamora, M. R. 2004. Cytomegalovirus and lung transplantation. Am. J. Transplant. 4:1219-1226. [DOI] [PubMed] [Google Scholar]