Abstract
A host genetic variant (−35C/T) correlates with increased human leukocyte antigen C (HLA-C) expression and improved control of HIV-1. HLA-C-mediated immunity may be particularly protective because HIV-1 is unable to remove HLA-C from the cell surface, whereas it can avoid HLA-A- and HLA-B-mediated immunity by Nef-mediated down-modulation. However, some individuals with the protective −35CC genotype exhibit high viral loads. Here, we investigated whether the ability of HIV-1 to replicate efficiently in the “protective” high-HLA-C-expression host environment correlates with specific functional properties of Nef. We found that high set point viral loads (sVLs) were not associated with the emergence of Nef variants that had acquired the ability to down-modulate HLA-C or were more effective in removing HLA-A and HLA-B from the cell surface. However, in individuals with the protective −35CC genotype we found a significant association between sVLs and the efficiency of Nef-mediated enhancement of virion infectivity and modulation of CD4, CD28, and the major histocompatibility complex class II (MHC-II)-associated invariant chain (Ii), while this was not observed in subjects with the −35TT genotype. Since the latter Nef functions all influence the stimulation of CD4+ T helper cells by antigen-presenting cells, they may cooperate to affect both the activation status of infected T cells and the generation of an antiviral cytotoxic T-lymphocyte (CTL) response. In comparison, different levels of viremia in individuals with the common −35TT genotype were not associated with differences in Nef function but with differences in HLA-C mRNA expression levels. Thus, while high HLA-C expression may generally facilitate control of HIV-1, Nef may counteract HLA-C-mediated immune control in some individuals indirectly, by manipulating T-cell function and MHC-II antigen presentation.
The accessory human immunodeficiency virus type 1 (HIV-1) Nef protein is required for the maintenance of high viral loads and thus accelerates disease progression (2, 22). Nef is a myristoylated protein of ∼27 kDa that facilitates viral immune evasion and enhances HIV-1 replication by a variety of functions. For example, Nef reduces the levels of CD4, major histocompatibility complex class I (MHC-I), CD28, and CXCR4 (CXCL12) cell surface expression by recruiting these molecules to the endocytic machinery or by rerouting them to lysosomes for degradation (29). Thus, Nef can modulate the responsiveness of HIV-1-infected T cells to stimulation, protect them against lysis by cytotoxic T lymphocytes (CTL), reduce their migration in response to stromal cell-derived factor 1 (SDF-1), prevent superinfection, and facilitate the release of fully infectious virions. Furthermore, Nef can interfere with MHC-II antigen presentation by up-modulating the invariant chain (Ii) associated with nonfunctional immature MHC-II complexes (39). Finally, Nef interacts with cell signaling pathways to modulate T-cell activation and viral replication and increases the infectivity of progeny virions (22). Thus, Nef is the most versatile of all HIV-1 accessory proteins.
Down-modulation of MHC-I is one of the best-defined Nef activities and protects primary HIV-1-infected T cells from CTL killing (8). Studies in the simian immunodeficiency virus (SIV)/macaque model demonstrated that Nef-mediated MHC-I downregulation provides a selective advantage for viral replication in vivo (27) and attenuates the CD8+ T-cell response (40). Nef also seems to limit the virus-specific CD8+ T-cell response in HIV-1 infection since unusually strong CTL responses have been demonstrated in humans infected with nef-defective HIV-1 strains (11). Furthermore, it has been reported that the ability of Nef to down-modulate MHC-I correlates with the breadth of the CTL response (24) and is impaired in late-stage AIDS patients when the selective pressure exerted by the immune response is reduced (4). Usually, MHC-I down-modulation should render virally infected cells susceptible to natural killer (NK) cells, which preferentially lyse cells lacking such molecules. However, Nef down-modulates only HLA-A and HLA-B, which are recognized by the majority of CTL, but not HLA-C and HLA-E, which also interact with inhibitory NK cell receptors (7). Thus, Nef facilitates viral evasion of both innate and adaptive immune responses by simultaneously preventing NK cell killing and reducing CTL recognition of HIV-1-infected cells. Not surprisingly, this selective nature of Nef-mediated MHC-I down-regulation is conserved among different groups of primate lentiviruses (10, 36).
While the role of Nef in viral immune evasion is well established, the influence of host genetic factors on its functions has not been investigated. Recently, a genome-wide association study identified a single nucleotide polymorphism (SNP) 35 kb upstream of the HLA-C gene (−35C/T) as a major determinant of the level of circulating virus in the plasma during the nonsymptomatic phase preceding the progression to AIDS, referred to here as set point viral loads (sVLs) (12, 13). Individuals homozygous for the minor allele C at this locus (−35CC) had sVLs that were on average 0.8 log lower than those of subjects homozygous for the major allele T (−35TT). A subsequent study showed that the −35C variant is associated with high levels of HLA-C cell surface expression and delayed progression to AIDS (41). Thus, the −35C SNP may be associated with improved control of HIV/AIDS because of improved HLA-C-mediated antigen presentation.
Although the average sVLs in individuals with the −35CC “high-HLA-C-expression” genotype are significantly lower than those in individuals with the −35TT genotype, the distributions overlap. This suggests that some individuals with the −35CC genotype are not able to mount effective HLA-C-mediated immune responses or that the virus “learns” to counteract them. Under normal circumstances, HIV-1 would be expected to be under strong selective pressure against HLA-C down-modulation to avoid NK lysis of infected target cells. However, this could be different in HIV-1-infected individuals with the protective −35CC genotype, who may be capable of mounting significantly enhanced innate as well as adaptive HLA-C-mediated immune responses. To test this hypothesis, we investigated whether high sVLs in individuals with the −35CC genotype are associated with an increased capability of Nef to down-modulate HLA-C. Our results revealed no evidence for such a scenario. Surprisingly, however, nef alleles from subjects with the protective −35CC genotype and high sVLs were significantly more effective in down-modulating CD4, CD28, and CXCR4 and in up-modulating Ii than those from −35CC subjects with low sVLs. No such associations between Nef function and sVLs were observed in individuals with the −35TT genotype. Interestingly, however, we found that individuals with the −35TT genotype and low sVLs expressed levels of HLA-C mRNA that were as high as those detected in individuals with the protective −35CC genotype. These results suggest that HLA-C expression levels are directly contributing to HIV-1 control and that the −35C/T variant represents only an imperfect tagging polymorphism because the levels of HLA-C expression in individuals with the protective −35CC and the susceptible −35TT genotype vary substantially and overlap. In addition, in a subset of individuals with the protective −35CC genotype, Nef seems to counteract immune control by HLA-C, at least to some extent, by indirect mechanisms, targeting MHC-II antigen presentation and CD4+ T-helper-cell function.
MATERIALS AND METHODS
Patient samples.
Plasma and peripheral blood mononuclear cells (PBMCs) were obtained from 49 HIV-1-infected individuals previously included in a genome-wide association study of major determinants for host control of HIV-1 (12). The criteria of patient selection and the definition of sVLs have been described previously (12, 13). In brief, patients were eligible for the study if they had valid seroconversion data and sVLs defined from multiple measurements in the first 3 years after seroconversion in the absence of antiretroviral therapy. All individual data were carefully assessed, and outlier VL data, such as those measured early in infection before the set point was reached or during the late phase of disease when the VL slope significantly increased, were eliminated (12, 13). HIV-1 sVL was defined as the average of all remaining VL results. Patients were further selected based on their genotype (−35TT or −35CC) and the presence of high or low sVLs. Viral load and CD4+ T-cell count data were available from the time point of sample collection. Five individuals were of self-reported African ancestry. To avoid systematic bias between groups due to the influence of protective HLA-I alleles on viral load, we excluded individuals carrying HLA-B*5701 as tagged by the HCP5 rs2395029 SNP described in reference 12. Direct HLA allelic typing results were obtainable for 36 of 49 subjects (see Table S1 in the supplemental material): two of the −35CC individuals had an HLA-B*27, one with low and one with high sVLs.
HLA-C mRNA levels.
Cellular RNA was extracted from uncultured PBMCs of 43 individuals using the RNeasy minikit (Qiagen) and reversely transcribed using the high-capacity cDNA reverse transcription kit (ABI). Raji cells were used to establish the standard curve. TaqMan gene expression assay HLA-C Hs 00762610_s1 (6-carboxyfluorescein [FAM]-minor groove binder [MGB]) and human β-actin endogenous control (VIC-MGB) were used in a real-time PCR for 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Expression analysis was done in triplicate from one cDNA per patient sample.
PCR amplification of nef alleles.
Genomic DNA was extracted from uncultured peripheral blood mononuclear cells (PBMCs) of 46 individuals by using the QIAamp DNA minikit (Qiagen). Viral RNA was extracted from the plasma of 3 individuals (one from each group except for −35TTlow) using the RNeasy minikit (Qiagen), and cDNA was synthesized using the high-capacity reverse transcription kit (ABI). Nef genes were amplified in bulk using nested PCR (HotStarTaq Master Mix kit; Qiagen) and degenerate oligonucleotide primers. Outer primers included SG2069 (5′-ATACCTASAMGAATMAGACARGG-3′) and SG2070 (5′-CTGCTTATATGCAGCATCTGAGGG-3′), while inner primers included SG2067 (5′-TAAMATGGGKRGCAAMTGGTC-3′) and SG2068 (5′-AGCAASYTCKRTGTCAGCAGT-3′) (standard abbreviations are used for positions of base ambiguity). The resulting PCR products were purified using the peqGold Cycle Pure kit (peqLab) and sequenced directly using the inner primers SG2067 and SG2068. Thus, the nef sequences should represent the predominant form present in the respective patients. The authenticity of all bulk nef sequences was confirmed by the analysis of individual nef alleles.
Proviral constructs.
NL4-3 proviral constructs containing primary HIV-1 nef genes followed by an internal ribosome entry site (IRES) and the enhanced green fluorescent protein (eGFP) gene were generated essentially as described previously (31, 34). Briefly, splice-overlap-extension PCR was used to replace the NL4-3 nef gene with the patient-derived HIV-1 nef alleles. PCR fragments containing the 3′ end of the NL4-3 env gene were fused to pools of primary nef genes and cloned into pBR-NL43-IRES-eGFP-nef+ using unique HpaI and MluI sites. Aliquots of transformed Escherichia coli were plated on Luria broth-ampicillin dishes to assess transformation efficiency, as well as being used for direct inoculation of medium-scale plasmid preparations. The percentage of the plasmid population containing a nef insert was estimated by restriction enzyme analysis. To confirm the integrity of the PCR-derived inserts, between 3 and 12 individual proviral HIV-1 NL4-3 nef-IRES-eGFP clones (a total of 224) per patient sample were subjected to sequence analysis.
Phylogenetic analysis.
The amino acids of the newly sequenced HIV-1 Nef alleles were aligned with NA7 (GenBank accession number DQ242535) using ClustalX (42). Sites with a gap in any sequence were eliminated. The tree was constructed using the neighbor-joining method implemented in ClustalX using Kimura's correction and 1,000 bootstrapped replicates.
Cell culture.
Jurkat, THP-1, and 293T cells were cultured as described previously (32, 33). 293T cells were maintained in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum. PBMCs from healthy human donors were isolated using lymphocyte separation medium (Biocoll separating solution; Biochrom), stimulated for 3 days with phytohemagglutinin (PHA; 2 μg/ml), and cultured in RPMI 1640 medium with 10% fetal calf serum (FCS) and 10 ng/ml interleukin-2 (IL-2) prior to infection. CD4+ T cells were isolated from whole blood using the human CD4+ T cell RosetteSep kit following the protocol provided by the manufacturer.
Virus stocks and transductions.
To generate viral stocks, 293T cells were cotransfected with the proviral HIV-1 constructs either alone (for infectivity or replication assays) or together with a plasmid (pHIT-G) expressing the vesicular stomatitis virus G protein (VSV-G) (31). The latter was used to achieve high initial infection levels for flow cytometric analysis. However, all HIV-1 constructs contained intact env genes and were thus replication competent following the first round of infection. The medium was changed after overnight incubation, and virus was harvested 24 h later. Residual cells in the supernatants were pelleted, and the supernatants were stored at −70°C. Virus stocks were quantified using a p24 antigen capture assay provided by the NIH AIDS Research and Reference Reagent Program. All experiments with infectious material were approved by the institutional biosafety officer and performed in a biosafety level 3 facility. Handling of wild-type and pseudotyped HIV-1 was performed under laminar flow hoods by well-trained laboratory personnel using special protective laboratory wear.
Infectivity assays.
Virus infectivity was determined using TZM-bl and P4-CCR5 cells as described previously (26). Briefly, the cells were plated in 96-well-dishes in a volume of 100 μl and infected after overnight incubation with virus stocks containing 0.5 or 5.0 ng of p24 antigen produced by transiently transfected 293T cells. Two days postinfection, viral infectivity was detected using the Gal screen kit from Tropix as recommended by the manufacturer. β-Galactosidase activities were quantified as relative light units per second (RLU/s) using the Orion Microplate luminometer.
Flow cytometric analysis.
CD4, CXCR4, MHC-I, CD28, and eGFP reporter expression in Jurkat T cells, human PBMCs, or purified CD4+ T cells and Ii surface expression by THP-1 cells transduced with HIV-1 (NL4-3) constructs coexpressing Nef and eGFP were measured as described previously (31, 33). CD8, IL-2 receptor (IL-2R), and CD69 expression were measured by standard fluorescence-activated cell sorting (FACS) staining, using CD8 (BD Pharmingen; clone RPA-T8), CD25 (BD Pharmingen; clone M-A251), and CD69 (BD Pharmingen; clone FN50) monoclonal antibodies (MAbs). For quantification of Nef-mediated modulation of specific surface molecules, the levels of receptor expression (red fluorescence) were determined for cells expressing a specific range of eGFP levels. The extent of up- or down-modulation (n-fold) was calculated by dividing the mean fluorescence intensity (MFI) obtained for cells infected with the nef-negative NL4-3 control viruses by the corresponding values obtained for cells infected with viruses coexpressing Nef and eGFP.
PBMC activation and apoptosis.
PBMCs were stimulated with PHA (1 μg/ml) for 3 days, infected with various HIV-1 eGFP/Nef constructs, and subsequently cultured in RPMI 1640 (10% FCS, 10 ng/ml IL-2) for another 2 days. At this time the PBMCs expressed very low levels of CD69 and IL-2R and hence had a resting phenotype. Thereafter, the PBMCs were treated a second time with PHA and CD69 and IL-2R expression levels were measured by FACS analysis 1, 2, or 3 days later. The frequency of virally infected apoptotic cells was determined using the annexin V (AnV) apoptosis detection kit (BD Bioscience) as recommended by the manufacturer.
NFAT induction.
Jurkat cells stably transfected with an NFAT (nuclear factor of activated T cells)-dependent reporter gene vector (16) were either left uninfected or transduced with HIV-1 Nef/eGFP constructs expressing various nef alleles. Except for those cells used as controls, cultures were treated with PHA (1 μg/ml; Murex). Luciferase activity was measured, and n-fold induction was determined by calculating the ratio between measured relative light units of treated samples and those of untreated samples as described previously (16, 32).
Statistical analysis.
The activities of nef alleles derived from HIV-1-infected individuals homozygous for the −35CC genotype and high (n = 19) or low (n = 6) VLs and the control group of patients with the −35TT genotype and high (n = 19) or low (n = 5) VLs were compared using a two-tailed Student t test. The PRISM package version 4.0 (Abacus Concepts, Berkeley, CA) was used for all calculations.
Nucleotide sequence accession numbers.
HIV-1 nef sequences have been submitted to GenBank, and accession numbers are HM244483 to HM244682.
RESULTS
High levels of HLA-C mRNA expression in −35TT individuals with low sVLs.
To investigate a possible association between Nef function and sVLs in subjects with different −35C/T genotypes, we selected 49 HIV-1-infected individuals who were homozygous for either the −35CC protective allele (n = 25) or the more common −35TT allele (n = 24) and amplified their respective nef genes from plasma viral RNA (n = 3) or proviral DNA (n = 46) by using nested PCR (here referred to as “−35CC” and “−35TT” Nefs). These nef alleles were further subdivided into those derived from subjects with high (−35CChi, 104.8 ± 0.3, n = 19, and −35TThi, 105.1 ± 0.6, n = 19) or low (−35CClow, 102.7 ± 0.4, n = 6, and −35TTlow, 102.0.3 ± 0.2, n = 5) sVLs defined as the log10-transformed average copy number of genomic viral RNA per ml plasma during the early chronic phase of infection (12, 13). On average, the sVLs differed by more than 2 orders of magnitude between the “high” and “low” groups (Fig. 1 A). Notably, the HIV-1-infected individuals analyzed are not representative because we included a high number of noncontrollers with the −35CC genotype to increase the chance of identifying nef alleles capable of down-modulating HLA-C. Usually, the majority (62.3%) of −35CC individuals but only a minority (15.1%) of −35TT subjects restrict the viral load to <2,000 RNA copies/ml (41).
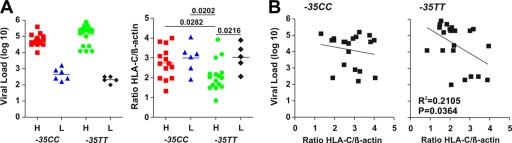
FIG. 1.
Genotype, set point viral loads, and HLA-C mRNA levels of study subjects. (A) Set point viral loads and HLA-C mRNA levels in HIV-1-infected individuals at the time of PBMC or plasma sampling for nef analysis. Patient samples were grouped based on their −35CC or −35TT genotype and the VLs (H, high; L, low) and are color coded: red, −35CChi; blue, −35CClow; green, −35TThi; black, −35TTlow. The levels of HLA-C mRNA represent averages of two measurements. The horizontal bars indicate average numbers per group. (B) Correlation between the sVLs and the levels of HLA-C mRNA expression. The levels of HLA-C transcripts were available for 43 of the 49 HIV-1-infected individuals analyzed.
To verify the genotype-dependent differences in the levels of HLA-C mRNA expression, we quantified HLA-C transcripts in peripheral blood mononuclear cells (PBMCs) of 43 of the 49 individuals by real-time PCR. Our results confirmed that the −35CC genotype is associated with increased levels of HLA-C mRNA expression (−35CC, 2.84 ± 0.16, n = 22, versus −35TT, 2.31 ± 0.18, n = 21; P = 0.0371). Unexpectedly, however, this difference was due only to the low levels of HLA-C transcripts in subjects carrying −35TT alleles with high sVLs (2.1 ± 0.19, n = 16) (Fig. 1A). In contrast, the average HLA-C mRNA levels in −35TT individuals who controlled HIV-1 replication were as high (3.0 ± 0.30, n = 5) as those found in individuals with the protective −35CC genotype. High levels of HLA-C mRNA expression correlated with low sVLs in the −35TT group (Fig. 1B). Thus, in a minority of cases, enhanced HLA-C-mediated immunity may also contribute to the control of HIV-1 in individuals with this most common genotype. In comparison, the levels of HLA-C expression in the high- and low-sVL groups of −35CC individuals were similar and did not correlate with the sVLs, probably because we preferentially selected subjects failing to control HIV-1 for analysis.
High sVLs in −35CC individuals are not associated with differences in Nef-mediated modulation of MHC-I molecules.
For functional analyses, nef amplicons from all 49 patient samples were cloned in bulk into replication-competent HIV-1 NL4-3-based IRES-eGFP constructs coexpressing Nef and eGFP from bicistronic RNAs (30). Bulk nef alleles were used because they reflect the circulating viral quasispecies more accurately than do individual clones. Nonetheless, it remains possible that the ratio of specific nef alleles in some proviral constructs may not be entirely representative for the respective HIV-1-infected individual if the number of viral templates in the initial PCR was low. For quality control, nef genes of 3 to 12 proviral clones (a total of 224) per patient sample were sequenced and phylogenetically analyzed. These studies confirmed that the majority (87%) of proviral constructs contained intact nef genes and that the sequences of the individual nef clones were closely related to those obtained by direct sequencing of the corresponding PCR fragments and formed patient-specific clades (see Fig. S1 in the supplemental material).
An alignment of the consensus Nef amino acid sequences from the 49 patients is shown in Fig. S2 in the supplemental material. As expected, the Nef sequences were highly variable but known functional domains, such as the N-terminal myristoylation signal, the acidic region, the P(xxP)3 motif, the ExxxLL endocytosis motif in the C loop, and a V1H interaction site (17), were conserved. The four groups of Nef alleles differed in the frequencies of specific amino acid residues at various positions but did not exhibit apparent group-specific sequence signatures. To ensure appropriate representation of the nef alleles present in vivo, viral stocks were derived from 50 or more independent transformants per patient sample. Western blot analysis of 293T cells transfected with these proviral constructs showed that all expressed the p24 core antigen, eGFP, and functional Nef proteins (data not shown).
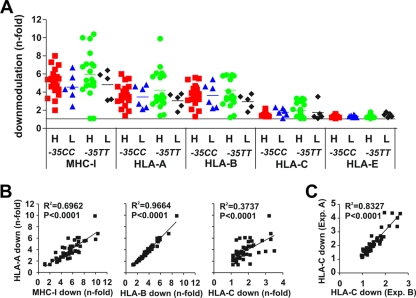
To determine whether nef alleles from the four subject groups differed in their abilities to down-modulate MHC-I, we transduced Jurkat T cells stably transfected with CD8 chimeras containing the intracytoplasmic domain of HLA-A, -B, -C, and -E molecules (10, 36) with the various Nef-expressing proviral constructs. Previous studies have shown that Nef selectively down-regulates HLA-A and HLA-B because of specific amino acid variations in the cytoplasmic domain of MHC-I (7) and that these cell lines represent a useful tool to assess the effect of Nef on the different HLA molecules (10, 36). Flow cytometric analysis showed that nef alleles from all subject groups efficiently down-modulated native MHC-I as well as CD8-HLA-A and CD8-HLA-B chimeras (Fig. 2A). All nef alleles that down-modulated CD8-HLA-A and -B fusions were also active in removing native MHC-I from the surface of Jurkat T cells (Fig. 2B) and PBMCs (see Fig. S3 in the supplemental material). None of the nef alleles reduced surface expression of CD8-HLA-E fusions. However, some Nef proteins were capable of down-modulating CD8-HLA-C, albeit with substantially lower efficiency (up to 3.5-fold; 1.82 ± 0.08, given as mean value ± standard error of the mean [SEM]) than HLA-A (up to 9.9-fold; 3.80 ± 0.23) and HLA-B (up to 8.8-fold; 3.80 ± 0.22). Surprisingly, all seven of these were derived from individuals carrying −35TT alleles who did not show unusually high levels of HLA-C mRNA expression. The modest effects on HLA-C fusions were highly reproducible (Fig. 2C) and confirmed using the 221 B-cell line expressing the Cw4 HLA-C allele (7) (data not shown). Overall, nef alleles from individuals with high sVLs tended to down-modulate MHC-I molecules with greater potency than did those from subjects with low sVLs (Fig. 2A). However, these differences failed to reach significance even when data for MHC-I, HLA-A, and HLA-B modulation were combined (4.48 ± 0.19, n = 114, versus 3.8 ± 0.24, n = 33; P = 0.0525). Thus, Nef proteins from HIV-1-infected individuals with high sVLs are generally capable of efficiently down-modulating HLA-A and -B, but not HLA-C, even in the high-expression −35CC host environment.
FIG. 2.
Modulation of HLA molecules by Nef alleles. (A) Nef-mediated down-modulation of the MHC-I and chimeric CD8 fusion proteins containing the intracytoplasmic domains of HLA-A, -B, -C, and -E on Jurkat cells. HIV-1 nef genes were grouped by VLs (H, high; L, low) and patient genotype and are color coded as in Fig. 1. Each symbol indicates the n-fold down-modulation (average of three measurements) of the indicated receptor molecule by one of the 49 NL4-3 proviral recombinants expressing bulk HIV-1 nef alleles. The horizontal bars indicate average activities per group. (B) Correlation between down-modulation of HLA-A and MHC-I (left), HLA-B (middle), and HLA-C (right). (C) Reproducibility of Nef-mediated down-modulation of CD8-HLA-C fusions in independent experiments.
High sVLs in −35CC individuals are associated with efficient modulation of CD4, CD28, and Ii by Nef.
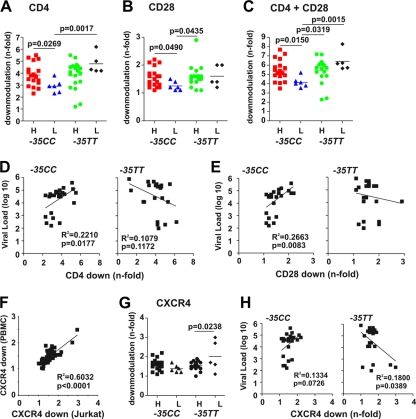
In addition to MHC-I, Nef modulates the surface expression of several other receptors involved in antigen presentation and the interaction of T cells and antigen-presenting cells (APCs). To examine whether the −35C/T SNP or different VLs are associated with differences in these Nef functions, we first examined down-modulation of CD4, one of the best-characterized Nef functions. For this analysis, we used proviral HIV-1 constructs defective in vpu and env since both gene products also reduce CD4 cell surface expression (23) and thus mask the effect of Nef. We found that nef alleles from the −35CChi group were significantly more active in CD4 down-modulation than were those derived from the −35CClow group (3.8 ± 0.2 versus 2.9 ± 0.2; P = 0.027) (Fig. 3A). This was not a general characteristic of subjects with low viral loads since nef alleles from −35TTlow individuals were all highly active in CD4 down-modulation. Similarly, nef alleles from the −35CChi group were also more effective in down-modulating the CD28 costimulatory factor of T-cell activation than were those derived from the −35CClow group (Fig. 3B), while the opposite was observed for nef alleles from subjects with the −35TT genotype. The strongest association between high sVLs, the −35CC genotype, and Nef function was seen when CD4 and CD28 down-modulation were analyzed in combination (Fig. 3C). Finally, the effect of the 49 nef alleles on CD4 and CD28 was also confirmed in PBMCs from three different donors (R2 generally > 0.8, P < 0.0001). In these experiments, the efficiencies of CD4 and CD28 down-modulation by Nef correlated with high sVLs in individuals with the −35CC genotype, whereas the opposite was observed in the −35TT group (Fig. 3D and E). These data suggest that efficient inhibition of costimulatory signals and T-cell activation by Nef is advantageous for effective viral persistence in individuals with the protective −35CC but not the susceptible −35TT genotype.
FIG. 3.
nef-mediated down-modulation of CD4, CD28, and CXCR4. (A to C) Quantitation of Nef-mediated down-modulation of CD4 (A), CD28 (B), and both receptors (C) on PBMCs infected with HIV-1 Nef/eGFP constructs. A vpu- and env-defective HIV-1 backbone was used to measure the effect of Nef on CD4 surface expression. HIV-1 nef genes were grouped by VLs (H, high; L, low) and patient genotype and are color coded as in Fig. 1. Each symbol represents n-fold down-modulation of the indicated receptor molecule by proviral recombinants expressing bulk HIV-1 nef alleles (average values of down-modulation derived from three independent experiments are shown). Panel C shows the sum of CD4 and CD28 down-modulation. The horizontal bars indicate average activities per group. (D and E) Correlation between Nef-mediated down-modulations of CD4 (D) and CD28 (E) and the sVLs in −35CC and −35TT individuals. (F) Correlation between Nef-mediated down-modulation of CXCR4 in Jurkat T cells and in PBMCs. (G) Functional activity of nef alleles derived from the different groups of HIV-1-infected individuals in modulating CXCR4 surface expression. (H) Correlation between Nef-mediated down-modulation of CXCR4 and VLs in −35CC or −35TT individuals.
Nef also down-modulates CXCR4 to inhibit T-cell migration in response to the chemokine stromal cell-derived factor 1 (SDF-1). In agreement with previous results (18), all HIV-1 nef alleles only weakly affected the levels of CXCR4 surface expression in both Jurkat cells and PBMCs (Fig. 3F and data not shown). Increased Nef-mediated down-modulation of CXCR4 was usually associated with low VLs in the −35TT group, whereas the opposite was observed in −35CC individuals (Fig. 3G and H). Although the overall effects of HIV-1 Nef proteins on CXCR4 surface expression were weak, these results indicate that suppression of T-cell migration may be more beneficial for viral persistence in the presence of the −35C allele.
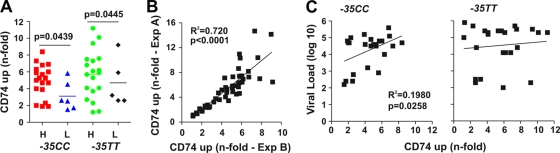
Nef not only manipulates T cells but may also impair antigen presentation by APCs by up-modulating the invariant chain (Ii, CD74) associated with immature MHC-II complexes. Previous studies have shown that up-modulation of Ii requires only low levels of Nef expression and is observed in HIV-1-infected macrophages (30, 34). Flow cytometric analysis showed that the great majority of Nef alleles enhanced Ii surface expression on HIV-1-infected monocytic THP-1 cells (43). Nef alleles from −35CClow individuals were significantly less active in up-modulating Ii than were those obtained from −35CChi subjects (4.8 ± 0.4 versus 3.1 ± 0.6; P = 0.0439) (Fig. 4A and B). Moreover, high sVLs in the −35CC, but not in the −35TT, host environment correlated with the efficiency of Nef-mediated up-modulation of Ii (Fig. 4C). Thus, an increased ability of Nef to impair MHC-II antigen presentation seems to be beneficial for viral persistence in individuals with the −35CC genotype.
FIG. 4.
Nef-mediated up-modulation of Ii. (A) Functional activity of nef alleles derived from different groups of HIV-1-infected individuals (as in Fig. 1) in up-modulating Ii (CD74) surface expression on THP-1 cells. Each symbol represents the average activity of one individual nef allele measured in three independent experiments. The horizontal bars indicate average activities per group. H, high sVL; L, low sVL. (B) Correlation between the efficiencies of Nef-mediated Ii (CD74) up-modulation in two independent experiments. (C) Correlation between the efficiencies of Nef-mediated up-modulation of Ii and VLs in −35CC and −35TT individuals.
High sVLs in −35CC individuals are associated with reduced NFAT activation.
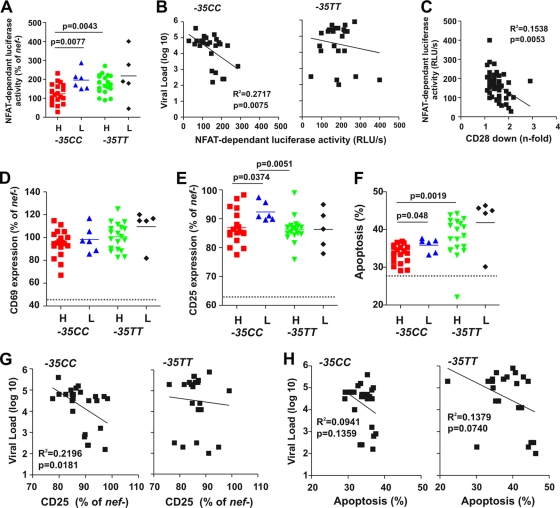
HIV-1 Nef proteins also interact with cellular factors involved in signal transduction and thus modulate the responsiveness of virally infected T cells to stimulation (15, 16, 35). Expression of some HIV-1 Nef proteins is associated with the hyperactivation of the nuclear factor of activated T cells (NFAT) (16), which regulates IL-2 gene expression, a hallmark of T-cell activation. The advantage for the virus is that an increased activation status of the infected cells is associated with increased viral gene expression. However, it may come at the cost of a reduced life span of the infected cell and increased viral antigen presentation and hence vulnerability to CTL lysis. To examine whether the nef alleles from the four subject groups differentially affected T-cell activation, we transduced Jurkat T cells stably transfected with a luciferase reporter gene under the control of an NFAT-dependent promoter (16) with the HIV-1 constructs and measured their responsiveness to activation by PHA. We found that T cells expressing nef alleles derived from −35CChi subjects expressed significantly lower levels of NFAT-dependent luciferase activity than did those derived from −35CClow individuals (124.1 ± 12.1, n = 19, versus 195.3 ± 20.8, n = 6; P = 0.0077) (Fig. 5A). Furthermore, low levels of NFAT correlated with high VLs in −35CC individuals (R2 = 0.2717; P = 0.0075) but not in −35TT subjects (Fig. 5B). Remarkably, primary HIV-1 nef alleles varied substantially in their ability to activate NFAT: the majority rendered the virus-infected T cells hyperresponsive to stimulation as reported elsewhere (16). However, 8 nef alleles from the −35CChi (n = 6) and −35TTlow (n = 2) groups suppressed NFAT activation. To further assess the activation status of the HIV-1-infected Jurkat cells, we also measured the levels of CD69 and CD25 surface expression, well-established markers for early and late T-cell activation. Overall the levels of CD69 and CD25 correlated with those of NFAT activity and inversely with the efficiency of CD28 (but not CD4 or MHC-I) down-modulation by Nef (Fig. 5C and data not shown). Thus, although HIV-1 Nef proteins down-modulate CD28 only up to 3-fold, this seems sufficient to suppress the responsiveness of HIV-1-infected T cells to stimulation. Altogether, these results suggest that reduced levels of NFAT and T-cell activation promote viral replication in −35CC but not in −35TT individuals, possibly because reduced viral gene expression and antigen presentation are advantageous in the protective high-HLA-C-expression environment.
FIG. 5.
Nef-mediated effects on NFAT expression, CD4+ T-cell activation, and apoptosis. (A) Induction of NFAT in T-cell cultures infected with HIV-1 variants expressing nef alleles from HIV-1-infected individuals with different sVLs (H, high; L, low) and the −35C/T genotype. Levels of NFAT-dependent luciferase reporter activity are the averages (±standard deviations [SD]) of triple infections. The horizontal bars indicate average activities per group. Similar results were obtained in two independent experiments. (B) Correlation between the levels of NFAT-dependent luciferase reporter activity and VLs in −35CC and −35TT individuals. (C) Correlation between Nef-mediated down-modulation of CD28 and PHA-induced levels of NFAT-dependent luciferase activities. (D to F) Expression of CD69 (D), CD25 (E), and levels of apoptosis (F) in CD4+ T cells infected with HIV-1 Nef/eGFP constructs. All values represent averages of duplicate experiments. Similar results were obtained in PBMCs. (G and H) Correlation between the levels of CD25 expression (G) and apoptosis (H) and the VLs in −35CC and −35TT individuals.
We also examined the effects of nef alleles on cellular activation and programmed death in human PBMCs infected with the eGFP-expressing HIV-1 constructs. Flow cytometric analysis showed that the levels of CD69 expression did not differ significantly between the four groups (Fig. 5D). In comparison, expression of nef alleles derived from −35CClow individuals was associated with enhanced levels of late T-cell activation, as indicated by increased surface expression of CD25 (Fig. 5E). All nef alleles had similar effects on CD69 and CD25 expression in PBMCs obtained from different donors and in the presence or absence of CD8+ T cells (R2 generally > 0.4; P < 0.0001; n = 4). Nef alleles from −35TT individuals were usually associated with higher percentages of apoptotic cells than were those derived from −35CC subjects (38.6% ± 1.2% versus 34.09% ± 0.5%; P = 0.0008). On average, expression of low-sVL nef genes resulted in higher levels of apoptosis than did those amplified from individuals with high sVLs (Fig. 5F). The levels of CD25 expression correlated with the percentages of apoptotic PBMCs (R2 = 0.3033; P < 0.0001) and higher levels of NFAT induction (R2 = 0.3116; P < 0.0001). It is noteworthy that all nef alleles suppressed the induction of CD69 induction in Jurkat T cells but not in PBMCs (see Fig. S4A in the supplemental material). These results are in agreement with previous data showing that HIV-1 Nef proteins suppress T-cell receptor (TCR) signaling in Jurkat T cells (19), while only HIV-2 and SIV Nef proteins which down-modulate CD3 block early events of T-cell activation in PBMCs (31). Finally, all nef alleles tested suppressed the induction of CD25 (Fig. S4B and data not shown). Altogether, our data showed that high sVLs in −35CC subjects (but not in −35TT subjects) were associated with Nef proteins, resulting in enhanced down-modulation of CD28 (R2 = 0.2264; P = 0.0162) and decreased expression of CD25 (R2 = 0.2196; P = 0.0181) in HIV-1-infected PBMCs (Fig. 5G and data not shown). Furthermore, we observed a nonsignificant trend for an association between high sVLs and reduced levels of apoptosis in both −35CC and −35TT individuals (Fig. 5H).
Nef alleles from −35CC individuals with high sVLs are particularly active in enhancing virion infectivity but not viral spread.
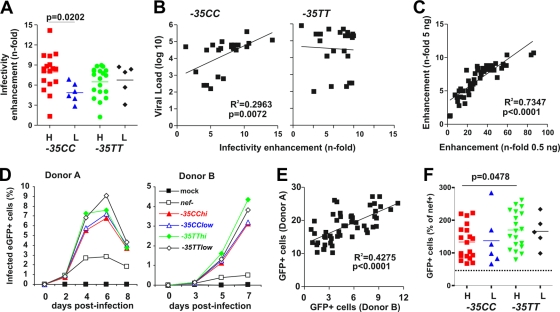
In addition to facilitating viral immune evasion and manipulating T-cell activation, Nef also enhances the infectivity of HIV-1 virions (6, 25, 37). To examine whether differences in sVLs or the −35T/C variant were associated with differences in this Nef function, we infected the HeLa-CD4/LTR-LacZ indicator cell line P4-CCR5 (5, 14) with p24 antigen-normalized viral stocks (5 ng) and determined the β-galactosidase activities 2 days later. All HIV-1 Nef proteins enhanced HIV-1 infectivity, albeit with differential efficiencies (Fig. 6A). On average, −35CChi nef alleles were more active in this function than were those derived from the remaining three groups (8.0 ± 0.7 versus 6.2 ± 0.4; P = 0.0214). Again, effective enhancement of virion infectivity was associated with high VLs in −35CC patients (R2 = 0.2961; P = 0.0073) but not in −35TT subjects (Fig. 6B). The potencies of the nef alleles in promoting HIV-1 infection were similar at a 10-fold-lower infectious dose, although the magnitude of Nef-mediated enhancement was substantially increased (Fig. 6C).
FIG. 6.
Nef-mediated enhancement of virion infectivity and viral spread in PBMCs. (A) P4-CCR5 indicator cells were infected with proviral constructs expressing bulk nef alleles. Infections were performed in triplicate with virus stocks containing 5 ng p24 antigen. Values represent the averages of 3 measurements compared to the infectivity of the virus expressing the NL4-3 Nef (100%). H, high sVL; L, low sVL. (B) Correlation between the infectivities of proviral constructs expressing primary Nef alleles in P4-CCR5 cells and VLs in the respective patients. (C) Correlation between the enhancements of virion infectivity at two different viral doses. (D) Percentages of virally infected GFP+ cell levels detected in PBMCs derived from two donors after infection with proviral constructs expressing nef alleles from the indicated groups of HIV-1-infected individuals. Shown are average values for the entire group of nef alleles (−35CChi, n = 19; −35CClow, n = 6; −35TThi, n = 19; −35TTlow, n = 5). (E) Correlation between the cumulative numbers of HIV-1-infected GFP+ cells detected in PBMC donors A and B. (F) The average cumulative numbers of HIV-1-infected GFP+ cells were measured for each of the 49 proviral constructs investigated. Values are shown relative to the proviral HIV-1 NL4-3 Nef (nef+) construct (100%).
Finally, we examined whether the four groups of nef alleles differ in their abilities to promote viral spread in infected PBMC cultures. The HIV-1 NL4-3 Nef/eGFP constructs are replication competent and have the advantage that both the numbers of virally infected cells and the levels of virus production can be readily determined. HIV-1 strains expressing primary nef alleles were usually spreading with about 3- to 5-fold-higher efficiency in the PBMC cultures than that of the control virus containing a disrupted nef gene (Fig. 6D). On average, nef alleles derived from −35TT individuals were slightly more active in promoting viral spread in PBMCs than those derived from −35CC subjects (169.2 ± 11.3 versus 134.4 ± 11.7, P = 0.0371) (Fig. 6F). This difference remained significant when only nef alleles from patients with high sVLs were included in the analysis (170.1 ± 13.2 versus 133.4 ± 12.1; P = 0.0478) (Fig. 6E). Further analyses showed that the efficiency of the 49 bulk nef alleles in promoting viral spread in PBMC cultures correlated with that of CD4 down-modulation (R2 = 0.1463; P = 0.0067) but not with the enhancement of virion infectivity or the VLs of the patients (data not shown).
DISCUSSION
In the present study we characterized nef genes from 49 HIV-1-infected individuals representing both “protective” −35CC and “susceptible” −35TT HLA-C genotypes who controlled their virus to various degrees. In particular, we examined the capacity of these nef alleles to modulate CD4, CD28, CXCR4, MHC-I, HLA-A, -B, -C, -E, and Ii cell surface expression, T-cell activation, programmed cell death, virion infectivity, and viral replication (summarized in Table S2 in the supplemental material). All nef genes were amplified and cloned in bulk to ensure that they were representative for each patient and examined in the context of a replication-competent HIV-1 provirus in different cell types, including primary PBMCs and CD4+ T cells. Given that functional nef genes are associated with viral loads higher by several orders of magnitude in SIVmac-infected macaques (20) and in HIV-1-infected human individuals (9, 21), we expected to find some associations between Nef function and the levels of viremia regardless of the −35 genotype. Second, we anticipated detecting nef alleles showing an increased capability of down-modulating HLA-C in −35CC subjects who failed to control their virus. Neither of these turned out to be the case. However, detailed functional analyses identified a set of Nef functions associated with high levels of viral replication in subjects with the protective −35CC genotype. Importantly, these Nef functions were not simply an indicator of high viral loads, since these associations were not found in the −35TT group. Moreover, the particular Nef phenotypes did not involve HLA-A and HLA-B down-modulation and thus general CTL control. Instead, they all centered around T-cell activation and APC interaction (indicated in Fig. 7), suggesting that in a subset of −35CC individuals Nef evolved to counteract HLA-C-mediated immune control, albeit by indirect mechanisms.
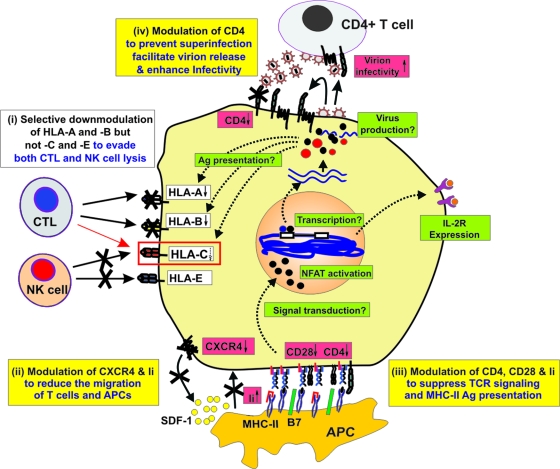
FIG. 7.
Schematic of Nef functions associated with high sVLs in −35CC individuals. Selective down-modulation of HLA-A and HLA-B, but not HLA-C and HLA-E, by Nef was maintained in −35CC people (i). However, high sVLs in the high-HLA-C-expression −35CC host environment were associated with increased modulation of CD4, CD28, CXCR4, and Ii that was most likely to suppress (ii) the migration of HIV-1-infected T cells and APCs and (iii) antigen-dependent T-cell activation via the immunological synapse. As a consequence −35CChi nef alleles may also be particularly effective in modulating downstream signaling events affecting NFAT activation and the transcription of the viral long terminal repeat (LTR) promoter, as well as of cellular genes and activation markers. Nef functions that are increased in −35CC individuals with high sVLs are indicated by red, and those that are not (or may be reduced) are indicated by green boxes. Reported biological consequences of Nef functions associated with high sVLs in the −35CC group are highlighted by yellow boxes. Please note that the proposed in vivo effects of these variations in Nef function are hypothetical.
We initially speculated that Nef variants capable of down-modulating HLA-C may emerge in some HIV-1-infected −35CC individuals to directly counteract HLA-C-induced CTL responses. As shown in Fig. 2, such variants did not arise in the −35CC group. Unexpectedly, however, nef alleles capable of moderate HLA-C down-modulation were observed in several subjects with the −35TT genotype (Fig. 2). Thus, our failure to find nef alleles capable of HLA-C down-modulation in the −35CC group is not due to the fact that Nef is entirely unable to acquire this function. Instead, our finding raises the possibility that reduction of HLA-C from the cell surface may enhance NK lysis of infected cells to an extent that—on balance—this is disadvantageous to the virus. It should be noted, however, that it is currently unclear whether the magnitude of our observed HLA-C modulation is sufficient to influence NK or CTL lysis in vivo.
We observed only a weak association between the efficiency of MHC-I modulation by Nef and high sVLs. At first glance, this seemed surprising since previous studies indicate a clear role of MHC-I down-modulation by Nef in viral immune evasion (1, 27, 40). However, our results are in agreement with the finding that the potency of Nef-mediated MHC-I down-modulation in vitro correlates with the breadth of the HIV-1-specific CTL response but not with the levels of plasma viremia in HIV-1-infected individuals (24). One likely explanation for this is that the selective pressures affecting Nef-mediated MHC-I modulation are rather complex. Whereas other Nef functions, such as down-modulation of CD4 or enhancement of viral infectivity, are most likely always advantageous, the impact of MHC-I modulation is dependent on the host's CTL response. For example, lack of Nef-mediated MHC-I down-modulation may be associated with high sVLs in the absence of an effective CTL response (4, 24, 28). However, Nef is also targeted by CTL (3) and escape mutations may reduce this protein's ability to remove MHC-I from the surface and increase the susceptibility of virus-infected cells to CTL lysis (1, 44). Many Nef proteins analyzed in the present study exhibited variations in known CTL epitopes (data not shown). Thus, ineffective Nef-mediated down-modulation of MHC-I may result from either effective or ineffective CTL responses and hence be associated with particularly effective or ineffective immune control and differential sVLs.
Although the protective −35CC host environment was not associated with enhanced down-modulation of HLA-C, we found that several Nef functions known to influence the interaction of APCs with CD4+ helper T cells were enhanced in −35CC (but not −35TT) individuals who failed to control their virus (Fig. 7). Specifically, high sVLs in the −35CC host genetic background were associated with increased Nef-mediated down-modulation of CD4, CD28, and CXCR4 and up-modulation of Ii. Down-modulation of CD4 may negatively affect APC-T-cell interaction because CD4 is associated with the TCR-CD3 complex and binds a section of the MHC molecule (45). Lack of the CD28 costimulatory signal will prevent lasting T-cell activation and may render the HIV-1-infected T cells anergic and hence nonresponsive to further stimulation. CXCR4 down-modulation inhibits T-cell migration in response to SDF-1 (17), and high surface levels of Ii (or CD74) impair MHC-II antigen presentation by APCs (39). Most nef alleles from the −35CC group that were highly active in CD4 down-modulation were also highly active in modulating CD28, CXCR4, and Ii and in promoting viral replication but not in enhancing virion infectivity or down-modulating MHC-I. Notably, the correlation with high sVLs in the −35CC group became substantially more significant when we used cumulative or average CD4, CD28, CXCR4, and Ii modulation values for calculation (Fig. 3C and data not shown). This is expected since the combined effect of Nef on all four receptors should be a better indicator of the functional alterations of APCs and T-cell interactions than the individual activities (schematically indicated in Fig. 7). Whether or not the cooperative effect of several modest differences in individual Nef functions significantly affects T-cell function, antigen presentation, and evasion of HLA-C-mediated immune control in vivo remains elusive. Nonetheless, our data suggest that the ability of Nef to interfere with the initiation and maintenance of an effective antiviral immune response as well as antigen-dependent T-cell stimulation is particularly important for the maintenance of high viral loads in the “high HLA-C” −35CC host environment. Thus, it is tempting to hypothesize that HIV-1 evades HLA-C-mediated immune control indirectly, by manipulation of immune functions critical for the development of cellular immune responses.
Other Nef features associated with high sVLs in −35CC individuals were reduced levels of NFAT activation and CD25 expression and increased virion infectivity. The first two phenotypes correlated inversely with Nef's ability to down-modulate CD28. Thus, they are likely a consequence of increased Nef-mediated suppression of T-cell activation. Lower levels of T-cell activation and NFAT expression should be associated with decreased viral gene expression (38). In agreement with this, nef genes from −35CChi individuals were on average less active in promoting viral spread in PBMC cultures, although as a group they enhanced virion infectivity with higher efficiency than did nef alleles from −35TT subjects (Fig. 6). The efficiency of virus spread in vivo depends on both the number and infectiousness of progeny virions produced by the infected cells. Thus, increased Nef-mediated enhancement of virion infectivity may be associated with high sVLs in the −35CC host background because it compensates for the reduced virus production, although larger sample sets need to be analyzed to further assess this possibility.
Enhanced modulation of CD4, CD28, CXCR4, and Ii was not associated with high sVLs in −35TT individuals. Instead, we found that the levels of HLA-C mRNA in −35TT individuals with low sVLs were significantly higher than those in subjects with the same genotype and high sVLs (Fig. 1). The finding that the average levels of HLA-C mRNA expression are higher in individuals with the −35CC genotype than in those with −35TT but overlap and vary substantially within each group is in agreement with published data (41). Notably, the individual in the −35TT high-sVL group showing the highest level of HLA-C mRNA expression most likely failed to control HIV-1 because he was homozygous at all three HLA class I loci (data not shown). Our results suggest that high levels of HLA-C expression may be protective irrespectively of the −35 genotype. Indeed, the −35 variant is unlikely to directly alter HLA-C expression but rather behaves as an imperfect proxy for HLA-C expression. It is conceivable that we did not observe a significant correlation between the levels of HLA-C expression and sVLs in the −35CC group because we preferentially analyzed the minority of individuals with this genotype who failed to control HIV. Usually, the majority (62.3%) of individuals with the −35CC “high-HLA-C-expression” genotype (but only 15.1% with −35TT) efficiently control HIV (41). Thus, only in exceptional cases does HIV-1 seem capable of evading improved HLA-C-mediated immune control by acquiring changes in Nef or elsewhere. Most likely, our selection criteria allowed identification both of the rare events that may allow HIV-1 (at least in part) to evade the improved immune control associated with high levels of HLA-C expression and of some individuals expressing high levels of HLA-C despite their “susceptible” −35TT genotype.
In conclusion, our results suggest that high levels of HLA-C expression may facilitate control of HIV irrespectively of the −35 genotype. Furthermore, we show that the impact of specific Nef functions on HIV-1 replication depends, at least in part, on the genetic properties of the infected host. In the “protective” high-HLA-C −35CC host environment effective immune evasion functions of Nef seem critical for efficient virus replication. Since HIV-1 cannot antagonize HLA-C-mediated immune control directly, it may achieve this (in some individuals) by indirect mechanisms, such as interfering with T-cell function (CD4, CD28, and CXCR4 down-modulation) and MHC-II antigen presentation (Ii up-modulation). In this context, it is tempting to speculate that the combined effects of Nef-mediated modulation of CD4, CD28, CXCR4, and Ii on T-cell and APC function in HIV-1-infected individuals in vivo are underestimated by measuring individual Nef functions in vitro. Finally, Nef variants capable of efficient down-modulation of HLA-C did not emerge even in a host environment expected to favor the selection of such variants. Effective Nef-mediated counteraction of improved HLA-C-mediated immunity by complex indirect mechanisms is obviously the exception because most individuals with the −35CC genotype efficiently control HIV (41). Thus, future AIDS vaccine approaches should be geared toward inducing potent HLA-C-mediated immune responses since they seem less susceptible to effective direct viral evasion strategies.
Supplementary Material
Acknowledgments
We thank Thomas Mertens for support, Nicola Schrott, Martha Mayer, Kerstin Regensburger, and Daniela Krnavek for excellent technical assistance, and Ingrid Bennett for critical readings of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft, the Swiss National Science Foundation, and Public Health Service grants AI067057 (F.K.), AI63993 (D.T.E.), AI71306 (D.T.E.), and AI067854 (B.H.H. and D.B.G.) from the National Institutes of Health. D.T.E. is an Elizabeth Glaser Scientist supported by the Elizabeth Glaser Pediatric AIDS Foundation. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 12 May 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ali, A., S. Pillai, H. Ng, R. Lubong, D. D. Richman, B. D. Jamieson, Y. Ding, M. J. McElrath, J. C. Guatelli, and O. O. Yang. 2003. Broadly increased sensitivity to cytotoxic T lymphocytes resulting from Nef epitope escape mutations. J. Immunol. 171:3999-4005. [DOI] [PubMed] [Google Scholar]
- 2.Ariën, K. K., and B. Verhasselt. 2008. HIV Nef: role in pathogenesis and viral fitness. Curr. HIV Res. 6:200-208. [DOI] [PubMed] [Google Scholar]
- 3.Brumme, Z. L., C. J. Brumme, D. Heckerman, B. T. Korber, M. Daniels, J. Carlson, C. Kadie, T. Bhattacharya, C. Chui, J. Szinger, T. Mo, R. S. Hogg, J. S. Montaner, N. Frahm, C. Brander, B. D. Walker, and P. R. Harrigan. 2007. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 3:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carl, S., T. C. Greenough, M. Krumbiegel, M. Greenberg, J. Skowronski, J. L. Sullivan, and F. Kirchhoff. 2001. Modulation of different human immunodeficiency virus type 1 Nef functions during progression to AIDS. J. Virol. 75:3657-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charneau, P. G., P. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 24:651-662. [DOI] [PubMed] [Google Scholar]
- 6.Chowers, M. Y., C. A. Spina, T. J. Kwoh, N. J. Fitch, D. D. Richman, and J. C. Guatelli. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J. Virol. 68:2906-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, G. B., R. T. Gandhi, D. M. Davis, O. Mandelboim, B. K. Chen, J. L. Strominger, and D. Baltimore. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661-671. [DOI] [PubMed] [Google Scholar]
- 8.Collins, K. L., B. K. Chen, S. A. Kalams, B. D. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 9.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. McPhee, A. L. Greenway, A. Ellett, C. Chatfield, V. A. Lawson, S. Crowe, A. Maerz, S. Sonza, J. Learmont, J. S. Sullivan, A. Cunningham, D. Dwyer, D. Dowton, and J. Mills. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988-991. [DOI] [PubMed] [Google Scholar]
- 10.DeGottardi, M. Q., A. Specht, B. Metcalf, A. Kaur, F. Kirchhoff, and D. T. Evans. 2008. Selective downregulation of rhesus macaque and sooty mangabey major histocompatibility complex class I molecules by Nef alleles of simian immunodeficiency virus and human immunodeficiency virus type 2. J. Virol. 82:3139-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyer, W. B., G. S. Ogg, M. A. Demoitie, X. Jin, A. F. Geczy, S. L. Rowland-Jones, A. J. McMichael, D. F. Nixon, and J. S. Sullivan. 1999. Strong human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte activity in Sydney Blood Bank Cohort patients infected with nef-defective HIV type 1. J. Virol. 73:436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fellay, J., K. V. Shianna, D. Ge, S. Colombo, B. Ledergerber, M. Weale, K. Zhang, C. Gumbs, A. Castagna, A. Cossarizza, A. Cozzi-Lepri, A. De Luca, P. Easterbrook, P. Francioli, S. Mallal, J. Martinez-Picado, J. M. Miro, N. Obel, J. P. Smith, J. Wyniger, P. Descombes, S. E. Antonarakis, N. L. Letvin, A. J. McMichael, B. F. Haynes, A. Telenti, and D. B. Goldstein. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fellay, J., D. Ge, K. V. Shianna, S. Colombo, B. Ledergerber, E. T. Cirulli, T. J. Urban, K. Zhang, C. E. Gumbs, J. P. Smith, et al. 2009. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 5:e1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenard, D., G. Lambeau, E. Valentin, J. C. Lefebvre, M. Lazdunski, and A. Doglio. 1999. Secreted phospholipases A2, a new class of HIV inhibitors that block virus entry into host-cells. J. Clin. Invest. 104:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenard, D., W. Yonemoto, C. de Noronha, M. Cavrois, S. A. Williams, and W. C. Greene. 2005. Nef is physically recruited into the immunological synapse and potentiates T cell activation early after TCR engagement. J. Immunol. 175:6050-6057. [DOI] [PubMed] [Google Scholar]
- 16.Fortin, J. F., C. Barat, Y. Beauséjour, B. Barbeau, and M. J. Tremblay. 2004. Hyper-responsiveness to stimulation of human immunodeficiency virus-infected CD4+ T cells requires Nef and Tat virus gene products and results from higher NFAT, NF-kappaB, and AP-1 induction. J. Biol. Chem. 279:39520-39531. [DOI] [PubMed] [Google Scholar]
- 17.Geyer, M., O. T. Fackler, and B. M. Peterlin. 2001. Structure-function relationships in HIV-1 Nef. EMBO Rep. 2:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrecka, K., T. Swigut, M. Schindler, F. Kirchhoff, and J. Skowronski. 2005. Nef proteins from diverse groups of primate lentiviruses down-modulate CXCR4 to inhibit migration to the chemokine stromal derived factor 1. J. Virol. 79:10650-10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iafrate, A. J., S. Bronson, and J. Skowronski. 1997. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 16:673-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kestler, H. W., D. J. Ringler, K. Mori, D. L. Panicali, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651-662. [DOI] [PubMed] [Google Scholar]
- 21.Kirchhoff, F., T. C. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Absence of intact nef sequences in a long-term, nonprogressing survivor of HIV-1 infection. N. Engl. J. Med. 332:228-232. [DOI] [PubMed] [Google Scholar]
- 22.Kirchhoff, F., M. Schindler, A. Specht, N. Arhel, and J. Münch. 2008. Role of Nef in primate lentiviral immunopathogenesis. Cell. Mol. Life Sci. 65:2621-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lama, J. 2003. The physiological relevance of CD4 receptor down-modulation during HIV infection. Curr. HIV Res. 1:167-184. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, M. J., A. Balamurugan, A. Ohno, S. Kilpatrick, H. L. Ng, and O. O. Yang. 2008. Functional adaptation of Nef to the immune milieu of HIV-1 infection in vivo. J. Immunol. 180:4075-4081. [DOI] [PubMed] [Google Scholar]
- 25.Miller, M. D., M. T. Warmerdam, I. Gaston, W. C. Greene, and M. B. Feinberg. 1994. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J. Exp. Med. 179:101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Münch, J., D. Rajan, M. Schindler, A. Specht, E. Rücker, F. J. Novembre, E. Nerrienet, M. C. Müller-Trutwin, M. Peeters, B. H. Hahn, and F. Kirchhoff. 2007. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J. Virol. 81:13852-13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Münch, J., N. Stolte, D. Fuchs, C. Stahl-Hennig, and F. Kirchhoff. 2001. Efficient class I major histocompatibility complex down-regulation by simian immunodeficiency virus Nef is associated with a strong selective advantage in infected rhesus macaques. J. Virol. 75:10532-10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel, P. G., M. T. Kimata, J. E. Biggins, J. M. Wilson, and J. T. Kimata. 2002. Highly pathogenic simian immunodeficiency virus mne variants that emerge during the course of infection evolve enhanced infectivity and the ability to downregulate CD4 but not class I major histocompatibility complex antigens. J. Virol. 76:6425-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roeth, J. F., and K. L. Collins. 2006. Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways. Microbiol. Mol. Biol. Rev. 70:548-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schindler, M., J. Münch, and F. Kirchhoff. 2005. Human immunodeficiency virus type 1 inhibits DNA damage-triggered apoptosis by a Nef-independent mechanism. J. Virol. 79:5489-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindler, M., J. Münch, O. Kutsch, H. Li, M. L. Santiago, F. Bibollet-Ruche, M. C. Müller-Trutwin, F. J. Novembre, M. Peeters, V. Courgnaud, E. Bailes, P. Roques, D. L. Sodora, G. Silvestri, P. M. Sharp, B. H. Hahn, and F. Kirchhoff. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055-1067. [DOI] [PubMed] [Google Scholar]
- 32.Schindler, M., J. Schmökel, A. Specht, H. Li, J. Münch, M. Khalid, D. L. Sodora, B. H. Hahn, G. Silvestri, and F. Kirchhoff. 2008. Inefficient Nef-mediated down-modulation of CD3 and MHC-I correlates with loss of CD4+ T cells in natural SIV infection. PLoS Pathog. 4:e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindler, M., S. Wildum, N. Casartelli, M. Doria, and F. Kirchhoff. 2007. Nef alleles from children with non-progressive HIV-1 infection modulate MHC-II expression more efficiently than those from rapid progressors. AIDS 21:1103-1107. [DOI] [PubMed] [Google Scholar]
- 34.Schindler, M., S. Würfl, P. Benaroch, T. C. Greenough, R. Daniels, P. Easterbrook, M. Brenner, J. Münch, and F. Kirchhoff. 2003. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J. Virol. 77:10548-10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrager, J. A., and J. W. Marsh. 1999. HIV-1 Nef increases T cell activation in a stimulus-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 96:8167-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Specht, A., M. Q. DeGottardi, M. Schindler, B. Hahn, D. T. Evans, and F. Kirchhoff. 2008. Selective down-modulation of HLA-A and -B by Nef alleles from different groups of primate lentiviruses. Virology 373:229-237. [DOI] [PubMed] [Google Scholar]
- 37.Spina, C. A., T. J. Kwoh, M. Y. Chowers, J. C. Guatelli, and D. D. Richman. 1994. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J. Exp. Med. 179:115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson, M. 2003. HIV-1 pathogenesis. Nat. Med. 9:853-860. [DOI] [PubMed] [Google Scholar]
- 39.Stumptner-Cuvelette, P., S. Morchoisne, M. Dugast, S. Le Gall, G. Raposo, O. Schwartz, and P. Benaroch. 2001. HIV-1 Nef impairs MHC class II antigen presentation and surface expression. Proc. Natl. Acad. Sci. U. S. A. 98:12144-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swigut, T., L. Alexander, J. Morgan, J. Lifson, K. G. Mansfield, S. Lang, R. P. Johnson, J. Skowronski, and R. Desrosiers. 2004. Impact of Nef-mediated downregulation of major histocompatibility complex class I on immune response to simian immunodeficiency virus. J. Virol. 78:13335-13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas, R., R. Apps, Y. Qi, X. Gao, V. Male, C. O'hUigin, G. O'Connor, D. Ge, J. Fellay, J. N. Martin, J. Margolick, J. J. Goedert, S. Buchbinder, G. D. Kirk, M. P. Martin, A. Telenti, S. G. Deeks, B. D. Walker, D. Goldstein, D. W. McVicar, A. Moffett, and M. Carrington. 2009. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat. Genet. 41:1290-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuchiya, S., M. Yamabe, Y. Yamaguchi, Y. Kobayashi, T. Konno, and K. Tada. 1980. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26:171-176. [DOI] [PubMed] [Google Scholar]
- 44.Ueno, T., C. Motozono, S. Dohki, P. Mwimanzi, S. Rauch, O. T. Fackler, S. Oka, and M. Takiguchi. 2008. CTL-mediated selective pressure influences dynamic evolution and pathogenic functions of HIV-1 Nef. J. Immunol. 180:1107-1116. [DOI] [PubMed] [Google Scholar]
- 45.Weiss, A., and D. R. Littman. 1994. Signal transduction by lymphocyte antigen receptors. Cell 76:263-274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.