Abstract
We have utilized electron paramagnetic resonance spectroscopy to study secondary structure, subunit interaction, and molecular orientation of vimentin molecules within intact intermediate filaments and assembly intermediates. Spectroscopy data prove α-helical coiled-coil structures at individual amino acids 316–336 located in rod 2B. Analysis of positions 305, 309, and 312 identify this region as conforming to the helical pattern identified within 316–336 and thus demonstrates that, contrary to some previous predictions, this region is in an α-helical conformation. We show that by varying the position of the spin label, we can identify both intra- and inter-dimer interactions. With a label attached to the outside of the α-helix, we have been able to measure interactions between positions 348 of separate dimers as they align together in intact filaments, identifying the exact point of overlap. By mixing different spin-labeled proteins, we demonstrate that the interaction at position 348 is the result of an anti-parallel arrangement of dimers. This approach provides high resolution structural information (<2 nm resolution), can be used to identify molecular arrangements between subunits in an intact intermediate filament, and should be applicable to other noncrystallizable filamentous systems as well as to the study of protein fibrils.
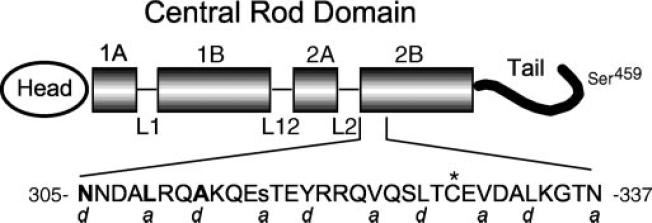
Several human diseases have been traced to point mutations in human IF1 genes (1–5). However, the mechanism(s) by which mutations produce disease has not been elucidated because the crystal structure has not been solved for any IF protein (6), although partial sequences have been solved (7, 8). Primary sequence analysis suggests that all IF proteins have a central rod domain composed principally of what is predicted to be an α-helical sequence flanked by variably sized head and tail domains (Fig. 1 shows a schematic of the IF protein vimentin) (9–11). In rare instances, the tail may be absent (12, 13). The central rod domain is relatively uniform in size (about 310 amino acids) and format, consisting of four coil domains separated by three short linker domains (14–16). The former are mainly characterized by a primary sequence containing a heptad repeat pattern (designated as residues a–g), a signature for an α-helical coiled-coil dimer (17, 18). In this motif, the a and d positions are usually occupied by hydrophobic side chains, which promote the coiled association between two parallel α-helices. Circular dichroism studies of IFs confirm the presence of substantial levels of α-helical content, but without a solved crystal structure the exact identification of α-helical and non-helical regions within the central rod domain is imprecise.
FIG.1. Vimentin schematic.
The predicted vimentin molecule is shown diagrammatically, with the central rod domain emphasized. α-helical rod subdomains 1A, 1B, 2A, and 2B are shown as shaded boxes. Hypothesized non-helical linker regions L1, L12, and L2 are drawn as thin lines. The region of rod subdomain 2B subject to study is expanded, and the sequence of this region is shown in single-letter amino acid abbreviations. Positions 305, 309, and 312 are bold; position 316 is lowercase. An asterisk marks the site of the single endogenous cysteine. Letters a and d below the amino acid sequence represent positions within individual heptad repeats in this area. The location of Ser459 is abstractly indicated near the end of the vimentin tail.
Extensive cross-linking studies have been conducted to determine the arrangement of proteins in IFs (19, 20). Although data have been generated that are consistent with the initial formation of an in-parallel and in-register dimer and the subsequent assembly of these dimers, the interpretation of such data is not always straightforward and must be made cautiously. For example, cross-links between Cys328 of vimentin have been interpreted as occurring between chains within a dimer (intra-dimer), as well as between chains of separate dimers (inter-dimer) (21, 22).
These same issues, protein solubility and the inability to achieve crystallization, have plagued efforts to illuminate the molecular structure and interactions of membrane proteins. However, discrete structural information of membrane protein architecture has been achieved through site-directed spin labeling (SDSL) and electron paramagnetic resonance (EPR). We report here the use of these techniques to ask specific questions about the architecture of IF proteins and the arrangement of proteins within IFs.
Generally, SDSL is performed by engineering a unique cysteine amino acid into the peptide chain at a chosen position and modifying the free sulfhydryl with a thiol-specific nitroxide spin label. Subsequent EPR spectra of the spin-labeled side chain can be interpreted to provide information relevant to the relationship of protein structure and function (23, 24). Side chain dynamics measured from the EPR line shape data reflect backbone and contact-limited motions, information used to elucidate local secondary structure as well as protein folding and assembly (25). Furthermore, the interaction between site-directed spin labels is used to determine spatial relationships in proteins, information required to solve structure at the tertiary and quaternary levels. Because of its sensitivity and capability for real-time measurements under physiological conditions, EPR spectroscopy is particularly attractive for investigating conformational dynamics and mapping protein associations (24).
The ocular lens is composed of a large central mass of fiber cells expressing the IF proteins vimentin, filensin, and phakinin (CP49) surrounded by a single layer of lens epithelial cells expressing the IF protein vimentin (26). Vimentin is homopolymeric, whereas filensin and CP49 are copolymers. A mouse vimentin knockout displayed no obvious phenotype, but mutations in the fiber cell-specific protein CP49 can cause cataracts in humans (27–29). As a precursor to the analysis of CP49 and filensin structure and assembly, we performed experiments with the homopolymeric IF protein vimentin as a model system. This report documents the utility of an SDSL-EPR-based approach to study IF assembly, documenting α-helical coiled-coil structures at individual amino acids and identifying arrangements of molecules within an intact filament.
MATERIALS AND METHODS
Vimentin was produced by bacterial expression using pET vectors and isopropyl-β-D-thiogalactopyranoside induction; the vimentin expression construct was generously provided by Roy Quinlan (University of Durham, Durham, UK). Mutants were created with a Stratagene QuikChange kit and verified by DNA sequencing. Inclusion bodies were purified from bacteria using lysozyme/DNase (30) and were subject to high and low salt washes. Purified inclusion bodies were dissolved in 8 M urea and chromatographed over a SuperDex gel filtration column using a fast protein liquid chromatography system (Amersham Biosciences); fractions were analyzed by SDS-PAGE, and peak fractions were pooled. Purified proteins were incubated in 100 micromolar TCEP (tris-(2-carboxyethyl)phosphine, hydrochloride; Molecular Probes, Eugene, OR) followed by spin labeling with 500 micromolar O-87500 ((1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate-d15[MTSL-d15]; Toronto Research Chemicals, Toronto, Canada). The spin-labeled protein was separated from unincorporated label by chromatography over a CM-Sepharose column. All purified proteins were stored at −80 °C. Filaments were assembled either by dialysis of labeled proteins in a stepwise fashion (31) and aliquots removed at different stages for EPR analysis or by single step dialysis overnight (32). EPR measurements were carried out in a JEOL X-band spectrometer fitted with a loop-gap resonator (33). An aliquot of purified, spin-labeled protein (5 μl) at a final concentration of approximately 100 μM protein was placed in a sealed quartz capillary contained in the resonator. Spectra of samples at room temperature (20–22 °C) were obtained by a single 60-s scan over 100 G at a microwave power of 2 milliwatts and a modulation amplitude optimized to the natural line width of 1 G, as described previously (34). Electron microscopy was performed as described by Quinlan and co-workers (31).
RESULTS
Structural Differences between the Central Rod and Carboxyl Tail Domains Revealed by EPR Spectra
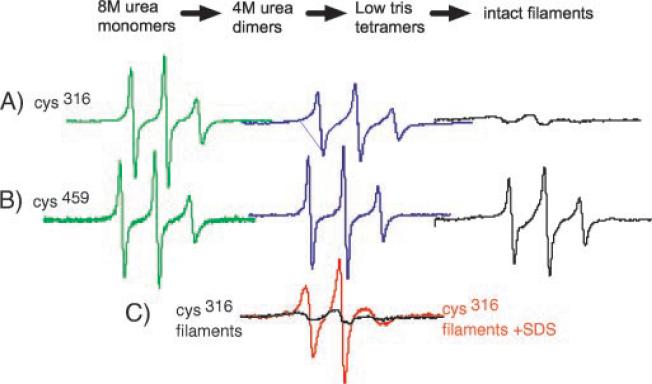
Fig. 1 shows the hypothesized domain structure of vimentin and the region studied. Initial experiments were performed to compare the spectra of samples labeled within the central rod or carboxyl terminal tail domains followed by assembly into filaments. Consistent with current models, EPR spectra recorded from mutant vimentin labeled either within the central rod domain at position 316 (Cys316) or the tail domain at position 459 (Cys459) reveal striking differences in the local structure of these distinct domains (Fig. 2). When unfolded in 8 M urea, the spectra from mutant Cys316 and Cys459 proteins are identical, with sharp narrow peaks (Fig. 2, green spectra). Upon reduction of urea concentration from 8 to 4 M, corresponding to the assembly of dimers, the spectrum of the label attached to the central rod domain (Cys316) becomes more anisotropic, reflecting the adoption of a more ordered structure (Fig. 2A, blue trace) (24, 35). In the assembled filament, the motional freedom of the Cys316 side chain is greatly reduced, as revealed by its highly anisotropic spectrum (Fig. 2A, black trace).
FIG.2. EPR analysis of assembly.
In vitro assembly of intermediate filaments occurs through various intermediates, starting with soluble monomers denatured in 8 M urea. EPR spectra can be recorded at any stage; shown are spectra recorded from samples in 8 M urea (green, left), 4 M urea (blue, middle), and assembled filaments (black, right). EPR spectra were recorded from vimentin samples spin-labeled within the central rod (Cys316, Ser328) (A) or near the end of the tail (Ser328, Cys459) (B). C shows the effect of SDS on the intermediate filament structure as reported by position 316. Each spectrum represents the identical number of spins from the same sample, pre- and post-SDS treatment. Interaction between spins in assembled filaments results in a spectrum that is nearly flat (black line). Addition of SDS destroys the IF structure and results in the spectrum with sharp peaks (red line).
In contrast, the spectra from vimentin labeled at Cys459 in the tail domain reveals a location that retains a high level of mobility, even in the assembled IF (Fig. 2B, black trace). The sharp peaks of the spectra clearly demonstrate a high level of spin label mobility, indicating a lack of rigid structure. Thus, these two distinct spin-labeled vimentin samples demonstrate that the EPR line shapes can 1) resolve distinct dynamic states associated with specific stages of the assembly process and 2) identify secondary structural features unique to specific IF domains.
The severe broadening of the Cys316 intact filament spectra shown in Fig. 2A suggests a magnetic interaction between probes within the assembled filament. Fig. 2C demonstrates how a substantial interaction between labels can be readily identified. The black line displays the EPR spectrum of intact filaments spin-labeled at position 316. The spectrum of the same sample was then collected following addition of SDS (red trace), which disrupts the assembled filament into detergent-solubilized subunits. The large differences in the spectra, where the protein concentration is identical, reveals the extent of interaction between labels. Integration of the SDS-treated sample provides an accurate measure of the total number of spin labels present in the sample. This value can then be used to normalize the intensities between samples according to the total number of spins determined in the presence of SDS.
Analysis of the Amino Terminus of Rod 2B
To test the hypothesis that the initial stage of vimentin assembly is a coiled-coil dimer and to establish whether this dimer was arranged in- or anti-parallel, in-register, or out of register, we engineered a series of single-cysteine mutants at each position from 323 to 336 (as shown in Fig. 1). This sequence spans two contiguous heptad repeats in a region predicted to be rod domain 2B (11, 14, 16). Each mutant contained a single introduced cysteine residue with the wild type cysteine replaced by serine. Mutant proteins were produced by bacterial expression, solubilized in urea, purified, and then spin-labeled. Filament assembly was performed either by single step dialysis overnight (32) or multistep dialysis over several days (31). Assembly into native-looking filaments was confirmed by electron microscopy (31).
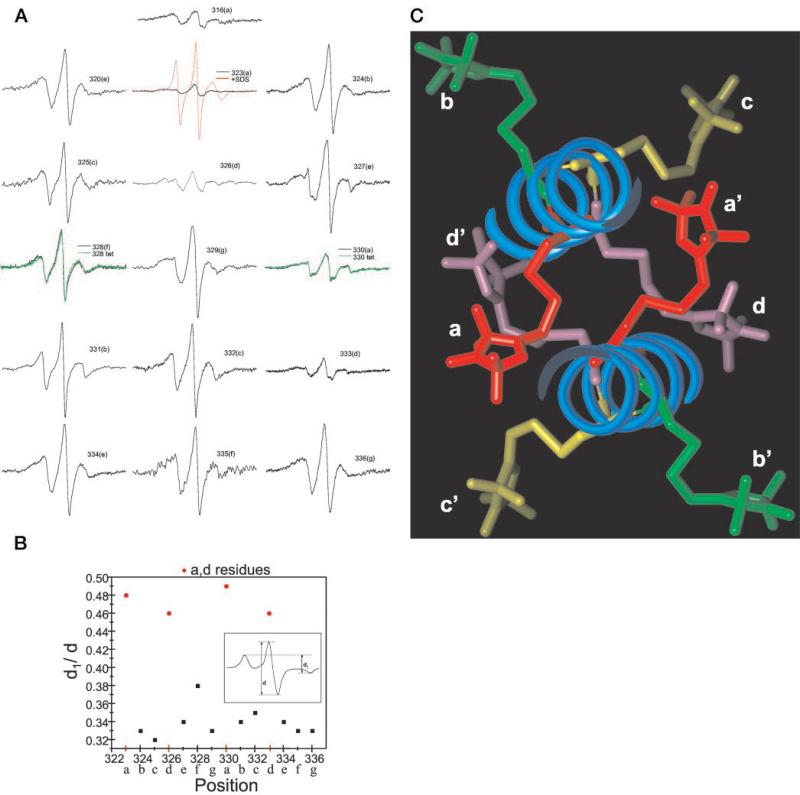
Fig. 3A shows the EPR spectra generated from these mutants. The spectrum from each of these mutants, normalized for the same number of spins, reveals that the spin label is in a non-random environment, indicative of a stable backbone and varying degrees of tertiary contact (36). Direct evidence of the coiled-coil structure of the rod domain is provided by the magnetic interactions between labels in close proximity, manifested by increased broadening (most easily seen as a loss in amplitude) of the spectrum when the label is placed in a or d positions (323, 326, 330, and 333) of the heptad repeat. Thus, these data identify residues that were predicted to appose one another in a coiled-coil configuration.
FIG.3. EPR characterization over two heptad repeats for vimentin spin-labeled at positions 323–336.
A, room temperature EPR spectra were recorded from assembled intermediate filaments from the indicated mutants. Letters in parenthesis indicate the position of the residue within the heptad repeat. Spectra are scaled according to the same number of spins. The total spins for each sample was determined by adding SDS (final of 2%) to each intermediate filament sample (see position 323, red line) and calculating the double integral, which was then normalized among all samples. The green spectra at positions 328 and 330 represent the normalized spectra for the mutants assembled under low ionic strength conditions that halt assembly at the tetrameric protofilament. B, spectral broadening in the absence of motion plotted over positions 323–336. The line-width ratio d1/d (inset) from spectra acquired at −100 °C reflects the dipolar interaction strength with a 1/r3 dependence (38). C, cross-section of a coiled-coil backbone showing the projections of spin-labeled cysteine replacements. The α-helical backbone of each monomer associated within a dimer is show as a blue coil. Shown are spin-labeled cysteine side chains at the a position (red), b position (green), c position (yellow), and d position (purple). The model was constructed with Insight II molecular modeling software (MSI, Inc.) with the solved crystal structure of a 100-amino acid region of cortexillin as the starting template.
Evaluation of the broadening as a function of only the distance-dependent dipolar interaction was performed by collecting the EPR spectra of frozen samples. In the absence of motion (i.e. frozen in liquid N2), semi-quantitative analysis of the extent of broadening caused by dipolar interaction can be obtained from the d1/d spectral ratio (see Fig. 3B, inset) (37, 38). Fig. 3B shows the dipolar broadening ratio versus sequence position. The difference between a,d positions and non-a,d positions is demonstrated as a binary distribution of points. The d1/d ratio for a,d positions clusters near 0.5, and the d1/d ratio for non-a,d positions all approach 0.33. These values translate into distances of 1.0 to 1.5 nm for a,d positions and >2 nm for all other positions. This analysis provides a measure of the separation between spin labels, revealing the periodicity of the distance separation along the positions of the heptad. From the data, we conclude that these two heptads are assembled in a coiled-coil dimer and that the individual monomers of this dimer are arranged in parallel and in exact register.
To illustrate the α-helical coiled-coil structure within this region of vimentin, we used the recently determined crystal structure of a 100-amino acid region of cortexillin (39) as a model. With this crystal structure as a template for vimentin amino acids 305–405, we used molecular modeling software to introduce spin-labeled cysteine amino acids to heptad positions a through g. Fig. 3C shows an end-on view of the coiled coil with the spin-labeled side chain of each amino acid position in a different color. The appropriate positions of one heptad are labeled a–d, and equivalent positions on the second protein are labeled a′–d′. Looking down the helix, positions a–d (and a′–d′) are labeled red, green, yellow, and purple, respectively. The red and purple moieties are more centrally located, whereas the yellow and green moieties occupy more exterior positions. Spin labels at a and d positions are close enough to interact within one dimer; positions b and c are farther separated and at the limit of spin-spin interaction detection. From this modeling and our EPR data of Fig. 3, A and B, the periodicity of the spectra is easily seen to coincide exactly with the helical nature of the coiled coil. Positions e, f, and g of the heptad can also be modeled and reveal the same exterior positions as positions b and c.
In summary, our examination of vimentin structure within the 323–336 span confirms a self-interaction between labels attached at either the a or d position. If the monomers were aligned in an anti-parallel manner, then at best, only one residue within the entire rod domain would have shown interaction. Had the monomers been in-parallel, but out of register, then no interaction would have been seen. Thus, SDSL-EPR is able to provide the first real-time data that IF monomers are aligned as in-parallel and in-register dimers, within native filaments, formed in physiologic conditions in a manner consistent with predictions of coiled-coil assembly.
Determination of the Structure of Positions 305–316
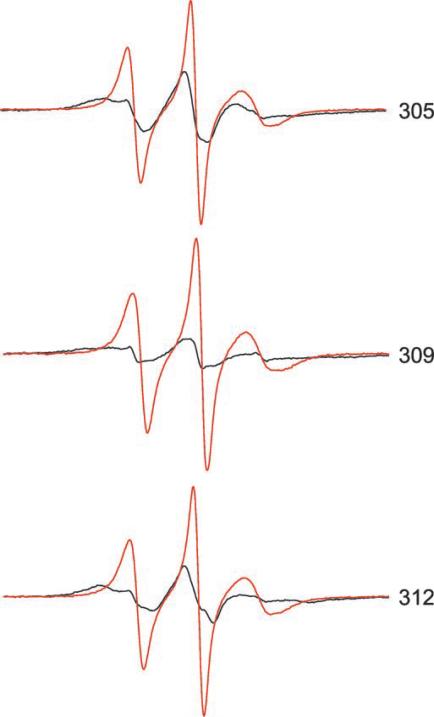
Having established the ability to identify coiled-coil structures at the level of individual amino acids, we sought to clarify the structure of vimentin positions 305–315, which have been assigned alternately to rod domain 2B or linker 2 (11, 14, 16). Within this sequence, amino acids 305, 309, and 312 were individually changed to cysteine, and the resulting proteins were subject to EPR analysis. As shown in the spectra of these positions (Fig. 4) 305, 309, and 312 resemble the a–d positions in line shape and extensive broadening. From these data, we conclude that the vimentin region 305–316 is in an α-helical coiled structure, revealing a heptad repeat pattern continuous with the region 323–336 discussed above. Thus, EPR analysis shows that rod domain 2B extends from at least position 305.
FIG.4. Room temperature EPR spectra from mutants 305, 309, and 312.
Spectra were collected from samples of intact filaments (black) and from SDS-treated filaments (red) at room temperature and normalized as described in the legend to Fig. 2A.
Use of EPR to Identify Inter-dimer Interactions
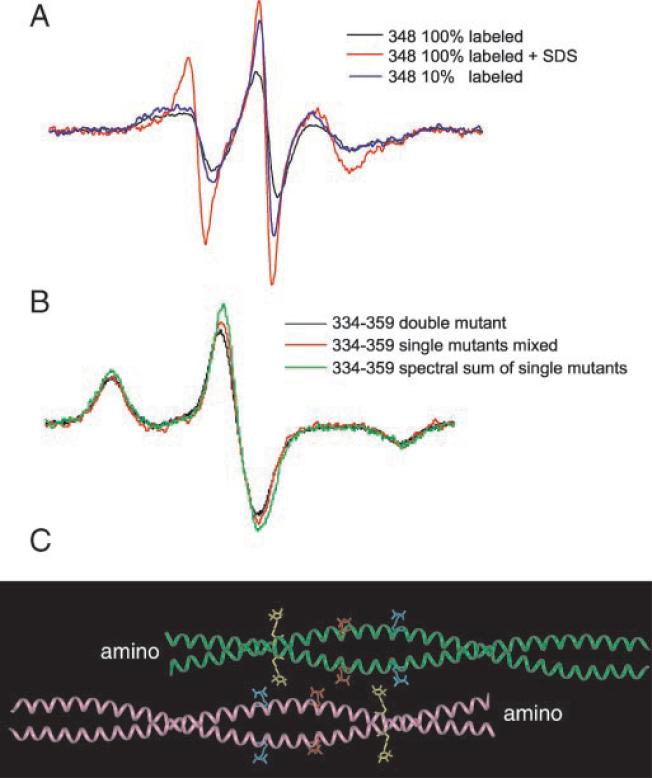
In the previous experiments, a,d residues were found to interact because they are positioned at apposing surfaces of two monomers. The lack of interaction of the non-a–d residues results from the greater separation between spin labels located on opposite sides of the exterior surface of the dimer. We hypothesized that spin labels such as these, on the surface of the dimer, would permit us to study the precise arrangement of dimers in native filaments. Therefore, we spin-labeled vimentin at multiple positions along the outer surface of the α-helix of the rod domain, assembled filaments, and recorded spectra, looking for evidence of spin label interaction. We found that EPR spectra from spin labels placed at positions 345, 348, and 349 showed evidence of interaction indicating physical proximity. Of these three positions, 348 provided strongest interaction, establishing this residue as a point of overlap between two dimers (Fig. 5A).
FIG.5. Alignment of vimentin dimers detected by spin interaction.
A, room temperature EPR spectra from filaments containing 100 and 10% labeled Cys348 vimentin. All three spectra reflect the same number of spins. Scaling was accomplished by the addition of 2% SDS to the 10 and 100% samples and by normalizing the resulting double integrals. The 100% labeled sample is clearly more broadened (black line) than the 10% labeled sample (blue line). The sample with 10% labeled Cys348 was prepared from a mixture of unlabeled wild type vimentin and 100% labeled Cys348 vimentin (9:1). B, the dipolar interaction between labels attached at positions 334 and 359 observed in the EPR spectra of frozen filaments. The 334–359 double mutant (black line) contains the two Cys substitutions within the same gene. The 334 + 359 mixture (red line) contains equal amounts spin-labeled Cys334 and Cys359 vimentin, which were prepared by isolating and spin labeling each protein separately. For experiments in A and B, protein concentrations were measured using a Pierce BCA assay kit, with bovine serum albumin as a standard. Appropriate amounts of protein were then mixed, and filament assembly was performed using a single step dialysis procedure. The amount of dipolar broadening relative to the spectral sum of single labeled 334 + single labeled 359 (green) was analyzed using a Pake deconvolution of normalized spectra (see “Results”). The three scans normalized to the same number of spins were obtained over 200 G at −100 °C (although only a 100 G portion is shown). C, ribbon diagram of an α-helical coiled coil indicating the locations of 334 (yellow), 348 (red), and 359 (blue) on the outside of the helices. Shown are spin-labeled Cys substitutions along the coiled-coil backbone structure obtained from the x-ray crystal structure of a 100-amino acid cortexillin fragment (39), which exists as an α-helical coiled coil. The molecular model was constructed using Insight II software.
Interaction between spin labels at position 348 is interpreted as resulting from an interaction between spin labels attached to separate dimers. Support for this conclusion is provided by comparing EPR spectra from samples with different proportions of spin label (25). A sample of vimentin containing 10% spin-labeled 348 and 90% wild type vimentin produces a spectrum that shows no evidence of spin-spin interaction; the spin labels are diluted and very rarely interact. The traces in Fig. 5A each represent the same number of spins as determined from the integration of the sample following addition of SDS. The spectrum from protein completely labeled at position 348 (100%, Fig. 5A, black line) is significantly more broadened than from the sample containing 10% labeled 348 (Fig. 5A, blue line). The spectrum of the 10% sample demonstrates that position 348 is a non-a,d residue of the heptad (compare the spectra to those in Fig. 3A), but the spectrum from the 100% labeled sample is broadened. Note that each dimer contains two spin-labeled positions, but only one would be predicted to interact with the second dimer. The second spin-labeled position of each dimer is on the opposite side and would be predicted not to interact unless tetramers assemble further.
Although the data in Fig. 5A show that position 348 is a site of interaction between dimers, these data do not identify whether these dimers are arranged in- or anti-parallel. Unambiguous demonstration of the anti-parallel alignment of dimers with a midpoint overlap at position 348 is provided by EPR spectra of mixtures of spin-labeled proteins (Fig. 5B). We selected position 334 as the starting point on the outside of the helix (an e position) and predicted that a site within the region of 355–365 would interact with 334 if dimers were arranged in an anti-parallel fashion. Therefore, mutants at positions 356(b heptad position), 359(e), 363(b), or 366(e) on the outside of the helix were prepared. Spin-labeled 334 was individually mixed equimolar with each spin-labeled 356, 359, 363, or 366 mutant. Assembly of each mixture into filaments was confirmed by electron microscopy, and EPR spectra were collected. To evaluate these combinations independently of line shape differences arising from differences in the rate of rotational averaging of hyperfine anisotropy, EPR analysis was performed on the samples frozen at −100 C°. Quantitative analysis of the spectra was achieved using a Pake deconvolution of broadening functions (40, 41), which optimizes results according to both the strength of the interaction and the fraction of interacting species. Only the mixture of the 334 and 359 spin-labeled proteins displayed evidence of interaction as seen by significant broadening of the spectra (Fig. 5B). From this mixture, the Pake deconvolution optimization of frozen spectra resulted in a mean interspin distance of 1.6 nm, with a non-interacting fraction of 74%. This labeling level coincides with a distribution of approximately one quarter of the spins interacting. Given an equal number of labeled 334 and 359 proteins introduced into the assembly mixture and a random pairing of these proteins during assembly, the expected population of spins paired at the tetramer level is 25%. Thus, these results provide strong evidence that vimentin dimers in intact filaments are associated in an anti-parallel manner, overlapping within 1.6 nm of position 348.
In observing the interaction between positions 334 and 359, more broadening would be predicted if spin labels were placed at both 334 and 359 within the same polypeptide chain. Hence we constructed a double mutant that codes for a Cys substitution at both position 334 and position 359. To simplify analysis, the assembly of the 334–359 double mutant was performed in low ionic strength Tris buffer; under these conditions, tetrameric protofilaments have been demonstrated to predominate (22, 42, 43). EPR spectra of the 334–359 double mutant (Fig. 5B, black line) is different from the spectral sum of the individual mutants (green line). A Pake deconvolution analysis of the double mutant collected at −100 C° was optimized with a mean interaction distance of 1.7 nm and a non-interacting population of 57%. The results correspond to a system in which approximately half of the spins are interacting, matching the alignment predicted by the experiments using a mixture of single-labeled proteins. Thus, EPR spectra provide evidence for the interaction of positions 334 and 359 within vimentin tetramers. Furthermore, quantitative analysis supports the hypothesis that vimentin in low ionic strength Tris is a relatively homogeneous population of tetramers arranged anti-parallel with rod 2B interacting.
Visualization of the side chain interactions identified between position 348 and itself, as well as 334 and 359, is provided in Fig. 5C. As in Fig. 2C, we have used the crystal structure of cortexillin as a model for vimentin dimer structure and have arranged two dimers in a structure consistent with our EPR data. Within the figure, the spin-labeled side chain of position 334 is shown in yellow; 348 is red, and 359 is blue. No other side chains are shown. As depicted, interactions between 348 and 348 as well as 334 and 359 can be seen.
DISCUSSION
The data presented conclusively show that assembly of vimentin intermediate filaments can be studied by site-directed spin labeling and EPR. In summary, our examination of vimentin structure within the 323–336 span confirms a self-interaction between labels attached at either the a or d position. If the monomers were aligned in an anti-parallel manner, then at best, only one residue within the entire rod domain would have shown interaction. Had the monomers been in parallel but out of register, then no interaction would have been seen. Spectral data provide precise structural information detailing the protein-protein contacts between IF proteins in intact filaments. Thus, SDSL-EPR is able to provide the first real-time data that IF monomers are aligned as in-parallel and in-register dimers, within native filaments formed in physiologic conditions in a manner consistent with predictions of coiled-coil assembly.
Our data reveal the α-helical coiled-coil nature of the vimentin rod domain and provide evidence that the region 305–336 is an α-helical coiled coil; this identification resolves a discrepancy in the literature (11, 14).
Two factors account for the broad spectra reported by the a,d positions. First, the wide splittings reflect a lack of motional averaging by side chains located at these positions. This indicates that the rotational freedom of the label is restricted by the protein structure. Second, the magnitude of broadening suggests a dipolar interaction among the labels when they are attached at either an a or d position. Thus the dynamic and spatial observations of spin-labeled positions within this span provide the first direct evidence of an α-helical coiled-coil arrangement with an exact register of the two peptide chains. Non-a,d positions, predicted to be on the exterior of the dimer, do not reveal such strong broadening, indicating that spin labels are farther apart, yet the overall structure is ordered and α-helical.
Beyond determination of structure, we have used SDSL-EPR to study assembly of IFs. We have been able to collect spectra from samples in low ionic strength Tris and subsequently from the same samples assembled into filaments. A surprising finding was that little change in rod domain structure is observed between normalized spectra obtained from fully assembled filaments and protofilaments formed in low ionic strength Tris. Under these conditions, higher order assembly beyond tetrameric protofilaments is inhibited, resulting in a predominantly tetramer population (22, 42, 43). Two examples of tetramer spectra are shown in Fig. 3A (green line). This suggests that vimentin assembly beyond the tetramer level does not generate strong tertiary contacts involving the rod domain examined here. Thus, we do not see evidence for a tight packing of neighboring tetramers via their rod domains. This is consistent with the fact that non-a,d positions display moderate motional freedom even in the intact filament.
As shown in Fig. 3C, non-a,d positions are widely separated on the outside of the helix. In a fortunate coincidence, this degree of separation is just slightly greater than the distance EPR can be used to measure. Thus, we have used spin labels placed on the outside of the helix to study interactions between dimers in filaments and assembly intermediates. With this strategy, we have identified an anti-parallel arrangement of dimers that is one of the configurations suggested by Steinert et al. from protein cross-link data (20). Our conclusions extend the data of Steinert et al. (20) and identify amino acid position 348 as the midpoint of overlap. Additionally, we have predicted an interaction between adjacent dimers and identified an interaction between positions 334 and 359 of adjacent dimers; this interaction thus proves the ability of EPR to identify interaction between the outer surfaces of two adjacent α-helices. This identification allows for the rational prediction of amino acids that interact between adjacent rods 2B in the anti-parallel arrangement.
Our data validate our experimental approach and provide a method for understanding the individual mechanisms behind the growing number of IF gene mutations identified as causing inherited genetic diseases. Because of the numbers and locations of identified mutations, each is likely to have subtle differences from the others. Abnormal assembly products can now be characterized by EPR at any stage during assembly, and the mechanisms behind such abnormal assembly can be elucidated. More broadly, these data establish the utility of SDSL-EPR to provide structural data on filament-forming proteins in general.
Acknowledgment
We thank Dr. Christian Altenbach for kindly providing the Pake deconvolution and simulation program.
This work was supported by National Institutes of Health Grant RO1EY08747 and Core Grant P30EY-12576 (both to P. G. F.) and by a UC Davis Health Sciences Research Fund grant (to J. F. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IF, intermediate filament; SDSL, site direct spin labeling; EPR, electron paramagnetic resonance; SDS, sodium dodecyl sulfate; PAGE, polyacrylamide gel electrophoresis.
REFERENCES
- 1.Lane EB, Rugg EL, Navsaria H, Leigh IM, Heagerty AH, Ishida-Yamamoto A, Eady RA. Nature. 1992;356:244–246. doi: 10.1038/356244a0. [DOI] [PubMed] [Google Scholar]
- 2.Irvine AD, Corden LD, Swensson O, Swensson B, Moore JE, Frazer DG, Smith FJ, Knowlton RG, Christophers E, Rochels R, Uitto J, McLean WH. Nat. Genet. 1997;16:184–187. doi: 10.1038/ng0697-184. [DOI] [PubMed] [Google Scholar]
- 3.Brenner M, Johnson AB, Boespflug-Tanguy O, Rodriguez D, Goldman JE, Messing A. Nat. Gen. 2001;27:117–120. doi: 10.1038/83679. [DOI] [PubMed] [Google Scholar]
- 4.Bonifas JM, Rothman AL, Epstein EH., Jr. Science. 1991;254:1202–1205. doi: 10.1126/science.1720261. [DOI] [PubMed] [Google Scholar]
- 5.Coulombe PA, Hutton ME, Letai A, Hebert A, Paller AS, Fuchs E. Cell. 1991;66:1301–1311. doi: 10.1016/0092-8674(91)90051-y. [DOI] [PubMed] [Google Scholar]
- 6.Fuchs E, Cleveland DW. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann H, Strelkov SV, Feja B, Rogers KR, Brettel M, Lustig A, Häner M, Parry DA, Steinert PM, Burkhard P, Aebi U. J. Mol. Biol. 2000;298:817–832. doi: 10.1006/jmbi.2000.3719. [DOI] [PubMed] [Google Scholar]
- 8.Strelkov SV, Herrmann H, Geisler N, Lustig A, Ivaninskii S, Zimbelmann R, Burkhard P, Aebi U. J. Mol. Biol. 2001;306:773–781. doi: 10.1006/jmbi.2001.4442. [DOI] [PubMed] [Google Scholar]
- 9.Hanukoglu I, Fuchs E. Cell. 1983;33:915–924. doi: 10.1016/0092-8674(83)90034-x. [DOI] [PubMed] [Google Scholar]
- 10.Steinert PM, Steven AC, Roop DR. Cell. 1985;42:411–420. doi: 10.1016/0092-8674(85)90098-4. [DOI] [PubMed] [Google Scholar]
- 11.Albers K, Fuchs E. Int. Rev. Cytol. 1992;134:243–279. doi: 10.1016/s0074-7696(08)62030-6. [DOI] [PubMed] [Google Scholar]
- 12.Bader BL, Magin TM, Hatzfeld M, Franke WW. Embo J. 1986;5:1865–1875. doi: 10.1002/j.1460-2075.1986.tb04438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hess JF, Casselman JT, FitzGerald PG. Curr. Eye Res. 1993;12:77–88. doi: 10.3109/02713689308999499. [DOI] [PubMed] [Google Scholar]
- 14.Conway JF, Parry DAD. Int. J. Biol. Macromol. 1988;10:79–98. [Google Scholar]
- 15.Steinert PM, Roop DR. Annu. Rev. Biochem. 1988;57:593–625. doi: 10.1146/annurev.bi.57.070188.003113. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs E, Weber K. Annu. Rev. Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 17.Crick FHC. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 18.Cohen C, Parry DAD. Trends Biol. Sci. 1986;11:245–248. [Google Scholar]
- 19.Steinert PM, Marekov LN, Fraser RD, Parry DA. J. Mol. Biol. 1993;230:436–452. doi: 10.1006/jmbi.1993.1161. [DOI] [PubMed] [Google Scholar]
- 20.Steinert PM, Marekov LN, Parry DA. J. Biol. Chem. 1993;268:24916–24925. [PubMed] [Google Scholar]
- 21.Quinlan RA, Hatzfeld M, Franke WW, Lustig A, Schulthess T, Engel J. J. Mol. Biol. 1986;192:337–349. doi: 10.1016/0022-2836(86)90369-4. [DOI] [PubMed] [Google Scholar]
- 22.Rogers KR, Herrmann H, Franke WW. J. Struct. Biol. 1996;117:55–69. doi: 10.1006/jsbi.1996.0069. [DOI] [PubMed] [Google Scholar]
- 23.Perozo E, Cortes DM, Cuello LG. Science. 1999;285:73–78. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 24.Hubbell WL, Cafiso DS, Altenbach C. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 25.Langen R, Isas JM, Luecke H, Haigler HT, Hubbell WL. J. Biol. Chem. 1998;273:22453–22457. doi: 10.1074/jbc.273.35.22453. [DOI] [PubMed] [Google Scholar]
- 26.Sandilands A, Masaki S, Quinlan RA. In: Intermediate Filaments Subcellular Biochemistry. Herrmann H, Harris JR, editors. Vol. 31. Plenum Press; New York: 1998. pp. 291–318. [PubMed] [Google Scholar]
- 27.Colucci-Guyon E, Portier MM, Dunia I, Paulin D, Pournin S, Babinet C. Cell. 1994;79:679–694. doi: 10.1016/0092-8674(94)90553-3. [DOI] [PubMed] [Google Scholar]
- 28.Conley YP, Erturk D, Keverline A, Mah TS, Keravala A, Barnes LR, Bruchis A, Hess JF, FitzGerald PG, Weeks DE, Ferrell RE, Gorin MB. Am. J. Hum. Genet. 2000;66:1426–1431. doi: 10.1086/302871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakobs PM, Hess JF, FitzGerald PG, Kramer P, Weleber RG, Litt M. Am. J. Hum. Genet. 2000;66:1432–1436. doi: 10.1086/302872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagai K, Thøgersen HC. Methods Enzymol. 1987;153:461–481. doi: 10.1016/0076-6879(87)53072-5. [DOI] [PubMed] [Google Scholar]
- 31.Carter JM, Hutcheson AM, Quinlan RA. Exp. Eye Res. 1995;60:181–192. doi: 10.1016/s0014-4835(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 32.Herrmann H, Hofmann I, Franke WW. J. Mol. Biol. 1992;223:637–650. doi: 10.1016/0022-2836(92)90980-x. [DOI] [PubMed] [Google Scholar]
- 33.Hubbell WL, Froncisz W, Hyde JS. Rev. Sci. Instrum. 1987;58:1879–1886. [Google Scholar]
- 34.Chomiki N, Voss JC, Warden CH. Eur. J. Biochem. 2001;268:903–913. doi: 10.1046/j.1432-1327.2001.01946.x. [DOI] [PubMed] [Google Scholar]
- 35.Hubbell WL, Gross A, Langen R, Lietzow MA. Curr. Opin. Struct. Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 36.Mchaourab H, Lietzow MA, Hideg K, Hubbell WL. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 37.Kokorin AI, Zamaraev KI, Grigorian GL, Ivanov VP, Rozantsev EG. Biofizika. 1972;17:34–41. [PubMed] [Google Scholar]
- 38.Likhtenshtein GI. Biophysical Labeling Methods in Molecular Biology. Cambridge University Press; New York: 1993. pp. 57–62. [Google Scholar]
- 39.Burkhard P, Kammerer RA, Steinmetz MO, Bourenkov GP, Aebi U. Structure Fold. Des. 2000;8:223–230. doi: 10.1016/s0969-2126(00)00100-3. [DOI] [PubMed] [Google Scholar]
- 40.Rabenstein MS, Shin Y-K. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altenbach C, Oh K-J, Trabanino RE, Hideg K, Hubbell WL. Biochemistry. 2001;40:15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 42.Meng JJ, Khan S, Ip W. J. Biol. Chem. 1994;269:18679–18685. [PubMed] [Google Scholar]
- 43.Potschka M, Nave R, Weber K, Geisler N. Eur. J. Biochem. 1990;190:503–508. doi: 10.1111/j.1432-1033.1990.tb15602.x. [DOI] [PubMed] [Google Scholar]