Abstract
Protein-arginine methyltransferase 1 (PRMT1) plays pivotal roles in various cellular processes. However, its role in megakaryocytic differentiation has yet to be investigated. Human leukemia K562 cells have been used as a model to study hematopoietic differentiation. In this study, we report that ectopic expression of HA-PRMT1 in K562 cells suppressed phorbol 12-myristate 13-acetate (PMA)-induced megakaryocytic differentiation as demonstrated by changes in cytological characteristics, adhesive properties, and CD41 expression, whereas knockdown of PRMT1 by small interference RNA promoted differentiation. Impairment of the methyltransferase activity of PRMT1 diminished the suppressive effect. These results provide evidence for a novel role of PRMT1 in negative regulation of megakaryocytic differentiation. Activation of ERK MAPK has been shown to be essential for megakaryocytic differentiation, although the role of p38 MAPK is still poorly understood. We show that knockdown of p38α MAPK or treatment with the p38 inhibitor SB203580 significantly enhanced PMA-induced megakaryocytic differentiation. Further investigation revealed that PRMT1 promotes activation of p38 MAPK without inhibiting activation of ERK MAPK. In p38α knockdown cells, PRMT1 could no longer suppress differentiation. In contrast, enforced expression of p38α MAPK suppressed PMA-induced megakaryocytic differentiation of parental K562 as well as PRMT1-knockdown cells. We propose modulation of the p38 MAPK pathway by PRMT1 as a novel mechanism regulating megakaryocytic differentiation. This study thus provides a new perspective on the promotion of megakaryopoiesis.
Keywords: Cell Differentiation, Hematopoiesis, MAPKs, p38 MAPK, Signal Transduction, Megakaryocytic Differentiation, PRMT1, Protein-arginine Methyltransferase 1
Introduction
Megakaryocytes arise from hematopoietic stem cells in bone marrow and ultimately release circulating platelets (1). They are thus regarded as a crucial source for treating thrombocytopenia, which affects a wide range of patients (1). Understanding the underlying molecular mechanisms would be important for developing new therapies for thrombocytopenia.
The human leukemia cell line K562 has been used as a model to study megakaryocytic differentiation (2). Phorbol 12-myristate 13-acetate (PMA)2 can induce K562 cells to differentiate in a manner that resembles the cytological and biochemical events observed during megakaryocytic differentiation in the bone marrow. These characteristics include enlarged cell size, expression of specific surface markers such as integrin αIIbβ3 (CD41/61) and integrin α2 (CD49b), acquisition of adhesive properties, cytoplasmic vacuolation, and the formation of a multilobed nucleus due to endomitosis (3–7). However, the molecular events that govern these events have not yet been completely elucidated.
Members of the MAPK family have been reported to participate in PMA-induced differentiation of K562 cells toward the megakaryocytic lineage. The ERK1/2 MAPK is activated (3, 4, 8), and the activation is essential as shown by using specific inhibitors or dominant negative MAPK/ERK kinase (MEK), the ERK upstream regulator (3–8). Several signaling molecules, including protein kinase C (6, 7) and Raf-1 (9), have been reported to mediate PMA-induced activation of the ERK MAPK pathway. The role of the other two members of the MAPK family, the JNK and the p38 MAPK, is less understood. In some reports, PMA treatment has been shown to activate JNK (6, 10), but its activation does not appear to be required for expression of the specific megakaryocyte marker CD49b (10). Previous studies did not find conclusive evidence for activation of p38 MAPK after PMA stimulation of K562 cells (6, 10, 11). The potential role of p38 MAPK is also controversial. Studies using inhibitors of p38 MAPK reported either stimulation (6) or no significant effects (10) on PMA-induced surface expression of megakaryocytic markers. In addition to K562 cells, the MAPK pathways have also been shown to play significant roles in megakaryopoiesis of primary progenitor cells, CD34+ hematopoietic cells (8, 12, 13), and other cell lines (8, 14, 15) induced by thrombopoietin (TPO), which is regarded as the primary growth factor that regulates megakaryopoiesis in vivo (1). The ERK pathway is activated and is essential for TPO-induced megakaryocytic differentiation (8, 12–15).
Protein arginine methylation mediated by protein-arginine methyltransferases (PRMTs) plays a pivotal role in numerous cellular functions (16, 17). PRMT1 was the first protein-arginine methyltransferase to be identified (18) and contributes to the majority of PRMT activity (>85%) in mammals (19, 20). The Prmt1 null mice are lethal at a very early embryonic stage (19), indicating an essential role of PRMT1 in embryonic development. Several lines of evidence suggest the potential involvement of PRMT1 in cell differentiation. Increased arginine methylation is reported during erythroid differentiation (21). In nerve growth factor-induced neuronal differentiation of PC12 cells, arginine methylation is shown to occur mainly at PRMT1 substrate sites (22). Overexpression of PRMT1 enhances retinoid-induced gene expression in myeloid cells (23). A recent report shows that knockdown of PRMT1 affects neurite outgrowth of Neuro2a cells (24). However, more studies are required to establish a direct link of PRMT1 in regulating cell differentiation.
In this study, we show that enforced expression of PRMT1 suppresses PMA-induced megakaryocytic differentiation of K562 cells. Conversely, knockdown of PRMT1 enhances differentiation, suggesting a novel role of PRMT1 in regulating megakaryocytic differentiation. Our data also clearly show that suppression or knockdown of p38 MAPK promotes PMA-induced megakaryocytic differentiation, supporting a negative regulatory role for p38 MAPK in this process. Further studies demonstrate that PRMT1 suppresses megakaryocytic differentiation through promoting activation of p38 MAPK without inhibiting the activation of ERK MAPK. Taken together, this study unveils a novel mechanism underlying regulation of megakaryocytic differentiation through PRMT1 and p38 MAPK.
EXPERIMENTAL PROCEDURES
Materials
Phorbol 12-myristate 13-acetate (PMA) was obtained from Sigma. S-Adenosyl-l-[methyl-3H]methionine ([3H]AdoMet) (63.6 Ci/mmol, NET-155H) and fluorographic enhancer, EN3HANCE, were obtained from PerkinElmer Life Sciences. The fluorescein isothiocyanate-conjugated anti-Plt-1 (CD41) antibody was obtained from Beckman Coulter, Inc. TPO, interleukin-3, stem cell factor, flk2/flt3 ligand, interleukin-6, granulocyte colony-stimulating factor, and granulocyte-macrophage colony-stimulating factor were obtained from PeproTech EC Ltd.
Plasmids
The pPCDNA3HA2-PRMT1 plasmid harboring rat PRMT1 cDNA (18) was used for ectopic expression of HA-PRMT1 in mammalian cells. A single amino acid mutation (G80R) was introduced in PRMT1 by PCR to impair its enzymatic activity (25). The human PRMT2 cDNA was amplified from K562 by reverse transcription-PCR and subcloned into a pPCDNA3HA2 plasmid. The human PRMT5 cDNA was excised from a pGEX-4T-1-PRMT5 plasmid, a gift from Dr. W. Y. Tarn (Institute of Biomedical Sciences, Academia Sinica, Taiwan), and inserted into a pPCDNA3HA2 plasmid for expression in mammalian cells. The mammalian p38α expression plasmid was a gift from Dr. J. Han (The Scripps Research Institute, La Jolla, CA). The pLKO.1-puro plasmid-based shRNAs, including shLuc (luciferase shRNA), TRCN0000035933 (PRMT1-sh1), TRCN0000000509 (p38α-sh1), and TRCN0000010052 (p38α-sh2), were obtained from the National RNAi Core Facility, Taiwan. The PRMT1-sh2 (26) was expressed from the pSUPER vector (Oligoengine) and used for transient knockdown of PRMT1. The PRMT1 sh-1 was used for selection of stable clones. To select stable p38α-knockdown clones, K562 cells were transfected simultaneously with both p38α-sh1 and p38α-sh2.
Cell Culture and Stable Clone Selection
The human chronic myelogenous leukemia K562 cell line was from the American Type Culture Collection (ATCC). Cells were cultivated in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, and 100 IU/ml streptomycin. Transfection was performed using LipofectamineTM 2000 reagent (Invitrogen). Stable clones expressing HA-PRMT1 or HA-PRMT1G80R were selected with G418 (0.5 mg/ml, Calbiochem). Stable clones expressing shRNAs were selected with puromycin (0.5 μg/ml, Calbiochem).
Cell Growth and Differentiation Analysis of K562 Cells
For cell cycle analysis, cells were fixed in 70% ethanol, stained with a solution containing propidium iodide (50 μg/ml) and DNase-free RNase A (1 μg/μl) for 1 h at room temperature, and analyzed by flow cytometry using the FACScan system and CellQuest software (BD Biosciences). For megakaryocytic differentiation, K562 cells were treated with 40 nm phorbol 12-myristate 13-acetate (PMA). Adherent cells with pseudopodia were examined by phase contrast light microscopy. For quantification, cells in suspension or loosely attached were first removed from the culture dish carefully, and adherent cells were then removed by trypsinization, collected, and counted. Cytological changes, including a multilobed nucleus and vacuolated cytoplasm, were examined by modified Wright-Giemsa staining. Three to four hundred cells were examined in each assay. To analyze the surface marker CD41, cells were first incubated in 2% bovine serum albumin for 30 min and then with fluorescein isothiocyanate-conjugated anti-Plt-1 (CD41) antibodies (1:80) in 1% bovine serum albumin for 30 min; cells were then analyzed by flow cytometry. Cell viability was measured by trypan blue exclusion.
Preparation of Cell Lysates
For the methylation assay, cells were resuspended in extraction buffer (50 mm Tris-HCl, pH 7.4; 0.5 mm EDTA; 0.5 mm EGTA; 10 μg/ml leupeptin; 10 μg/ml aprotinin; 10 μg/ml pepstatin; 1 mm phenylmethylsulfonyl fluoride; 0.5 mm dithiothreitol; 5% glycerol) and disrupted by homogenization with a glass tissue grinder. The homogenates were centrifuged at 10,000 × g for 30 min, and the supernatants were stored at −80 °C. For Western blot analysis, cells were lysed with RIPA buffer (50 mm Tris-HCl, pH 7.4; 150 mm NaCl; 1% Triton X-100; 0.1% SDS; 1 mm edta; 1 mm phenylmethylsulfonyl fluoride; 10 μg/ml aprotinin; 10 μg/ml leupeptin; 10 μg/ml pepstatin; 1% sodium deoxycholate; 1 mm sodium fluoride; 1 mm sodium orthovanadate; 25 mm β-glycerophosphate).
Methylation Analysis
The thioredoxin-fused hnRNP K proteins were expressed and purified as described previously (27). Cell homogenates (4 μg) and hnRNP K proteins (5 μg) were incubated in the presence of 1.65 μCi of [3H]AdoMet and 25 mm Tris-HCl, pH 8.0, in a final volume of 30 μl at 30 °C for 30 min. Reactions were stopped by the addition of SDS sample buffer and then subjected to SDS-PAGE. After staining and de-staining, gels were soaked in the fluorographic enhancer EN3HANCE, dried, and then exposed to x-ray film (Kodak) at −70 °C for fluorographic analysis.
Western Blot Analysis
Western blot was performed with the following antibodies: anti-HA (1:1000, HA.11, Covance); anti-PRMT1 (1:1000, Sigma); anti-asymmetric dimethylarginine (1:500, ASYM24, Millipore); anti-ERK2 (1:2500, Santa Cruz Biotechnology); anti-phospho-p44/42 (p-ERK) (1:1250, Cell Signaling); anti-p38 MAPK (1:1000, Cell Signaling); anti-phospho-p38 (p-p38) (1:1000, Cell Signaling); anti-actin (1:15,000, Chemicon); and anti-glyceraldehyde-3-phosphate dehydrogenase (1:15,000, Abcam). Detections were performed using the ECLTM Western blotting detection reagents (GE Healthcare). The levels of phosphorylated and total p38 were quantified using a laser scanning densitometer (GE Healthcare). Levels of phosphorylated p38 were normalized to total p38 levels to measure the extent of activation of p38.
Protein Kinase Assay
To analyze the kinase activity, active p38 MAPK was immunoprecipitated with an immobilized anti-phospho-p38 antibody, and phosphorylation of a specific substrate (ATF-2 peptides) was measured according to the manufacturer's instructions (nonradioactive p38 MAPK assay kit, Cell Signaling).
Isolation of CD34+ Cells and Analysis of Differentiation
CD34+ cells were derived from human umbilical cord blood with consent from the mother and were collected and processed according to governmental regulations (“Guidelines for Collection and Use of Human Specimens for Research.” Department of Health, Taiwan) and after approval from the scientific committees of the Food Industry Research and Development Institute, Taiwan. Isolation and expansion of CD34+ cells were performed as described previously (28). Briefly, the CD34+ cells were purified with CD34 microbeads by a Miltenyi VarioMACS device (Miltenyi Biotec, Bergisch Gladbach, Germany) and cultivated in serum-free Iscove's modified Dulbecco's medium (HyClone) supplemented with serum substitutes (1.5 g/liter bovine serum albumin, 4.4 μg/ml insulin, 60 μg/ml transferrin, and 25.9 μm 2-mercaptoethanol) and a cytokine mixture (8.5 ng/ml TPO, 4.1 ng/ml interleukin-3, 15 ng/ml stem cell factor, 6.7 ng/ml flk2/flt3 ligand, 0.8 ng/ml interleukin-6, 3.2 ng/ml granulocyte colony-stimulating factor, and 1.3 ng/ml granulocyte-macrophage colony-stimulating factor) for 6 to 7 days. To induce megakaryocytic differentiation, the expanded CD34+ cells were cultivated (5 × 104 cells/ml) in media described above without cytokine mixture for 6 h. TPO (100 ng/ml) was added to induce differentiation. Expression of CD41 was analyzed by flow cytometry 15 days after TPO stimulation.
The TAT-mediated protein transduction system can deliver proteins into cells rapidly, efficiently, and noninvasively (29). Recombinant TAT-fused HA-PRMT1 and control TAT-fused HA-GFP proteins were expressed in Escherichia coli and purified using Ni+-nitrilotriacetic acid-agarose (Qiagen). Endotoxins were removed using Detoxi-GelTM endotoxin removing gel (Pierce) according to the manufacturer's instructions. For differentiation study, recombinant TAT-fused proteins were added to CD34+ cells 6 h before TPO stimulation and again at the time of TPO addition.
Statistical Analysis
The Student's t test was used for statistical analysis. Values are presented as means ± S.E. All experiments were performed at least three times. p < 0.05 was considered statistically significant.
RESULTS
Ectopic Expression of HA-PRMT1 Suppresses PMA-induced Megakaryocytic Differentiation of K562 Cells
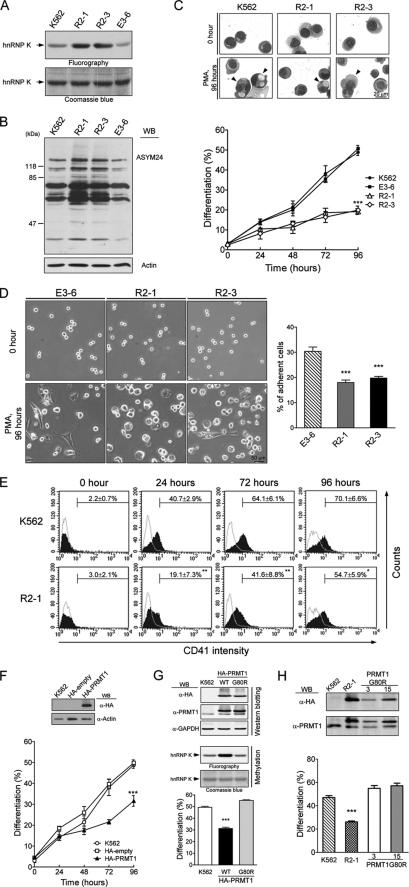
The human K562 cell line is a multipotent leukemia cell line that undergoes megakaryocytic differentiation upon PMA treatment. To investigate whether protein arginine methylation plays a role in differentiation, we established cell clones that stably expressed HA-PRMT1. These stable clones, R2-1 and R2-3, exhibited elevated PRMT1 activity as measured by the methylation of hnRNP K, a known PRMT1 substrate (Fig. 1A) (27) or as measured by the intracellular levels of asymmetric dimethylarginine, a product of PRMT1 catalytic activity (Fig. 1B). The E3-6 cells were an empty vector control that exhibited PRMT1 activity similar to the parental K562 cells. When immunoprecipitated from R2-1 and R2-3 cells, the HA-PRMT1 protein was enzymatically active and methylated wild type hnRNP A2 and hnRNP K proteins but not mutants lacking the methyl acceptor glycine- and arginine-rich motifs (supplemental Fig. S1).
FIGURE 1.
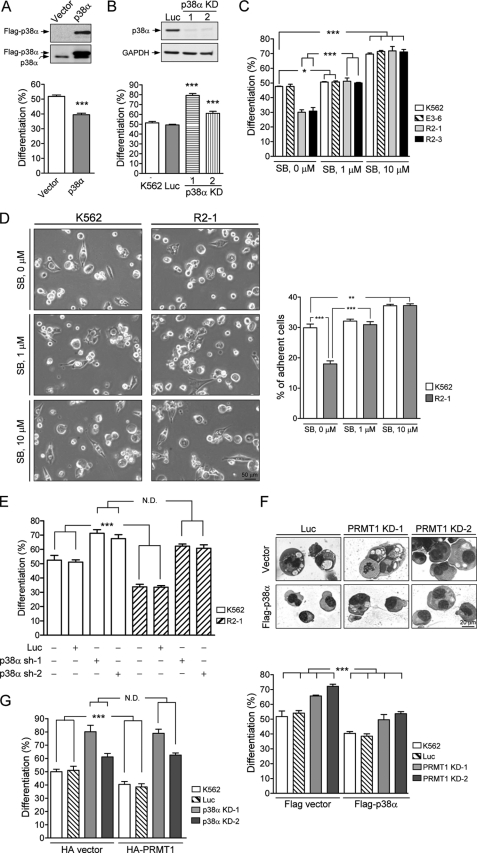
Ectopic expression of HA-PRMT1 suppresses PMA-induced megakaryocytic differentiation of K562 cells. K562 cells were stably transfected with pPCDNA3HA2-PRMT1 plasmids (R2-1 and R2-3) or empty vectors (E3-6). The methyltransferase activity of PRMT1 in cell homogenates was assayed using the known substrate hnRNP K and detected by fluorography (A). Levels of asymmetric dimethylarginine, a product of PRMT1 activity, in cell lysates were detected by Western blot (WB) analysis using a specific antibody (ASYM24) (B). Various K562 cell clones were treated with PMA (40 nm). Megakaryocytic differentiation was detected by modified Wright-Giemsa staining for cytological changes (C), by phase contrast microscopy for examination of adherent cells with pseudopodia (D), or by flow cytometry for the expression of the specific surface marker CD41 (E). Cells with enlarged and lobed nuclei and microvesicles are marked by arrowheads (C, upper panel). The suspended and adherent cells were collected and quantified (D, right panel). Dashed lines in E are isotypic controls. Ectopic expression of HA-PRMT1, R2-1, and R2-3 significantly suppressed differentiation. Transient transfection of K562 cells with pPCDNA3HA2-PRMT1 led to similar effects (F). Inset shows expression of HA-PRMT1 in transfected cells. The enzymatically impaired mutant PRMT1G80R, when transiently expressed in K562 cells, did not suppress differentiation as the wild type (WT) enzyme (G, lower panel). Ectopic expression of mutant PRMT1 did not increase the methyltransferase activity in cell homogenates when assayed with a known PRMT1 substrate hnRNP K (G, middle panel). Expression levels of wild type and mutant HA-PRMT1 proteins were similar (G, upper panel). Stable clones of mutant PRMT1G80R also did not show suppressive effects on differentiation (H, lower panel). Expression of wild type and mutant HA-PRMT1 proteins were examined by Western blot (H, upper panel). All experiments were performed at least three times, and data are presented as means ± S.E.; *, p < 0.05; **, p < 0.01; ***, p < 0.005 as compared with K562 parental cells.
Megakaryopoiesis can be characterized by cytological changes and expression of lineage-specific markers such as CD41 (3–7). After PMA treatment, K562 cells exhibited enlarged and lobed nuclei and multiple microvesicles, which were readily detected by modified Wright-Giemsa staining (Fig. 1C, upper panel, K562). About 50% of the E3-6 and the K562 cells exhibited characteristics of megakaryocytes 96 h after PMA treatment (Fig. 1C, lower panel). In addition, we observed an increased number of adherent cells, about 30% at 96 h (Fig. 1D, E3-6). Only about 20% of R2-1 and R2-3 cells were identified as megakaryocytes by modified Wright-Giemsa staining (Fig. 1C, lower panel), indicating that ectopic expression of HA-PRMT1 significantly suppressed differentiation. The expression of the megakaryocyte-specific marker CD41 on the cell surface was also significantly decreased, from 70 to 55% at 96 h, in HA-PRMT1-expressing cells (Fig. 1E). CD41 is a subunit of integrin αIIbβ3, which is essential for the acquisition of adherent properties. Consistent with reduced CD41 expression in R2-1 and R2-3 cells, fewer adherent cells were observed in these cells, about 17–20% at 96 h (Fig. 1D, R2-1 and R2-3). When transiently overexpressed in K562 cells, HA-PRMT1 also suppressed PMA-induced megakaryocytic differentiation (Fig. 1F). These results together suggest that the enforced expression of PRMT1 suppresses PMA-induced megakaryocytic differentiation in K562 cells. When tested in another leukemia cell line HEL, transient expression of HA-PRMT1 also suppressed megakaryocytic differentiation (supplemental Fig. S2), suggesting that the suppressive effect of PRMT1 may be a common event in PMA-induced megakaryocytic differentiation of leukemia cell lines.
A single amino acid mutation (G80R) in the conserved AdoMet binding domain of PRMT1 has been shown to impair its methyltransferase activity (25). Transient expression of mutant PRMT1G80R in K562 cells did not suppress differentiation (Fig. 1G, lower panel). The PRMT1 methyltransferase activity did not increase in PRMT1G80R-overexpressing cell homogenates (Fig. 1G, middle panel) either. In contrast, overexpression of wild type PRMT1 significantly enhanced methyltransferase activity and suppressed differentiation (Fig. 1G). The expression level of these two proteins was similar as detected by Western blot analysis (Fig. 1G, upper panel). In addition, the mutant PRMT1G80R stable clones, G80R-3 and G80R-15, were also unable to suppress megakaryocytic differentiation (Fig. 1H). These results suggest that the methyltransferase activity of PRMT1 is essential for its suppressive effect on megakaryocytic differentiation.
Reduced Levels of Endogenous PRMT1 Enhance PMA-induced Megakaryocytic Differentiation of K562 Cells
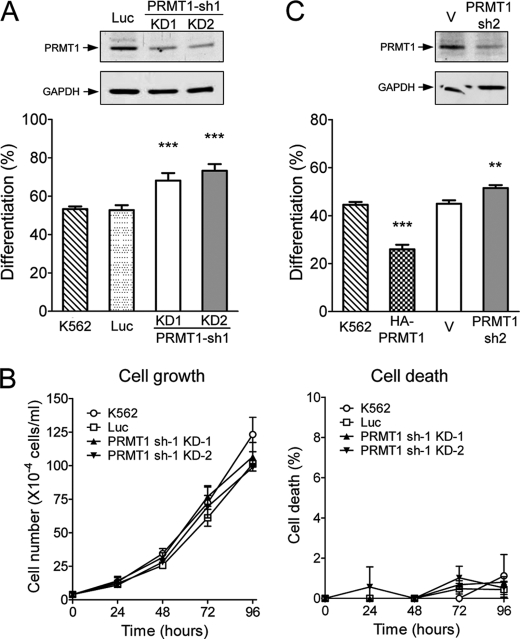
We used RNA interference to investigate whether endogenous PRMT1 plays a role in megakaryocytic differentiation. Cell clones that stably expressed shRNA (PRMT1-sh1) were selected. The protein levels of endogenous PRMT1 in stable clones 1 and 2 were significantly reduced (Fig. 2A, upper panel). Upon PMA stimulation, these knockdown clones exhibited significantly higher levels of megakaryocytic differentiation (∼70% as compared with ∼50%; Fig. 2A, lower panel). Cells transfected with the control luciferase shRNA displayed differentiation levels similar to the parental K562 (Fig. 2A). The stable knockdown of PRMT1 did not affect cell growth (Fig. 2B, left panel) or viability (Fig. 2B, right panel) under normal culture conditions. These results suggest that a reduced level of endogenous PRMT1 favors PMA-induced megakaryocytic differentiation. This enhancement was also observed when endogenous PRMT1 was transiently knocked down using a different shRNA (PRMT1-sh2) (Fig. 2C). These results are consistent with the suppressive effect of transient expression of HA-PRMT1 (Figs. 1F and 2C) and suggest that endogenous PRMT1 plays a negative role in the regulation of PMA-induced megakaryocytic differentiation of K562 cells.
FIGURE 2.
Reduced levels of endogenous PRMT1 enhance PMA-induced megakaryocytic differentiation of K562 cells. KD-1 and KD-2 cell clones were stably transfected with PRMT1 shRNA (sh-1). Western analysis using an anti-PRMT1 antibody showed reduced expression of PRMT1 proteins (A, upper panel). Megakaryocytic differentiation was analyzed 96 h after PMA treatment (A, lower panel). Luc, luciferase shRNA. The growth and viability of these cell clones were similar under regular culture conditions (B). Effects of PRMT1 knockdown were also assayed by transient transfection with a different shRNA (sh-2). PRMT1 protein levels were detected by Western blot analysis (C, upper panel) and megakaryocytic differentiation was detected by modified Wright-Giemsa staining (C, lower panel). Luciferase shRNA (Luc) and the pSUPER empty vector (V) were used as controls. All experiments were performed at least three times, and data are presented as means ± S.E.; **, p < 0.01; ***, p < 0.005 as compared with K562 parental cells.
Differential Effects of PRMT1 on PMA-induced Growth Arrest and Differentiation
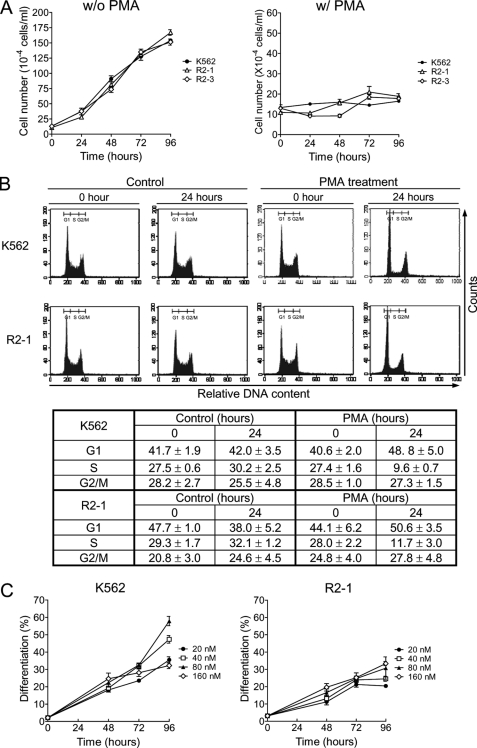
Because cell growth arrest is a prerequisite for cell differentiation, we examined whether PRMT1 interferes with this process. In K562 parental cells, PMA treatment caused a cessation of cell growth (Fig. 3A, right panel) and a significant decrease in the number of S phase cells, from 27.4 to 9.6%, after 24 h (Fig. 3B). Similar effects were observed in HA-PRMT1-expressing cells. These results indicate that HA-PRMT1-expressing cells still retain the ability to respond to PMA-induced growth arrest. The growth rate (Fig. 3A, left panel) and viability (supplemental Fig. S3) of these PRMT1 clones were similar to parental and empty control cells under normal culture conditions. We further examined the response of these cell clones to different concentrations of PMA. PMA stimulated megakaryocytic differentiation of parental K562 cells in a dose-dependent manner, from ∼35 to ∼57% at concentrations ranging from 20 to 80 nm (Fig. 3C, K562). However, HA-PRMT1-expressing R2-1 cells responded only slightly to the increasing concentrations of PMA (Fig. 3C, R2-1). An increase of PMA to 160 nm did not further promote differentiation in K562 cells or R2-1 cells (Fig. 3C), suggesting that the suppressive effect of PRMT1 cannot be overcome with higher concentrations of PMA.
FIGURE 3.
Differential effects of PRMT1 on PMA-induced growth arrest and differentiation. Under regular culture conditions, the growth rate of HA-PRMT1-expressing cells, R2-1 and R2-3, were similar to parental cells (A, left panel). PMA treatment (40 nm) caused a cessation of growth (A, right panel) and cell cycle arrest at G1 phase (B) in all cell clones. To examine the dose response of these cells to PMA, K562 and R2-1 cells were treated with increasing concentrations of PMA as indicated, and megakaryocytic differentiation was analyzed by modified Wright-Giemsa staining (C). All experiments were performed at least three times and data are presented as means ± S.E. w/o, without; w/, with.
Modulation of the MAPK Pathways by PRMT1
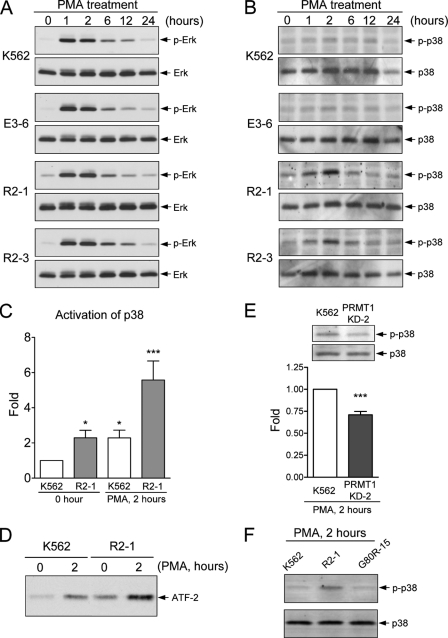
Activation of the ERK MAPK has been shown to be essential for megakaryocytic differentiation in K562 cells and hematopoietic stem cells (8). MAPKs are activated through phosphorylation of specific threonine and tyrosine residues by upstream MAPK kinases (30). To investigate whether PRMT1 blocks this pathway, we examined the activation of ERK MAPK by Western blot using antibodies that specifically recognize phosphorylated threonine and tyrosine residues. The ERK MAPK was significantly activated 1 h after PMA stimulation, and the activation was gradually decreased after 2 h in parental and control cells (Fig. 4A, K562 and E3-6). Notably, in R2-1 and R2-3 cells, ERK MAPK was also activated in a similar pattern (Fig. 4A), suggesting that the ectopic expression of HA-PRMT1 does not block this signaling pathway, which is essential for differentiation. In contrast to the remarkable activation of ERK MAPK upon PMA stimulation, p38 MAPK was only slightly activated in control cells (Fig. 4B, K562 and E3-6). Interestingly, p38 MAPK was more significantly activated in R2-1 and R2-3 cells as compared with that in parental cells; this activation peaked 2 h after PMA treatment (Fig. 4B). In R2-1 cells, p38 MAPK activation increased ∼4–5-fold at its peak, although in K562 cells, activation increased by only ∼2-fold at its peak as compared with unstimulated K562 cells (Fig. 4C). Furthermore, the basal level of active p38 MAPK in untreated R2-1 cells was already higher than basal levels in parental cells (∼2.5-fold) (Fig. 4C). Activation of p38 MAPK was further demonstrated directly by kinase assays. The active form of p38 MAPK was immunoprecipitated, and kinase activity was assayed by phosphorylation of its substrate ATF-2 (Fig. 4D). Consistent with the results obtained with Western blot analysis (Fig. 4C), both the basal and stimulated activities of p38 MAPK in R2-1 were higher than those observed in parental K562 cells (Fig. 4D). These results suggest that expression of HA-PRMT1 potentiates activation of the p38 MAPK. Consistently, knockdown of endogenous PRMT1 decreased PMA-stimulated activation of p38 (Fig. 4E). Furthermore, the methyltransferase-impaired HA-PRMT1G80R mutant could not promote activation of p38 MAPK (Fig. 4F), suggesting that the methyltransferase activity of PRMT1 is required to promote activation of the p38 MAPK.
FIGURE 4.
Modulation of the MAPK pathways by PRMT1. Parental, empty control (E3-6), and HA-PRMT1-expressing R2-1 and R2-3 cells were treated with PMA (40 nm), collected, and lysed in RIPA buffer. Activation of ERK (A) and p38 (B) was detected with antibodies against the specific phosphorylated forms. Activation of p38 2 h after PMA treatment was quantified and normalized to the total amount of p38 protein (C). The active form of p38 was immunoprecipitated, and its kinase activity was assayed by phosphorylation of its substrate ATF-2 (D). The kinase activity of p38 was significantly activated in PRMT1-overexpressing cells (C and D). Knockdown of endogenous PRMT1 decreased the activation of p38 (E). Abrogation of PRMT1 enzyme activity (G80R-15 mutant) could no longer promote activation of p38 (F). All experiments were performed at least three times, and data are presented as means ± S.E.; *, p < 0.05; ***, p < 0.005 as compared with K562 parental cells.
p38 MAPK Negatively Regulates Megakaryocytic Differentiation
To further elucidate the role of p38 MAPK in megakaryocytic differentiation, K562 cells were first transiently transfected with plasmids expressing p38α and then treated with PMA. Megakaryocytic differentiation was reduced (40% versus 52%) in p38α-transfected cells as detected by the modified Wright-Giemsa staining (Fig. 5A). In addition, fewer adhesive cells were observed in p38α-transfected cells, 30% versus 15% in parental and p38α-transfected cells (supplemental Fig. S4). We then knocked down endogenous p38α by generating cell clones that stably expressed p38α shRNAs (Fig. 5B, upper panel). In these clones, we found that more cells were induced to differentiate toward the megakaryocyte lineage (Fig. 5B, lower panel). Consistent with these results, suppression of kinase activity with SB203580, a specific inhibitor of p38 MAPK, led to an increase in megakaryocytic differentiation (70% versus 48% at 96 h after PMA treatment) in parental and empty vector cell clones; this increase was found to occur in a dose-dependent manner (Fig. 5C, K562). Similar enhancement was observed in adhesive properties of these cells after treatment with the inhibitor (Fig. 5D, K562). Taken together, these results provide solid evidence that p38α MAPK plays a negative regulatory role in PMA-induced megakaryocytic differentiation of K562 cells.
FIGURE 5.
PRMT1-mediated suppression of megakaryocytic differentiation is dependent on activation of the p38 MAPK. K562 cells were transiently transfected with FLAG-p38α and empty vectors and analyzed for megakaryocytic differentiation 96 h after PMA treatment (A). Stable cell clones (p38α KD) transfected simultaneously with two p38α shRNAs (sh-1 and sh-2) were selected, and the protein levels were examined by Western blot analysis (B, upper panel). Luc, luciferase shRNA; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Reduced expression of p38α enhanced megakaryocytic differentiation (B, lower panel). Treatment with the p38 inhibitor SB203580 (SB) greatly enhanced megakaryocytic differentiation in both parental K562 and HA-PRMT1-expressing R2-1 and R2-3 cells when analyzed 96 h after PMA treatment (C). The specific inhibitor of p38 MAPK, SB203580, was added 30 min before stimulation with PMA. Adherent cells were examined by phase contrast microscopy 96 h after PMA treatment (D, left panel) and quantified (D, right panel). Transient knockdown of p38α with either p38α sh-1 or p38α sh-2 shRNAs enhanced megakaryocytic differentiation in both K562 and R2-1 cells (E). Ectopic expression of p38α by transient transfection suppressed megakaryocytic differentiation in both K562 and PRMT1-knockdown cells (PRMT1 KD-1 and KD-2) (F). Cytological changes were detected by modified Wright-Giemsa staining (F, upper panel) and quantified (F, lower panel). HA-PRMT1 was transiently expressed in stable p38α-knockdown clones (p38α KD-1 and p38α KD-2) (G). Ectopic expression of HA-PRMT1 could no longer suppress megakaryocytic differentiation in p38α-deficient cells. Cells stably transfected with the luciferase shRNA (Luc) were used as a negative control. All experiments were performed at least three times, and data are presented as means ± S.E.; *, p < 0.05; **, p < 0.01; ***, p < 0.005; N.D. means no difference.
PRMT1 Suppresses Differentiation through Activation of p38α MAPK
To investigate the involvement of p38α in PRMT1-mediated suppression of megakaryocytic differentiation, we knocked down p38α in R2-1 cells by transient transfection with p38α shRNAs, (p38α sh-1 or p38α sh-2). We not only observed enhanced PMA-induced differentiation of K562 parental cells (65–70% versus 50–55%; Fig. 5E, K562) but also enhanced R2-1 cell differentiation (62% versus 30–35%; Fig. 5E, R2-1). The differentiation levels were similar in K562 and R2-1 cells after p38α were knocked down, suggesting that p38α silencing could reverse PRMT1-mediated suppression. The control luciferase shRNA did not affect differentiation in either parental or R2-1 cells (Fig. 5E). Likewise, treatment with SB203580 also led to an increase in PMA-induced megakaryocytic differentiation of R2-1 and R2-3 cells, similar to that observed in control E3-6 and parental K562 cells, as detected by modified Wright-Giemsa staining (Fig. 5C) and cell adherent properties (Fig. 5D). Taken together, suppression of p38α, by either RNA interference or a specific kinase inhibitor, enhanced differentiation in PRMT1-overexpressing cells to a level comparable with that of parental cells. These results indicate that p38α MAPK mediates the suppressive effect of PRMT1 on PMA-induced differentiation.
Transient expression of FLAG-tagged p38α suppressed differentiation not only in parental and control cells but also in stable PRMT1-knockdown clones, PRMT1 KD-1 and KD-2 (Fig. 5F). Cytological changes were examined by modified Wright-Giemsa staining (Fig. 5F, upper panel) and quantified (Fig. 5F, lower panel). Together with the results in Fig. 4, E and F, and Fig. 5E, these data suggest that p38 functions downstream of PRMT1. This notion was further supported by ectopic expression of HA-PRMT1 in p38α-knockdown cell clones (p38α KD-1 and KD-2). The PRMT1-mediated suppression of differentiation was almost completely diminished when p38α was knocked down (Fig. 5G). Taken together, these results indicate that p38α MAPK mediates the suppressive effect of PRMT1 on PMA-induced differentiation.
Enforced Expression of PRMT1 in Human CD34+ Hematopoietic Cells Suppresses TPO-induced Megakaryocytic Differentiation
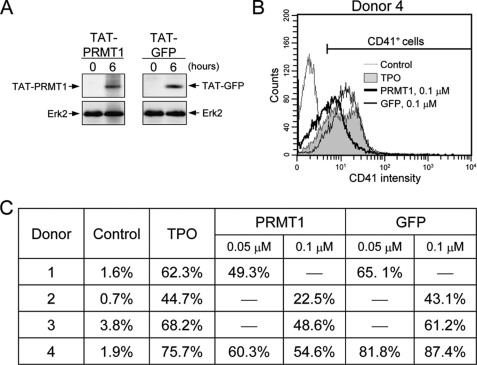
To investigate the effect of PRMT1 in more physiologically relevant conditions, HA-PRMT1 was introduced into human CD34+ hematopoietic cells by a TAT-mediated protein transduction system, which delivers conjugated proteins into cells in a noninvasive and highly efficient manner (29). Both the recombinant TAT-conjugated HA-PRMT1 protein and the HA-GFP control protein were detected in cells by Western blot analysis (Fig. 6A). TPO induces CD34+ hematopoietic cells to undergo differentiation toward the megakaryocytic lineage as detected by the expression of the specific surface marker CD41. Flow cytometric analysis of donor 4 is shown in Fig. 6B. TPO-induced CD41 expression ranged from 45 to 76% in four different donors at day 15 after TPO treatment (Fig. 6C). Transduction of TAT-PRMT1 proteins significantly suppressed TPO-induced differentiation (23–55% with 0.1 μm TAT-PRMT1; Fig. 6C). The suppression occurred in a dose-dependent manner (Fig. 6C, donor 4). These results suggest that PRMT1 may negatively regulate TPO-induced megakaryocytic differentiation of CD34+ hematopoietic cells.
FIGURE 6.
Enforced expression of PRMT1 in human CD34+ hematopoietic cells suppresses TPO-induced megakaryocytic differentiation. Recombinant TAT-HA-PRMT1 and TAT-HA-GFP (control) proteins were added to CD34+ cells. Entrance of the TAT-fused proteins into cells was examined by Western blot using anti-HA antibodies (A). ERK2 was used as a loading control. To examine the effect on megakaryocytic differentiation, TAT-fused proteins were added into cells 6 h before TPO treatment and again at the time of TPO treatment. After 15 days, cells were analyzed for expression of CD41 by flow cytometry (B and C). A representative result from donor 4 is shown in B.
Effects of PRMT2 and PRMT5 on PMA-induced Megakaryocytic Differentiation
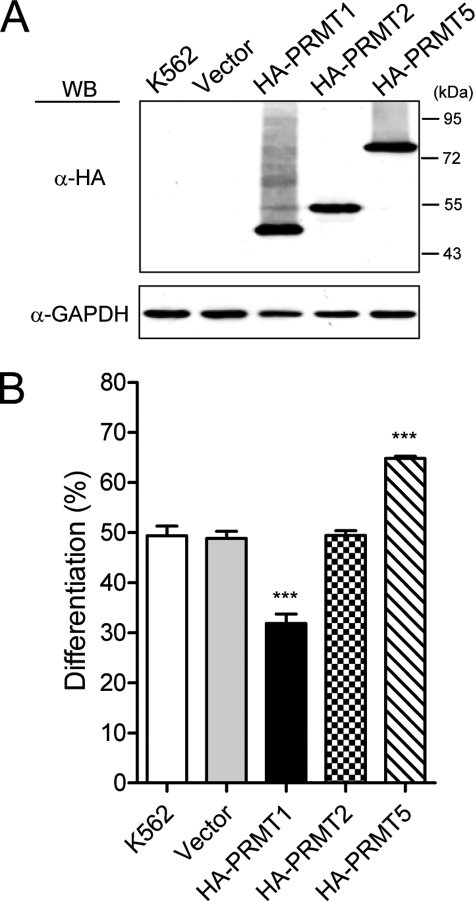
In mammals, PRMTs are classified according to the products of their enzymatic activity. The type I enzymes produce asymmetric ω-NG,NG-dimethylarginine and type II enzymes form symmetric ω-NG,N′G-dimethylarginine (16, 17). PRMT1 is the predominant type I enzyme (19, 20). To test whether other PRMTs play roles in N′G megakaryocytic differentiation, we further examined the effects of PRMT2 and PRMT5 by transient transfection. PRMT2 has been recently identified as a type I enzyme (31). Both PRMT1 and PRMT2 can function as a coactivator in regulating gene expression (32–35). In contrast to PRMT1, which suppressed differentiation, ectopic expression of PRMT2 had no apparent effect on PMA-induced differentiation (Fig. 7B). PRMT5 is a well characterized type II methyltransferase (16, 17). Interestingly, ectopic expression of PRMT5 exhibited a stimulatory effect on differentiation (Fig. 7B). These PRMTs were expressed at a similar level (Fig. 7A). These results suggested a unique role of PRMT1 in suppression of megakaryocytic differentiation.
FIGURE 7.
Effect of PRMT2 and PRMT5 on megakaryocytic differentiation. K562 cells were transiently transfected with pPCDNA3HA2 plasmids expressing either HA-tagged PRMT1, PRMT2, or PRMT5 proteins that were detected by Western blot (WB) using anti-HA antibodies (A). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. Megakaryocytic differentiation was analyzed by modified Wright-Giemsa staining 96 h after PMA treatment (B). All experiments were performed at least three times, and data are presented as means ± S.E.; ***, p < 0.005.
DISCUSSION
PRMT1 plays critical roles in various cellular processes (16, 17). We show in this study that PRMT1 negatively modulates differentiation toward the megakaryocytic lineage in the multipotent leukemia cell lines K562 and HEL. Similar results are also observed in human CD34+ hematopoietic cells (Fig. 6). These results are consistent with our observation that PRMT1 activity is down-regulated during megakaryocytic differentiation (supplemental Fig. S5). We also show that the p38 pathway is required for the suppressive effect of PRMT1, as suppression of megakaryocytic differentiation no longer occurs in the presence of the p38 inhibitor SB203580 or p38 shRNAs (Fig. 5). Consistent with these results, ectopic expression of PRMT1 potentiates p38 activation; however, knockdown of PRMT1 reduces its activation (Fig. 4). Previously, the role of p38 MAPK in megakaryocytic differentiation was ambiguous and controversial (6, 10, 13, 36–39). Here, we clearly show that the reduction of p38α greatly promotes differentiation (Fig. 5, B and E), whereas overexpression of p38α suppresses differentiation (Fig. 5, A and F). Pathologically, constitutively activated p38 MAPK has been found to be associated with poor differentiation in megakaryocytes and other hematopoietic cells in patients with myelodysplastic syndromes (40, 41). Inhibition of p38 enhances hematopoiesis in progenitors of myelodysplastic syndromes (40). These reports support the clinical relevance of our findings.
PRMT1 may function either directly on p38 MAPK or indirectly on upstream regulatory molecules. A preliminary examination using anti-asymmetric dimethylarginine antibodies did not detect modification of p38 MAPK (data not shown), so the direct target(s) are likely upstream molecules. The p38 MAPK is activated via phosphorylation at threonine and tyrosine residues by dual kinases MKK3, MKK4, and MKK6 (30). So far, to the best of our knowledge, these MAPK kinases are not reported to be modified in arginine residues. Members of the small GTPase Rho family, including Rac1 and Cdc42, are also upstream regulators of the p38 MAPK pathway. In TCR/CD28 costimulated T cells, the GDP-releasing factor Vav protein regulates the activity of Rac-1 and thus activates p38 MAPK (42). This costimulation also induces arginine methylation of Vav1 and its selective localization to the nucleus (43). Although the methyltransferase responsible for Vav1 arginine methylation has not yet been identified, this study indicates that arginine methylation of upstream molecules may play a role in modulation of p38 MAPK. A number of signaling molecules are shown to be modified by arginine methylation. Estrogen receptor α is transiently methylated by PRMT1, which then triggers its interaction with phosphatidylinositol 3-kinase and Src (44). PRMT1 also methylates the FOXO1 transcription factor and blocks its phosphorylation by Akt (45). These reports show that PRMT1-mediated methylation can lead to changes in activity, intracellular localization, or protein-protein interaction of the modified molecules. Further work is needed to determine the direct target(s) of PRMT1 in the p38 MAPK pathway.
Cell differentiation requires a balance between positive and negative regulatory events. ERK MAPK activation is essential for megakaryocytic differentiation of various cell lines and of primary CD34+ hematopoietic cells using different inducers (1, 8), suggesting its common role in megakaryocytic differentiation. Here, we have confirmed a negative role for p38α MAPK in PMA-induced megakaryocytic differentiation in K562 cells. Numerous studies have shown that ERK and p38 MAPKs can differentially regulate the same cellular events. For example, expression of cyclin D1 is positively regulated by the ERK MAPK pathway and negatively regulated by the p38 MAPK pathway in serum-stimulated fibroblasts (46). In K562 cells, cyclin D1 is up-regulated during PMA-induced megakaryocytic differentiation via the ERK MAPK pathway (47). In addition, overexpression of cyclin D1 is shown to be associated with the formation of polyploidy of megakaryocytes in transgenic mice (48). These observations raise the possibility that the activated p38 MAPK may counteract the effect of ERK MAPK on the expression of cyclin D1 and thus affect megakaryocytic differentiation in K562 cells. Furthermore, p38 MAPK has been reported to negatively regulate ERK under various stimulation conditions (49, 50). Insulin-stimulated induction of Egr-1 and Krox20 transcription is dependent on activation of ERK MAPK (49). Inhibition of the p38 MAPK pathway with specific inhibitors augments and prolongs ERK MAPK activation and further enhances induction of both genes (49). We did not observe inhibition of ERK MAPK activation upon overexpression of PRMT1 (Fig. 4A); however, PMA-induced megakaryocytic differentiation is suppressed, suggesting that p38 MAPK may act downstream of ERK MAPK. The activated ERK MAPK translocates to the nucleus where it activates transcription of specific genes (4). This process may be suppressed by p38. In monocytic THP-1 cells, nuclear translocation of activated ERK MAPK induced by PMA is modulated by a p38-specific inhibitor (51). These results support that p38 MAPK can potentially modulate the downstream events after ERK is activated to regulate megakaryocytic differentiation.
Expression of the surface marker CD41 is detected in hematopoietic progenitor cells (52) and is further stimulated during megakaryocytic commitment (13). In unstimulated CD34+ hematopoietic cells, RUNX1 positively regulates CD41 transcription, potentially as a result of arginine methylation of RUNX1 by PRMT1 (53). However, PMA-stimulated transcription of CD41 in K562 cells requires the cooperation of transcription factors GATA1, RUNX1, and CBFβ (54). Our results reveal that PRMT1 negatively regulates surface expression of CD41 in K562 cells simulated by PMA (Fig. 1E) and in CD34+ cells stimulated by TPO (Fig. 6), suggesting that PRMT1 may also regulate the activity of transcription factors responsible for stimulating expression of CD41 or for the presentation of CD41 on cell surface. The p38 and ERK MAPKs can either positively or negatively regulate GATA-1 activity in different cell contexts (55, 56). Whether GATA-1 activity can be regulated by PRMT1 through MAPK-dependent phosphorylation is of interest to be investigated.
TPO, alone or in combination with other cytokines, regulates various steps in megakaryopoiesis, including survival, expansion, and differentiation (1). Multiple pathways, including JAK/STATs, phosphatidylinositol 3-kinase/Akt, and ERK MAPK, are activated by TPO via its receptor Mpl (1, 8). The functional role of p38 MAPK is less studied and still elusive. The p38 MAPK is activated and essential for TPO-induced megakaryocytic differentiation in UT-7 cells expressing the Mpl receptor (57) and also for TPO-induced Hox4B expression and proliferation (58). However, treatment of SB203580 shows no effects on CD41 expression in human CD34+ hematopoietic progenitor cells (13) or polyploidy in murine CD41-positive cells (38). In primary hematopoietic progenitors, p38 is shown to mediate the suppressive effect of type I interferons on hematopoiesis (59). TPO may stimulate different pathways under different conditions such as developmental stages of cells used, cytokine combinations, and treatment protocols. The concentrations and specificity of inhibitors used could potentially lead to the discrepancy in the role of p38. Whether p38 MAPK mediates the suppressive effect of PRMT1 on differentiation of CD34+ hematopoietic cells requires more extensive investigation.
Because megakaryocytes are the precursors of platelets, searching for regulators to promote megakaryopoiesis for potential therapeutic treatment for thrombocytopenia has been an active area. In addition to activation of positive regulators, suppression of negative regulators can often contribute to further promotion of the cellular processes that they control. Our findings that both PRMT1 and p38 MAPK are negative regulators of megakaryocytic differentiation not only reveal new scientific insights into megakaryocytic differentiation but also provide a new perspective for a potential therapeutic application to promote megakaryopoiesis.
Supplementary Material
Acknowledgment
We thank K.-L. Tang for technique assistance.
This work was supported by National Science Council, Taiwan, Grant NSC 95-2311-B-010-002 (to W.-J. L.), University System of Taiwan Grant 97DFA2200014 (to W.-J. L.), and a grant from the Ministry of Education, Aim for the Top University Plan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- PMA
- phorbol 12-myristate 13-acetate
- PRMT1
- protein-arginine methyltransferase 1
- TPO
- thrombopoietin
- MAPK
- mitogen-activated protein kinase
- [3H]AdoMet
- S-adenosyl-l-[methyl-3H]methionine
- HA
- hemagglutinin
- ERK
- extracellular signal-regulated kinase
- shRNA
- short hairpin RNA
- JNK
- c-Jun NH2-terminal kinase
- hnRNP
- heterogeneous nuclear ribonucleoprotein.
REFERENCES
- 1.Kaushansky K. (2005) J. Clin. Invest. 115, 3339–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alitalo R. (1990) Leuk. Res. 14, 501–514 [DOI] [PubMed] [Google Scholar]
- 3.Racke F. K., Lewandowska K., Goueli S., Goldfarb A. N. (1997) J. Biol. Chem. 272, 23366–23370 [DOI] [PubMed] [Google Scholar]
- 4.Whalen A. M., Galasinski S. C., Shapiro P. S., Nahreini T. S., Ahn N. G. (1997) Mol. Cell. Biol. 17, 1947–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herrera R., Hubbell S., Decker S., Petruzzelli L. (1998) Exp. Cell Res. 238, 407–414 [DOI] [PubMed] [Google Scholar]
- 6.Jacquel A., Herrant M., Defamie V., Belhacene N., Colosetti P., Marchetti S., Legros L., Deckert M., Mari B., Cassuto J. P., Hofman P., Auberger P. (2006) Oncogene 25, 781–794 [DOI] [PubMed] [Google Scholar]
- 7.Goldfarb A. N., Delehanty L. L., Wang D., Racke F. K., Hussaini I. M. (2001) J. Biol. Chem. 276, 29526–29530 [DOI] [PubMed] [Google Scholar]
- 8.Séverin S., Ghevaert C., Mazharian A. (2010) J. Thromb. Haemost. 8, 17–26 [DOI] [PubMed] [Google Scholar]
- 9.Delehanty L. L., Mogass M., Gonias S. L., Racke F. K., Johnstone B., Goldfarb A. N. (2003) Blood 101, 1744–1751 [DOI] [PubMed] [Google Scholar]
- 10.Eriksson M., Arminen L., Karjalainen-Lindsberg M. L., Leppä S. (2005) Exp. Cell Res. 304, 175–186 [DOI] [PubMed] [Google Scholar]
- 11.Tseng C. P., Huang C. H., Tseng C. C., Lin M. H., Hsieh J. T., Tseng C. H. (2001) Biochem. Biophys. Res. Commun. 285, 129–135 [DOI] [PubMed] [Google Scholar]
- 12.Fichelson S., Freyssinier J. M., Picard F., Fontenay-Roupie M., Guesnu M., Cherai M., Gisselbrecht S., Porteu F. (1999) Blood 94, 1601–1613 [PubMed] [Google Scholar]
- 13.Miyazaki R., Ogata H., Kobayashi Y. (2001) Ann. Hematol. 80, 284–291 [DOI] [PubMed] [Google Scholar]
- 14.Rouyez M. C., Boucheron C., Gisselbrecht S., Dusanter-Fourt I., Porteu F. (1997) Mol. Cell. Biol. 17, 4991–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia J., de Gunzburg J., Eychène A., Gisselbrecht S., Porteu F. (2001) Mol. Cell. Biol. 21, 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bedford M. T., Clarke S. G. (2009) Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf S. S. (2009) Cell. Mol. Life Sci. 66, 2109–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin W. J., Gary J. D., Yang M. C., Clarke S., Herschman H. R. (1996) J. Biol. Chem. 271, 15034–15044 [DOI] [PubMed] [Google Scholar]
- 19.Pawlak M. R., Scherer C. A., Chen J., Roshon M. J., Ruley H. E. (2000) Mol. Cell. Biol. 20, 4859–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang J., Kao P. N., Herschman H. R. (2000) J. Biol. Chem. 275, 19866–19876 [DOI] [PubMed] [Google Scholar]
- 21.Bakker W. J., Blázquez-Domingo M., Kolbus A., Besooyen J., Steinlein P., Beug H., Coffer P. J., Löwenberg B., von Lindern M., van Dijk T. B. (2004) J. Cell Biol. 164, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cimato T. R., Tang J., Xu Y., Guarnaccia C., Herschman H. R., Pongor S., Aletta J. M. (2002) J. Neurosci. Res. 67, 435–442 [DOI] [PubMed] [Google Scholar]
- 23.Balint B. L., Szanto A., Madi A., Bauer U. M., Gabor P., Benko S., Puskás L. G., Davies P. J., Nagy L. (2005) Mol. Cell. Biol. 25, 5648–5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyata S., Mori Y., Tohyama M. (2008) Neurosci. Lett. 445, 162–165 [DOI] [PubMed] [Google Scholar]
- 25.Kwak Y. T., Guo J., Prajapati S., Park K. J., Surabhi R. M., Miller B., Gehrig P., Gaynor R. B. (2003) Mol. Cell 11, 1055–1066 [DOI] [PubMed] [Google Scholar]
- 26.Rezai-Zadeh N., Zhang X., Namour F., Fejer G., Wen Y. D., Yao Y. L., Gyory I., Wright K., Seto E. (2003) Genes Dev. 17, 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiou Y. Y., Lin W. J., Fu S. L., Lin C. H. (2007) Protein J. 26, 87–93 [DOI] [PubMed] [Google Scholar]
- 28.Yao C. L., Feng Y. H., Lin X. Z., Chu I. M., Hsieh T. B., Hwang S. M. (2006) Stem Cells Dev. 15, 70–78 [DOI] [PubMed] [Google Scholar]
- 29.Gump J. M., Dowdy S. F. (2007) Trends Mol. Med. 13, 443–448 [DOI] [PubMed] [Google Scholar]
- 30.Platanias L. C. (2003) Blood 101, 4667–4679 [DOI] [PubMed] [Google Scholar]
- 31.Lakowski T. M., Frankel A. (2009) Biochem. J. 421, 253–261 [DOI] [PubMed] [Google Scholar]
- 32.Qi C., Chang J., Zhu Y., Yeldandi A. V., Rao S. M., Zhu Y. J. (2002) J. Biol. Chem. 277, 28624–28630 [DOI] [PubMed] [Google Scholar]
- 33.Meyer R., Wolf S. S., Obendorf M. (2007) J. Steroid Biochem. Mol. Biol. 107, 1–14 [DOI] [PubMed] [Google Scholar]
- 34.Wang H., Huang Z. Q., Xia L., Feng Q., Erdjument-Bromage H., Strahl B. D., Briggs S. D., Allis C. D., Wong J., Tempst P., Zhang Y. (2001) Science 293, 853–857 [DOI] [PubMed] [Google Scholar]
- 35.Klinge C. M., Jernigan S. C., Mattingly K. A., Risinger K. E., Zhang J. (2004) J. Mol. Endocrinol. 33, 387–410 [DOI] [PubMed] [Google Scholar]
- 36.Jiang F., Jia Y., Cohen I. (2002) Blood 99, 3579–3584 [DOI] [PubMed] [Google Scholar]
- 37.Meshkini A., Yazdanparast R. (2008) Toxicol. In Vitro 22, 1503–1510 [DOI] [PubMed] [Google Scholar]
- 38.Mazharian A., Watson S. P., Séverin S. (2009) Exp. Hematol. 37, 1238–1249.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conde I., Pabon D., Jayo A., Lastres P., Gonzalez-Manchon C. (2010) Eur. J. Haematol. 84, 430–440 [DOI] [PubMed] [Google Scholar]
- 40.Navas T. A., Mohindru M., Estes M., Ma J. Y., Sokol L., Pahanish P., Parmar S., Haghnazari E., Zhou L., Collins R., Kerr I., Nguyen A. N., Xu Y., Platanias L. C., List A. A., Higgins L. S., Verma A. (2006) Blood 108, 4170–4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahjahan M., Dunphy C. H., Ewton A., Zu Y., Monzon F. A., Rice L., Chang C. C. (2008) Am. J. Clin. Pathol. 130, 635–641 [DOI] [PubMed] [Google Scholar]
- 42.Salojin K. V., Zhang J., Delovitch T. L. (1999) J. Immunol. 163, 844–853 [PubMed] [Google Scholar]
- 43.Blanchet F., Cardona A., Letimier F. A., Hershfield M. S., Acuto O. (2005) J. Exp. Med. 202, 371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Romancer M., Treilleux I., Leconte N., Robin-Lespinasse Y., Sentis S., Bouchekioua-Bouzaghou K., Goddard S., Gobert-Gosse S., Corbo L. (2008) Mol. Cell 31, 212–221 [DOI] [PubMed] [Google Scholar]
- 45.Yamagata K., Daitoku H., Takahashi Y., Namiki K., Hisatake K., Kako K., Mukai H., Kasuya Y., Fukamizu A. (2008) Mol. Cell 32, 221–231 [DOI] [PubMed] [Google Scholar]
- 46.Lavoie J. N., L'Allemain G., Brunet A., Müller R., Pouysségur J. (1996) J. Biol. Chem. 271, 20608–20616 [DOI] [PubMed] [Google Scholar]
- 47.Lee C. H., Yun H. J., Kang H. S., Kim H. D. (1999) IUBMB Life 48, 585–591 [DOI] [PubMed] [Google Scholar]
- 48.Sun S., Zimmet J. M., Toselli P., Thompson A., Jackson C. W., Ravid K. (2001) Haematologica 86, 17–23 [PubMed] [Google Scholar]
- 49.Keeton A. B., Bortoff K. D., Bennett W. L., Franklin J. L., Venable D. Y., Messina J. L. (2003) Endocrinology 144, 5402–5410 [DOI] [PubMed] [Google Scholar]
- 50.Ding X. Z., Adrian T. E. (2001) Biochem. Biophys. Res. Commun. 282, 447–453 [DOI] [PubMed] [Google Scholar]
- 51.Numazawa S., Watabe M., Nishimura S., Kurosawa M., Izuno M., Yoshida T. (2003) J. Biochem. 133, 599–605 [DOI] [PubMed] [Google Scholar]
- 52.Emambokus N. R., Frampton J. (2003) Immunity 19, 33–45 [DOI] [PubMed] [Google Scholar]
- 53.Zhao X., Jankovic V., Gural A., Huang G., Pardanani A., Menendez S., Zhang J., Dunne R., Xiao A., Erdjument-Bromage H., Allis C. D., Tempst P., Nimer S. D. (2008) Genes Dev. 22, 640–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elagib K. E., Racke F. K., Mogass M., Khetawat R., Delehanty L. L., Goldfarb A. N. (2003) Blood 101, 4333–4341 [DOI] [PubMed] [Google Scholar]
- 55.Towatari M., Ciro M., Ottolenghi S., Tsuzuki S., Enver T. (2004) Hematol. J. 5, 262–272 [DOI] [PubMed] [Google Scholar]
- 56.Stassen M., Klein M., Becker M., Bopp T., Neudörfl C., Richter C., Heib V., Klein-Hessling S., Serfling E., Schild H., Schmitt E. (2007) Mol. Immunol. 44, 926–933 [DOI] [PubMed] [Google Scholar]
- 57.Tang Y. S., Zhang Y. P., Xu P. (2008) Leukemia 22, 1018–1025 [DOI] [PubMed] [Google Scholar]
- 58.Kirito K., Fox N., Kaushansky K. (2003) Blood 102, 3172–3178 [DOI] [PubMed] [Google Scholar]
- 59.Verma A., Deb D. K., Sassano A., Uddin S., Varga J., Wickrema A., Platanias L. C. (2002) J. Biol. Chem. 277, 7726–7735 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.