Abstract
The lipid raft protein Flotillin-1 was previously shown to be required for cell proliferation. Here we show that it is critical for the maintenance of the levels of the mitotic regulator Aurora B. Knockdown of Flotillin-1 induced aberrant mitotic events similar to those produced by Aurora B depletion and led to a marked decline in Aurora B levels and activity. Transfection of wild-type full-length Flotillin-1 or forms directed to the nucleus increased Aurora B levels and activity. Flotillin-1 interacted with Aurora B directly through its SPFH domain in a complex distinct from the chromosomal passenger protein complex, and the two proteins co-purified in nuclear, non-raft fractions. These observations are the first evidence for a function of Flotillin-1 outside of lipid rafts and suggest its critical role in the maintenance of a pool of active Aurora B.
Keywords: Cell Cycle, Lipid Raft, Mitosis, Protein-Protein Interactions, RNA Silencing, Aurora B, Flotillin
Introduction
As a signaling regulatory molecule, Flotillin-1 tethers protein complexes to lipid rafts through interactions with the sorbin homology domains of the recruited proteins (1, 2). Flotillins oligomerize to form microdomain scaffolds that regulate the assembly of glycosylphosphatidylinositol-anchored and transmembrane proteins, favoring the communication with intracellular signal-transducing molecules (1, 3). Several membrane-resident receptor kinases, upon triggering by their cognate growth factors, form a ternary complex with the receptor-phosphorylated Cbl-associated protein and the ubiquitin ligase c-Cbl, and localize to lipid rafts via interaction with flotillins to initiate signaling (4, 5). In addition to its cell membrane-associated signaling functions, Flotillin-1 regulates clathrin-independent endocytic pathways, phagosome traffic, and actin cytoskeleton dynamics (6–9). An indication for a function of Flotillin-1 in non-membrane compartments was provided by the observation that it can translocate to the nucleus upon mitogenic stimuli (10). In that study, nuclear localization was shown to be required for Flotillin-1 to induce cell proliferation. Here we report that Flotillin-1 regulates the abundance and activity of the mitotic regulator Aurora B kinase.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfections, and Reagents
HeLa and PC3 cells were obtained from the American Type Culture Collection (Rockville, MD). Cells transduced with lentiviral particles were selected with 5 μg/ml puromycin (Sigma). siRNA5 duplexes for Flotillin-1 (Dharmacon, Lafayette, CO) or Aurora B (Qiagen) were transfected using TransIT HeLa Monster Transfection Reagent (Mirus Bio Corp., Madison, WI). Cell proliferation was measured daily by trypsinization and counting. Nocodazole (1 μm) treatments were carried out overnight. Antibodies and sources were as follows: Flotillin-1 and Aurora B (mouse) from BD Transduction Laboratories (Lexington, KY); hemagglutinin from Roche Diagnostics (Mannheim, Germany); β-tubulin and Aurora B (rabbit) from Sigma; phospho-histone H3 (Ser-10) from Upstate (Billerica, MA); actin and borealin from Santa Cruz Biotechnology (Santa Cruz, CA); INCENP from Cell Signaling (Denvers, MA); and survivin from Abcam (Cambridge, UK). Serum from a patient with CREST syndrome was kindly provided by O. Viñas (Hospital Clínic, Barcelona, Spain). Epoxomicin and lactacystin were from Sigma.
siRNAs and shRNAs Sequences
siRNAs F92, F93, and F94 were directed to sequences at the 3′-untranslated end of endogenous Flotillin-1 mRNA and are not present in exogenous HA-Flotillin-1 or GFP-NLS-Flotillin-1 constructs. The sequences targeted for RNA interference duplex or shRNA knockdown are shown in Table 1.
TABLE 1.
RNA interference target sequences
| siRNA/shRNA | Target sequence | Vendor |
|---|---|---|
| FLOT1-F92 | ACCUCACACUGCUAUGAUUdTdT | Qiagen |
| FLOT1-F93 | GAAUAUUUUCCUGACCAAGdTdT | Qiagen |
| FLOT1-F94 | UGUCCAUUGACAGUGAGGdTdT | Qiagen |
| FLOT1-shRNA-1 | GCAGAGAAGTCCCAACTAATT | Sigma |
| FLOT1-shRNA-2 | CCAGGACTATTTGCACTCTTT | Sigma |
| AURKB | AGGGAUCCCUUCUUUCCdTdT | Qiagen |
Plasmids
Plasmids HA-Flotillin-1, HA-Flot1ΔC, and Flot1-HA have been described (10). pGST-Aurora B and pGST-Survivin were kindly provided by S. P. Wheatley (University of Sussex, Falmer, Brighton, UK). The Myc-tagged Aurora B plasmid was kindly provided by E. Nigg (Max Planck Institute of Biochemistry, Martinsried, Germany). The FLOTILLIN-1 domain and the SPFH domain of Flotillin-1 were subcloned in-frame with the hemagglutinin tag into the EcoRI and BglII sites, and the BglII and KpnI sites of pCMV-HA, to yield plasmids HA-FLOT and HA-SPFH. pGST-Flotillin-1 was generated by cloning the EcoRI-XhoI fragment containing the Flotillin-1 full-length cDNA into pGEX-4T. The nuclear localization sequence from the SV40 T antigen was inserted in-frame downstream of the GFP sequence into pGFP-Flot1 (10).
Real-time Reverse Transcription-PCR
RNA was isolated from cells with the RNeasy Mini Kit (Qiagen). After reverse transcription using random primers (Invitrogen) and Moloney murine leukemia virus reverse transcriptase, the reaction products were analyzed by PCR with SYBR Green incorporation. The sequences of the primers used are shown in Table 2. Two independent experiments with triplicate determinations each were performed. The ΔΔCt method was applied to estimate relative transcript levels, normalized for ribosomal RPS14 transcript amplification.
TABLE 2.
Primers used for real-time reverse transcription-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| FLOT1 | GGCAGAAATTCTCAGAACAG | GTGCAAATAGTCCTGGTCAT |
| AURKB | TCCTCTTCAAGTCCCAGATA | GTTGTAGAGACGCAGGATGT |
| INCENP | CAAGAAGACTGCCGAAGAGC | TCAGGAGCCTCTCCAGGTAA |
| RPS14 | GGCAGACCGAGATGAATCCTCA | CAGGTCCAGGGGTCTTGGTC |
Immunofluorescence
Immunofluorescence was performed as described before (10). DNA was stained with Hoechst 33258 (Sigma). Fluorescent images were captured with a confocal-spectral microscope (FV1000, Olympus, Tokyo, Japan) and quantified with Olympus Fluorview software. In all quantifications, at least 300 cells were scored in duplicate experiments.
Flow Cytometry
Cells were fixed and washed in 70% ethanol in phosphate-buffered saline at −20 °C for 1 h, washed several times with cold phosphate-buffered saline, treated with RNase A (50 μg/ml) and propidium iodide (25 μg/ml), and analyzed on an Epics XL flow cytometer (Coulter, Miami, FL) for DNA content (filter set at 675 nm). Ten thousand events were analyzed. Cells were assigned to specific cell cycle phases by applying the Multicycle cell cycle analysis software (Phoenix Flow Systems, San Diego, CA).
Immunoprecipitation and Pulldown Assays
Co-immunoprecipitation experiments were performed as described (10). For pulldown assays, GST-tagged proteins were affinity-purified from bacteria with glutathione-Sepharose 4B beads (Sigma), eluted with 10 mm reduced glutathione and frozen until use. Cell lysates were incubated with glutathione-Sepharose-immobilized GST-tagged proteins, washed, eluted with Laemmli loading buffer, and subjected to immunoblotting.
TUNEL Assays
TUNEL assays were performed using the In Situ Cell Detection Kit, TMR Red (Roche Applied Science). Samples were analyzed by immunofluorescence.
Direct Interaction of Aurora B and Flotillin-1 Proteins
In vitro transcribed and translated [35S]Met Flotillin-1 was synthesized using pCDNA3-Flot1 and the T7 TnT-coupled transcription/translation system (Promega, Madison, WI). Translated reactions were incubated with GST-Aurora B pre-bound on glutathione-Sepharose beads in binding buffer (10 mm Tris-HCl (pH 8.0), 150 mm NaCl, 10% glycerol, 0.2% Triton X-100, 2 mm sodium vanadate, 10 mm NaF, protease inhibitor mixture, and 200 μg/ml of RNase A) at 4 °C for 1 h. Following washes, bound proteins were separated by SDS-PAGE, and gels were fluorographed, dried, and exposed to film.
Sucrose Density Gradient Centrifugation
Cells were disrupted with a Dounce homogenizer, and nuclear and non-nuclear fractions were separated by centrifugation as described (10). The non-nuclear fractions were further fractionated on discontinuous sucrose gradients. Fractions were collected and analyzed by Western blotting.
RESULTS
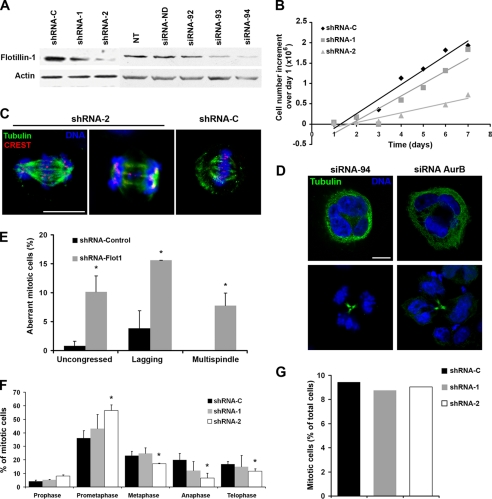
We had previously observed that Flotillin-1 depletion causes a significant inhibition of cell proliferation (10). To further investigate this effect, we tested siRNA duplexes (siRNA-92, -93, and -94) or shRNAs (shRNA-1 and -2) that target five distinct sequences on the Flotillin-1 mRNA and deplete protein levels with different efficiencies (Fig. 1A). Of these, siRNA-94 and shRNA-2 reproducibly inhibited Flotillin-1 expression at significant levels and were used in subsequent experiments (Figs. 1 and 2). The ensuing proliferative inhibition showed a good correlation with the degree of Flotillin-1 depletion, obtained with two different shRNA sequences (shRNA-1 and -2, Fig. 1B). Growth inhibition was accompanied with the accumulation of a significant fraction of cells with aberrant mitotic events, including uncongressed and lagging chromosomes, multispindle cells (Fig. 1, C–E), multinucleated and apoptotic cells (Fig. 1D, and supplemental Fig. S1). Cells treated with control siRNA or shRNA duplexes presented normal mitotic morphologies and correct chromosome congression at the metaphase plate, with <5% showing aberrant mitoses in all cases. Knockdown of Flotillin-1 caused an accumulation of cells in prometaphase and a decrease in the proportion of cells distributed in metaphase, anaphase, and telophase (Fig. 1F), without affecting the mitotic index (Fig. 1G) or the cell cycle phase distribution (supplemental Fig. S2).
FIGURE 1.
Knockdown of Flotillin-1 inhibits cell proliferation and induces mitotic defects. A, specific depletion of Flotillin-1 protein in HeLa cells by distinct siRNAs (100 nm) or shRNAs, each targeting different sequences on the Flotillin-1 mRNA, analyzed by Western blotting. Controls siRNA-ND and shRNA-C do not target any human gene-specific sequences. NT, non-transfected cells. Actin is used as a control for protein loading. B, different degrees of Flotillin-1 knockdown by shRNA-1 and -2 inhibit HeLa cell proliferation at different levels. Cells, transduced with shRNA, were grown in complete medium and triplicate wells counted every day after trypsinization. C, aberrant mitotic figures associated with Flotillin-1 depletion. Cells were stained for tubulin (green), kinetochores (red), and DNA (blue, Hoechst 33258), and images were captured by confocal microscopy. Scale bar, 10 μm. D, depletion of Flotillin-1 and Aurora B by RNA interference cause multipolar and multinucleated figures. Cells were stained for tubulin and DNA, and images were captured by confocal microscopy. Scale bar, 10 μm. E, cells with uncongressed and lagging chromosomes and multispindle figures were scored as a fraction of the total number of cells in mitosis and are indicated as “Aberrant mitotic cells.” At least 500 mitotic cells were counted per each condition. *, p < 0.05 (Student's t test). F, distribution of cells in different mitotic phases after Flotillin-1 depletion by two distinct shRNAs. Cells in different stages of mitosis were counted after staining for lamin A/C and DNA, and are represented over the total number of mitotic cells. *, p < 0.05. At least 300 cells were counted in each determination in two independent experiments. G, mitotic index is not affected by depletion of Flotillin-1. The number of mitotic cells for each shRNA was scored and is represented over the total number of cells. At least 300 cells were counted per condition.
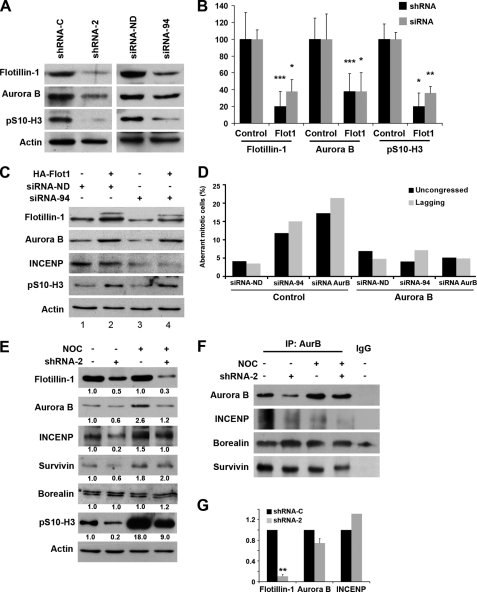
FIGURE 2.
Depletion of Flotillin-1 causes the down-regulation of Aurora B. A, representative image of the depletion of Flotillin-1 by different shRNAs or siRNAs causing a significant decline of Aurora B levels and of phosphorylated histone H3 (Ser-10). After lentiviral infection or transfection of the indicated shRNA or siRNA, cells were analyzed by Western blotting. Actin signals did not show significant variations between samples in these experiments and were used as indicators of protein loading and normalization. B, quantification of normalized Western blotting signals by scanning densitometry, shown as the average of three independent experiments similar to that in A. *, p < 0.05; **, p < 0.005; ***, p < 0.001. C, exogenous HA-Flotillin-1 rescues the down-regulation of Aurora B induced by RNA interference-mediated depletion of endogenous Flotillin-1. Cells were transfected with siRNA-94 specifically targeting endogenous Flotillin-1, or control siRNA, and HA-Flotillin-1, resistant to siRNA-94, or a control plasmid (2 μg) and analyzed by Western blotting 48 h after transfection. Flotillin-1 antibody also identifies exogenous HA-Flot1 (slower migrating band). Signal for actin was used as a control for protein loadings. D, transfection of Myc-Aurora B reverts the mitotic phenotypes observed after depletion of Flotillin-1 or Aurora B. Cells were transfected with the indicated siRNA and either control or Myc-Aurora B plasmids (2 μg). After 48 h, cells were fixed and stained for tubulin and DNA. The number of mitotic cells harboring uncongressed and lagging chromosomes was scored relative to the total number of cells in mitosis and is indicated as “Aberrant mitotic cells.” At least 500 mitotic cells were counted per condition. E, Flotillin-1 depletion causes a decrease in Aurora B in mitotic cells. Equal amounts of protein extracts (50 μg) from Flotillin-1-depleted cells, enriched by overnight nocodazole treatment (1 μm), or control untreated cells, were analyzed by Western blotting with the indicated antibodies. *, slower specific band. Actin was used as a control for protein loadings and normalization. Numbers at the bottom of each lane indicate relative levels of expression normalized to actin and to control untreated cells. F, the formation of the CPC is compromised in cells depleted of Flotillin-1. Equal amounts of protein extracts (300 μg) from control and Flotillin-1-depleted cells, without and with nocodazole (1 μm, overnight) were immunoprecipitated with Aurora B antibody (3 μg/sample). Immunoprecipitated proteins were analyzed by Western blotting with the indicated antibodies. IgG, control antibody used for immunoprecipitation. The faster migrating band in the Western blot for Borealin is nonspecific. G, Flotillin-1 knockdown does not significantly affect Aurora B or INCENP transcript levels. Real-time reverse transcription-PCR was used to quantify the levels of Aurora B, INCENP, and Flotillin-1 transcripts in HeLa cells treated with shRNA-2. **, p < 0.005.
These effects are reminiscent of those caused by depletion of the chromosomal passenger complex (CPC) protein Aurora B (11). Indeed, RNA interference depletion of Aurora B in HeLa cells led to a similar failure of chromosomes to congress, failure to complete cytokinesis, and the accumulation of multinucleated cells (Fig. 1D; see also Fig. 2D).
Because of the above phenotype, we hypothesized the occurrence of a functional association between Flotillin-1 and Aurora B. Depletion of Flotillin-1 by siRNA-94 or shRNA-2 was followed by a significant decline in Aurora B protein levels (Fig. 2, A and B). The decrease in Aurora B was paralleled by a marked decrease in the phosphorylation of histone H3 at serine 10, a major substrate of Aurora B activity (12, 13) (Fig. 2 (A and B) and supplemental Fig. S3). Likewise, depletion of Flotillin-1 caused a reduction in the levels of INCENP, a protein tightly associated with Aurora B (Fig. 2, C and E). The stability and relative abundance of Aurora B and INCENP are strongly interdependent (14), and therefore the down-regulation of INCENP after Flotillin-1 knockdown could be an indirect consequence of the depletion of Aurora B. Because INCENP is an activator of Aurora B (11), its concomitant decrease after Flotillin-1 knockdown might also contribute to the significant reduction of histone H3 (Ser-10) phosphorylation observed. Transfection of cells depleted of endogenous Flotillin-1 with HA-Flotillin-1, resistant to siRNA-94 and specifically targeting the endogenous protein, re-established the levels of Aurora B and the levels of phosphorylation of histone H3 at serine 10 (Fig. 2C). Similarly, expression in these cells of an Myc-tagged form of Aurora B led to the complete correction of the chromosomal abnormalities caused by Flotillin-1 depletion (Fig. 2D). These results suggest that the mitotic phenotypes caused by knockdown of Flotillin-1 are due to a down-regulation of Aurora B. Supportive of these results, the depletion of Flotillin-1 and the concomitant down-regulation of Aurora B and phospho-histone H3 (Ser-10) levels were also detected in PC3 prostate cancer cells (supplemental Fig. S4).
The best described function of Aurora B is in mitosis. As expected, in mitotically enriched nocodazole-treated cells, depletion of Flotillin-1 clearly diminished the levels of Aurora B and phosphorylated histone H3 (Ser-10) (Fig. 2E). In addition, INCENP levels also diminished in both mitotic and asynchronous cells depleted of Flotillin-1, although changes in other CPC components in mitotic cells were not significant. The decrease in Aurora B levels that follows knockdown of Flotillin-1 might reflect a decrease in the amount of holo-CPC formed in mitosis, as also suggested by the altered mitotic phenotypes observed. To explore this possibility, equal amounts of proteins from cells depleted of Flotillin-1 and treated, or not, with nocodazole were immunoprecipitated with Aurora B antibody (Fig. 2F). The amount of Aurora B and co-immunoprecipitated INCENP and survivin was decreased in Flotillin-1-depleted cells, suggesting a depletion of both the Aurora B/INCENP subcomplex and the holo-CPC in cells depleted of Flotillin-1 (11, 14, 15).
Knockdown of Flotillin-1 by shRNAs did not significantly affect the levels of Aurora B or INCENP transcripts, as shown by quantitative real-time reverse transcription-PCR (Fig. 2G), ruling out a direct effect on their mRNA levels and suggesting that the down-regulation of Aurora B and INCENP proteins in Flotillin-1-depleted cells is a post-transcriptional effect. Treatment of cells with the proteasome inhibitors lactacystin or epoxomicin restored to control levels the Aurora B protein levels that were down-regulated upon Flotillin-1 depletion, suggesting that Flotillin-1 depletion might affect the stability of Aurora B (supplemental Fig. S5). However, the half-life of the kinase was not significantly different in Flotillin-1-depleted from control cells (supplemental Fig. S6), suggesting that mechanisms other than protein stability might be involved in the regulation of Aurora B levels by Flotillin-1.
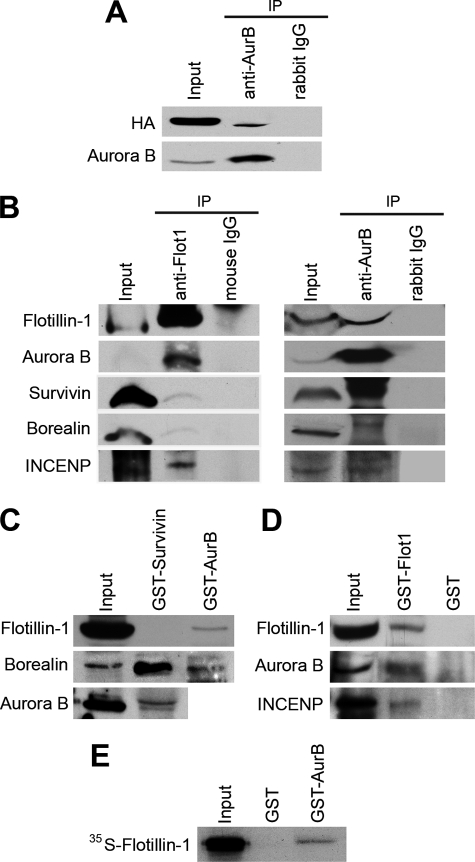
Next, we ascertained if the functional association observed between Flotillin-1 and Aurora B involved physical interactions between these proteins. Both endogenous and transfected Flotillin-1 co-immunoprecipitated with endogenous Aurora B and INCENP but not with borealin or survivin (Fig. 3(A and B) and supplemental Fig. S7). In reciprocal experiments, anti-Aurora B brought down Flotillin-1 and, as expected, also survivin, borealin and INCENP (Fig. 3B and supplemental Fig. S7). Moreover, GST-Aurora B, but not GST-survivin, pulled down endogenous and exogenous Flotillin-1 (Figs. 3C and 4A). As controls, both GST-Aurora B and GST-survivin pulled down the CPC protein borealin (Fig. 3C). Conversely, GST-Flotillin-1 pulled down endogenous Aurora B and INCENP from whole cell extracts (Fig. 3D). In vitro translated Flotillin-1 specifically interacted with immobilized GST-Aurora B, evidencing a direct interaction between the two proteins (Fig. 3E). These results support the occurrence of a complex between Flotillin-1 and Aurora B distinct from the canonical CPC. Although Aurora B forms a tight tetrameric complex with the CPC proteins in mitotic cells (15), a substantial pool of Aurora B is found in other complexes (16–18), including an Aurora B/INCENP subcomplex devoid of survivin or borealin (16), which does not target to any defined structure during mitosis and might have functions distinct from the CPC (15). This subcomplex has been suggested to be directly responsible for the phosphorylation of histone H3 at Ser-10 (13, 19). Consistent with its association with this non-CPC Aurora B/INCENP complex, Flotillin-1 did not localize to kinetochores during mitosis (supplemental Fig. S8), and, in mitotic cells depleted of Flotillin-1, no changes were seen in the expected localization of the CPC proteins Aurora B, survivin, and INCENP (supplemental Fig. S9).
FIGURE 3.
Flotillin-1 interacts with Aurora B in vivo and in vitro. A, transfected HA-Flotillin-1 and endogenous Flotillin-1 (B) co-immunoprecipitate with endogenous Aurora B. Endogenous Flotillin-1 associated with Aurora B and INCENP but not borealin or survivin. In reciprocal experiments, Aurora B co-immunoprecipitated with Flotillin-1, survivin, borealin, and INCENP. C, GST-Aurora B, but not GST-survivin, pulled down endogenous Flotillin-1. HeLa cell lysates were incubated with beads loaded with GST-survivin or GST-Aurora B for 1 h at 4 °C and washed, and eluted proteins were analyzed by Western blotting. D, GST-Flotillin-1 pulled down endogenous Flotillin-1, Aurora B, and INCENP. HeLa cell lysates were incubated with GST-Flotillin-1 or GST beads for 1 h at 4 °C and washed, and eluted proteins were analyzed by Western blotting. E, Flotillin-1 interacts in vitro with GST-Aurora B. pCDNA3-Flot1 was transcribed and translated in vitro using [35S]Met and reticulocyte extracts. Translated proteins were incubated with GST-Aur B or control GST, for 1 h at 4 °C, washed, and analyzed by SDS-PAGE and fluorography.
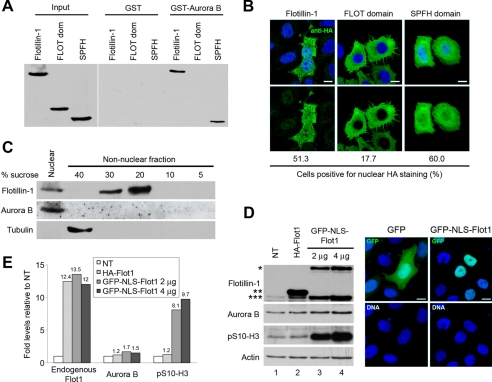
FIGURE 4.
Nuclear localization of Flotillin-1 is relevant for its action on Aurora B. A, Flotillin-1 interacts with Aurora B through its SPFH domain. GST-Aurora B was used in pulldown assays performed with lysates of cells transfected with HA-Flotillin-1, HA-FLOT-domain, or HA-SPFH-domain, visualized by Western blotting with anti-HA antibody. B, the SPFH domain, but not the FLOTILLIN domain, is sufficient for nuclear localization of Flotillin-1. Confocal images are representative of subdomains localization: anti-HA antibody identifies transfected cells. DNA was stained with Hoechst 33258. Quantification of nuclear localization, scored in at least 300 cells, is shown at the bottom. C, Aurora B localizes to nucleus and does not partition to lipid rafts. Nuclear and non-nuclear fractions were separated, and the latter was fractionated on discontinuous sucrose gradients. Lipid rafts were fractionated at 20% sucrose, as shown by enrichment of Flotillin-1. Reactivity with tubulin indicates a correct fractionation. D, overexpression of GFP-NLS-Flotillin-1 induces increased Aurora B protein levels and greatly induces the phosphorylation of histone H3 at Ser-10. Western blotting of cells transfected with pGFP (2 μg, lane 1), HA-Flotillin-1 (2 μg, lane 2), or GFP-NLS-Flot1 (2 μg, lane 3; and 4 μg, lane 4). Antibody to Flotillin-1 identifies GFP-NLS-Flot1 (*), HA-Flot1 (**), and the endogenous protein (***). Actin levels were used as protein loading controls and as normalization standards for the Western blotting signals. Quantifications of proteins levels are shown in panel E. Right panel, confocal microscopy images from cells transfected with GFP-NLS-Flotillin-1 localized exclusively to the nucleus, whereas GFP alone distributed to both nuclear and cytoplasmic compartments. DNA was stained with Hoechst 33258. Scale bar, 10 μm. E, graph shows relative levels of endogenous Flotillin-1, Aurora B, and phosphorylated histone H3 (Ser-10) compared with levels in GFP-transfected cells obtained by scanning densitometry of the Western blot signals in D, after normalization to actin levels.
Flotillin-1 contains two major functional and structural domains, the SPFH domain, also present in stomatin, prohibitin, podocin, and erlin, and the coiled-coil FLOTILLIN domain, shared only with the closely related paralog Flotillin-2 (20). Pulldown experiments with lysates from cells transfected with HA-Flot1-SPFH, expressing only the SPFH domain, or with HA-Flot1-FLOT, expressing only the FLOTILLIN coiled-coil domain, showed that the SPFH domain, but not the FLOTILLIN domain of Flotillin-1, was sufficient to associate with Aurora B (Fig. 4A). Immunofluorescent analysis of cells transfected with full-length or deletion variants showed that the SPFH domain of Flotillin-1, but not its coiled-coil domain, retained the capacity for nuclear localization, comparable to that of the full-length protein (Fig. 4B). Furthermore, subcellular fractionation and sucrose gradient centrifugation showed that, unlike Flotillin-1, Aurora B did not sediment in the lipid raft fraction, whereas both Flotillin-1 and Aurora B co-purified in the nuclear fraction (Fig. 4C). These observations suggest that Flotillin-1 and Aurora B interact outside of lipid rafts and mainly in the nucleus, through the SPFH domain of Flotillin-1.
To study the effects of overexpression and nuclear localization of Flotillin-1 on Aurora B protein levels, HeLa cells were transfected either with HA-Flotillin-1 or a form forcefully directed to the nucleus through the inclusion of a nuclear localization signal at the N terminus of the protein, GFP-NLS-Flot1. Transfection of increasing amounts of the latter plasmid, but not a control plasmid expressing GFP alone, resulted in corresponding increases in the levels of Aurora B (Fig. 4, D and E). Transfection of GFP-NLS-Flot1 induced a large enhancement of histone H3 (Ser-10) phosphorylation that was significantly superior to the increase in Aurora B levels observed in the same experiments (Fig. 4, D and E), suggesting that Flotillin-1 regulates not only the protein levels but also the enzymatic activity of the kinase. In contrast, transfection of Flotillin-1-HA or HA-Flotillin-1ΔC, forms that are deficient in nuclear translocation and are unable to induce cell proliferation (10), did not induce increased levels of Aurora B (supplemental Fig. S10). In these experiments, the levels of endogenous Flotillin-1 were significantly increased after transfection of HA-Flot1 or GFP-NLS-Flot1 (Fig. 4, D and E; see also Fig. 2C), suggesting that overexpression of exogenous Flotillin-1 leads to the accumulation of the endogenous protein. It has been shown that the formation of homo- and hetero-oligomers stabilizes Flotillin-1 (25). These observations, together with the significant decline in Aurora B protein levels upon depletion of Flotillin-1, reinforce the hypothesis of a direct correlation between levels and nuclear localization of Flotillin-1 with levels and activity of Aurora B.
DISCUSSION
Aurora B kinase activity peaks in mitosis, when it promotes the destabilization and release of microtubules that are incorrectly attached to kinetochores (11). After metaphase, the kinase either associates with the central spindle in anaphase or is inactivated by PP2A (21). Termination of Aurora B is also achieved by APC/C ubiquitination and proteasome degradation (22, 23). Inhibition of Aurora B leads to mono-orientated and mal-orientated chromosomes (24, 25) and impairs cytokinesis with appearance of multinucleated cells (11, 12, 14). Aurora B also has functions outside of mitosis, as exemplified by its regulation of mammalian target of rapamycin activity whereby it is required for the G1- to S-phase transition (18).
Our observations suggest that Aurora B protein levels and activity are dependent on the levels of Flotillin-1. In our experiments, knockdown of Flotillin-1 caused a decline in cell proliferation that was associated with a decrease in the levels and function of Aurora B. Conversely, overexpression of Flotillin-1 was associated with increased Aurora B levels and cell proliferation and significantly stimulated phosphorylation of its major substrate histone H3 (Ser-10).
Our evidence also suggests that Flotillin-1 forms a complex with Aurora B and INCENP that does not contain survivin or borealin, and it is therefore different from the chromosomal passenger complex (14, 17). In agreement with its association with a non-CPC Aurora B/INCENP complex, Flotillin-1 failed to localize to kinetochores in mitosis. Flotillin-1 interacted with Aurora B directly through its SPFH domain and, when it was directed to the nucleus, greatly induced the activity of the kinase on its substrate histone H3 (Ser-10). Our observation that only nuclear Flotillin-1 participates in the regulation of Aurora B is in agreement with our previous findings showing that only forms of Flotillin-1 capable of nuclear translocation are able to stimulate cell proliferation (10). Although Flotillin-1 does not interact with the CPC at mitosis, it might affect its function by determining the amount of Aurora B available to enter the complex, as suggested by our observation that depletion of Flotillin-1 was accompanied with a concomitant depletion of holo-CPC.
The requirement for Flotillin-1 to maintain Aurora B function and holo-CPC abundance represents a new function for Flotillin-1, possibly related to the chaperone functions found for other SPFH domain-containing proteins (26, 27). Prohibitins show a partial homology with GroEL/Hsp60 that has suggested a chaperone function for oligomerized prohibitins, defined as “holdases,” for the assembly of the respiratory chain complexes in mitochondria (28, 29). Similarly, the notion that flotillins form scaffolds for protein assemblies that are involved in signaling across the plasma membrane is supported by several evidences (1, 30). Flotillin-1 and Flotillin-2 associate in hetero- or homo-oligomers that stabilize the oligomerized proteins, such that the proteasome-dependent degradation of Flotillin-1 is accelerated in the absence of Flotillin-2 (26). Our observation that the overexpression of exogenous HA-Flot1 and GFP-NLS-Flot1 significantly increased the levels of endogenous Flotillin-1 may be related to the formation of stable homo-oligomers between endogenous and exogenous Flotillin-1.
Currently we do not know the mechanism by which Flotillin-1 depletion causes a down-regulation of Aurora B. Our observations, that Flotillin-1 depletion induces a down-regulation of Aurora B but does not affect its transcript levels or its protein half-life, might suggest regulation at levels other than transcription or protein degradation, possibly by modulating the rate of translation of Aurora B. On the other hand, overexpression of forms of Flotillin-1 that can translocate to the nucleus increases the levels of Aurora B and strongly enhances its activity, as deduced from hyperphosphorylation of histone H3 (Ser-10). Flotillin-2, a closely related Flotillin-1 paralog, does not translocate to the nucleus (10), and therefore it would not be expected to participate in the nuclear Flotillin-1/Aurora B complex observed here.
To our knowledge, our observations are the first evidence that Flotillin-1 plays relevant physiological roles beyond its previously known functions in cell and organelle membranes, namely the regulation of the overall abundance of Aurora B for a correct and timely cell cycle progression. These results are compatible with a model in which Flotillin-1 is released from its membrane localization in response to extracellular cues, such as serum growth factor stimulation (10), followed by migration to the nucleus where it enhances Aurora B kinase activity and facilitates CPC function. It remains to be explored if the nuclear function of Flotillin-1 observed in the present study is specific for a subset of proteins with which it associates, such as Aurora B, or if it involves more general mechanisms that may regulate additional proteins involved in cell cycle control. Because increased levels of Aurora B are frequently associated with the development of cancer (31), targeting Flotillin-1 might represent a novel approach to modulate Aurora B activity and a potentially useful avenue for investigation in cancer therapy.
Supplementary Material
Acknowledgment
We thank V. Plans for help with immunofluorescence.
This work was supported by the Ministry of Science and Education (Grants SAF2005-05848, SAF2008-03936, and SAF2008-04136-C02-01), the Ministry of Science and Technology (Grant SAF2005-05109-C02-01), the Fundació Marató TV3 (Grants 052610 and 053130), the Institut de Recerca Vall d'Hebrón, and the Centre de Referència en Biotecnologia de Catalunya.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S10.
- siRNA
- small interference RNA
- shRNA
- short hairpin RNA
- GST
- glutathione S-transferase
- CMV
- cytomegalovirus
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- CPC
- chromosomal passenger complex
- TUNEL
- terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling.
REFERENCES
- 1.Langhorst M. F., Reuter A., Stuermer C. A. (2005) Cell Mol. Life Sci. 62, 2228–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simons K., Toomre D. (2000) Nat. Rev. Mol. Cell Biol. 1, 31–39 [DOI] [PubMed] [Google Scholar]
- 3.Stuermer C. A., Lang D. M., Kirsch F., Wiechers M., Deininger S. O., Plattner H. (2001) Mol. Biol. Cell 12, 3031–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann C. A., Ribon V., Kanzaki M., Thurmond D. C., Mora S., Shigematsu S., Bickel P. E., Pessin J. E., Saltiel A. R. (2000) Nature 407, 202–207 [DOI] [PubMed] [Google Scholar]
- 5.Limpert A. S., Karlo J. C., Landreth G. E. (2007) Mol. Cell. Biol. 27, 5686–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dermine J. F., Duclos S., Garin J., St-Louis F., Rea S., Parton R. G., Desjardins M. (2001) J. Biol. Chem. 276, 18507–18512 [DOI] [PubMed] [Google Scholar]
- 7.Glebov O. O., Bright N. A., Nichols B. J. (2006) Nat. Cell Biol. 8, 46–54 [DOI] [PubMed] [Google Scholar]
- 8.Liu J., Deyoung S. M., Zhang M., Dold L. H., Saltiel A. R. (2005) J. Biol. Chem. 280, 16125–16134 [DOI] [PubMed] [Google Scholar]
- 9.Neumann-Giesen C., Fernow I., Amaddii M., Tikkanen R. (2007) J. Cell Sci. 120, 395–406 [DOI] [PubMed] [Google Scholar]
- 10.Santamaría A., Castellanos E., Gómez V., Benedit P., Renau-Piqueras J., Morote J., Reventós J., Thomson T. M., Paciucci R. (2005) Mol. Cell. Biol. 25, 1900–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruchaud S., Carmena M., Earnshaw W. C. (2007) Nat. Rev. Mol. Cell Biol. 8, 798–812 [DOI] [PubMed] [Google Scholar]
- 12.Giet R., Glover D. M. (2001) J. Cell Biol. 152, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams R. R., Maiato H., Earnshaw W. C., Carmena M. (2001) J. Cell Biol. 153, 865–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda R., Körner R., Nigg E. A. (2003) Mol. Biol. Cell 14, 3325–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeyaprakash A. A., Klein U. R., Lindner D., Ebert J., Nigg E. A., Conti E. (2007) Cell 131, 271–285 [DOI] [PubMed] [Google Scholar]
- 16.Gassmann R., Carvalho A., Henzing A. J., Ruchaud S., Hudson D. F., Honda R., Nigg E. A., Gerloff D. L., Earnshaw W. C. (2004) J. Cell Biol. 166, 179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange B. M., Rebollo E., Herold A., González C. (2002) EMBO J. 21, 5364–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song J., Salek-Ardakani S., So T., Croft M. (2007) Nat. Immunol. 8, 64–73 [DOI] [PubMed] [Google Scholar]
- 19.Vagnarelli P., Earnshaw W. C. (2004) Chromosoma 113, 211–222 [DOI] [PubMed] [Google Scholar]
- 20.Browman D. T., Hoegg M. B., Robbins S. M. (2007) Trends Cell Biol. 17, 394–402 [DOI] [PubMed] [Google Scholar]
- 21.Sun L., Gao J., Dong X., Liu M., Li D., Shi X., Dong J. T., Lu X., Liu C., Zhou J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7153–7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart S., Fang G. (2005) Cancer Res. 65, 8730–8735 [DOI] [PubMed] [Google Scholar]
- 23.Nguyen H. G., Chinnappan D., Urano T., Ravid K. (2005) Mol. Cell Biol. 25, 4977–4992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C. L., Peters J. M. (2003) J. Cell Biol. 161, 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lampson M. A., Renduchitala K., Khodjakov A., Kapoor T. M. (2004) Nat. Cell Biol. 6, 232–237 [DOI] [PubMed] [Google Scholar]
- 26.Solis G. P., Hoegg M., Munderloh C., Schrock Y., Malaga-Trillo E., Rivera-Milla E., Stuermer C. A. (2007) Biochem. J. 403, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winter A., Kämäräinen O., Hofmann A. (2007) Proteins 68, 353–362 [DOI] [PubMed] [Google Scholar]
- 28.Nijtmans L. G., de Jong L., Artal Sanz M., Coates P. J., Berden J. A., Back J. W., Muijsers A. O., van der Spek H., Grivell L. A. (2000) EMBO J. 19, 2444–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatsuta T., Model K., Langer T. (2005) Mol. Biol. Cell 16, 248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann-Giesen C., Falkenbach B., Beicht P., Claasen S., Lüers G., Stuermer C. A., Herzog V., Tikkanen R. (2004) Biochem. J. 378, 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carvajal R. D., Tse A., Schwartz G. K. (2006) Clin. Cancer Res. 12, 6869–6875 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.