Abstract
The single transmembrane domain serine/threonine kinase activin receptor type IIB (ActRIIB) has been proposed to bind key regulators of skeletal muscle mass development, including the ligands GDF-8 (myostatin) and GDF-11 (BMP-11). Here we provide a detailed kinetic characterization of ActRIIB binding to several low and high affinity ligands using a soluble activin receptor type IIB-Fc chimera (ActRIIB.Fc). We show that both GDF-8 and GDF-11 bind the extracellular domain of ActRIIB with affinities comparable with those of activin A, a known high affinity ActRIIB ligand, whereas BMP-2 and BMP-7 affinities for ActRIIB are at least 100-fold lower. Using site-directed mutagenesis, we demonstrate that ActRIIB binds GDF-11 and activin A in different ways such as, for example, substitutions in ActRIIB Leu79 effectively abolish ActRIIB binding to activin A yet not to GDF-11. Native ActRIIB has four isoforms that differ in the length of the C-terminal portion of their extracellular domains. We demonstrate that the C terminus of the ActRIIB extracellular domain is crucial for maintaining biological activity of the ActRIIB.Fc receptor chimera. In addition, we show that glycosylation of ActRIIB is not required for binding to activin A or GDF-11. Together, our findings reveal binding specificity and activity determinants of the ActRIIB receptor that combine to effect specificity in the activation of distinct signaling pathways.
Keywords: Growth Factors, Protein Chimeras, Receptor Modification, Receptor Structure-Function, Surface Plasmon Resonance (SPR), ActRIIB, Activin Receptor Type IIB, GDF-11, SPR, Myostatin
Introduction
The cytokine transforming growth factor β (TGF-β)2 and its homologs, including bone morphogenic proteins (BMPs), activins, and growth and differentiation factors (GDFs), comprise a large superfamily that controls many major cellular processes, including proliferation, differentiation, apoptosis, angiogenesis, and steroid synthesis (1–4). TGF-β superfamily members (ligands) form covalently and non-covalently linked homo- and heterodimers that bind two type I and two type II serine/threonine kinase receptors at the same time. Both receptor types consist of an extracellular ligand-binding domain, a single transmembrane span, and a cytoplasmic serine/threonine kinase domain. Formation of the hexameric receptor-ligand complex causes the constitutively active type II receptor kinase to phosphorylate type I receptor. Thus, activated type I receptors phosphorylate Smad proteins, which subsequently translocate into the nucleus and control expression of different genes (2, 5, 6). Five type II receptors have been identified: ActRIIA, ActRIIB, BMPRII, TGFβRII, and MISRII. The ActRIIB receptor is of particular interest because it binds multiple ligands from the activin, GDF, and BMP subgroups.
ActRIIB extracellular domain (ECD) sequence is exceptionally conserved, with only one amino acid difference between mice and humans and ∼90% identity between species as divergent as chickens and humans. Although ActRIIB-deficient mice develop to term, most animals (∼70%) die shortly after birth (4). Disruption of ActRIIB expression leads to cardiac and kidney malformation, defects in axial patterning, and disturbance of left-right asymmetry in mice (7).
Four different isoforms of ActRIIB were found in mice and humans (ActRIIB1, ActRIIB2, ActRIIB3, and ActRIIB4). The ECDs of ActRIIB1 and ActRIIB2 contain an insertion in the C-terminal portion of the ECD that is absent in isoforms ActRIIB3 and ActRIIB4. The biological significance of the different isoforms remains unclear. The ActRIIB2 isoform is most predominant in humans. It was previously suggested that the longer isoforms are the most potent. For example, the longer isoforms ActRIIB1 and ActRIIB2 have been shown to have a 3–4-fold higher affinity for activin A than the shorter isoforms ActRIIB3 and ActRIIB4 (8).
ActRIIB binds to a diverse group of TGF-β family members, including activin A, BMP-2, BMP-7, GDF-8 (growth and differentiation factor 8 or myostatin), and GDF-11. Activin A, one of the most abundant proteins of the TGF-β/BMP family, is thought to be a negative regulator of bone formation and other tissues (9); BMP-2 has been associated with ectopic bone formation and periarticular ossification (10); BMP-7 has been associated with bone homeostasis (11) and kidney development (12); and GDF-8 and GDF-11 are associated with negative regulation of skeletal muscle mass (13). Moreover, activins and BMPs are known also to use different signaling pathways. Thus, recruitment of BMPs or activins leads to activation of different Smad signaling events. For example, activin binding to ActRIIB leads to activation of the Smad2/3 pathway, whereas binding to BMP-2 results in activation of the Smad1/5/8 pathway (14).
GDF-8 (myostatin) and the closely related GDF-11 are essential for proper regulation of skeletal muscle growth. For example, mice lacking GDF-8 show a dramatic increase in skeletal muscle mass (15), and naturally occurring mutations in the Gdf-8 gene show increased muscle mass in cattle (16). Mice carrying a targeted mutation in the Gdf-8 gene have about twice the normal muscle mass as a result of a combination of muscle fiber hyperplasia and hypertrophy. Similar inactivating mutations have been shown to cause increased muscle mass in other species, including cattle (16–18), sheep (19), dogs (20), and humans (21). Blocking GDF-8 activity may therefore be effective for treatment of various muscle-wasting diseases. Indeed, loss of GDF-8 function has been shown to have beneficial effects in degenerative muscle and metabolic disease models (22–24). GDF-11 is highly related to GDF-8 in the mature region of the protein and is also expressed in skeletal muscle (25–27). Moreover, there is compelling biochemical and genetic evidence suggesting that GDF-11 signals through the same type II receptors as GDF-8. Co-immunoprecipitation studies carried out with Xenopus embryo have shown that, similar to GDF-8, GDF-11 is capable of binding to ActRIIA and ActRIIB, and, as in the case of GDF-8, GDF-11 appears to bind more strongly to ActRIIB than to ActRIIA (28). In addition, the phenotype of GDF-11 knock-out mice (25) is remarkably similar to that of mice deficient in activin type II receptor (7, 29).
In this paper, we performed a systematic analysis of several deletions in the C-terminal region of ActRIIB.Fc ECD and examined the effect of such truncations on the biological activity of the receptor. To answer key questions concerning specificity of signaling, we determined the binding affinities of different ligands to ActRIIB and assessed various residues on the interface and in the ligand binding pocket of ActRIIB ECD. We further investigated the contribution of glycosylation and isoform differences on the biological activity with GDF-11 and activin A. Finally, we identified a point mutation in ActRIIB that abrogates binding of the receptor to activin A without appreciably affecting its interaction with GDF-8 or GDF-11.
EXPERIMENTAL PROCEDURES
Materials
The Biacore 3000, Biacore T100, research grade CM5 chips, amine coupling reagents, HBS-EP buffer, and anti-hFc IgG capture kit were purchased from Biacore (GE Healthcare). The Infinite M200 luminometer was obtained from Tecan. Activin A, BMP-2, BMP-7, GDF-11, and GDF-8 were obtained from R&D Systems. FuGENE6 transfection reagent was obtained from Roche Applied Science. Goat anti-human IgG and goat anti-human IgG-AP for the enzyme-linked immunosorbent assay were from Southern Biotech. Novex 4–12% gels and other electrophoresis reagents were from Invitrogen. Restriction enzymes were purchased from New England Biolabs.
Construction of ActRIIB Plasmids and Mutants
Human ActRIIB cDNA was obtained from GeneCopoeia, and human IgG1 cDNA was from Invitrogen. The plasmid pAID4T-ActRIIB.Fc encoding the fusion molecule containing the ECD region of ActRIIB (residues 19–134; see Fig. 1A) was constructed as follows. The ECD ActRIIB region was made using ActRIIB cDNA as the template and primers (supplemental Table 1A) incorporating a 5′ SfoI site on the N terminus of ActRIIB and a 3′ AgeI site on the C terminus. The Fc domain was cloned by PCR using human IgG cDNA as the template and primers encoding a 5′ AgeI and 3′ EcoRI endonuclease site. A three-way ligation was set up using the vector pAID4T cut with SfoI and EcoRI. When ligated into pAID4T, ActRIIB is preceded by the tissue plasminogen activator signal sequence and fused in-frame to the human IgG1 Fc domain.
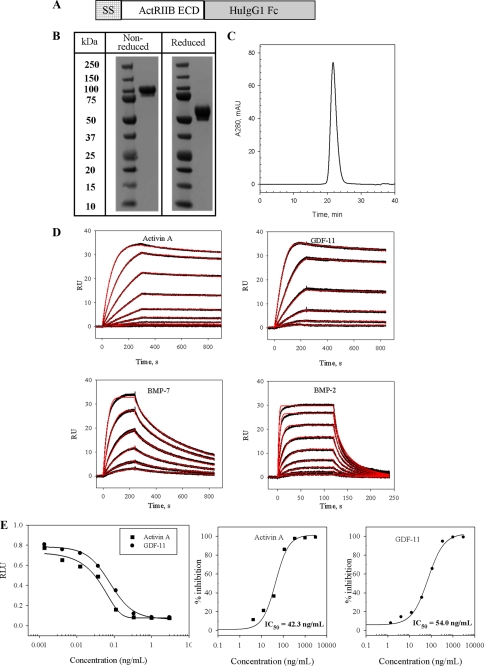
FIGURE 1.
Construction, purification, and activity of ActRIIB.Fc chimera. A, schematic representation of ActRIIB.Fc featuring (from left to right) the tissue plasminogen activator signal sequence (SS), the ActRIIB extracellular region (ActRIIB ECD) (residues 19–134) or a mutant thereof, and the human IgG1 Fc domain, including the hinge, CH2, and CH3 subdomains (HuIgG1Fc). B, SDS-PAGE of purified ActRIIB.Fc under reducing and non-reducing conditions. 10 μg of ActRIIB.Fc was diluted in NuPAGE gel-loading buffer with or without β-mercaptoethanol and boiled for 5 min. The eluates were run on a NuPAGE 4–12% BisTris gel in MES SDS running buffer. The gel was stained with SimplyBlue (Invitrogen). C, size exclusion chromatography of ActRIIB.Fc performed on a Superose 12 10/300 GL column. D, kinetic characterization of GDF-11, activin A, BMP-2, and BMP-7 binding to ActRIIB.Fc measured by SPR. The experiment was performed on the Biacore T100 instrument at 25 °C. Purified receptor was captured on the anti-hFc IgG chip at ∼140 response units (RU), and different concentrations of ligands were injected over captured receptor. Experiments were performed in duplicate. Sensograms (black lines) are overlaid with fits to a 1:1 interaction model with mass transport limitations (red lines). Kinetic data are presented in Table 1. E, activity of ActRIIB.Fc in dual luciferase reporter assay in A204 cells. Luciferase activity was measured at each concentration of ActRIIB.Fc (n = 2) tested in the presence of either GDF-11 or activin A. The experimental sample value was normalized to the control value and expressed as the ratio of relative luciferase units (RLU). The IC50 for each curve was calculated using XLfit.
All ActRIIB mutants were made by PCR-based site-directed mutagenesis (BD Biosciences Advantage-HF2). C terminus truncations of the molecule were made using primers as shown in supplemental Table 1B. Mutations within the mature ActRIIB sequence were introduced by utilizing an overlapping PCR approach and incorporating a 5′ SfoI site and a 3′ AgeI site (supplemental Table 1C). These unique sites were used to subclone the PCR product into pAID4T-ActRIIB.Fc, which had been cut with SfoI and AgeI to remove the wild-type ActRIIB insert. This resulted in the ActRIIB mutants ligated in-frame to the human IgG Fc domain. All constructs were confirmed by DNA sequencing analysis.
COS Expression of ActRIIB.Fc Molecules
SV40-transformed green monkey kidney (COS-1) cells (ATCC) were seeded at 3 × 106 cells/10-cm2 plate in Dulbecco's modified Eagle's medium (high glucose; supplemented with 10% fetal calf serum) and incubated overnight at 37 °C, 5% CO2. The cells were transfected with 10 μg of experimental plasmids or vector control using FuGENE6 reagent. After an overnight incubation, the cells were washed once in phosphate-buffered saline, and 4 ml of serum-free Dulbecco's modified Eagle's medium was added to each plate. Approximately 48 h post-transfection, the serum-free conditioned medium (CM) was harvested and centrifuged to remove cellular debris. The expressed fusion proteins in the CM were quantified by an enzyme-linked immunosorbent assay, using goat anti-human IgG as the capture antibody and detection with goat anti-human IgG-AP. A standard curve was run using purified ActRIIB.Fc.
Purification of ActRIIB.Fc and Variants
Conditioned medium containing expressed receptor was purified using MAB SelectSure (Protein A) and Q-Sepharose column chromatography (GE Healthcare Life Sciences). The purified receptor was dialyzed into 10 mm Tris-buffered saline, pH 7.2. Protein purity was analyzed by SDS-PAGE under reducing and non-reducing conditions. Size exclusion chromatography on a Superose 12 10/300 GL (GE Healthcare Life Sciences) column was performed to determine the purity of the protein and to check for aggregation.
Deglycosylation of ActRIIB.Fc
PNGase F, O-glycanase, and sialidase A (PROzyme enzymatic deglycosylation kit GK80115) were used for the enzymatic removal of oligosaccharides on ActRIIB.Fc. The digests were performed under non-denaturing conditions and incubated for 3 days at 37 °C. To assess the removal of carbohydrates, ActRIIB.Fc treated with different deglycosylation enzymes was evaluated on a silver-stained SDS-polyacrylamide gel under reducing conditions and compared with untreated and mock-treated (digest buffer minus enzyme) ActRIIB.Fc. The activity of the deglycosylated samples was also evaluated in the A204 (human rhabdomyosarcoma cell line) reporter gene assay and Biacore surface plasmon resonance (SPR) binding assay.
Reporter Gene Assay
A204 cells (ATCC) were seeded in 48-well plates at 1 × 105 cells/well in McCoy's 5A medium (Invitrogen) supplemented with 10% fetal calf serum and incubated overnight at 37 °C, 5% CO2. The next day, each well was transiently co-transfected with experimental luciferase reporter plasmid pGL3(CAGA)12 (200 ng) and control luciferase reporter plasmid pRL-CMV (10 ng). In separate plates, serial dilutions of the COS CM containing 0.0014–3 μg/ml ActRIIB.Fc or corresponding mutant receptor were incubated with 5 ng/ml GDF-11 or activin A (R&D Systems) for 45 min at 37 °C, 5% CO2. The dilutions of COS CM and ligands were made in serum-free McCoy's 5A medium containing 0.1% bovine serum albumin. The mix was then added to the transfected A204 cells and incubated for 6 h at 37 °C, 5% CO2. The cells were lysed and assayed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Chemiluminescence was measured using the Infinite M200 plate reader. The luciferase activity of the experimental reporter was normalized by the luciferase activity obtained from control reporter.
SPR Assay
Receptor-ligand binding affinities were determined by SPR using the Biacore 3000 and Biacore T100. Goat anti-human Fc antibody (anti-hFc IgG) was immobilized onto four flow cells of research grade CM5 chip using standard amino coupling chemistry. 100–150 RU of the ActRIIB.Fc and variants were captured on the experimental flow cells by injecting either purified receptor or CM expressing corresponding receptor. Another flow cell was used as a reference (control) to subtract for nonspecific binding, drift, and bulk refractive index. For kinetic analysis, a concentration series of different ligands (activin A, BMP-2, BMP-7, GDF-11, or GDF-8) was injected over experimental and control flow cells at either room temperature or 25 °C at a flow rate of ≥40 μl/min to minimize mass transport limitations. The antibody surface was regenerated between binding cycles by injection of 3 m MgCl2. HBS-EPS buffer (0.01 m HEPES, 0.5 m NaCl, 3 mm EDTA, 0.005% (v/v) surfactant P20, pH 7.4) containing 0.5 mg/ml bovine serum albumin was used as running buffer. All sensograms were processed by double referencing. To obtain kinetic rate constants, the corrected data were fitted to a 1:1 interaction model with mass transport term using BiaEvaluation software (Biacore). The equilibrium binding constant KD was determined by the ratio of binding rate constants kd/ka.
RESULTS
Production, Purification, and Characterization of ActRIIB.Fc Fusion Protein
The ActRIIB.Fc fusion protein, composed of two disulfide-linked monomers, each containing the ECD of ActRIIB and human Fc (Fig. 1A), was produced by transient transfection of the expression construct in COS cells and purified from conditioned medium by sequential column chromatography as described under “Experimental Procedures.” A Coomassie-stained SDS-polyacrylamide gel showed a broad protein band (due to glycosylation) with a molecular mass of ∼100 kDa under non-reducing conditions and of ∼60 kDa under reducing conditions (Fig. 1B). Size exclusion chromatography showed that under non-reducing conditions, the purified protein contained ∼98% ActRIIB.Fc dimer, and no aggregation was observed (Fig. 1C).
The binding affinity of purified ActRIIB.Fc for GDF-11, activin A, BMP-2, and BMP-7 as determined by SPR is shown in Fig. 1D. In order to mimic native conditions and to allow the receptor molecule to adopt a homogeneous orientation, purified ActRIIB was captured on an anti-hFc IgG Biacore chip to a low density of ∼140 response units, and different concentrations of ligand were injected over captured receptor. A 1:1 interaction model with mass transport term provided an excellent fit to the data, as shown by the overlay of the simulated binding responses (Fig. 1D, red lines). Activin A is known to bind to ActRIIB with high affinity. Accordingly, the apparent equilibrium dissociation constant KD measured by SPR was determined to be 35.7 pm (Table 1). Moreover, ActRIIB.Fc complexed with activin A is extremely stable, with kd of 1.48 × 10−4 s−1.
TABLE 1.
Analysis of ActRIIB.Fc binding to different ligands by SPR
Purified ActRIIB.Fc was captured on a Biacore sensor chip by immobilized anti-human Fc antibody. Different concentrations of ligands were injected over captured ActRIIB.Fc in duplicates. Kinetic analysis of activin A, GDF-11, BMP-2, and BMP-7 was performed on a Biacore T100 instrument at 25 °C. Kinetic analysis of binding to GDF-8 was performed on a Biacore 3000 instrument. Data were globally fit to a 1:1 binding model with mass transfer term using BIAevaluation software. Representative sensograms and fits are given in Fig. 1D.
| Ligand | ka | kd | KD |
|---|---|---|---|
| m−1s−1 | s−1 | pm | |
| Activin A | 4.14 × 106 | 1.48 × 10−4 | 35.7 |
| GDF-8 | 3.69 × 105 | 3.45 × 10−5 | 93.5 |
| GDF-11 | 4.55 × 106 | 1.38 × 10−4 | 30.3 |
| BMP-2 | 1.47 × 107 | 1.2 × 10−1 | 8215 |
| BMP-7 | 1.05 × 107 | 4.38 × 10−3 | 418 |
It was previously suggested that ActRIIB also has a high affinity for myostatin (GDF-8) and the highly related ligand GDF-11. Using SPR, we showed that GDF-11 forms a very stable complex with ActRIIB.Fc, with an affinity of 30.3 pm (Fig. 1D and Table 1). Myostatin and GDF-11 share a high degree of homology (∼90%) and are thought to have similar functions. Thus, it is not surprising that GDF-8 also binds to ActRIIB.Fc with picomolar affinity (Table 1) and kinetic parameters comparable with those of GDF-11.
According to the literature, BMP-2 and BMP-7 have low binding affinities for the ECD of ActRIIB (30). Our SPR studies indicated that binding of BMP-2 to ActRIIB.Fc was 270- and 230-fold weaker compared with the high affinity ligands GDF-11 and activin A, respectively. Although there was only ∼3-fold difference in the rate of receptor-ligand complex formation, we observe a striking difference in the stability of the complex. Thus, kd for the BMP-2/ActRIIB.Fc interaction was 0.12 s−1, whereas kd for the high affinity ligands GDF-11 and activin A was 1.38 × 10−4 s−1 and 1.48 × 10−4 s−1, respectively. We also found that BMP-7 binds to ActRIIB.Fc with much higher affinity than BMP-2, mainly due to a slower off-rate (4.38 × 10−3 s−1).
The kinetic parameters that we provide in this paper should be considered apparent because they have been obtained for the ECD of human ActRIIB fused to an immunoglobulin Fc domain expressed in mammalian cells. Nevertheless, we show selectivity of ActRIIB for different ligands under the same experimental conditions.
To evaluate the ability of ActRIIB.Fc to antagonize Smad signaling induced by GDF-11, GDF-8, or activin A, we performed a reporter gene assay using a human rhabdomyosarcoma cell line (A204) transfected with pGL3(CAGA)12-luc, a Smad2/3-responsive construct, and pRL-CMV-luc, as internal control. The cells were treated with corresponding ligand in the presence or absence of ActRIIB.Fc and assayed for luciferase activity after 6 h. ActRIIB.Fc inhibited signaling by GDF-11 (Fig. 1E), GDF-8 (data not shown), and activin A (Fig. 1E) in a dose-dependent manner with an inhibitory concentration (IC50) of 54.0 ng/ml for GDF-11, 26.0 ng/ml for GDF-8 (data not shown), and 42.3 ng/ml for activin A. These data demonstrate that the ActRIIB.Fc chimeric fusion protein inhibited signaling by GDF-11, GDF-8, and activin A via the Smad pathway. Additional testing using a BMP-specific cell-based reporter assay (i.e. T98G cells containing the BRE-luciferase reporter gene) did not show inhibition of BMP-2 and BMP-7 signaling by ActRIIB.Fc (data not shown).
Due to the similarity of the above results for GDF-8 and GDF-11, which is consistent with their high degree of homology and similarity of function, in addition to the consistent quality of commercially available GDF-11 compared with GDF-8, we chose to use GDF-11 alone for the subsequent studies described below.
Effect of Deglycosylation on the Activity of ActRIIB.Fc
The ActRIIB.Fc protein contains three potential N-glycosylation sites: two in the extracellular domain of ActRIIB and one in the Fc portion of the fusion molecule. In order to assess the role of carbohydrates in the activity of ActRIIB.Fc, we performed enzymatic deglycosylation of the protein. SDS-PAGE analysis of ActRIIB treated with deglycosylation enzymes is presented in Fig. 2A. Upon PNGase F digestion, the band corresponding to ActRIIB.Fc migrated faster compared with the untreated ActRIIB.Fc due to removal of N-linked carbohydrates (Fig. 2A, lane N-Gly). No band indicative of incomplete digestion of ActRIIB.Fc was observed. Peptide mapping of PNGase F-treated ActRIIB.Fc allowed for the assignment of the three tryptic peptides containing Asn23, Asn46, and Asn193, confirming the predicted location of N-linked oligosaccharides (data not shown).
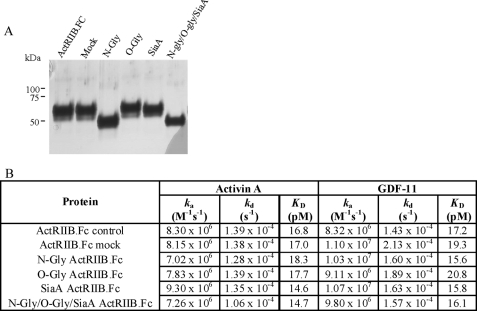
FIGURE 2.
Effect of deglycosylation of ActRIIB. A, SDS-PAGE (reduced) analysis of ActRIIB.Fc treated with deglycosylation enzymes. ActRIIB.Fc was treated by PNGase F (lane N-Gly); O-glycanase (lane O-Gly), sialidase A (lane SiaA), or the combination of all three enzymes (lane N-Gly/O-Gly/SiaA) at 37 °C. Control ActRIIB.Fc (lane ActRIIB.Fc) and ActRIIB.Fc incubated at 37 °C without the addition of the enzymes (lane Mock) are shown for comparison. B, effect of deglycosylation on the activity of ActRIIB.Fc. Purified ActRIIB.Fc was treated with deglycosylation enzymes as described under “Experimental Procedures.” Deglycosylated proteins along with the control ActRIIB.Fc were captured on a Biacore sensor chip by immobilized anti-human Fc antibody; different concentrations of activin A and GDF-11 were injected over captured receptors in duplicates. Analysis was performed on a Biacore 3000 instrument at room temperature. Data were globally fit to a 1:1 binding model with mass transfer term using BIAevaluation software.
Upon combined treatment with three deglycosylation enzymes (PNGase F, O-glycanase, and sialidase A), the diffuse band of ActRIIB.Fc collapsed into a single band whose mobility was consistent with the theoretical molecular mass of ∼39 kDa, suggesting complete deglycosylation of the protein (Fig. 2A, lane N-Gly/O-Gly/SiaA). The deglycosylated ActRIIB.Fc was able to bind both activin A and GDF-11 with the same kinetic parameters as untreated and mock ActRIIB.Fc (Fig. 2B). We also tested the effect of deglycosylation on the ability of ActRIIB.Fc to inhibit activation of promoters responsive to GDF-11 and activin A. In the A204 reporter gene assay, enzyme-treated ActRIIB.Fc retained its ability to inhibit signaling by GDF-11, suggesting that the carbohydrate side chains are not required for binding to this ligand (data not shown).
Effect of C-terminal Deletions
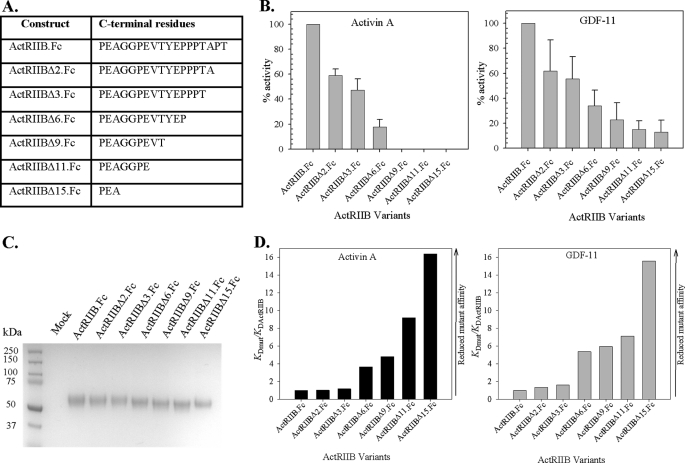
In order to characterize the role of the C terminus of the ECD, we created an ActRIIB.Fc variant with the 15 C-terminal amino acids deleted from the ECD of the receptor. The relative potencies of ActRIIB.Fc (full-length ECD chimera) and ActRIIBΔ15.Fc mutant with activin A and GDF-11 were investigated. We found that the biological activity of the Δ15 mutant was dramatically decreased for both activin A and GDF-11 (Fig. 3). Thus, in the A204 assay with GDF-11, ActRIIBΔ15.Fc showed ∼10% activity (i.e. inhibited ∼10% of the signaling by GDF-11 compared with 100% inhibition by non-truncated ActRIIB.Fc). In the A204 assay with activin A, no activity of ActRIIBΔ15.Fc was detected (Fig. 3B). SDS-PAGE analysis indicated that the quality of the chimeric truncated version of ActRIIB and the quality of the full-length ECD chimera were comparable (Fig. 3C). In order to confirm that the observed effect was not due to misfolding of the protein, we further tested the ability of purified ActRIIBΔ15.Fc to bind both GDF-11 and activin A using SPR (Fig. 3D). The equilibrium binding constant (KD) of the truncated protein ActRIIBΔ15.Fc was compared with the KD of ActRIIB.Fc (i.e. the chimera containing the full-length ECD). Fig. 3D presents the relative binding data, expressed as KD(mut)/KD(ActRIIB). ActRIIBΔ15.Fc showed ∼16-fold increased KD compared with ActRIIB.Fc. Moreover, the percentage of correctly folded protein, as estimated from the theoretical maximum response term (Rmax) obtained from SPR, was significantly decreased for ActRIIBΔ15.Fc (data not shown).
FIGURE 3.
Effect of ActRIIB C-terminal deletions. A, nomenclature of ActRIIB.Fc fusion proteins containing ActRIIB ECD C-terminal truncations. B, effect of C-terminal truncations on biological activity of ActRIIB.Fc on activin A (left) and GDF-11 (right) signaling measured in the A204 reporter gene assay. The average of the IC50s from at least three independent experiments was calculated for each experimental protein. The experiment was performed with proteins expressed in COS CM without additional purification. The percentage activity was defined as relative activity of each protein in the presence of deletion variant versus ActRIIB.Fc. C, SDS-polyacrylamide gel analysis of ActRIIB.Fc mutants under reducing conditions. Concentration of mutants in COS CM was quantified by an enzyme-linked immunosorbent assay. 8 μg of each protein was precipitated by Protein A-Sepharose, washed, and eluted by NuPAGE gel-loading buffer with β-mercaptoethanol and boiled for 5 min. The eluates were run on a NuPAGE 4–12% BisTris gel in MES SDS running buffer. The gel was stained with SimplyBlue. D, effect of C-terminal truncations of ActRIIB on binding activity to activin A (left) and GDF-11 (right) measured by SPR. Mutant proteins were captured by anti-hFc IgG on a CM5 chip, and different concentrations of activin A or GDF-11 were injected over captured receptors at room temperature. Equilibrium binding constants were obtained by fitting the data to 1:1 binding model with mass transport term. Data are presented as ratios of KD(mutant)/KD(ActRIIB).
To investigate systematically the role of the C terminus of ActRIIB, we prepared several mutants whose C termini are truncated: ActRIIBΔ11.Fc, ActRIIBΔ9.Fc, ActRIIBΔ6.Fc, ActRIIBΔ3.Fc, and ActRIIBΔ2.Fc (Fig. 3A). The biological activity of these proteins as well as their kinetic properties were tested in conditioned medium without purification. We found that deletion of 2 and 3 amino acids from the C terminus of the protein resulted in a 2-fold difference in KD between truncated ActRIIBΔ3.Fc, ActRIIBΔ2.Fc, and ActRIIB.Fc with respect to activin A and GDF-11 binding. The ligand-receptor interactions weakened dramatically upon further truncation of the C terminus. This effect is attributable both to impaired recognition of the complex, as judged by decreased ka, and to the decreased stability of the receptor-ligand complex, as attested by increased kd (data not shown). These data correlate well with cell-based activity measurements (Fig. 3B). Deletions have a more pronounced effect on the biological activity of activin A than GDF-11. In order to confirm that these differences were not due to decreased stability of the mutant proteins, we performed additional experiments to verify the level of expression. Fig. 3C shows an SDS-PAGE analysis of the mutants, which suggests that the quality of all of the mutant and wild-type ActRIIB.Fc proteins was comparable.
Ligand Binding Pocket Mutations
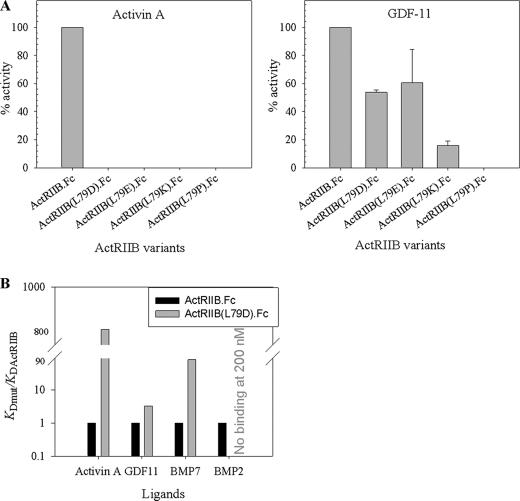
The crystal structure of the truncated ECD domain of ActRIIB in complex with activin A and BMP-2 provided information about the residues involved in the interactions with the high affinity ligand activin A and the low affinity ligand BMP-2 (30, 31). The activin A/ActRIIB interface involves hydrophobic, charged, and polar residues on both ActRIIB and activin A (Fig. 4). The concave ActRIIB interface consists of residues on fingers 1, 2, and 3 and comprises an ∼634 Å2 surface (Fig. 4A). Leu79 (shown in red in Fig. 4A) accounts for approximately one-sixth (96/634 Å2) of the total ActRIIB surface buried in the complex with activin A (31). Substitution of this residue with proline previously was reported to reduce dramatically the binding of ActRIIB to activin A and to a lesser extent substitution with alanine (30). To determine the relative contributions of Leu79 to binding of activin A and GDF-11, a series of ActRIIB.Fc mutants containing amino acid substitutions at position 79 were constructed and tested in the A204 assay for their ability to inhibit signaling by these ligands (Fig. 5A). Unexpectedly, all substitutions at this site effectively eliminated activity (i.e. the ability of ActRIIB.Fc to inhibit signaling by activin A in the A204 assay (Fig. 5A, left)), whereas inhibition of GDF-11 signaling was observed in the presence of three of the four ActRIIB.Fc variants (Fig. 5A, right). Approximately 50% activity was observed with the L79D and L79E variants. Even substitution with a large, charged residue (L79K) did not fully eliminate inhibition of GDF-11 signaling, whereas a substitution expected to induce strain on the polypeptide chain conformation (L79P) effectively eliminated inhibition. SDS-PAGE analysis demonstrated that the quality of all of the variant and wild-type ActRIIB.Fc proteins was comparable (data not shown). The kinetic rate constants for activin A and GDF-11 binding to these variants are presented in Table 2. We observed a dramatic effect of the substitutions on the activin A interactions with mutated receptors. Thus, the KD for negatively charged substitution in the ActRIIB(L79D).Fc mutant was ∼13,500 pm, and KD for positively charged substitution in ActRIIB(L79K).Fc was ∼5920 pm, compared with 35.7 pm for wild-type ActRIIB.Fc. This difference in the equilibrium dissociation constants is mainly due to the striking differences in the dissociation rate constants between Leu79 variants and wild-type ActRIIB.Fc receptors. Due to limitations of Biacore instrumentation, accurate determination of the kd for Leu79 mutants was not possible; however we could estimate that they were faster than 0.1 s−1, compared with kd of 1.48 × 10−4 s−1 for ActRIIB.Fc. This suggests that Leu79 plays a significant role in the stability of the receptor-activin A complex. No measurable binding of ActRIIB(L79P).Fc to activin A was observed. In contrast, the KD for GDF-11 was modestly (1.5–4.3-fold) affected by the substitutions at Leu79, mainly due to an increase in the kd rate. In order to assess the importance of Leu79 for the ActRIIB.Fc interactions with other ligands, we have chosen the ActRIIB(L79D).Fc variant, which was purified and further characterized by SPR. Fig. 5B shows that the affinity of ActRIIB(L79D).Fc mutant relative to wild-type ActRIIB.Fc for activin A, GDF-11, and BMP-7 was decreased ∼850-, 3-, and 83-fold, respectively, and binding to BMP-2 was entirely abolished.
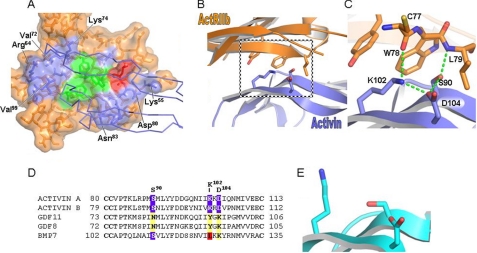
FIGURE 4.
Structure of ActRIIB in complex with activin A. A, surface representation of ActRIIB ECD. ActRIIB is colored orange, with Leu79 colored red, the cluster of aromatic amino acids (ActRIIB residues Tyr60, Trp78, and Phe101) at the center of the ActRIIB-activin A interface colored green, and the remaining interface residues colored blue. Activin A is shown as a blue Cα trace (31). B, ribbon diagram of the ActRIIB-activin A complex (31). This view is orthogonal to the orientation in A, with ActRIIB ECD colored orange and activin A colored blue. Interface residues proximal to ActRIIB Leu79 are also shown. C, magnified view of B showing ActRIIB Leu79 contributions to activin A binding. ActRIIB Leu79 combines with ActRIIB residues Tyr60 and Trp78 to orient activin A residues Ser90, Lys102, and Asp104, thereby generating a cluster of hydrogen bonds (green dotted line) central to the complex interface. D, sequence alignment of activin A, activin B, GDF-11, GDF-8, and BMP-7. The alignment was performed by ClustalW. Only amino acids corresponding to activin A residues 80–113 are shown. GDF-8 and GDF-11 sequences are divergent at positions corresponding to activin A residues Ser90, Lys102 and Asp104. E, ribbon diagram of activin A in its unbound state (40). Activin A is colored cyan. Residues that form the hydrogen bond cluster in the complex, activin A Ser90, Lys102, and Asp104, are also shown.
FIGURE 5.
Effect of mutations of Leu79 on the specificity of ActRIIB.Fc protein. A, effect of amino acid substitution of Leu79 by aspartic acid, glutamic acid, lysine, and proline on biological activity of ActRIIB.Fc with activin A (left) and GDF-11 (right) was measured in the A204 reporter gene assay. The average of the IC50 values from at least three independent experiments was calculated for each experimental protein. The percentage activity was defined as activity of each protein relative to ActRIIB.Fc. B, effect of mutation substitution of Leu79 by aspartic acid on the binding of ActRIIB.Fc to different ligands was tested by SPR. Purified ActRIIB(L79D).Fc and control ActRIIB.Fc were captured on the anti-hFc IgG chip, and different concentrations of ligands were injected over the captured receptors at room temperature. Data are presented as ratios of KD(mutant)/KD(ActRIIB). Error bars, S.D.
TABLE 2.
Mutational analysis of the ligand binding interface measured by SPR
ActRIIB.Fc mutants expressed in COS conditioned medium were captured on an anti-hFc IgG chip. Activin A or GDF-11 at different concentrations was injected over captured receptors. The experiment was performed on a Biacore 3000 instrument at room temperature. Data were globally fit to a 1:1 binding model with mass transfer term using BIAevaluation software.
| ActRIIB mutants | Ligands |
|||||
|---|---|---|---|---|---|---|
| Activin A |
GDF-11 |
|||||
| ka | kd | KD | ka | kd | KD | |
| m−1s−1 | s−1 | pm | m−1s−1 | s−1 | pm | |
| ActRIIB.Fc | 4.14 × 106 | 1.48 × 10−4 | 35.7 | 8.80 × 106 | 4.95 × 10−4 | 56.0 |
| ActRIIB(L79D).Fc | 13,500a | 8.83 × 106 | 9.96 × 10−4 | 109.0 | ||
| ActRIIB(L79E).Fc | 9390a | 7.55 × 106 | 1.00 × 10−3 | 133.0 | ||
| ActRIIB(L79K).Fc | 5920a | 7.21 × 106 | 8.28 × 10−4 | 115.0 | ||
| ActRIIB(L79P).Fc | No binding | 6.87 × 106 | 1.65 × 10−3 | 240.0 | ||
a Due to extremely fast dissociation, it was impossible to accurately measure the kinetic constant; thus, KD was determined using affinity fit.
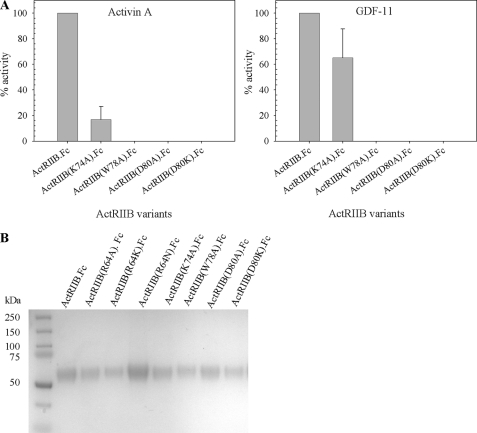
A cluster of three aromatic amino acids, Tyr60, Trp78, and Phe101, lie at the center of the receptor-ligand (ActRIIB-activin A) interface and contribute ∼20–25% of the total buried surface of the receptor (Fig. 4A). These residues are highly conserved between ActRIIB and another activin A binding receptor, ActRIIA, with Trp78 and Phe101 being 100% conserved and Tyr60 being replaced by phenylalanine in ActRIIA. Individual mutations of Tyr60, Trp78, and Phe101 in the ECD of ActRIIA eliminated receptor binding to activin A (32). Substitution of Trp78 and, to a lesser extent, substitution of Tyr60 dramatically reduced binding of ActRIIB to low affinity ligand BMP-2 and the high affinity ligand activin A (30). To check if the same amino acids are involved in ActRIIB interactions with GDF-11, we have made a series of point mutations in the ActRIIB ECD in which residues 78 and 101 were replaced by alanine. The ability of these ActRIIB.Fc variants to inhibit signaling through the GDF-11 pathway was measured in the A204 assay. Fig. 6A shows that the point mutation W78A at the hydrophobic interface eliminates activity (i.e. no inhibition of signaling by activin A was observed). The same effect was observed with the F101A mutant (data not shown). Because these mutants did not show detectable biological activity in the A204 assay, we verified the level of expression. Fig. 6B shows SDS-PAGE analysis of mutants in conditioned medium, which suggests that the quality of the W78A variant and the quality of the wild-type ActRIIB.Fc proteins were comparable. Both mutations of Trp78 and Phe101 completely eliminated binding to GDF-11 and activin A in the Biacore binding assay (data not shown). Thus, the same hydrophobic residues on ActRIIB are important for binding to both activin A and GDF-11.
FIGURE 6.
Effect of mutations in ligand binding pocket. A, biological activity of the ActRIIB.Fc mutants with activin A (left) and GDF-11 (right) was measured in the cell-based A204 assay. B, expression of the mutants in COS CM was visualized by SDS-PAGE under reducing conditions. 8 μg of protein was precipitated by Protein A-Sepharose, washed and eluted by NuPAGE gel-loading buffer with β-mercaptoethanol, and boiled for 5 min. The eluate was run on a NuPAGE 4–12% BisTris gel in MES SDS running buffer. The gel was stained with SimplyBlue. Error bars, S.D.
In addition to the hydrophobic residues in the binding pocket, several residues (Lys55, Lys74, and Asp80) identified as constituents of the ionic/polar binding interface (31) were investigated. Replacement of Lys55 with Ala only somewhat reduced binding affinity of the ActRIIB(K55A).Fc for GDF-11 or activin A (data not shown). We also found that the K74A substitution decreased the ability of ActRIIB(K74A).Fc to inhibit GDF-11 or activin A activation of the reporter gene in the A204 assay by ∼35 and ∼83%, respectively, whereas substitutions of Asp80 by alanine or lysine effectively abolished this biological activity of ActRIIB(D80A).Fc or ActRIIB(D80K).Fc in the A204 assay. The level of protein expression of these mutants was confirmed as shown in Fig. 6B. Another residue of ActRIIB, Glu39, makes a minor (16 Å2) contribution to the ligand interface and also is involved in intramolecular contacts with Lys74. As expected, the ActRIIB(E39K).Fc variant failed to bind either activin A or GDF-11 (data not shown).
Effect of Amino Acid Substitution at Position 64 of ECD of ActRIIB
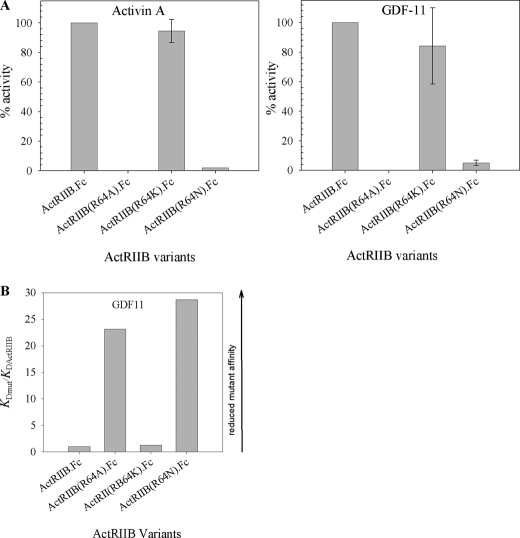
Residue 64, which is at the outer edge of the ligand binding interface also was investigated. We replaced Arg64 in ActRIIB.Fc with alanine to mimic the reported naturally occurring human mutation and lysine in order to preserve positive charge in this position; asparagine was chosen as an alternative amino acid substitution. We further assessed the effect of these mutations on the biological activity of ActRIIB.Fc using the cell-based potency assay (Fig. 7A) and the ability of the fusion protein to bind GDF-11 using SPR (Fig. 7B). Mutation of Arg64 to alanine or asparagine completely eliminated the ability of ActRIIB.Fc to inhibit both activin A (Fig. 7A, left) and GDF-11 (Fig. 7A, right) signaling in the A204 assay. It also had a marked effect on the GDF-11 binding affinity, with interactions weakened 24- and 29-fold for ActRIIB(R64A).Fc and ActRIIB(R64N).Fc, respectively (Fig. 7B). In the related ActRIIA receptor, there is a lysyl residue in the corresponding position. Consequently, we replaced residue 64 with lysine, thereby preserving positive charge at position 64 (ActRIIB(R64K).Fc mutant). This mutation did not have a significant effect on either the biological activity of the protein in the A204 assay or the affinity of the receptor for GDF-11.
FIGURE 7.
Effect of Arg64 mutation on activin A and GDF-11 binding activity. A, effect of amino acid substitution at position 64 on the biological activity with activin A (left) and GDF-11 (right) was measured in the A204 reporter gene assay. B, binding of different mutants to GDF-11 (right) measured by SPR. Mutants in COS CM were captured by anti-hFc IgG on the CM5 chip, and different concentrations of GDF-11 were injected over captured ActRIIB.Fc. Experiments were performed on the Biacore 3000 instrument at room temperature. Equilibrium binding constants were obtained by fitting the data to a 1:1 binding model that included mass transport term. Data are reported as ratios of KD(mutant)/KD(ActRIIB).
DISCUSSION
ActRIIB has been proposed to be the major type II receptor for GDF-11 and GDF-8 signaling. ActRIIB has also been shown to bind additional ligands with varying affinities (e.g. high affinity for activin A and low affinity for BMP-2 and BMP-7) (30). To understand the mechanism of ActRIIB selectivity, we performed a detailed kinetic analysis of GDF-11, activin A, BMP-2, and BMP-7 binding to the soluble ECD of ActRIIB and variants thereof fused to human Fc (hFc). These constructs produced a stable receptor dimer that could be expressed readily in COS cells and purified to homogeneity in two simple steps. This protein is glycosylated and thus closely represents the natural form of the extracellularly accessible portion of ActRIIB.
Previous studies have determined affinities of ActRIIB for different ligands (30, 33, 34); however, these studies did not result in any consensus because data varied greatly, depending on the assay design, experimental conditions, source of ligands and receptors, etc. To determine the binding parameters of different ligands and ActRIIB, we used SPR. The use of a human Fc as part of the ActRIIB fusion protein has allowed us to use a capture format, where receptors were captured on the chip with covalently immobilized anti-hFc IgG. Our approach has an advantage over the direct coupling approach, where molecules of receptor or ligand are chemically cross-linked to the surface in random orientation, because our approach creates a homogeneous population of receptors that have the same orientation on the chip surface. Our results indicate that we produced a functional ActRIIB molecule, as measured by its ability to bind the high affinity ligand activin A as well as the low affinity ligands BMP-7 and BMP-2 with expected relative affinities, and in addition validate our observations of high affinity (picomolar magnitude) binding of ActRIIB.Fc to GDF-8 and GDF-11 (i.e. comparable with ActRIIB.Fc binding to activin A). Thus, ActRIIB.Fc is a highly specific receptor for GDF-8/GDF-11 as well as activin A.
The affinity of ActRIIB for activin A has been determined previously. For example, the affinity of membrane-bound ActRIIB was reported as 100–380 pm (8, 35). The affinity of mouse ActRIIB ECD for activin A determined by SPR with receptor or activin A being covalently coupled on the Biacore chip varied between 0.244 and 6.87 nm (33). Our data show that full-length ECD of ActRIIB fused to Fc is capable of binding activin A with low picomolar affinity. Conjoining ActRIIB ECD with Fc does not affect ligand binding, as was shown previously by del Re et al. (34). Previously reported binding affinities of ActRIIB to activin A possibly underestimate the actual binding affinities because measurements were made between ligands and isolated ECD. Receptors function in situ as membrane-bound dimers whose orientations are thus restricted. Therefore, affinities for activin A and GDF-11 reported here are more likely to represent physiological values.
Glycosylation can play a significant role in receptor ligand recognition by members of the TGF-β receptor family. N-Glycosylation of BMP-6 is crucial for recognition by activin receptor type I, as shown by total diminishment of BMP-6 signaling through this receptor upon removal of the carbohydrate moiety (36). In another report, unglycosylated TGFβRII-ECD showed a 1000-fold lower affinity compared with the intact cell surface receptor (37). The ActRIIB.Fc used in our study is glycosylated at two N-glycosylation sites in the receptor portion and at one N-glycosylation site in the Fc region. Commercial preparations of ActRIIB available from R&D Systems produced in insect and mammalian cells are described as having a 10-fold difference in activity, suggesting that differences in glycosylation might affect the biological activity of ActRIIB. We show here that enzymatic deglycosylation of ActRIIB.Fc with PNGase F, O-glycanase, and sialidase A does not affect binding of the receptor to either GDF-11 or activin A.
The effect of ActRIIB C-terminal deletions on the binding affinity of the fusion protein for activin A and GDF-11 is of interest because it has been shown that in mice alternative splicing of mouse ActRIIB mRNA gives rise to four different isoforms (ActRIIB1, ActRIIB2, ActRIIB3, and ActRIIB4). The ECDs of isoforms ActRIIB1 and ActRIIB2 contain a three-proline cluster surrounded by five flanking amino acids in the C-terminal portion of the ECD, a feature that is absent in isoforms ActRIIB3 and ActRIIB4. Interestingly, ActRIIB2 is the predominant isoform in mice. Although the biological relevance of these isoforms remains unclear, in mice, the ActRIIB1 and ActRIIB2 isoforms were suggested to bind activin A with higher affinity than ActRIIB3 and ActRIIB4 variants, indicating that alternative splicing in the C terminus of the ECD can have a significant effect on ligand binding (8). The structure of the ECD of mouse ActRIIB lacking the 14 C-terminal amino acids purified from yeast has been solved (38). Using analytical centrifugation, it was concluded that the 14 C-terminal residues are not required for complex formation with activin. However, no information about the effect of the C-terminal portion for the kinetics of interactions was provided. In a study of the highly homologous ActRII protein (mouse), Greenwald et al. (39) concluded that the C-terminal portion of the receptor does not affect binding to activin A. In this investigation, we demonstrate that deletion of 15 C-terminal residues of ECD (i.e. elimination of the proline cluster) dramatically reduces binding of both GDF-11 and activin A to the receptor and also has a significant effect on biological activity. Although a chief function of the C-terminal portion is tethering the ECD to the cell membrane, it cannot be ruled out that these residues may also directly impact the binding interaction. Consistent with the findings of Attisano et al. (8), even deletion of as few as 2 or 3 residues from the C terminus of the ECD caused a measurable decrease in the potency of the ActRIIB.Fc protein as an inhibitor of GDF-11 and activin A.
Analysis of the ligand binding interface from the ActRIIB-activin A crystal structure has identified roles for the hydrophobic residues Trp78 and Phe101 and the ionic/polar residues Lys74 and Glu39 of ActRIIB in binding to both activin A and GDF-11. These findings largely corroborate those of other investigators concerning the key role played by respective residues at receptor-ligand interfaces within the TGF-β superfamily. In addition, however, we also show that Leu79 plays an important role in activin A binding, which is very sensitive to amino acid substitution of this residue, whereas GDF-11 binding is considerably less so. The unique group of amino acid residues in GDF-8 and GDF-11 that are in close proximity to Leu79 of ActRIIB may help distinguish their role and the dynamics of their signaling mechanisms within the TGF-β superfamily of ligands. This finding also demonstrates that the ligand specificity of soluble forms of ActRIIB can readily be modified to potentially help understand and distinguish the relative physiological effects of its native ligands. Assessment of cell-based assays wherein the signaling via endogenous receptors is replaced by full-length (i.e. membrane-bound) versions of these modified forms of ActRIIB as a means to further confirm our in vitro findings is the subject of ongoing studies.
The ActRIIB-activin structure identifies three residues in activin A (Pro88, Asp104, and Ser90) that are in close proximity with Leu79 of ActRIIB (Fig. 4, B and C). The Pro88 and Ser90 residues are conserved to a high degree within the TGF-β superfamily, although residues such as alanine, threonine, glycine, and valine also are represented. Notably, only GDF-8, GDF-11, and inhibin have an asparagine in the sequence location corresponding to Ser90 of activin A. GDF-8 and GDF-11 also have a lysine at the site corresponding to Asp104 of activin A (Fig. 4D). We hypothesize that the lysyl residue at this site may have more flexibility to accommodate the removal of the hydrophobic Leu79 of ActRIIB, and/or the aliphatic component of the lysine may provide additional stability to the bound versus unbound ligand states that compensates for the loss of the hydrophobic Leu79 contribution from the receptor (see Fig. 4, C and E, for the bound (31) versus unbound (40) conformations of activin A). The Asn83 residue of GDF-11 also may play a role because BMP-7, which has the corresponding lysine of GDF-11 but retains the serine similar to activin A, exhibits intermediate affinity for the ActRIIB(L79D).Fc variant.
There are a few potential salt bridges and hydrogen bonds between ActRIIB and activin A that surround the hydrophobic center of the interface. Thus, Lys55 is involved in hydrogen bonding with Glu111 of activin A, and Asp80 contacts Arg87 of activin A. Our finding that the K55A mutation had only a relatively minor effect on activin A or GDF-11 binding as judged by SPR and cell-based assays is consistent with a previously reported finding (32) that mutation of the corresponding residue in the ActRIIA receptor did not drastically affect receptor binding to activin A.
Asp80 was identified as a key residue contributing to binding to activin A through both ionic and polar interactions with Arg87 of activin A (31). Although mutation of the corresponding aspartyl residue in the ActRIIA receptor had a marginal effect on activin A binding (32), it is somewhat surprising that our mutations eliminated binding to both activin A and GDF-11 because GDF-11 has a serine at the position corresponding to the arginine in activin A. We tentatively ascribe the loss of binding to a defect in protein folding.
A residue with amphiphilic properties, Lys74, is involved in hydrophobic contacts across the interface and intramolecular ionic/polar contact with Glu39. As expected, the ActRIIB(E39K).Fc variant failed to bind either activin A or GDF-11, which is attributable to loss of stable orientation required for ligand binding.
The primary sequence of ActRIIB ECD is highly conserved. Thus, human ActRIIB ECD shares 99% sequence identity with mouse, rat, and bovine ActRIIB and 78% sequence identity with Xenopus ActRIIB. Nevertheless, there exists a report of a natural allelic variation of arginine or alanine at residue 64 (41). We demonstrate that both R64A and R64N mutations have a devastating effect on the biological activity of the receptor as well as on its binding to GDF-11 but that maintaining positive charge with either arginine (native protein) or lysine (introduced mutation) at this position does not significantly affect the activity. These observations raise questions concerning the frequency and sustainability of an Ala64 allele in the human population.
In summary, in this investigation using soluble ActRIIB.Fc chimeras, we have demonstrated that both GDF-8 and GDF-11 bind the extracellular domain of ActRIIB with high affinities comparable with those of activin A, whereas the affinities of BMP-2 and BMP-7 for ActRIIB are lower by orders of magnitude. The binding of ActRIIB to GDF-11 and activin A are distinguishable by substitutions in ActRIIB Leu79 that effectively abolish ActRIIB binding to activin A but not to GDF-11. We also have demonstrated that the C terminus of the ActRIIB extracellular domain is required for maintaining biological activity of the ActRIIB.Fc receptor chimera. In addition, we show that glycosylation of ActRIIB.Fc is not required for binding to activin A or GDF-11. These findings reveal binding specificity and activity determinants of the ActRIIB receptor that help explain specificity in the activation of distinct signaling pathways.
Supplementary Material
Acknowledgments
We thank Tim Ahern, a medical writer supported by Acceleron Pharma, who provided editorial assistance during preparation of the manuscript. We also thank John Knopf and John Quisel for critical review of the manuscript and all members of Acceleron Pharma's Cell Biology and Protein Purification Groups for contributions in support of this work.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- TGF-β
- transforming growth factor β
- ActRIIB
- activin receptor type IIB
- BMP
- bone morphogenic protein
- CM
- conditioned medium
- ECD
- extracellular domain
- SPR
- surface plasmon resonance
- GDF
- growth and differentiation factor
- PNGase F
- peptide:N-glycanase F
- hFc
- human Fc
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Sun P. D., Davies D. R. (1995) Annu. Rev. Biophys. Biomol. Struct. 24, 269–291 [DOI] [PubMed] [Google Scholar]
- 2.Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 3.Watabe T., Miyazono K. (2009) Cell Res. 19, 103–115 [DOI] [PubMed] [Google Scholar]
- 4.Lee S. J., Reed L. A., Davies M. V., Girgenrath S., Goad M. E., Tomkinson K. N., Wright J. F., Barker C., Ehrmantraut G., Holmstrom J., Trowell B., Gertz B., Jiang M. S., Sebald S. M., Matzuk M., Li E., Liang L. F., Quattlebaum E., Stotish R. L., Wolfman N. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18117–18122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuchida K., Nakatani M., Uezumi A., Murakami T., Cui X. (2008) Endocr. J. 55, 11–21 [DOI] [PubMed] [Google Scholar]
- 6.Harrison C. A., Gray P. C., Vale W. W., Robertson D. M. (2005) Trends Endocrinol. Metab. 16, 73–78 [DOI] [PubMed] [Google Scholar]
- 7.Oh S. P., Li E. (1997) Genes Dev. 11, 1812–1826 [DOI] [PubMed] [Google Scholar]
- 8.Attisano L., Wrana J. L., Cheifetz S., Massagué J. (1992) Cell 68, 97–108 [DOI] [PubMed] [Google Scholar]
- 9.Pearsall R. S., Canalis E., Cornwall-Brady M., Underwood K. W., Haigis B., Ucran J., Kumar R., Pobre E., Grinberg A., Werner E. D., Glatt V., Stadmeyer L., Smith D., Seehra J., Bouxsein M. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7082–7087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddi A. H. (1998) Clin. Orthop. Relat. Res. 355, S66–S72 [DOI] [PubMed] [Google Scholar]
- 11.ten Dijke P. (2006) Curr. Med. Res. Opin. 22, Suppl. 1, S7–S11 [DOI] [PubMed] [Google Scholar]
- 12.Patel S. R., Dressler G. R. (2005) Trends Mol. Med. 11, 512–518 [DOI] [PubMed] [Google Scholar]
- 13.Wolfman N. M., McPherron A. C., Pappano W. N., Davies M. V., Song K., Tomkinson K. N., Wright J. F., Zhao L., Sebald S. M., Greenspan D. S., Lee S. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15842–15846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y. G., Hata A., Lo R. S., Wotton D., Shi Y., Pavletich N., Massagué J. (1998) Genes Dev. 12, 2144–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McPherron A. C., Lawler A. M., Lee S. J. (1997) Nature 387, 83–90 [DOI] [PubMed] [Google Scholar]
- 16.McPherron A. C., Lee S. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12457–12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grobet L., Poncelet D., Royo L. J., Brouwers B., Pirottin D., Michaux C., Ménissier F., Zanotti M., Dunner S., Georges M. (1998) Mamm. Genome 9, 210–213 [DOI] [PubMed] [Google Scholar]
- 18.Kambadur R., Sharma M., Smith T. P., Bass J. J. (1997) Genome Res. 7, 910–916 [DOI] [PubMed] [Google Scholar]
- 19.Clop A., Marcq F., Takeda H., Pirottin D., Tordoir X., Bibé B., Bouix J., Caiment F., Elsen J. M., Eychenne F., Larzul C., Laville E., Meish F., Milenkovic D., Tobin J., Charlier C., Georges M. (2006) Nat. Genet. 38, 813–818 [DOI] [PubMed] [Google Scholar]
- 20.Mosher D. S., Quignon P., Bustamante C. D., Sutter N. B., Mellersh C. S., Parker H. G., Ostrander E. A. (2007) PLoS Genet. 3, e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuelke M., Wagner K. R., Stolz L. E., Hübner C., Riebel T., Kömen W., Braun T., Tobin J. F., Lee S. J. (2004) N. Engl. J. Med. 350, 2682–2688 [DOI] [PubMed] [Google Scholar]
- 22.Bogdanovich S., Krag T. O., Barton E. R., Morris L. D., Whittemore L. A., Ahima R. S., Khurana T. S. (2002) Nature 420, 418–421 [DOI] [PubMed] [Google Scholar]
- 23.Wagner K. R., McPherron A. C., Winik N., Lee S. J. (2002) Ann. Neurol. 52, 832–836 [DOI] [PubMed] [Google Scholar]
- 24.McPherron A. C., Lee S. J. (2002) J. Clin. Invest. 109, 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McPherron A. C., Lawler A. M., Lee S. J. (1999) Nat. Genet. 22, 260–264 [DOI] [PubMed] [Google Scholar]
- 26.Gamer L. W., Wolfman N. M., Celeste A. J., Hattersley G., Hewick R., Rosen V. (1999) Dev. Biol. 208, 222–232 [DOI] [PubMed] [Google Scholar]
- 27.Nakashima M., Toyono T., Akamine A., Joyner A. (1999) Mech. Dev. 80, 185–189 [DOI] [PubMed] [Google Scholar]
- 28.Oh S. P., Yeo C. Y., Lee Y., Schrewe H., Whitman M., Li E. (2002) Genes Dev. 16, 2749–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song J., Oh S. P., Schrewe H., Nomura M., Lei H., Okano M., Gridley T., Li E. (1999) Dev. Biol. 213, 157–169 [DOI] [PubMed] [Google Scholar]
- 30.Weber D., Kotzsch A., Nickel J., Harth S., Seher A., Mueller U., Sebald W., Mueller T. D. (2007) BMC Struct. Biol. 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson T. B., Woodruff T. K., Jardetzky T. S. (2003) EMBO J. 22, 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray P. C., Greenwald J., Blount A. L., Kunitake K. S., Donaldson C. J., Choe S., Vale W. (2000) J. Biol. Chem. 275, 3206–3212 [DOI] [PubMed] [Google Scholar]
- 33.Greenwald J., Vega M. E., Allendorph G. P., Fischer W. H., Vale W., Choe S. (2004) Mol. Cell 15, 485–489 [DOI] [PubMed] [Google Scholar]
- 34.del Re E., Sidis Y., Fabrizio D. A., Lin H. Y., Schneyer A. (2004) J. Biol. Chem. 279, 53126–53135 [DOI] [PubMed] [Google Scholar]
- 35.Mathews L. S., Vale W. W. (1993) Receptor 3, 173–181 [PubMed] [Google Scholar]
- 36.Saremba S., Nickel J., Seher A., Kotzsch A., Sebald W., Mueller T. D. (2008) FEBS J. 275, 172–183 [DOI] [PubMed] [Google Scholar]
- 37.Goetschy J. F., Letourneur O., Cerletti N., Horisberger M. A. (1996) Eur. J. Biochem. 241, 355–362 [DOI] [PubMed] [Google Scholar]
- 38.Greenwald J., Fischer W. H., Vale W. W., Choe S. (1999) Nat. Struct. Biol. 6, 18–22 [DOI] [PubMed] [Google Scholar]
- 39.Greenwald J., Le V., Corrigan A., Fischer W., Komives E., Vale W., Choe S. (1998) Biochemistry 37, 16711–16718 [DOI] [PubMed] [Google Scholar]
- 40.Harrington A. E., Morris-Triggs S. A., Ruotolo B. T., Robinson C. V., Ohnuma S., Hyvönen M. (2006) EMBO J. 25, 1035–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hildén K., Tuuri T., Erämaa M., Ritvos O. (1994) Blood 83, 2163–2170 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.