Abstract
Measles virus (MV), an enveloped negative-strand RNA virus, remains a major cause of morbidity and mortality in developing countries. MV predominantly infects immune cells by using signaling lymphocyte activation molecule (SLAM; also called CD150) as a receptor, but it also infects polarized epithelial cells, forming tight junctions in a SLAM-independent manner. Although the ability of MV to infect polarized epithelial cells is thought to be important for its transmission, the epithelial cell receptor for MV has not been identified. A transcriptional repressor, Snail, induces epithelial-mesenchymal transition (EMT), in which epithelial cells lose epithelial cell phenotypes, such as adherens and tight junctions. In this study, EMT was induced by expressing Snail in a lung adenocarcinoma cell line, II-18, which is highly susceptible to wild-type MV. Snail-expressing II-18 cells lost adherens and tight junctions. Microarray analysis confirmed the induction of EMT in II-18 cells and suggested a novel function of Snail in protein degradation and distribution. Importantly, wild-type MV no longer entered EMT-induced II-18 cells, suggesting that the epithelial cell receptor is down-regulated by the induction of EMT. Other polarized cell lines, NCI-H358 and HT-29, also lost susceptibility to wild-type MV when EMT was induced. However, the complete formation of tight junctions rather reduced MV entry into HT-29 cells. Taken together, these data suggest that the unidentified epithelial cell receptor for MV is involved in the formation of epithelial intercellular junctions.
Keywords: Cell Junctions, Epithelial Cell, Tight Junction, Virus Entry, Virus, Epithelial-Mesenchymal Transition, Measles Virus
Introduction
Measles is an acute febrile disease transmitted via aerosol droplets and remains a major cause of infant death in developing countries (1). Measles virus (MV),2 the causative agent of the disease, is an enveloped RNA virus belonging to the genus Morbillivirus in the family Paramyxoviridae. MV has two envelope glycoproteins, the hemagglutinin (H) and fusion (F) proteins, both of which are necessary for MV entry and syncytium formation. The H protein has the ability to bind to a host cell receptor, whereas the F protein causes the fusion of the viral envelope with a cell membrane upon the H protein's binding to a receptor. MV infects immune cells by using human signaling lymphocyte activation molecule (SLAM; also called CD150) as a receptor (2). Vaccine strains of MV use CD46, which is expressed on all human cells except for red blood cells, as an alternate receptor (3, 4). Although wild-type (WT) MV cannot use CD46 as a receptor, pathological data from humans and experimentally infected monkeys showed that MV also infects SLAM-negative epithelial tissues in various organs, such as the skin, oral mucosa, pharynx, trachea, esophagus, intestines, and urinary bladder (5–12). In addition, WT MV induces large syncytia in primary human small airway epithelial cells (13). Recent studies showed that MV infects polarized epithelial cell lines in a SLAM- and CD46-independent manner (14, 15). Thus, there must be another receptor for MV, which is different from SLAM and CD46. However, this epithelial cell receptor for MV has not been identified. MV buds from the apical surface of polarized epithelial cells (15, 16). Furthermore, Leonard et al. (16) reported that a recombinant MV that cannot use the epithelial cell receptor still infects rhesus monkeys and causes a rash and anorexia after intranasal infection but is not shed in the airways. These data suggested that infection of polarized epithelial cells is important for the spread of MV to a new host.
Epithelial-mesenchymal transition (EMT) is a process by which epithelial cells lose their cell junctions and acquire a mesenchymal cell-like phenotype (17). EMT is observed in some developmental processes as well as cancer invasion and metastasis. Snail, a transcriptional repressor, plays a central role during EMT (17, 18). Although the functions of Snail remain to be elucidated, some targets of Snail have been reported. For example, Snail inhibits expression of the gene encoding E-cadherin, which constitutes the adherens junction (19, 20). Snail also acts as a transcriptional repressor of genes encoding tight junction-related molecules, such as occludin, claudins, and Crumbs3 (21–24), and of other genes (25, 26). Furthermore, Ohkubo et al. reported that Snail down-regulates claudin-1 at the posttranscriptional level (22).
In this study, we show that a lung adenocarcinoma cell line II-18 (27), which forms tight junctions, is highly susceptible to WT MV. When Snail was transiently expressed in II-18 cells, they lost adherens and tight junctions. Microarray analysis also confirmed the induction of EMT in II-18 cells. Importantly, WT MV no longer entered EMT-induced II-18 cells, which suggests that the epithelial cell receptor is down-regulated by the induction of EMT. Other polarized cell lines, NCI-H358 and HT-29, also lost susceptibility to WT MV when EMT was induced. However, the complete formation of tight junctions rather reduced MV entry into HT-29 cells. Taken together, these data suggest that the unidentified epithelial cell receptor for MV may be involved in the formation of epithelial intercellular junctions.
EXPERIMENTAL PROCEDURES
Cells
II-18 cells (RCB2093, Riken Bioresource Center Cell Bank (Tsukuba, Japan)) were maintained in RPMI medium (ICN Biomedicals (Aurora, OH)) supplemented with 7.5% fetal bovine serum (FBS). PLAT-gp cells, kindly provided by M. Shimojima and T. Kitamura, are retrovirus-packaging cells, which contain the retroviral gag and pol genes (28). PLAT-gp cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 7.5% FBS and 10 μg/ml blasticidin (InvivoGen (San Diego, CA)). The characteristics and culture conditions used for the following cell lines were as described previously: Vero/hSLAM (29), NCI-H358 (30), HT-29 (31), and A549 (32).
Viruses
All full-length genome plasmids were derived from the p(+)MV323 plasmid encoding the antigenomic full-length cDNA of the IC-B WT strain of MV (33). The p(+)MV323-EGFP and p(+)MV323-Luci plasmids, which contain an additional transcriptional unit of enhanced green fluorescent protein (EGFP) and the Renilla luciferase gene, respectively, were reported previously (14, 34). p(+)MV/Ed-H-EGFP and p(+)MV/Ed-H-Luci contain the Edmonston tag H gene in place of the IC-B WT H gene (14, 35). p(+)MV323/H(Y543S)-EGFP has a mutation in the H gene resulting in the amino acid substitution Y543S; the mutant H protein is incapable of interacting with the putative epithelial cell receptor (15). Recombinant MV strains generated from p(+)MV323-EGFP, p(+)MV323-Luci, p(+)MV/Ed-H-EGFP, p(+)MV/Ed-H-Luci, and p(+)MV323/H(Y543S)-EGFP were named IC323-EGFP, IC323-Luci, IC323/Ed-H-EGFP, IC323/Ed-H-Luci, and IC323/H(Y543S)-EGFP, respectively. They were generated as reported previously (36, 37).
Plasmid Constructions
pCVSV-G is an expression plasmid that was made by cloning a cDNA encoding the vesicular stomatitis virus G protein into the expression vector pCAGGS (38). pMX-GFP, kindly provided by M. Shimojima and T. Kitamura, is a plasmid for a Moloney murine leukemia virus-based retrovirus vector expressing GFP (39, 40). pMXs-IP is a plasmid for a retrovirus vector with multiple cloning sites (41). To generate a plasmid for a retrovirus vector expressing Snail, total RNA was extracted from A549 cells, the cDNA encoding Snail was amplified by reverse transcription and PCR, and pMXs-IP-Snail was generated by inserting Snail cDNA into the EcoRI and NotI sites of pMXs-IP. The eukaryotic expression vectors pCA7 (36, 42) and pCA7ps (15) are derivatives of pCAGGS (38). pCA7ps-ICH, pCA7ps-EdH, and pCA7-ICF, which encode the IC-B H protein, the Edmonston tag H protein, and the IC-B F protein, respectively (15, 35), have been described previously.
Retrovirus Vector
PLAT-gp cells were cultured in collagen-coated 6-well cluster plates (Iwaki Glass Co. Ltd., Chiba, Japan). pMX-GFP (4 μg), pMXs-IP (4 μg), or pMXs-IP-Snail (4 μg) was transfected, together with pCVSV-G (0.4 μg), into PLAT-gp cells, using Lipofectamine 2000 (Invitrogen). After 2 or 3 days posttransfection, supernatant was harvested and filtered through a 0.45-μm filter (Millipore, Bedford, MA). The retrovirus vector expressing GFP (MX-GFP) was used for titration. II-18 cells were cultured in collagen-coated 48-well cluster plates (Corning Glass) and infected by serially diluted MX-GFP. After 48 h postinfection (p.i.), the number of GFP-expressing cells was counted under a fluorescence microscope. Cells were infected with the retrovirus vector expressing Snail in medium containing 4 μg/ml Polybrene at a multiplicity of infection (MOI) of 50–100. A retrovirus vector derived from pMXs-IP was used for control experiments.
FACS Analysis
Cells were harvested and fixed with phosphate-buffered saline (PBS) containing 4% paraformaldehyde for 20 min at room temperature. After being washed with PBS, cells were permeabilized in PBS containing 0.05% Triton X-100 for 5 min at room temperature. Cells were then incubated with the mouse polyclonal antibody against Snail (Abnova (Taipei, Taiwan)) for 1 h on ice, followed by incubation with Alexa Fluor 488-conjugated anti-mouse secondary antibody (Molecular Probes, Inc. (Eugene, OR)). Cells were analyzed by flow cytometry and ModFit software (version 3.0; Verity Software House).
Western Blot Analysis
Cells were lysed in a radioimmunoprecipitation assay buffer (150 mm NaCl, 10 mm Tris-HCl, pH 7.4, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS), and polypeptides in cells were separated by SDS-PAGE and blotted onto polyvinylidene difluoride membranes (Hybond-P; Amersham Biosciences). The membranes were then incubated with primary antibodies for 16 h. A mouse monoclonal antibody (mAb) against E-cadherin (Invitrogen), occludin (Invitrogen), or actin (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)) and rabbit polyclonal antibody against claudin-1 (Invitrogen) or ZO-1 (Invitrogen) were used. The membranes were washed with Tris-buffered saline containing 0.05% Tween 20 and incubated with peroxidase-conjugated goat anti-mouse (Bio-Rad) or anti-rabbit IgG antibody (Zymed) for 1 h at room temperature. After membranes were washed with Tris-buffered saline, 0.05% Tween 20, they were treated with the ECL Plus reagent (Amersham Biosciences), and chemiluminescent signals were detected and visualized using a VersaDoc 3000 imager (Bio-Rad).
Immunofluorescence Staining and Confocal Microscopy
Cells were cultured on 35-mm Petri dishes with thin bottoms (μ-Dish 35 mm, high; Ibidi (Munich, Germany)) or on collagen-coated coverslips. After the treatments described in the respective procedures, cells were fixed and permeabilized with PBS containing 2.5% formaldehyde and 0.5% Triton X-100. Cells were then washed with PBS and incubated with a mAb against ZO-1 (Invitrogen) or E-cadherin (Invitrogen) for 1 h at 37 °C, followed by incubation with Alexa Fluor 488-conjugated anti-mouse secondary antibody (Molecular Probes). Nuclear DNA was stained with propidium iodide (Sigma) at 10 μg/ml. Images of cells were obtained sequentially from the top to the bottom of the cells by using a confocal microscope (Radiance 2100, Bio-Rad) and merged together by using Lasersharp software (Bio-Rad).
cDNA Microarray Analysis
Total RNA was isolated from cells using the RNeasy Plus Mini kit (Qiagen (Valencia, CA)). RNA was amplified and labeled using the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX). HumanWG-6 version 3.0 BeadChip (Illumina, Hayward, CA) was hybridized with labeled cRNA. Data were submitted to the GEO database, accession number GSE19679, and analyzed using BeadStudio (Illumina) and KeyMolnet software (IMMD Inc., Tokyo, Japan).
Virus Titration
Monolayers of Vero/hSLAM cells on 12-well cluster plates were infected with serially diluted virus samples and incubated for 1 h at 37 °C. After being washed with PBS, the cells were overlaid with DMEM containing 7.5% FBS and 1% methylcellulose. At 4 days p.i., the titers of EGFP-expressing viruses were determined by counting the numbers of plaques under a fluorescence microscope. The number of plaques of luciferase-expressing viruses was counted after neutral red staining. Cell infectious units (CIU) of EGFP-expressing viruses were also determined as described previously (34, 42).
Virus Growth
II-18 cells cultured in 12-well cluster plates were infected with IC323-EGFP at an MOI of 0.01. At various days p.i., both cells and media were collected to determine the titers in them.
Measurement of Infectivity
Cells were cultured in collagen-coated 48-well cluster plates (Corning Glass). After the treatments described in the respective procedures, cells were incubated by Renilla luciferase-expressing MVs at an MOI of 0.01. After 3 h p.i., DMEM supplemented with 7.5% FBS and 100 μg/ml fusion block peptide (Z-d-Phe-Phe-Gly) (43) (Peptide Institute Inc. (Osaka, Japan)) was added to each well to block the second round of infection by progeny viruses. After 24 h p.i., cells were lysed in the Renilla luciferase assay lysis buffer. The Renilla luciferase activity in the cells was then analyzed using a Renilla luciferase assay system (Promega (Madison, WI)), according to the manufacturer's instructions. Chemiluminescence was measured using a Mithras LB940 plate reader (Berthold Technologies (Pforzheim, Germany)). Luciferase activity of control samples was set to 100% in each experiment.
Fusion Assay
CHO cells cultured in 6-well cluster plates were cotransfected with pCA7ps-ICH or pCA7ps-EdH (2 μg) plus pCA7-ICF (2 μg), using Lipofectamine 2000 (Invitrogen). Two days after transfection, the cells were cocultured with II-18 cells. After 2 days of incubation, the cells were observed after Giemsa staining under a light microscope.
Modulation of the Tight Junction Formation
HT-29 cells were seeded at 15% confluence on collagen-coated coverslips or 6-well plates. At various time intervals (1, 3, 10, and 16 days after plating), cells were used for virus infection or immunostaining. Alternatively, the culture media of HT-29 cells 8–12 days after plating were changed to calcium-free media (Invitrogen) or DMEM (Invitrogen). After 24 h, cells were used for virus infection or immunostaining. After virus infection, DMEM supplemented with 7.5% FBS and 100 μg/ml fusion block peptide was added to each well. At 36 h p.i., the number of EGFP-expressing cells was counted under a fluorescence microscope.
RESULTS
SLAM- and CD46-independent MV Infection of II-18 Cells
WT MV infects and replicates in polarized epithelial cell lines, such as NCI-H358, HT-29, Calu-3, Caco-2, and T84 (14, 15). However, these cell lines are not as easy to propagate as other common cell lines because of their slow growth and a requirement for special medium. After screening many cell lines, we found that a human lung adenocarcinoma cell line, II-18, is not only highly susceptible to WT MV but also grows fast in RPMI medium.
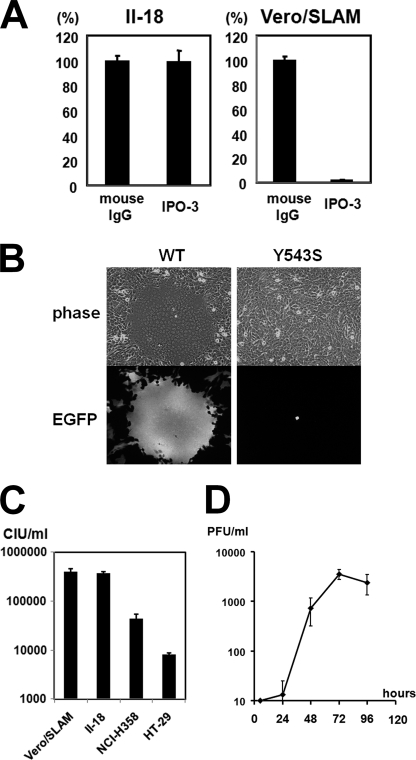
To examine whether MV infects II-18 cells in a SLAM-independent manner, II-18 and Vero/hSLAM cells were infected with IC323-Luci, a recombinant virus encoding a reporter luciferase based on a WT strain of MV, in medium containing a mAb against SLAM (IPO-3) (44) (Kamiya Biomedical (Seattle, WA)) or mouse control IgG. IPO-3 prevented IC323-Luci entry into Vero/hSLAM cells but not II-18 cells (Fig. 1A). An epithelial cell receptor-blind MV IC323/H(Y543S)-EGFP, which has a Y543S substitution on the receptor-binding H protein and cannot utilize the epithelial cell receptor, was reported previously (15). II-18 cells were infected with IC323-EGFP and IC323/H(Y543S)-EGFP and observed under a fluorescence microscope at 2 days p.i. Infection with IC323-EGFP induced syncytium formation in II-18 cells, whereas infection with IC323/H(Y543S)-EGFP did not (Fig. 1B). WT MV does not use CD46 as a receptor. All of these results indicate that MV infects II-18 cells in a SLAM- and CD46-independent manner. The infectious titer of the same stock of IC323-EGFP was measured in Vero/SLAM, II-18, NCI-H358, and HT-29 cells. The titer in II-18 cells was comparable with that in Vero/SLAM cells (Fig. 1C), indicating that WT MV infects II-18 and SLAM-positive cells at almost the same efficiency. The titers in NCI-H358 and HT-29 cells were much lower than that in II-18 cells. These data indicate that II-18 cells are suitable for studying an unidentified epithelial cell receptor. To investigate whether II-18 cells can support multiple cycles of MV growth, a growth curve of IC323-EGFP in II-18 cells was examined (Fig. 1D). Although II-18 cells supported multiple cycles of MV growth, the peak titer was lower than that in other polarized cell lines reported previously (14, 15). There may be postentry defects of MV replication in II-18 cells.
FIGURE 1.
SLAM-independent WT MV infection of II-18 cells. A, monolayers of II-18 and Vero/hSLAM cells were incubated with medium containing control mouse IgG or IPO3 (anti-SLAM mAb) and infected with IC323-Luci at an MOI of 0.01. At 3 h p.i., the fusion block peptide was added to medium, and at 24 h p.i., the Renilla luciferase activity was measured. The activity in cells treated with control mouse IgG was set to 100%. B, monolayers of II-18 cells were infected with IC323-EGFP or IC323/H(Y543S)-EGFP. At 2 days p.i., the monolayers were observed under a phase-contrast or fluorescence microscope. C, monolayers of Vero/hSLAM, II-18, NCI-H358, and HT-29 cells were infected with serially diluted virus samples of the same stock of IC323-EGFP. Titers were measured by counting the numbers of EGFP-expressing cells under a fluorescence microscope. D, monolayers of II-18 cells were infected with IC323-EGFP at an MOI of 0.01. At various days p.i., virus was harvested, and its plaque-forming units were measured in Vero/hSLAM cells. Error bars, S.D.
Destruction of Adherens and Tight Junctions in II-18 Cells by Snail Expression
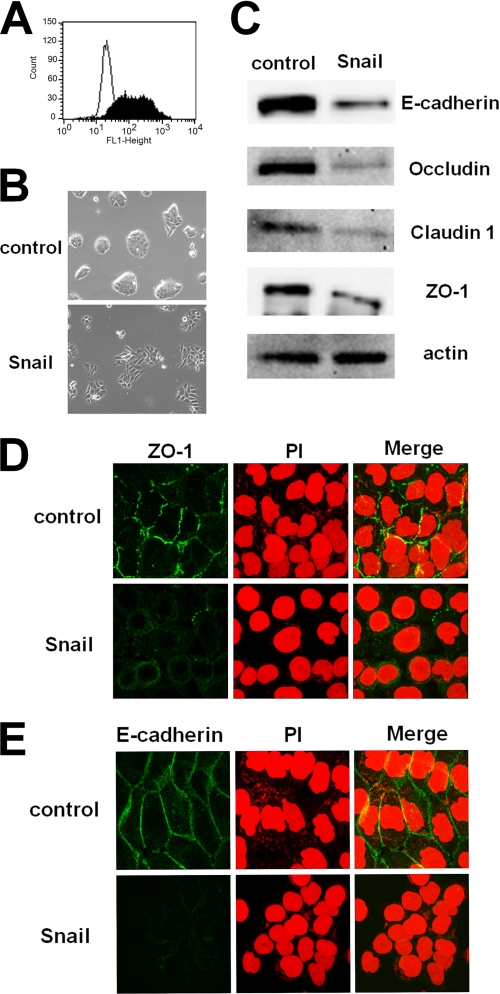
Snail is a transcriptional repressor that induces EMT (17). EMT-induced cells lose epithelial intercellular junctions, such as adherens and tight junctions (21, 22). To examine whether MV infection via the epithelial cell receptor is influenced by EMT, II-18 cells were infected with the retrovirus vector expressing Snail at an MOI of 50–100. Expression of Snail was confirmed by fluorescence-activated cell sorting analysis (Fig. 2A). After 2 days p.i., the morphology of II-18 cells was observed under a phase-contrast imaging microscope. II-18 cells expressing Snail seemed to have weak contacts with each other as compared with control cells (Fig. 2B). After 5 days p.i., expression of E-cadherin, occludin, claudin-1, and ZO-1 was examined by Western blot analysis. E-cadherin is a component of the adherens junction, and occludin, claudin-1, and ZO-1 are components of the tight junction. They are used as epithelial markers (17). All of these proteins were down-regulated in II-18 cells expressing Snail (Fig. 2C). To investigate whether the formation of adherens and tight junctions was affected by Snail expression in II-18 cells, immunofluorescence staining of E-cadherin and ZO-1 was performed. The immunofluorescent staining pattern of ZO-1 indicated that tight junctions were formed in control II-18 cells, but they were destroyed by Snail expression (Fig. 2D). Similarly, adherens junctions were formed between neighboring II-18 cells, but they were destroyed after the induction of EMT (Fig. 2E). These results indicate that Snail expression causes II-18 cells to lose the epithelial phenotype.
FIGURE 2.
Induction of EMT in II-18 cells by expressing Snail. A, II-18 cells were infected with the retrovirus vector expressing Snail or control vector at an MOI of 50–100. At 5 days p.i., fluorescence-activated cell sorting analysis was performed to detect the expression of Snail. The expression of Snail was detected by using mouse polyclonal antibody against Snail. The empty profile represents control II-18 cells, and the solid profile represents Snail-expressing II-18 cells. B, at 2 days p.i., control and Snail-expressing II-18 cells were observed under a phase-contrast microscope. C, at 5 days p.i., cells were harvested, and expressions of E-cadherin, occludin, claudin-1, ZO-1, and actin were detected by Western blot analysis. D and E, at 5 days p.i., cells were permeabilized, fixed, and stained with a mAb against ZO-1 (D) or E-cadherin (E), followed by incubation with Alexa Fluor 488-conjugated anti-mouse secondary antibody. Nuclear DNA was stained with propidium iodide (PI). The cells were observed using a confocal microscope.
Modulation of mRNA Transcription Levels in II-18 Cells by Snail
Next, microarray analysis was performed to comprehensively investigate the influence of Snail expression on mRNA levels in II-18 cells. II-18 cells were infected with the retrovirus vector expressing Snail or the control vector at an MOI of 50–100. After 5 days p.i., total RNA was isolated from control or Snail-expressing II-18 cells, amplified, and labeled (n = 3). The human-WG-6 version 3.0 BeadChip (Illumina), which has 48,803 probes, was used for hybridization. The signals for 192 probes (corresponding to 181 genes) were decreased more than 2-fold by Snail expression (supplemental Table 1), whereas the signals for 63 probes (corresponding to 55 genes) were increased more than 2-fold (supplemental Table 2). The list of down-regulated genes includes the E-cadherin gene, which is a well known target of Snail. In addition, some genes related to cell junctions were down-regulated (Table 1). Claudin-3, -7, and -8 are components of tight junctions. These data indicate that EMT was largely regulated at the mRNA level. Genes associated with protein trafficking and degradation were also affected by Snail expression (Table 2). Caveolin-1 and cathepsin L1, which have functions in the endocytosis pathway (45, 46), were up-regulated. Cathepsin L1 is a cysteine proteinase for protein degradation in the lysosome, and cystatin E/M, an inhibitor of cysteine proteinases including cathepsin L1 (47), was down-regulated. In addition, some down-regulated genes were associated with vesicular transport, especially the endosomal recycling pathway (Table 2). Rab proteins are monomeric small GTPases that regulate transport between organelles (48). The Rab family contains more than 60 members in humans. The list of down-regulated genes includes five Rab-associated genes, four of which encode the proteins located in the recycling endosome (RAB17, RAB25, RAB40C, and RAB11FIP1) (49–53). Furthermore, MAL2 is associated with apical sorting in polarized cells (54), and other down-regulated genes involved in the protein transport are shown in Table 2. These data suggest that Snail not only represses target genes related to cell junctions but also influences the expression of genes involved in protein metabolism and distribution, thereby regulating EMT in II-18 cells.
TABLE 1.
Down-regulated genes related to cell junctions
| Symbol | Definition | Snail/Controla |
|---|---|---|
| CEACAM6 | Carcinoembryonic antigen-related cell adhesion molecule 6 | 0.05 |
| CD9 | CD9 molecule | 0.34 |
| CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | 0.37 |
| CLDN3 | Claudin 3 | 0.11 |
| CLDN7 | Claudin 7 | 0.21 |
| CLDN8 | Claudin 8 | 0.23/0.34b |
a Signals in Snail-expressing cells divided by those in control cells.
b Down-regulated as examined with two different CLDN8 probes.
TABLE 2.
Up- or down-regulated genes related to endocytosis or protein trafficking
| Symbol | Definition | Snail/Controla |
|---|---|---|
| Endocytosis | ||
| Up-regulated | ||
| CAV1 | Caveolin 1, caveolae protein, 22 kDa | 2.38 |
| CTSL1 | Cathepsin L1 | 2.26/2.20b |
| Down-regulated | ||
| CST6 | Cystatin E/M | 0.20/0.25c |
| Protein trafficking | ||
| Down-regulated | ||
| RAB17 | RAB17, member RAS oncogene family | 0.20 |
| RAB25 | RAB25, member RAS oncogene family | 0.24 |
| RAB38 | RAB38, member RAS oncogene family | 0.29 |
| RAB40C | RAB40C, member RAS oncogene family | 0.29 |
| RAB11FIP1 | RAB11 family-interacting protein 1 | 0.41 |
| MAL2 | MAL, T-cell differentiation protein 2 | 0.23 |
| MALL | MAL, T-cell differentiation protein-like | 0.40 |
| STX3 | Syntaxin 3 | 0.40 |
| SCIN | Scinderin | 0.42 |
| MYO5C | Myosin VC | 0.45 |
a Signals in Snail-expressing cells divided by those in control cells.
b Up-regulated as examined with two different CTSL1 probes.
c Down-regulated as examined with two different probes of CST6.
Loss of MV Infection of EMT-induced Polarized Epithelial Cells
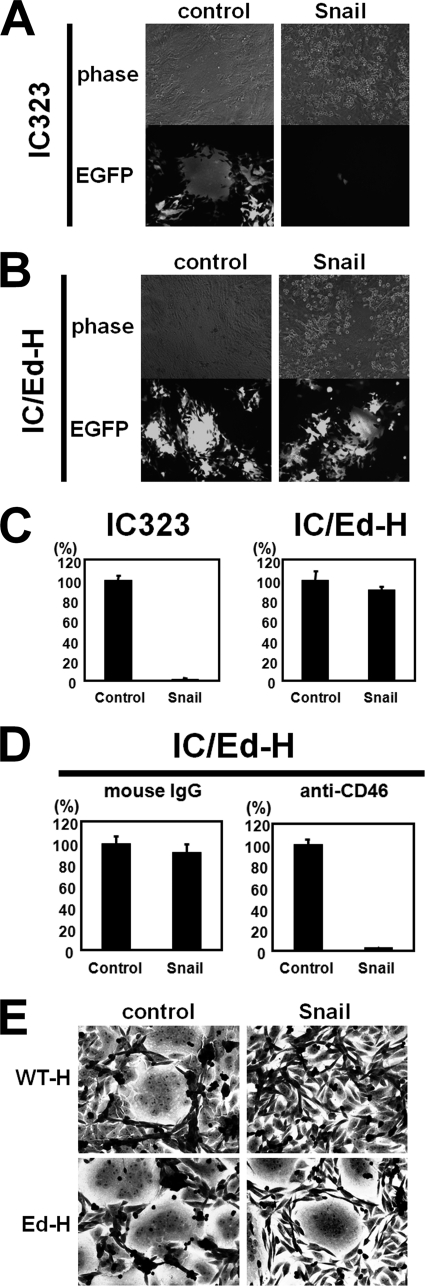
We then examined whether MV infection via the epithelial cell receptor is affected by EMT. In these experiments, not only IC323-EGFP or -Luci but also IC/EdH-EGFP or -Luci were used to infect cells. The latter viruses have the H protein of the Edmonston vaccine strain, which can use CD46 as a receptor in addition to SLAM and the epithelial cell receptor. Control and EMT-induced II-18 cells were infected with IC323-EGFP and observed under a fluorescence microscope (Fig. 3A). IC323-EGFP infected control cells efficiently and induced cell-cell fusion. By contrast, the number of EGFP-expressing cells was decreased, and syncytium formation was not observed in EMT-induced II-18 cells. CD46-using IC/EdH-EGFP infected both control and EMT-induced cells and induced cell-cell fusion in both (Fig. 3B). To quantify the levels of infection, the infectivities of IC323-Luci and IC/EdH-Luci were determined in control and EMT-induced II-18 cells (Fig. 3C). IC/EdH-Luci infected both cell types efficiently, and the infectivity was not affected by the induction of EMT. By contrast, the infectivity of IC323-Luci was greatly reduced in EMT-induced II-18 cells compared with control cells. IC/EdH-Luci infected EMT-induced II-18 cells via CD46, because M75, a mAb against CD46, almost completely blocked IC/Ed-H-Luci infection of those cells (Fig. 3D). These results clearly indicate that WT MV infection of EMT-induced II-18 cells is abolished at the entry step, very likely due to the absence of a receptor, because CD46-dependent entry and postentry replication of IC/EdH-Luci were not affected at all in EMT-induced II-18 cells. We also performed a fusion assay to obtain additional evidence that MV infection is inhibited at the entry step by the induction of EMT. CHO cells transiently expressing H and F proteins of MV were overlaid onto II-18 cells. CHO cells expressing the H and F proteins can fuse with cells with the corresponding receptors, which can interact with the H protein. Membrane fusion was not observed in EMT-induced II-18 cells when CHO cells expressing the H protein of WT MV were used, but it was observed in those cells when CHO cells expressing the H protein of the MV Edmonston strain were used (Fig. 3E). Thus, the H protein of WT MV does not support membrane fusion in EMT-induced II-18 cells, indicating that a receptor for WT MV does not exist in those cells.
FIGURE 3.
Loss of MV infection of EMT-induced II-18 cells. A and B, II-18 cells were infected with the retrovirus vector expressing Snail or control vector at an MOI of 50–100. Control and EMT-induced II-18 cells were infected with IC323-EGFP (A) or IC/Ed-H-EGFP (B). At 2 days p.i., the cells were observed under a phase-contrast or fluorescence microscope. C, control and EMT-induced II-18 cells were infected with IC323-Luci or IC/Ed-H-Luci. At 3 h p.i., the fusion block peptide was added to medium, and at 24 h p.i., the Renilla luciferase activity was measured. The activity in control cells was set to 100%. D, control and EMT-induced II-18 cells were incubated with medium containing control mouse IgG or a mAb against CD46 (M75) and infected with IC323-Luci. The procedures after MV infection were the same as those in C. E, CHO cells were transfected with pCA7ps-ICH or pCA7ps-EdH plus pCA7-ICF, using Lipofectamine 2000. Two days after transfection, the cells were harvested and cocultured with II-18 cells. After 2 days of incubation, the cells were subjected to Giemsa staining and observed under a light microscope. Error bars, S.D.
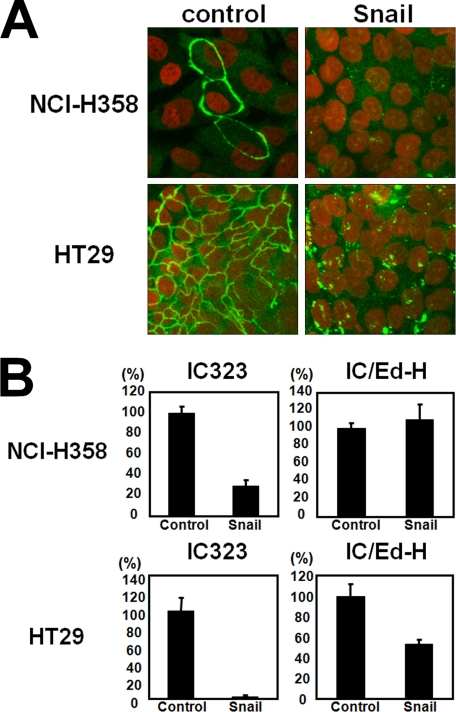
We next examined whether EMT also inhibits MV infection in other polarized epithelial cell lines, namely NCI-H358 and HT-29. By expressing Snail, the tight junctions of NCI-H358 and HT-29 cells were destroyed, as revealed by staining for ZO-1 (Fig. 4A). At 5 days after the induction of EMT with the retrovirus vector encoding Snail, NCI-H358 and HT-29 cells were infected with 1.0 × 103 plaque-forming units of IC323-Luci or IC323/Ed-H-Luci. The infectivity of IC323-Luci, but not IC/Ed-H-Luci, was reduced in EMT-induced NCI-H358 cells (Fig. 4B). The infectivity of IC323-Luci was almost completely abolished in EMT-induced HT-29 cells, whereas that of IC/Ed-H-Luci was only moderately reduced (Fig. 4B). Thus, the induction of EMT in polarized epithelial cells is correlated with the loss of infectivity of WT MV. These results indicate that the putative MV receptor on polarized epithelial cells disappears upon the induction of EMT.
FIGURE 4.
Loss of MV infection of EMT-induced NCI-H358 and HT-29 cells. A, NCI-H358 and HT-29 cells were infected with the retrovirus vector expressing Snail or control vector. At 7–10 days p.i., control and Snail-expressing cells were permeabilized, fixed, and stained with a mAb against ZO-1, followed by incubation with Alexa Fluor 488-conjugated anti-mouse secondary antibody. Nuclear DNA was stained with propidium iodide. The cells were observed using a confocal microscope. B, control and Snail-expressing cells were infected with IC323-Luci or IC/Ed-H-Luci. At 3 h p.i., the fusion block peptide was added to medium, and at 24 h p.i., the Renilla luciferase activity was measured. The activity in control cells was set to 100%. Error bars, S.D.
The Formation of Junction Structures per Se Is Not Required for WT MV Infection
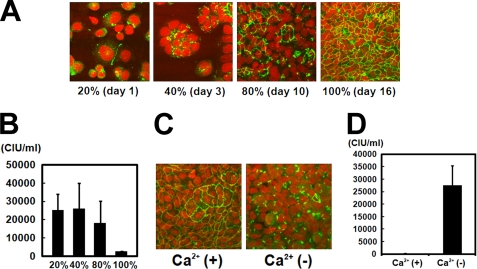
Last, experiments were performed to investigate whether the formation of junction structures, such as tight and adherens junctions, is necessary for MV to enter polarized epithelial cells. Polarized cells form tight junctions when growing densely in monolayers (55). HT-29 cells have the ability to form complete monolayers with the formation of tight junctions and can be propagated uniformly (Fig. 4A), unlike most other human polarized epithelial cell lines. HT-29 cells were seeded on culture plates to fill ∼15% of the culture area (day 0). On days 1 and 3, HT-29 cells filled ∼20 and ∼40% of the culture area, respectively, and poorly formed tight junctions (Fig. 5A). When the cells filled ∼80% of the culture area, on day 10, about half of the cells formed tight junctions (Fig. 5A). On day 16, cells filled the culture area almost entirely (100%), forming tight junctions well (Fig. 5A). These monolayers cultured in different densities were infected with 2.0 × 105 CIU of IC323-EGFP. To determine the initial infection of these monolayers by IC323-EGFP, the second round of infection was blocked by a fusion block peptide (34, 43). When the numbers of EGFP-expressing cells were counted at 2 days p.i., they were 2.5 × 104, 2.6 × 104, 1.8 × 104, and 2.3 × 103 in monolayers at densities of ∼20, 40, 80, and 100%, respectively (Fig. 5B). These data indicate that, when cells form tight junctions completely, the infection efficiency of WT MV becomes lower compared with that in cells having incomplete tight junctions.
FIGURE 5.
WT MV infection of HT-29 cells with or without tight junctions. A, HT-29 cells were seeded on culture plates to fill ∼15% of the culture area. At day 1, 3, 10, and 16 (when growing cells filled 20, 40, 80, and 100% of the culture area, respectively), cells were permeabilized, fixed, and stained with a mAb against ZO-1, followed by incubation with Alexa Fluor 488-conjugated anti-mouse secondary antibody. Nuclear DNA was stained with propidium iodide. The cells were observed using a confocal microscope. B, HT-29 cells grown under the same conditions as those in A were infected with 2.0 × 105 CIU of IC323-EGFP for 2 h (CIUs were determined on Vero/hSLAM cells). After 2 h of incubation, cells were washed with PBS and incubated in a standard culture medium with the fusion block peptide. Numbers of EGFP-expressing cells were counted at 2 days p.i. C, at 8–12 days after plating, HT-29 cells were fed with Ca2+-free medium or medium containing Ca2+. After 24 h of incubation, cells were used for immunostaining as in A. D, HT-29 cells grown under the same conditions as those in C were infected with IC323-EGFP, and CIUs were determined at 2 days p.i. Error bars, S.D.
We also analyzed the role of junction structure formation in WT MV infection in a different way. Ca2+ is a critical component for the formation of tight junctions (55). When Ca2+ is depleted from culture media, cells no longer form tight junctions (55). Confluent monolayers of HT-29 cells forming tight junctions were prepared. Before infection with MV, some monolayers were cultured in DMEM containing Ca2+ and others in Ca2+-free medium. After 24 h of incubation, the latter lost tight junctions (Fig. 5C), as reported previously (55). These monolayers with or without tight junctions were infected with 1 × 105 CIU of IC323-EGFP and cultured in the presence of the fusion block peptide. When counted at 2 days p.i., the number of EGFP-expressing cells was more than 100 times greater in monolayers without tight junctions than in those retaining the junctions (Fig. 5D). These data indicate that the formation of tight junctions per se is not required for WT MV infection and rather restricts MV infection.
DISCUSSION
MV predominantly infects immune cells in vivo by using SLAM as a receptor (56, 57), explaining the lymphotropism and immunosuppressive nature of MV. However, MV also infects polarized epithelial cells independently of SLAM (14–16). The ability of MV to infect polarized epithelial cells may not be required to cause disease symptoms but appears to be important for its transmission (16). Thus, the unknown epithelial cell receptor should be identified and investigated to fully understand MV pathogenesis. In this study, we have shown that the induction of EMT almost completely abolishes the susceptibility of polarized epithelial cell lines to WT MV, shedding light on the properties of the epithelial cell receptor.
We found that a lung adenocarcinoma cell line II-18 is very useful to study the mechanisms underlying MV infection of epithelial cells. II-18 cells are easily cultured in a standard culture medium, unlike other human polarized epithelial cell lines, and are highly susceptible to WT MV independently of SLAM and CD46. Furthermore, they are readily induced to undergo EMT by expressing the transcriptional repressor Snail in them. Upon expression of Snail, II-18 cells lost intercellular adherens and tight junctions, accompanied by the up- and down-regulation of a number of genes, as revealed by microarray analysis.
Previous studies reported that Snail affects gene expression of molecules involved in the cell junctions, cytoskeleton, cell metabolism, transcription, and cell signaling (26, 58, 59). Our results showed that Snail also affects the expression of genes responsible for protein degradation and trafficking. The list of up-regulated genes included those encoding caveolin-1 and cathepsin L1, which function in the endocytosis pathway (45, 46). By contrast, the gene encoding cystatin E/M, an inhibitor of cysteine proteinases, such as cathepsin L1 (47), was down-regulated. Furthermore, the list of down-regulated genes contained those encoding proteins involved in protein trafficking, especially protein recycling. De Craene et al. (26) also reported that RAB25 is repressed by Snail. Endocytosed proteins escape from degradation in the lysosome if they are transported to the recycling endosome and returned to the plasma membrane (60). The recycling endosome is also associated with apical/basolateral sorting, which maintains the polarity of the cell membrane (60). Thus, it is likely that Snail inhibits the recycling pathway and enhances protein degradation, thereby affecting the distribution of proteins. Together with down-regulation of genes encoding cell junction molecules, this may contribute to Snail-mediated EMT in II-18 cells. Similar observations have been reported. E-cadherin was found to be endocytosed and degraded in lysosomes when EMT was induced by TGF-β (61). Ohkubo et al. (22) showed that the tight junction constituent claudin-1 is down-regulated by Snail expression at the posttranscriptional level, although another group reported a different observation (23).
All cell lines that WT MV has been shown to infect in a SLAM-independent manner are polarized epithelial cells forming tight junctions. Therefore, we thought that EMT might affect the susceptibility of II-18 cells to MV. Beyond our initial expectations, EMT almost completely abolished MV infection of II-18 cells via the epithelial cell receptor. Inhibition of MV infection was at the entry step, because entry and membrane fusion mediated by CD46-using MV were not affected by EMT. The same findings were obtained in other polarized cell lines, namely NCI-H358 and HT-29. These data indicate that the unidentified epithelial cell receptor for MV is a polarized epithelial cell-specific molecule that disappears from the plasma membrane of II-18 cells upon the induction of EMT. It is also reported that EMT inhibits adenovirus infection through down-regulation of the coxsackie and adenovirus receptor (62), which is a component of the tight junction (63).
Despite the above findings, the formation of junction structures was not required for MV to enter polarized epithelial cells. On the contrary, the complete formation of tight junctions restricted MV entry into HT-29 cells. This indicates that the access of MV to the epithelial cell receptor is hindered when junction structures are completely formed. Infection by hepatitis C virus, which utilizes the tight junction molecules claudin and occludin as receptors, is reported to be restricted by cell polarization (64). The epithelial cell receptor for MV may also be a component of tight or adherens junctions. Alternatively, it may be a molecule strictly present at the basolateral surface of epithelial cells.
In an attempt to identify the putative epithelial cell receptor for MV, we knocked down, by using small interfering RNA, the molecules whose expressions are down-regulated by induced EMT. Although we have thus far tried small interfering RNAs for more than a dozen molecules, including CD9, CDH1, CLDN3, CLDN7, MAL2, MALL, and STX3 (Tables 1 and 2), the infectivity of MV is not affected in treated cells.
Identification of the epithelial cell receptor for MV will contribute to better understanding of MV pathogenesis, because the use of the epithelial cell receptor is important for MV transmission (15, 16). Thus, we continue to make efforts to identify this receptor. Furthermore, the present study suggests that susceptibility to MV may be used to monitor EMT or mesenchymal-epithelial transition (65).
Supplementary Material
Acknowledgments
We thank Y. Fujinaga for HT-29 cells; T. Seya for M75 hybridoma cells; M. Shimojima and T. Kitamura for PLAT-gp cells, pMX, pMX-GFP, pMXs-IP, and pCVSV-G; M. A. Billter for Edmonston tag cDNA; and K. Takayama for various human cell lines. We also thank the staff of the Research Support Center (Faculty of Medicine, Kyushu University) for technical support of microarray analysis.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology and the Ministry of Health, Labor, and Welfare of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
- MV
- measles virus
- WT
- wild-type
- EMT
- epithelial-mesenchymal transition
- GFP
- green fluorescent protein
- EGFP
- enhanced green fluorescent protein
- MOI
- multiplicity of infection
- p.i.
- postinfection
- H protein
- hemagglutinin protein
- F protein
- fusion protein
- SLAM
- signaling lymphocyte activation molecule
- hSLAM
- human SLAM
- FBS
- fetal bovine serum
- DMEM
- Dulbecco's modified Eagle's medium
- PBS
- phosphate-buffered saline
- mAb
- monoclonal antibody
- CIU
- Cell infectious units
- Z-
- benzyloxycarbonyl
- CHO
- Chinese hamster ovary.
REFERENCES
- 1.Bryce J., Boschi-Pinto C., Shibuya K., Black R. E. (2005) Lancet 365, 1147–1152 [DOI] [PubMed] [Google Scholar]
- 2.Tatsuo H., Ono N., Tanaka K., Yanagi Y. (2000) Nature 406, 893–897 [DOI] [PubMed] [Google Scholar]
- 3.Dörig R. E., Marcil A., Chopra A., Richardson C. D. (1993) Cell 75, 295–305 [DOI] [PubMed] [Google Scholar]
- 4.Naniche D., Varior-Krishnan G., Cervoni F., Wild T. F., Rossi B., Rabourdin-Combe C., Gerlier D. (1993) J. Virol. 67, 6025–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craighead J. E. (2000) in Pathology and Pathogenesis of Human Viral Disease (Craighead J. E. ed) pp. 397–410, Elsevier, London, UK [Google Scholar]
- 6.Kimura A., Tosaka K., Nakao T. (1975) Arch. Virol. 47, 295–307 [DOI] [PubMed] [Google Scholar]
- 7.Lightwood R., Nolan R. (1970) J. Pediatr. 77, 59–64 [DOI] [PubMed] [Google Scholar]
- 8.Moench T. R., Griffin D. E., Obriecht C. R., Vaisberg A. J., Johnson R. T. (1988) J. Infect. Dis. 158, 433–442 [DOI] [PubMed] [Google Scholar]
- 9.Nii S., Kamahora J., Mori Y., Takahashi M., Nishimura S., Okuno Y. (1964) Biken J. 6, 271–297 [PubMed] [Google Scholar]
- 10.Nommensen F. E., Dekkers N. W. (1981) J. Med. Virol. 7, 157–162 [DOI] [PubMed] [Google Scholar]
- 11.Olding-Stenkvist E., Bjorvatn B. (1976) J. Infect. Dis. 134, 463–469 [DOI] [PubMed] [Google Scholar]
- 12.Sakaguchi M., Yoshikawa Y., Yamanouchi K., Sata T., Nagashima K., Takeda K. (1986) Microbiol. Immunol. 30, 1067–1073 [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi K., Miyajima N., Nagata N., Takeda M., Tashiro M. (2003) Virus Res. 94, 11–16 [DOI] [PubMed] [Google Scholar]
- 14.Takeda M., Tahara M., Hashiguchi T., Sato T. A., Jinnouchi F., Ueki S., Ohno S., Yanagi Y. (2007) J. Virol. 81, 12091–12096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahara M., Takeda M., Shirogane Y., Hashiguchi T., Ohno S., Yanagi Y. (2008) J. Virol. 82, 4630–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leonard V. H., Sinn P. L., Hodge G., Miest T., Devaux P., Oezguen N., Braun W., McCray P. B., Jr., McChesney M. B., Cattaneo R. (2008) J. Clin. Invest. 118, 2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiery J. P., Sleeman J. P. (2006) Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 18.Barrallo-Gimeno A., Nieto M. A. (2005) Development 132, 3151–3161 [DOI] [PubMed] [Google Scholar]
- 19.Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García De Herreros A. (2000) Nat. Cell Biol. 2, 84–89 [DOI] [PubMed] [Google Scholar]
- 20.Cano A., Pérez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. (2000) Nat. Cell Biol. 2, 76–83 [DOI] [PubMed] [Google Scholar]
- 21.Ikenouchi J., Matsuda M., Furuse M., Tsukita S. (2003) J. Cell Sci. 116, 1959–1967 [DOI] [PubMed] [Google Scholar]
- 22.Ohkubo T., Ozawa M. (2004) J. Cell Sci. 117, 1675–1685 [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Estrada O. M., Cullerés A., Soriano F. X., Peinado H., Bolós V., Martínez F. O., Reina M., Cano A., Fabre M., Vilaró S. (2006) Biochem. J. 394, 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whiteman E. L., Liu C. J., Fearon E. R., Margolis B. (2008) Oncogene 27, 3875–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guaita S., Puig I., Franci C., Garrido M., Dominguez D., Batlle E., Sancho E., Dedhar S., De Herreros A. G., Baulida J. (2002) J. Biol. Chem. 277, 39209–39216 [DOI] [PubMed] [Google Scholar]
- 26.De Craene B., Gilbert B., Stove C., Bruyneel E., van Roy F., Berx G. (2005) Cancer Res. 65, 6237–6244 [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi S., Okada S., Inaba H., Syoji W., Hasumi T., Sato N., Fujimura S. (1990) Kokenshi 42, 73–79 [Google Scholar]
- 28.Morita S., Kojima T., Kitamura T. (2000) Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 29.Ono N., Tatsuo H., Hidaka Y., Aoki T., Minagawa H., Yanagi Y. (2001) J. Virol. 75, 4399–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brower M., Carney D. N., Oie H. K., Gazdar A. F., Minna J. D. (1986) Cancer Res. 46, 798–806 [PubMed] [Google Scholar]
- 31.Fogh J., Trempe G. (1975) in Human Tumor Cells in Vitro (Fogh J. ed) pp. 115–160, Plenum Publishing Corp., New York [Google Scholar]
- 32.Giard D. J., Aaronson S. A., Todaro G. J., Arnstein P., Kersey J. H., Dosik H., Parks W. P. (1973) J. Natl. Cancer Inst. 51, 1417–1423 [DOI] [PubMed] [Google Scholar]
- 33.Takeda M., Takeuchi K., Miyajima N., Kobune F., Ami Y., Nagata N., Suzaki Y., Nagai Y., Tashiro M. (2000) J. Virol. 74, 6643–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashimoto K., Ono N., Tatsuo H., Minagawa H., Takeda M., Takeuchi K., Yanagi Y. (2002) J. Virol. 76, 6743–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tahara M., Takeda M., Seki F., Hashiguchi T., Yanagi Y. (2007) J. Virol. 81, 2564–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda M., Ohno S., Seki F., Hashimoto K., Miyajima N., Takeuchi K., Yanagi Y. (2005) Virus Res. 108, 161–165 [DOI] [PubMed] [Google Scholar]
- 37.Nakatsu Y., Takeda M., Kidokoro M., Kohara M., Yanagi Y. (2006) J. Virol. Methods 137, 152–155 [DOI] [PubMed] [Google Scholar]
- 38.Niwa H., Yamamura K., Miyazaki J. (1991) Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 39.Kitamura T., Morikawa Y. (2000) Methods Mol. Biol. 134, 143–152 [DOI] [PubMed] [Google Scholar]
- 40.Onishi M., Kinoshita S., Morikawa Y., Shibuya A., Phillips J., Lanier L. L., Gorman D. M., Nolan G. P., Miyajima A., Kitamura T. (1996) Exp. Hematol. 24, 324–329 [PubMed] [Google Scholar]
- 41.Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. (2003) Exp. Hematol. 31, 1007–1014 [PubMed] [Google Scholar]
- 42.Takeda M., Ohno S., Seki F., Nakatsu Y., Tahara M., Yanagi Y. (2005) J. Virol. 79, 14346–14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson C. D., Scheid A., Choppin P. W. (1980) Virology 105, 205–222 [DOI] [PubMed] [Google Scholar]
- 44.Sidorenko S. P., Clark E. A. (1993) J. Immunol. 151, 4614–4624 [PubMed] [Google Scholar]
- 45.Mayor S., Pagano R. E. (2007) Nat. Rev. Mol. Cell Biol. 8, 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turk V., Turk B., Turk D. (2001) EMBO J. 20, 4629–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng T., Hitomi K., van Vlijmen-Willems I. M., de Jongh G. J., Yamamoto K., Nishi K., Watts C., Reinheckel T., Schalkwijk J., Zeeuwen P. L. (2006) J. Biol. Chem. 281, 15893–15899 [DOI] [PubMed] [Google Scholar]
- 48.Zerial M., McBride H. (2001) Nat. Rev. Mol. Cell Biol. 2, 107–117 [DOI] [PubMed] [Google Scholar]
- 49.Hunziker W., Peters P. J. (1998) J. Biol. Chem. 273, 15734–15741 [DOI] [PubMed] [Google Scholar]
- 50.Zacchi P., Stenmark H., Parton R. G., Orioli D., Lim F., Giner A., Mellman I., Zerial M., Murphy C. (1998) J. Cell Biol. 140, 1039–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casanova J. E., Wang X., Kumar R., Bhartur S. G., Navarre J., Woodrum J. E., Altschuler Y., Ray G. S., Goldenring J. R. (1999) Mol. Biol. Cell 10, 47–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Gabin A. G., Almazan G., Larocca J. N. (2004) J. Neurosci. Res. 76, 758–770 [DOI] [PubMed] [Google Scholar]
- 53.Hales C. M., Griner R., Hobdy-Henderson K. C., Dorn M. C., Hardy D., Kumar R., Navarre J., Chan E. K., Lapierre L. A., Goldenring J. R. (2001) J. Biol. Chem. 276, 39067–39075 [DOI] [PubMed] [Google Scholar]
- 54.de Marco M. C., Martín-Belmonte F., Kremer L., Albar J. P., Correas I., Vaerman J. P., Marazuela M., Byrne J. A., Alonso M. A. (2002) J. Cell Biol. 159, 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siliciano J. D., Goodenough D. A. (1988) J. Cell Biol. 107, 2389–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Swart R. L., Ludlow M., de Witte L., Yanagi Y., van Amerongen G., McQuaid S., Yüksel S., Geijtenbeek T. B., Duprex W. P., Osterhaus A. D. (2007) PLoS Pathog. 3, e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanagi Y., Takeda M., Ohno S., Hashiguchi T. (2009) Curr. Top. Microbiol. Immunol. 329, 13–30 [DOI] [PubMed] [Google Scholar]
- 58.Moreno-Bueno G., Cubillo E., Sarrió D., Peinado H., Rodríguez-Pinilla S. M., Villa S., Bolós V., Jordá M., Fabra A., Portillo F., Palacios J., Cano A. (2006) Cancer Res. 66, 9543–9556 [DOI] [PubMed] [Google Scholar]
- 59.Vetter G., Le Béchec A., Muller J., Muller A., Moes M., Yatskou M., Al Tanoury Z., Poch O., Vallar L., Friederich E. (2009) Biochem. Biophys. Res. Commun. 385, 485–491 [DOI] [PubMed] [Google Scholar]
- 60.Grant B. D., Donaldson J. G. (2009) Nat. Rev. Mol. Cell Biol. 10, 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Janda E., Nevolo M., Lehmann K., Downward J., Beug H., Grieco M. (2006) Oncogene 25, 7117–7130 [DOI] [PubMed] [Google Scholar]
- 62.Lacher M. D., Tiirikainen M. I., Saunier E. F., Christian C., Anders M., Oft M., Balmain A., Akhurst R. J., Korn W. M. (2006) Cancer Res. 66, 1648–1657 [DOI] [PubMed] [Google Scholar]
- 63.Cohen C. J., Shieh J. T., Pickles R. J., Okegawa T., Hsieh J. T., Bergelson J. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 15191–15196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mee C. J., Grove J., Harris H. J., Hu K., Balfe P., McKeating J. A. (2008) J. Virol. 82, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saito R. A., Watabe T., Horiguchi K., Kohyama T., Saitoh M., Nagase T., Miyazono K. (2009) Cancer Res. 69, 2783–2791 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.