Abstract
Endoplasmic reticulum oxidation 1 (ERO1) is a conserved eukaryotic flavin adenine nucleotide-containing enzyme that promotes disulfide bond formation by accepting electrons from reduced protein disulfide isomerase (PDI) and passing them on to molecular oxygen. Although disulfide bond formation is an essential process, recent experiments suggest a surprisingly broad tolerance to genetic manipulations that attenuate the rate of disulfide bond formation and that a hyperoxidizing ER may place stressed cells at a disadvantage. In this study, we report on the development of a high throughput in vitro assay for mammalian ERO1α activity and its application to identify small molecule inhibitors. The inhibitor EN460 (IC50, 1.9 μm) interacts selectively with the reduced, active form of ERO1α and prevents its reoxidation. Despite rapid and promiscuous reactivity with thiolates, EN460 exhibits selectivity for ERO1. This selectivity is explained by the rapid reversibility of the reaction of EN460 with unstructured thiols, in contrast to the formation of a stable bond with ERO1α followed by displacement of bound flavin adenine dinucleotide from the active site of the enzyme. Modest concentrations of EN460 and a functionally related inhibitor, QM295, promote signaling in the unfolded protein response and precondition cells against severe ER stress. Together, these observations point to the feasibility of targeting the enzymatic activity of ERO1α with small molecule inhibitors.
Keywords: Disulfide, Endoplasmic Reticulum (ER), Enzyme Inhibitors, High Throughput Screening (HTS), Protein Folding, Unfolded Protein Response
Introduction
Disulfide bonds stabilize the tertiary structures of secreted proteins (1), and disulfide formation is essential in all known life forms. The oxidative process that generates disulfide bonds in eukaryotes is accelerated by the formation of mixed disulfides between ER resident oxidoreductases, such as protein disulfide isomerases (PDIs)3 and reduced cysteine residues on newly translocated client proteins. Productive resolution of these mixed disulfides transfers the disulfide to the client protein and leaves the PDIs in a reduced state (2). Reduced PDIs are enzymatically reoxidized by an ER oxidase, ERO1, conserved from yeast to mammals (reviewed in Refs. 3, 4).
Yeast ERO1 contains a flavin adenine dinucleotide (FAD) tightly bound to a four helical bundle motif also found in other sulfhydryl oxidases (5). A series of disulfide exchange reactions, first between reduced PDI and a shuttle disulfide on ERO1 and then to an ERO1 disulfide lying adjacent to the isoalloxazine ring, leads to the reduction of the flavin and the eventual transfer of a pair of electrons to molecular oxygen (4). This disulfide relay results in the generation of one molecule of H2O2 for each disulfide bond formed in the ER (6).
In addition to the shuttle and active-site cysteines, ERO1 has several regulatory cysteines that form disulfide bonds that restrain the enzymatic activity of the fully oxidized enzyme (7, 8). ERO1 genes are also under transcriptional control, and their mRNA levels increase with the load of unfolded reduced proteins in the ER, a phenomenon mediated by the unfolded protein response (9, 10).
ERO1 is an essential enzyme in yeast, and its dysfunction leads to a rapid decline in oxidative protein folding, strong activation of the unfolded protein response, and marked loss of viability (11, 12). Interestingly, more modest attenuation of ERO1 activity improves the fitness of yeast challenged with high levels of protein misfolding in their ER (ER stress) (13). A similar situation prevails in worms; the single ero1 gene is essential, yet its partial inactivation enhances survival of worms exposed to the ER stress-inducing glycosylation inhibitor tunicamycin (14). Furthermore, compromise of ero1 in the adult also extends the life span of worms that are not exposed to conditions that promote unusual levels ER stress (15). These observations suggest potential benefits for partial inhibition of ERO1.
Mammals have two ERO1 isoforms encoded by separate genes (10). ERO1α is broadly expressed, whereas ERO1β is substantially restricted to the endocrine pancreas (16, 17). Interestingly, mice homozygous for nearly complete loss-of-function mutations in both isoforms are viable and have a very mild kinetic defect in oxidative protein folding in explanted cells (17). These observations point to the wide latitudes allowed for ERO1 activity in mammals and hold the prospect of targeting the enzyme with inhibitors. Here, we report on the application of a high throughput assay for ERO1α activity to the discovery of small molecule inhibitors and on the biochemical and cellular characterization of an inhibitor with a surprising mechanism for attaining specificity.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
A fusion protein of GST-SMT3-mouse ERO1α (residues 23–464) was expressed in the Rosetta (DE3) bacterial strain (Novagen). After growth at 37 °C to an A600 of 0.6–0.8, the culture was shifted to 18 °C, and isopropyl-β-d-thiogalactoside was added to 0.5 mm for an overnight incubation. Bacteria were harvested by centrifugation, resuspended in lysis buffer (20 mm Tris pH 7.5, 100 mm NaCl, 1 mm EDTA, 10 mm imidazole) supplemented with protease inhibitors (200 μm phenylmethylsulfonyl fluoride, 1 mg/ml pepstatin A), and lysed by high pressure cell disruption. To remove cell debris, the lysate was centrifuged for 30 min at 13,500 rpm, and the supernatant was subjected to glutathione affinity chromatography (5 ml of GSTrap 4B, GE Healthcare). Bacterially expressed and purified Ulp1p protease (18) was added to the purified protein and incubated overnight at 4° C to cleave the GST-SMT3 from the ERO1α (residues 23–464) prior to further gel filtration on a HiLoad 16/60 Superdex-75 prep grade column. The cleaved ERO1α (residues 23–464) protein was reapplied to glutathione beads, and the flow through was collected. The concentration of the purified protein was determined spectroscopically at 280 nm and 446 nm in 6 m guanidine HCl and 10 mm phosphate buffer (pH 6.8) assuming an extinction coefficient of 90,630 m−1 cm−1 (apo-ERO1α) and 11,300 m−1 cm−1 (FAD). All protein concentrations reported throughout this manuscript were determined similarly.
Where indicated, 500 μl aliquots of TrxA in buffer A (65 mm NaCl, 20 mm sodium phosphate buffer, pH 7.4, 1 mm EDTA) were reduced with a 100-fold excess of dithiothreitol (DTT) for 30 min at room temperature, and excess reductant was removed by size exclusion using a PD-10 gel filtration (GE Healthcare) equilibrated with buffer A. Fractions of 500 μl were collected, and the thiol titer for pooled reduced thioredoxin (TrxAred) fractions was determined using aliquots diluted into 1 mm 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB, Sigma) and measuring the average absorbance at 412 nm.
Amplex Ultra Red Fluorescence
ERO1α (residues 23–464), between 10 nm and 200 nm, and bacterially expressed TrxAred were combined in a 20-μl reaction in buffer A in a 384-well black round bottom plate (Corning catalog no. 3677) with 0.1 units/ml horseradish peroxidase (Worthington), 5 μm Amplex Ultra Red (AUR; Invitrogen) at room temperature and read kinetically at 535 ± 20 nm excitation and 590 ± 20 nm emission on a TECAN F500 fluorescent plate reader. The inhibitors were as follows: EN460 2-chloro-5-[4,5-dihydro-5-oxo-4-[(5-phenyl-2-furanyl)methylene]-3-(trifluoromethyl)-1H-pyrazol-1-yl], (TimTec, catalog no. ST05611) and QM295 (5(4H)-isoxazolone, 4-[(4-hydroxy-3-methoxyphenyl)methylene]-3-phenyl; Chembridge, catalog no. 5904135) were constituted to 10 mm in 100% dimethyl sulfoxide and added where indicated.
Oxygen Consumption
Oxygen consumption was measured using an Oxygraph Clark-type oxygen electrode (Hansatech Instruments, Pentney, Kings Lynn, UK). All experiments were done at room temperature in air-saturated buffer at sea level (∼250 μm O2 in buffer A) as described (6). Oxygen levels were monitored until a linear baseline was established, and catalytic oxygen consumption was initiated by the addition of TrxAred (50 μm) into a 1-ml reaction mixture containing 1 μm ERO1α (residues 23–464) with or without the indicated compounds (50 μm) with a final dimethyl sulfoxide concentration of 2%.
Ellman's Assay
200 nm ERO1α, 20 μm TrxAred, and 200 μm DTT with the indicated concentration of inhibitor were combined in a 384-well format white-walled clear flat bottom plate (Falcon 3963) in a 20-μl total reaction volume. At end point, 0.5 mm DTNB was added, and the absorbance at 405 nm was read on a TECAN F500 plate reader, essentially as described (19).
Mobility Shift Analysis of ERO1α Oxidative Status
ERO1α (1 μm) was reacted with TrxAred (10 μm) for the indicated amount of time ± inhibitors (25 μm) before quenching with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) (2 mm) and SDS (4%) for 1 h in the dark as described (7). To detect ERO1α, samples were resolved by nonreducing SDS-PAGE (10%) and immunoblotted with rabbit polyclonal serum raised to ERO1α (residues 23–464) protein. To detect TrxA, samples were resolved by nonreducing Tris-Tricine-PAGE (16.5%) and stained with Coomassie Blue.
Absorbance Spectroscopy
The absorbance spectra of compound EN460 and the indicated proteins were measured in a diode array (Agilent HP 8453) using a quartz cuvette with a path length of 1 cm. Absorbance of EN460 (100 μm) following reaction with DTT, reduced glutathione (GSH), tris(hydroxypropyl)phosphine (THP), or tris[2-carboxyethyl]phosphine (TCEP) (all 500 μm) was measured kinetically in 1 ml of buffer A (pH 6.5) and again following the addition of N-ethyl maleimide (NEM, 4 mm).
FAD Fluorescence
ERO1α (150 μm), TrxA (50 μm), EN460 (250 μm), and DTT (2 mm) were combined in 100 μl of buffer A for 60 min at room temperature in the dark, followed by gel filtration (G50 Sephadex, GE Healthcare). Samples were divided into three aliquots of 35 μl and exposed to THP (2 mm), FAD (1 mm), or both, where indicated, for 2 h at room temperature in the dark followed by a second gel filtration. ERO1α activity was measured by AUR assay. Where indicated, FAD concentration was determined by first denaturing in 8 m guanidine HCl, followed by measuring FAD fluorescence at 450ex and 535em at pH 2.
Effect of Inhibitors on ERO1α Redox Status in Vivo
Mouse embryonic fibroblast (MEF) cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and plated the day before the experiment at 70% confluence in 6-well dishes. Cells were challenged with DTT (10 mm) and/or the indicated ERO1α inhibitors (50 μm) where indicated for 30 min. In the washout experiment, cells were washed free of the DTT and incubated with fresh media ± inhibitor. At harvest, phosphate-buffered saline containing NEM (10 mm) was added and the cells subsequently kept on ice for 10 min in lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, protease inhibitors, and 10 mm NEM). Protein concentrations were estimated by Lowry assay, and comparable amounts of protein were run on a nonreducing SDS-PAGE and immunoblotted with antiserum to ERO1α.
Induction of an Unfolded Protein Response
293T cells stably transfected with the ATF6-UPRE-luciferase reporter (a gift of Michael Bassett and Jonathan Weissman, University of California, San Francisco) were treated overnight in low serum-containing media (5%) with the indicated concentration of tunicamycin (Calbiochem) or inhibitors. Cells were lysed in 25 ml of lysis buffer (25 mm glycylglycine, 15 mm MgSO4, 4 mm EGTA, 1 mm DTT, 1% Triton X-100), and luciferase was developed by addition of 25 ml of luciferase assay reagent (25 mm Gly-Gly, 15 mm MgSO4, 4 mm EGTA, 2 mm DTT, 11.7 mm potassium phosphate, 1.6 mm ATP, 0.2 mg/ml CoA, 500 μm luciferin), and luminescence was read in a 96-well white plate (Costar catalog no. 3912) on a TECAN F500 plate reader.
Cell Survival Assay
The effect of ERO1α inhibitors on the survival of cells exposed to tunicamycin were measured as described (20). Briefly, Perk−/− mouse fibroblasts (21) were plated at a density of 5000 cells/well in 24-well plates. A day later, the medium was replaced with complete medium with or without the compounds at 24 μm. The cells were incubated with or without compound for 7 h and then exposed to the indicated concentration of tunicamycin for another 24 h in the continued presence of compound. The cells were then washed with phosphate-buffered saline and returned to complete medium. After an additional 12 days of culture, the medium was replaced with fresh medium containing 0.05 mg/ml WST-1 (Dojindo) and 0.05 mg/ml phenazine methosulfate (Sigma), and the A450 minus the OD650 of 100 μl of medium from each well was measured after a 2–4-h incubation period.
RESULTS
A Homogenous High Throughput Assay for Mammalian ERO1α Activity
ERO1α catalyzes the transfer of a pair of electrons from reduced substrates to molecular oxygen with the formation of H2O2. A homogenous assay of ERO1α activity was developed in which reduced bacterial thioredoxin (TrxAred, a good in vitro surrogate for the natural substrates of ERO1α) is reacted with purified recombinant mouse ERO1α in the presence of oxygen. The H2O2 produced by this reaction drives the horseradish peroxidase (HRP)-mediated oxidation of AUR generating a fluorescent signal (Fig. 1A).
FIGURE 1.
A kinetic in vitro assay for mammalian ERO1α activity. A, scheme of assay: ERO1α activity is measured by a coupled fluorescence assay that detects the production of H2O2 upon oxidation of reduced substrate (thioredoxin A, TrxA; a surrogate for PDI), by recombinant mouse ERO1α (purified from Escherichia coli). HRP uses the H2O2 produced to oxidize the nonfluorescent AUR to a fluorescent molecule. B, shown is a time course of AUR fluorescence in reactions containing the indicated concentration of TrxAred and ERO1α (200 nm). RFU, relative fluorescence unit. THP (20 μm) was included as an electron donor. Inset, a plot of the initial rate of AUR fluorescence as a function of TrxAred concentration is shown. Shown are representative experiments reproduced more than three times. C, time course of AUR fluorescence in reactions containing the indicated concentration of ERO1α and TrxAred (20 μm). D, ERO1α turnover number as calculated by plotting the rate of H2O2 production (extracted from the initial rate of the change in AUR fluorescence in C) against the concentration of ERO1α in the reaction shown in C. Shown are representative experiments reproduced more than three times.
When reactions were performed in the presence of a large excess of HRP, the rate of development of the fluorescent signal was limited by the concentration of TrxAred and by the amount of enzyme in the reaction. Under these conditions, the Km for TrxAred was 3.1 μm (Fig. 1B). At 20 μm TrxAred the enzyme showed a turnover number of five disulfide bonds generated per minute with initial rates that were linearly dependent on ERO1α concentration (Fig. 1, C and D).
This assay was applied to a screen for small molecules that inhibit the development of the fluorescent signal. The assay performed well with Z values between 0.75 and 0.9 (supplemental Fig. 1). Among 210,965 compounds in the primary screen (supplemental Table 1), we identified 629 compounds that lowered AUR fluorescence by >2-fold. Inhibitors were counterscreened for their activity against HRP in an assay that employed H2O2, HRP, and AUR. Compounds that passed this test were rescreened for their ability to inhibit ERO1α in an orthogonal, colorimetric end point assay that detects the depletion of free thiols by ERO1α in the presence of TrxAred and DTT by the loss of reactivity to the Ellman reagent, DTNB (19). Because the latter assay is performed at high concentrations of DTT (200 μm), it also selects against nonspecific thiol-reactive compounds.
Identification of ERO1α Inhibitors
Compounds were stratified according to potency at the secondary screen level and restocked from powder. Employing these selective criteria (supplemental Fig. 2) left us with two candidate inhibitors: benzoic acid, 2-chloro-5-[4,5-dihydro-5-oxo-4-[(5-phenyl-2-furanyl)methylene]-3-(trifluoromethyl)-1H-pyrazol-1-yl], which we named EN460 in light of its enone group and molecular mass of 460 Da, and 5(4H)-isoxazolone,4-[(4-hydroxy-3-methoxyphenyl)methylene]-3-phenyl, which we named QM295, in view of its quinone methide group and molecular mass of 295 Da, (Fig. 2A). Both compounds inhibited the development of AUR fluorescence in a kinetic assay (Fig. 2B). The compound concentration needed to inhibit 50% of AUR fluorescence was nearly identical for both compounds, 1.9 μm (Fig. 2C).
FIGURE 2.
ERO1α inhibitors identified. A, structure of the inhibitors EN460 and QM295. B, a time course of AUR fluorescence in reactions containing the indicated concentration of each inhibitor. Shown are representative experiments reproduced more than three times. C, plot of percent inhibition as a function of compound concentration extracted from the initial velocity of the change in AUR fluorescence in the reactions shown in B. Data fitting with Sigma plot (n = 4). D, time-dependent changes in oxygen concentration in reactions containing ERO1α (1 μm) and TrxAred (50 μm) with or without EN460 or QM295 (50 μm). Shown is an experiment reproduced twice. E, immunoblot of ERO1α (1 μm) reacted for the indicated time with TrxAred (10 μm) in the absence or presence of EN460 or QM295 (25 μm) followed by quenching with AMS and separating each time point on a nonreducing SDS-PAGE. F, Coomassie-stained Tris-Tricine gel of TrxAred (10 μm) reacted for the indicated time with ERO1α (1 μm) in the absence or presence of EN460 (25 μm). Shown is an experiment reproduced twice. std error, S.E.
Both compounds also inhibited oxygen consumption by purified ERO1α in a reaction fed by TrxAred (Fig. 2D). As neither assay could distinguish between inhibitors that bound TrxAred and those that targeted ERO1α, we challenged purified ERO1α with TrxAred in the presence or absence of compounds and monitored the ERO1α redox state by labeling the free thiols with AMS. Like yeast ERO1, the cysteines in the mammalian enzyme mostly were oxidized in the basal state, and the protein demonstrated a high mobility in SDS-PAGE. Exposure to TrxAred lead to reduction of the regulatory disulfides, and a new steady state was attained whereby the enzyme was reduced, activated, and AMS-reactive, as demonstrated by its lower mobility on SDS-PAGE (Fig. 2E, compare lanes 9 and 10). In the absence of inhibitor in the assay, ERO1α returned to its oxidized, AMS-nonreactive, high mobility state following oxidation of the TrxAred, as reported in case of the yeast enzyme (7).
Although the inhibitors did not affect the reduction of ERO1α by TrxAred, they appeared to block the ability of ERO1α to completely oxidize TrxAred in the assay (shown for EN460 in Fig. 2F, compare lanes 2 and 3 with lanes 4 and 5) and to return ERO1α to its high mobility state (Fig. 2E, compare lanes 4, 8 and 12). This inability to return to the high mobility state likely indicates a defect in ERO1α reoxidation rather than a covalent modification by the compound that affects mobility, as the position of ERO1α on SDS-PAGE was unaffected by the compound alone (i.e. when AMS was omitted from the reaction, data not shown). Together, these experiments suggest that the inhibitors function not by interfering with the transfer of electrons from TrxAred to ERO1α but rather by targeting the reduced enzyme such that further oxidation of dithiol substrates by molecular oxygen is compromised.
A similar phenomenon was observed in vivo. At steady state, the ERO1α in mouse embryonic fibroblasts was mostly oxidized with high mobility on SDS-PAGE. Exposure of the cells to DTT (10 mm) lead to reduction of the disulfides in ERO1α and imparted a lower mobility on the protein in SDS-PAGE. Exposure of cells to EN460 or QM295 resulted in the accumulation of an ERO1α form with lower mobility, indicative of the reduced state (Fig. 3A). At steady state, QM295 had a less prominent effect of ERO1α oxidation than EN460 (compare lanes 3 and 4 in Fig. 3A). To confirm the inhibitory effect of QM295, we monitored ERO1α reoxidation following its reduction with DTT in cells and washout of the reductant. The presence of QM295 led to a marked delay in ERO1α reoxidation following DTT washout (Fig. 3B). Together, these observations suggest that both compounds were able to inhibit the enzyme in vivo as well as in vitro.
FIGURE 3.
Inhibition of endogenous ERO1α in vivo. A, immunoblot of endogenous ERO1α in lysates of untreated MEFs or MEFs exposed to DTT (10 mm, 30 min), EN460 or QM295 (50 μm, 30 min). MEFs were washed and lysed in the presence of NEM, and SDS-solubilized proteins were resolved on a nonreducing SDS-PAGE. ERO1ox, oxidized ERO1. B, immunoblot of endogenous ERO1α in lysates prepared as in A. The MEFs were exposed to a 30-min “pulse” of DTT (10 mm) followed by DTT-free “chase” for the indicated time in the absence or presence of QM295 (50 μm). ERO1ox, oxidized ERO1. Shown is an experiment reproduced twice. C, relative luciferase activity from an unfolded protein response reporter (ATF6::luciferase) in the 293T cell line after 16 h of exposure to the indicated concentrations of EN460 or QM295 or tunicamycin as a positive control. RFU, relative fluorescence unit. Values shown are the mean ± S.D. (n = 3) (*, p < 0.05, **, p < 0.01, two-tailed unpaired Student's t test compared with the untreated sample). D, survival of ER stress-hypersensitive Perk−/− MEFs that received no treatment (no tmt) or were treated with the indicated concentrations of EN460 or QM295 or TGD37BZ (TGD, a known protective compound, 20), followed by subsequent challenge with the indicated concentration of tunicamycin (Tm) for 24 h. Survival is expressed as relative amount of WST-1 reduced by tunicamycin-exposed cells compared with unexposed cells (arbitrarily set to 100%). Values shown are the mean ± S.D. (n = 3) (*, p < 0.05, **, p < 0.01, two-tailed unpaired Student's t test, relative to no treatment cells at each concentration of tunicamycin).
Blocking ERO1 activity in yeast and worms activates the unfolded protein response (11, 12). EN460 and QM295 were also found to activate an unfolded protein response reporter in cultured 293T cells (Fig. 3C). EN460 was a more potent activator, but its action was limited by toxicity at the highest concentrations used (data not shown), whereas QM295 had a more shallow and sustained dose response.
Lowered levels of ERO1 activity can protect against severe ER stress in worms, yeast, and cultured mammalian cells (13, 14, 17). Consistent with these observations, we found that continuous exposure to low concentrations of EN460 (and to a lesser degree QM295) protected hypersensitive Perk−/− mouse embryonic fibroblasts (22) from subsequent exposure to tunicamycin (Fig. 3D). Protection by ERO1α inhibitors was relatively modest, compared with a known protectant like TGD31BZ (20) and was likely limited by toxicity. Nonetheless, these observations point to the potential for ERO1α inhibitors to protect against the consequences of severe ER stress in mammalian cells.
Mechanism of Action of EN460
Hereafter, we described studies on the mechanism of action of the more potent inhibitor EN460.
A lag phase in EN460 inhibition of H2O2 production in the fluorescent assay (data not shown) suggested the hypothesis that the enone functionality of the inhibitor (Fig. 2A) reacts with cysteine residues uncovered during the reductive activation of ERO1α. To test for the irreversible inhibition predicted by this mechanism of action, we first exposed ERO1α (20 μm) to compound (250 μm) in the presence or absence of reduced substrate. We then diluted the mixture of enzyme and inhibitor 100-fold (to 200 nm and 2.5 μm, respectively) and compared the enzymatic activity in an assay that is sensitive to thiol depletion by ERO1α (Fig. 4A). When added to this assay at 2.5 μm, EN460 had a very modest inhibitory effect (Fig. 4B, compare lanes 8 and 9). In contrast, when diluted to a 2.5 μm final concentration after having been allowed to first react with the enzyme at a 100-fold higher concentration, the inhibition was far more conspicuous (Fig. 4B, compare lanes 8 and 12). Furthermore, the degree of inhibition that persisted after dilution was greater in the sample in which ERO1α was first activated by reducing substrate when exposed to EN460 than in the sample in which ERO1α was oxidized and inactive when exposed to EN460 (Fig. 4B, compare lanes 10 and 12).
FIGURE 4.
Sustained inhibition of ERO1α by EN460. A, schema of the experimental design to test the reversibility of ERO1α inhibition by EN460, using a colorimetric end point Ellman assay. The color-coding of the experimental arms is maintained in B, below. RT, room temperature. B, absorbance (Abs) of the Ellman reagent (DTNB) added 60 min after ERO1α, TrxA, and DTT were combined to allow ERO1α-mediated oxidation of free thiols. Retention of full ERO1α activity is signaled by the depletion of substrate thiols (as in B, lane 9). The indicated concentration of EN460 was included during the 60-min incubation in samples 5–8, whereas in samples 10 and 12, the same final concentration of EN460 and ERO1α as in lane 8 were attained by diluting a 100-fold concentrated solution of enzyme and inhibitor that were previously allowed to react for 60 min in a preincubation step (PI) in the absence of DTT (nonreducing conditions, NR; samples 10 and 11) or in its presence (reducing conditions, R; samples 12 and 13, **, p < 0.01, two tailed unpaired Student's t test). C, schema of the experimental design to test the reversibility of ERO1α inhibition by EN460, using the AUR fluorescence assay. The color-coding of the experimental arms in the cartoon corresponds to those in D. RT, room temperature. D, ERO1α activity measured by time-dependent AUR florescence. RFU, relative fluorescence unit. Where indicated ERO1α had previously been exposed to EN460 under reducing (+DTT) or non-reducing (−DTT) conditions for 60 min before gel filtration to separate the enzyme from free EN460. Shown is a representative experiment reproduced three times.
In a related, confirmatory experiment, ERO1 and EN460 were allowed to react in the presence or absence of a reduced substrate, and the enzyme was then separated from the small molecules in the reaction mix by gel filtration and assayed kinetically using the AUR fluorescent assay (Fig. 4C). ERO1α exposed to EN460 in the absence of reduced substrate retained most of its enzymatic activity following the removal of excess EN460 by gel filtration, whereas the sample that was exposed to inhibitor in the presence of reduced substrate showed very low activity following gel filtration (Fig. 4D). Together, these experiments suggested that EN460 is an activity-dependent, poorly reversible, or irreversible inhibitor of ERO1.
EN460 is colored red and has a broad absorbance peak between 450-550 nm both in aqueous solutions and in dimethyl sulfoxide. This chromophore is rapidly bleached by reductants, such as the thiols in DTT, GSH, and phosphine-based reagents THP and TCEP. A rapid decrease in visible absorbance is accompanied by a corresponding increase at 290 nm (Fig. 5, A and B). Consistent with the enone reactive functionality of EN460, DTT (Fig. 5C), and GSH (supplemental Fig. 3) form adducts with these thiols. These adducts are reversible as addition of excess NEM restored the absorbance peak at 470 nm and led to a loss of chromophore at 290 nm. In contrast, the phosphines generate reduced forms of EN460 that are reversed only slightly by NEM (Fig. 5C, upper panel).
FIGURE 5.
Spectroscopic evidence that EN460 reacts reversibly with free thiols. A, absorbance (Abs) spectra of EN460 (200 μm) before and after the addition of 200 μm DTT (black and red lines, respectively). B, time dependence of change in absorbance at 470 nm (upper panel) and 290 nm (lower panel) during the reaction of EN460 (100 μm) with DTT, reduced GSH, THP, and TCEP (all at 500 μm). C, time dependence of change in absorbance at 470 nm (upper panel) and 290 nm (lower panel) of EN460 that had been reacted with DTT, THP, or TCEP (as above) and subsequently exposed to NEM (10 mm) at t = 0. Shown is a representative experiment reproduced three times.
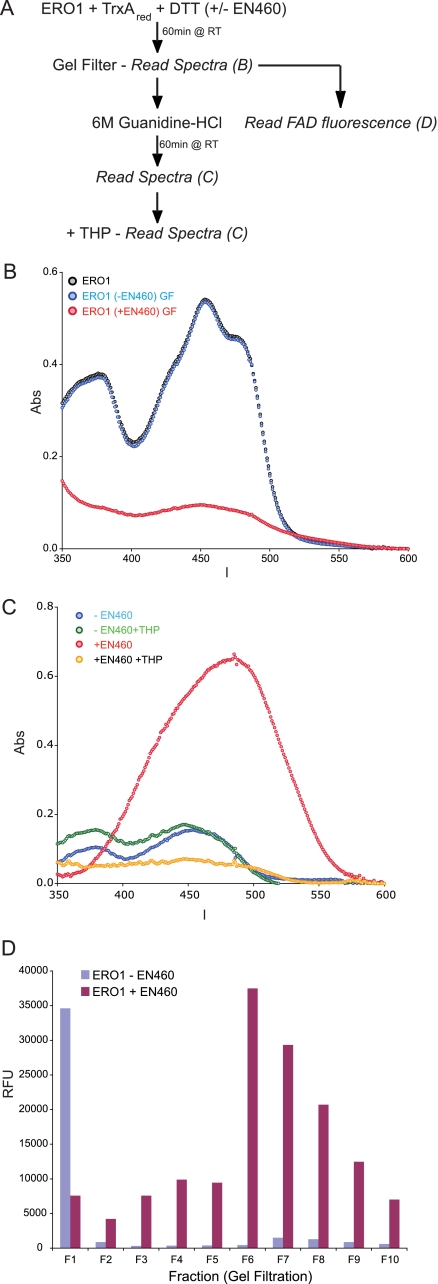
Like its yeast counterpart, mammalian ERO1α exhibits a bright yellow color due to the presence of FAD bound tightly but noncovalently within the active site of the oxidase (Fig. 6A, blue trace). The protein envelope contributes an additional absorbance band centered at 280 nm. In the experiments to follow, the subsequent modulation of the absorbance of ERO1α by covalent binding of EN460 was revealed after removing the spectral contribution of excess free reagent by gel filtration. When ERO1α was incubated with EN460 in the presence of TrxAred and DTT, the protein emerging from the gel filtration column showed notable additional absorbance at 290 nm and longer wavelengths (Fig. 6A, red trace). This feature was dominated by the covalent modification of the ERO1α protein because incubation of EN460 with reduced TrxAred and DTT in the absence of the oxidase did not generate this absorbance (Fig. 6A, compare gray and red traces). Adduct formation between EN460 and reduced ERO1α was accompanied by considerable loss of the flavin absorbance envelope. In contrast, flavin was retained when reduced ERO1α was gel-filtered without prior exposure to the inhibitor (Fig. 6A, purple trace).
FIGURE 6.
Spectroscopic evidence that the EN460 interaction with ERO1α thiols is stabilized by the protein structure. A, comparison of the spectrum of protein samples after gel filtration: ERO1α alone, with TrxA and DTT, and after including EN460 in the reductive incubation. A further control sample contained TrxA, DTT, and EN460 but not ERO1α. B, schema of the experimental design to test the reversibility of the EN460 interaction with ERO1α thiols by absorbance spectra. The color-coding of the experimental arms is maintained in C. RT, room temperature. C, time-dependent change in absorbance (Abs) of ERO1α that had been reacted with EN460 under reducing conditions and then gel-filtered to remove unbound small molecules, followed by denaturation in guanidine HCl (GDN), alkylation with NEM or a combination of both (all at t = 0). Shown is a representative experiment reproduced three times.
Before addressing the fate of the flavin prosthetic group upon reductive inactivation of ERO1α, we first examined the nature of the adduct between the prereduced oxidase and EN460. When inactivated ERO1α was denatured with guanidine HCl in the presence of the alkylating agent NEM, a rapid release of the original EN460 chromophore was seen (signaled by the increase in absorbance at 470 nm and a corresponding disappearance of the 290 absorbance; Fig. 6C, bottom and top panels, respectively). Overall, the loss of the 470 nm absorbance of EN460 upon reaction with reduced ERO1α, the appearance of a new band at 290 nm, and the reversal of this absorbance pattern in the presence of NEM are all consistent with the formation of a reversible thiol adduct (supplemental Fig. 3A). However, unlike the model thiols discussed earlier, release of EN460 from ERO1α adducts was only efficiently accomplished following denaturation of the oxidase (Fig. 6C, red trace).
To determine whether the loss of flavin absorbance observed in the gel-filtered samples in Fig. 6A involved a direct modification of the flavin chromophore by EN460 or indirect disruption of the FAD binding site, again unmodified and modified ERO1α were separated from excess small molecules by gel filtration (Fig. 7A) and attenuation of the FAD peak was confirmed (Fig. 7B). Next, purified ERO1α was exposed to NEM in the presence of guanidine HCl, which lead to release of the bound EN460 and appearance of a conspicuous absorbance peak at 470 nm, as expected (Fig. 7C, red trace). The subsequent addition of THP to this solution bleached the enone absorbance but revealed very low residual flavin absorbance (Fig. 7C, yellow trace). Control experiments showed that in the absence of EN460, neither exposure to THP nor guanidine HCl destroyed the FAD chromophore of ERO1α (Fig. 7C, blue and green traces). Together, these observations suggest that exposure to EN460 leads to loss of the FAD from the holoenzyme.
FIGURE 7.
Reaction of ERO1α with EN460 displaces the bound FAD. A, schema of the experimental design to measure the FAD absorbance (Abs) and fluorescence after exposure of ERO1α to EN460. RT, room temperature. B, absorbance (in the visible region of the spectra) of ERO1α, first reacted with EN460 (in the presence of TrxA and DTT) and then gel-filtered (GF; to remove unbound small molecules). Note the substantial disappearance of the FAD absorption spectrum in the sample that had been exposed to EN460. C, visible absorbance of denatured and alkylated ERO1α that had or had not been previously reacted with EN460. Where indicated, THP was added to reduce the released EN460 and reveal any underlying FAD absorbance. D, FAD fluorescence of fractions from the gel filtration of ERO1α that had or had not been reacted with EN460 under reducing conditions. The fractions were denatured and adjusted to pH 2, before measuring FAD fluorescence. Shown is a representative experiment that was reproduced three times.
Data presented in Fig. 7D provide further evidence that reductive inactivation of ERO1α by EN460 leads to weakened binding of FAD. As a control, untreated ERO1α was subject to gel filtration, and eluted fractions were analyzed for their FAD content by diluting them into guanidine HCl at pH 2 and measuring the fluorescence of the free FAD (which is conspicuous at low pH). ERO1α eluted in fraction F1 and the FAD fluorescence observed in the untreated sample reflected the expected release of the flavin from the denatured ERO1α. Subsequent fractions (F2–F10) exhibited very low residual flavin fluorescence, consistent with tight binding of FAD to apo-ERO1α. In contrast, F1 from EN460-inactivated ERO1α showed a residual flavin content of only ∼20% of the control, and the bulk of the FAD fluorescence was distributed in the low molecular mass fractions (F2–F10) that emerge after the protein peak (Fig. 7D). Thus, ERO1α that elutes after reductive inactivation with EN460 is substantially lacking in FAD and is derivatized by attachment of EN460 to a protein thiol.
The recovery of EN460 absorbance at 470 nm following denaturation and alkylation of ERO1, provides a basis for estimating the stoichiometry of the interaction of ERO1 and EN460; the concentration of ERO1 estimated from the protein absorbance and independently from the bound FAD absorbance of the starting material was 70 μm and 78 μm, respectively. After adjusting for the losses and dilution of the gel filtration procedure, the estimated concentration of ERO1 in the experiment shown in Fig. 7 is 46 μm. The extinction coefficient of EN460 at 470 nm was measured as 32 mm−1 cm−1, and the concentration of the EN460 therefore was ∼20 μm. Given that about one-third of the FAD absorption remained associated with ERO1, the concentration of the inhibited enzyme is in the order of 30 μm, suggesting a 1:1 complex with EN460.
Given the reversibility of EN460-thiol interactions, we wanted to know whether any of the inhibition of ERO1α by EN460 could be reversed by disruption of the enone-protein adduct and by allowing FAD to rebind to the apoprotein. We exposed the gel-filtered, EN460-inhibited ERO1α to THP alone, FAD alone, and THP + FAD for 2 h at room temperature, purified the protein away from small molecules by a second round of gel filtration, and assayed its enzymatic activity by AUR fluorescence (Fig. 8A). Interestingly, FAD alone had little affect, and THP alone led to some recovery, but the combination of THP and FAD led to significant recovery in enzymatic activity (Fig. 8B). Next, we denatured the protein in each sample and measured the amount of released FAD fluorescence; it was conspicuous only in the sample that had been exposed to THP and FAD prior to the terminal gel filtration (Fig. 8C). These observations suggested that EN460 labeling of ERO1α promoted loss of FAD, but removal of the enone by irreversible derivatization with THP allowed FAD rebinding and the recovery of some oxidase activity.
FIGURE 8.
Reconstitution of EN460-inhibited ERO1α activity with exogenous FAD. A, schema of the experimental design to test the role of FAD in reconstitution of inhibited ERO1α. RT, room temperature. B, time-dependent change of AUR fluorescence in an assay performed in the presence of uninhibited ERO1α and EN460-inhibited ERO1α that had been subsequently incubated with THP, FAD, or both, followed by gel filtration to remove unbound small molecules. C, FAD fluorescence of the protein peak (fraction 1) of gel-filtered ERO1α from the three experimental arms (as in Fig. 7D). GDN, guanidine HCl. RFU, relative fluorescence unit.
DISCUSSION
Here, we describe the application of a high throughput assay of ERO1α enzymatic activity to the identification of small molecule enzyme inhibitors and report on the characterization of two such inhibitors. Predictably, the inhibitors promoted an unfolded protein response and provided protection against otherwise lethal levels of ER stress in susceptible cultured cells. These findings point to the feasibility of targeting ERO1α with small molecules and to the potential utility of this strategy for protecting ER-stressed cells.
The most interesting and important lesson of our study concerns the mechanism of action of EN460. Multiple lines of evidence suggest that at least one cysteine residue, generated during activation and/or catalytic turnover of ERO1α, is a target of EN460. This reaction leads to inactivation of the enzyme. Clearly, the enone function of EN460 is a potent Michael acceptor for a range of thiols including DTT and glutathione. The enone functionality is required for inhibition, as irreversible reduction by phosphines inactivates EN460. Thiol adduct formation is signaled by loss of the characteristic red color of EN460 and is accompanied by the appearance of a strong new UV absorbance at 290 nm. This UV feature is also observed in ERO1α labeled with EN460 after reductive activation of the enzyme.
Thiol-mediated EN460 adducts can be reversed rapidly upon addition of the thiol-specific maleimide NEM. We have documented this reaction with EN460 adducts of DTT and GSH and found that it is also observed with the comparable ERO1α adduct, providing that the protein is first denatured. The reversibility of this enone-thiol adduct suggests that the interaction of EN460 and ERO1α is stabilized by other, presumably noncovalent interactions between the protein and the compound. It is likely that the instability of the ERO1α-EN460 adduct that follows denaturation has frustrated our effort to map adduct-containing peptides on ERO1α. Although it is formally possible that the conformational changes underlying the allosteric activation of ERO1α by reduction of regulatory disulfide bonds exposes a nonthiol nucleophile that reacts with EN460, this mechanism is disfavored by the potency of the thiol-selective enone NEM in competing with EN460. Finally, phosphine reductants are largely irreversible reductants of EN460, and their reactivity consumes any enone released from these Michael adducts. Overall, EN460 forms adducts with activated ERO1α that can be reversed by reagents that either compete with protein thiols or that consume the inhibitor itself.
Despite its nonselective reactivity with free thiols, EN460 exhibits surprising specificity toward ERO1α. Thus, in vitro, micromolar concentrations of EN460 can inhibit ERO1α even in the presence of a vast molar excess of competing thiols. In vivo, nonselective reactivity with free thiols may explain the leftward shift in the dose-response curve (compared with the in vitro situation). Nonetheless EN460 is able to inhibit its target in vivo, apparently in the presence of millimolar concentrations of competing thiols in the cell.
Adduct formation to ERO1α also leads to a significant weakening of the binding of the FAD prosthetic group. Weakening of FAD binding is not observed when reduced and activated ERO1α is exposed to high concentrations of the nonselective thiol-reactive inhibitor NEM (data not shown), suggesting the possibility that elements of the ring structure of EN460 may serve to displace flavin from ERO1α, contributing to loss of enzyme activity and helping to explain the avidity with which EN460 captures ERO1α.
Despite the presence of several potentially reactive thiols in ERO1 (7), the stoichiometry of the enzyme inhibitor complex is close to 1:1. This finding is consistent with the idea that the adduct is stabilized by a limited set of conformations and may possibly be restricted to a limited subset of the free thiols of ERO1.
The mechanism for EN460, outlined here, may be shared by other inhibitors whose structures suggest the ability to react with free thiols. For example, the quinone methide functionality of QM295 is a powerful Michael acceptor (23). Similarly, the newly described erodoxin (24) is predicted to form thiol adducts by substitution nucleophilic aromatic reaction SNAr chemistry (supplemental Fig. 4). Interestingly, erodoxin, which is a potent inhibitor of yeast ERO1, has weak activity against mouse ERO1α (IC50 > 400 μm, data not shown) Assuming that the reactive thiol groups are conserved in the yeast and mammalian enzyme, this observation points to the importance of noncovalent interactions in enzyme inhibition. The mechanism proposed for EN460 selectivity also differs from that of other thiol-reactive selective enzyme inhibitors. For example, the 6- or 7-acrylamido-4-anilinoquinazolines, PD160678 and PD168393, irreversibly inhibit the tyrosine kinase activity of the epidermal growth factor receptor by stably interacting with cysteine 773. However these compounds appear to have little promiscuous reactivity with other thiols (25).
Neither EN460 nor QM295 are sufficiently selective to be of value as physiological probes into ER redox poise. Their relative low potency in vivo and their toxicity profile suggest the costs of promiscuous interactions with free thiols. Nonetheless, our study suggests that compounds with stronger non-covalent interactions and the ability to form otherwise reversible bonds with free thiols could emerge as potent inhibitors of enzymes like ERO1.
Supplementary Material
Acknowledgments
We are indebted to Michael Bassick and Jonathan Weissman (University of California, San Francisco) for the gift of the ATF6::LUC reporter 293T cell line and Chris Lima (Memorial Sloan-Kettering Cancer Center) for the SMT3 and ULP1 plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants DK075311, DK47119, and ES08681 (to D. R.) and GM26643 (to C. T.). This work was also supported by fellowships from the National Cancer Institute of Canada and the Canadian Institute of Health Research (to J. D. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1S–4S.
- PDI
- protein disulfide isomerase
- AUR
- Amplex Ultra Red
- DTNB
- 5,5′-dithiobis(2-nitrobenzoic acid)
- DTT
- dithiothreitol
- GST
- glutathione S-transferase
- TrxAred
- reduced thioredoxin
- AMS
- 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- GSH
- glutathione
- NEM
- N-ethyl maleimide
- MEF
- mouse embryonic fibroblast
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- ERO1
- endoplasmic reticulum oxidation 1
- FAD
- flavin-adenine dinucleotide
- TCEP
- tris(2-carboxyethyl)phosphine
- THP
- Tri(hydroxypropyl)phosphine.
REFERENCES
- 1.Anfinsen C. B., Redfield R. R. (1956) Adv. Protein Chem. 11, 1–100 [DOI] [PubMed] [Google Scholar]
- 2.Appenzeller-Herzog C., Ellgaard L. (2008) Biochim. Biophys. Acta. 1783, 535–548 [DOI] [PubMed] [Google Scholar]
- 3.Tu B. P., Weissman J. S. (2004) J. Cell Biol. 164, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sevier C. S., Kaiser C. A. (2008) Biochim. Biophys. Acta. 1783, 549–556 [DOI] [PubMed] [Google Scholar]
- 5.Gross E., Kastner D. B., Kaiser C. A., Fass D. (2004) Cell 117, 601–610 [DOI] [PubMed] [Google Scholar]
- 6.Gross E., Sevier C. S., Heldman N., Vitu E., Bentzur M., Kaiser C. A., Thorpe C., Fass D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sevier C. S., Qu H., Heldman N., Gross E., Fass D., Kaiser C. A. (2007) Cell 129, 333–344 [DOI] [PubMed] [Google Scholar]
- 8.Appenzeller-Herzog C., Riemer J., Christensen B., Sørensen E. S., Ellgaard L. (2008) EMBO J. 27, 2977–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. (2000) Cell 101, 249–258 [DOI] [PubMed] [Google Scholar]
- 10.Pagani M., Fabbri M., Benedetti C., Fassio A., Pilati S., Bulleid N. J., Cabibbo A., Sitia R. (2000) J. Biol. Chem. 275, 23685–23692 [DOI] [PubMed] [Google Scholar]
- 11.Pollard M. G., Travers K. J., Weissman J. S. (1998) Mol. Cell 1, 171–182 [DOI] [PubMed] [Google Scholar]
- 12.Frand A. R., Kaiser C. A. (1999) Mol. Cell 4, 469–477 [DOI] [PubMed] [Google Scholar]
- 13.Haynes C. M., Titus E. A., Cooper A. A. (2004) Mol. Cell 15, 767–776 [DOI] [PubMed] [Google Scholar]
- 14.Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., Ron D. (2004) Genes. Dev. 18, 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curran S. P., Ruvkun G. (2007) PLoS. Genet. 3, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dias-Gunasekara S., Gubbens J., van Lith M., Dunne C., Williams J. A., Kataky R., Scoones D., Lapthorn A., Bulleid N. J., Benham A. M. (2005) J. Biol. Chem. 280, 33066–33075 [DOI] [PubMed] [Google Scholar]
- 17.Zito E., Chin K. T., Blais J., Harding H. P., Ron D. (2010) J. Cell Biol. 188, 821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mossessova E., Lima C. D. (2000) Mol. Cell 5, 865–876 [DOI] [PubMed] [Google Scholar]
- 19.Ellman G. L. (1959) Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 20.Harding H. P., Zhang Y., Khersonsky S., Marciniak S., Scheuner D., Kaufman R. J., Javitt N., Chang Y. T., Ron D. (2005) Cell Metabolism. 2, 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harding H. P., Zeng H., Zhang Y., Jungries R., Chung P., Plesken H., Sabatini D. D., Ron D. (2001) Mol. Cell 7, 1153–1163 [DOI] [PubMed] [Google Scholar]
- 22.Harding H. P., Zhang Y., Bertolotti A., Zeng H., Ron D. (2000) Mol. Cell 5, 897–904 [DOI] [PubMed] [Google Scholar]
- 23.Rokita S. E. (2009) Quinone Methides: Reactive Intermediates in Chemistry and Biology (Rokita S. E. ed) Vol. 1, Ch. 9, pp. 297–356, John Wiley, New York [Google Scholar]
- 24.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St. Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010) Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fry D. W., Bridges A. J., Denny W. A., Doherty A., Greis K. D., Hicks J. L., Hook K. E., Keller P. R., Leopold W. R., Loo J. A., McNamara D. J., Nelson J. M., Sherwood V., Smaill J. B., Trumpp-Kallmeyer S., Dobrusin E. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12022–12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.