Abstract
Following transcription, mRNA is processed, packaged into messenger ribonucleoprotein (mRNP) particles, and transported through nuclear pores (NPCs) to the cytoplasm. At the NPC cytoplasmic face, Dbp5 mediates mRNP remodeling and mRNA export factor dissociation, releasing transcripts for translation. In Saccharomyces cerevisiae, the conserved poly(A) RNA-binding protein, Nab2, facilitates NPC targeting of transcripts and also modulates poly(A) tail length. Dbp5 removes Nab2 from mRNPs at the cytoplasmic face of the pore and, importantly, a Nab2 RNA-binding mutant suppresses the thermosensitive rat8-2 (dbp5) mutant. GFD1 is a multicopy suppressor of rat8-2 (dbp5), and Gfd1 interacts physically with both Dbp5 and the Nab2 N-terminal domain (Nab2-N). Here, we present a structural and functional analysis of the Gfd1/Nab2-N interaction. Crystallography, supported by solution NMR, shows that Gfd1 residues 126–150 form an α-helix when bound to Nab2-N. Engineered Nab2-N and Gfd1 mutants that inhibit this interaction in vitro were used to probe its function in vivo using the genetic interaction between GFD1 and NAB2. Although GFD1 is not essential for viability, its deletion severely impairs growth of rat8-2 (dbp5) cells. Moreover, although Gfd1 overexpression suppresses rat8-2 (dbp5), Gfd1 mutants that do not bind Nab2 only partially suppress rat8-2 (dbp5). Furthermore, rat8-2 (dbp5) cells that express nab2-Y34A, in which binding to Gfd1 is impaired, show a synthetic growth phenotype and nuclear accumulation of poly(A) RNA. These data support the importance of the Gfd1/Nab2 interaction for Dbp5 activity and provide further molecular details of the interactions that facilitate Dbp5-mediated mRNP remodeling in the terminal step of mRNA export.
Keywords: Crystal Structure, MRNA, Nuclear Transport, Protein Structure, Protein-Protein Interactions, RNA Transport, X-ray Crystallography
Introduction
The nuclear export of mRNA requires the coordination of multiple cellular processes and involves proteins within both the nucleus and the cytoplasm. Before being exported to the cytoplasm, nascent transcripts have to progress through a series of co-transcriptional modifications, including 5′-capping (1), intron splicing (2), 3′-cleavage, and polyadenylation (1, 3). These processes are mediated by a host of mRNA-binding proteins (4). In common with other nuclear trafficking pathways, mRNA export appears to be mediated by a thermal ratchet mechanism that involves three principal steps: formation of an export-competent messenger ribonucleoprotein (mRNP);3 translocation of this mRNP through nuclear pore complexes (NPCs); and disassembly of the export complex in the cytoplasm (5). There is a general consensus that the majority of the export machinery is loaded onto mRNA co-transcriptionally, linking the pre-mRNA processing with mRNA export (6), and 3′-end processing is thought to play a major role in recruiting the mRNA export machinery (7). In addition, there are surveillance mechanisms to ensure that only mature and functional mRNPs (which consist of mRNA and heterogeneous nuclear ribonucleoproteins) are exported from the nucleus (8–10). The actual export of bulk mRNA through NPCs is mediated primarily by the evolutionarily conserved Mex67·Mtr2 heterodimer in Saccharomyces cerevisiae (11, 12) that promotes mRNA export through interactions with both mRNPs and nuclear pore proteins (13). Following export, disassembly of the mRNP export complex at the cytoplasmic face of NPCs is performed by a key RNA-remodeling protein, Dbp5, a putative ATP-dependent DEAD box RNA helicase (5, 14). A temperature-sensitive mutant of Dbp5, rat8-2 (dbp5), shows impaired growth and nuclear mRNA accumulation at the non-permissive temperature (15, 16). This terminal mRNP disassembly step in mRNA export is critical because it prevents the return of the mRNA to the nucleus and facilitates the release of the mRNA for translation (5, 17, 18). Other conserved yeast mRNA export factors that have been implicated in this mRNP disassembly step and/or modulating Dbp5 activity include the polyadenosine RNA-binding protein, Nab2 (14, 19), the export factors, Gle1 (20) and Gle2/Rae1 (21), and the nuclear pore protein, Nup159 (22).
The heterogeneous RNP, Nab2, is an essential conserved yeast poly(A) RNA-binding protein that is required for proper regulation of RNA poly(A) tail length and mRNA export (23, 24). Nab2 localizes to the nucleus at steady state (19) but shuttles between the nucleus and the cytoplasm (23). Nab2 contains two key functional domains: a unique N-terminal domain (Nab2-N) and a C-terminal poly(A) RNA-binding domain that contains seven tandem CCCH zinc fingers. The N-terminal domain is required for efficient export of both mRNA and Nab2 from the nucleus (25, 26), and the zinc finger domain specifically binds to polyadenosine RNA (19, 27). Consistent with a function for Nab2 as a polyadenosine RNA-binding protein, recent studies provide evidence that Nab2 forms complexes with numerous RNA processing/export factors and apparently associates with a broad population of cellular RNAs (28, 29).
A Nab2 zinc finger mutant that exhibits weaker binding to poly(A) RNA efficiently suppresses the temperature sensitivity of the rat8-2 (dbp5) mutant (14), linking Nab2 function to Dbp5 activity in the terminal mRNP disassembly step. The N-terminal domain of Nab2 is critical for cell function, and a Nab2 mutant that lacks this domain, nab2-ΔN, shows a severe growth phenotype and strong nuclear accumulation of poly(A) RNA (26). The structure of Nab2-N is based on a five-α-helix bundle (30). Furthermore, Nab2-N interacts with at least two proteins, Mlp1, a nucleoporin-attached protein located on the nuclear face of the NPC (31) and Gfd1 (32). A hydrophobic patch on the surface of Nab2-N mediates a direct interaction with Mlp1 (30). This Nab2-N/Mlp1 interaction is hypothesized to contribute to targeting of transcripts to the nuclear pores and quality control of mRNA export from the nucleus (25). Nab2-N also interacts with Gfd1, a non-essential protein implicated in mRNA export (33).
Gfd1 is one of several factors that, although not essential for mRNA export, enhance the efficiency of the process, either by facilitating integration of different steps in the gene expression pathway or by increasing the rate of key steps. Gfd1 was initially identified in screens for high copy suppressors of the Dbp5 mutant, rat8-2 (dbp5) (33), and a mutant of the RNA export factor, Gle1, gle1-8 (34). Gfd1 interacts with a number of components of the mRNA export machinery in yeast two-hybrid assays (32–35). Most notably, Gfd1 interacts with the Dbp5-activating protein, Gle1, the cytoplasmic nucleoporin Nup42/Rip1, the putative RNA helicase, Dbp5, and a protein implicated in mRNA export, Zds1 (33–35). Gfd1 localizes to the cytoplasm and nuclear rim, a pattern similar to that observed for both Dbp5 and Gle1 (33). These observations led to the hypothesis that Gfd1 may be present in a complex together with Gle1, Dbp5, and Nup42/Rip1 on the cytoplasmic face of the NPCs and participate in the terminal stages of mRNA export (33, 34). In addition, Gfd1 forms a complex with Nab2 both in vitro and in vivo (32) in which Gfd1 binds to the N-terminal domain of Nab2. Although Gfd1 has been linked to mRNA export, its precise role in this export process remains unclear. The GFD1 gene is not essential, and its deletion does not alter Nab2 export from the nucleus and has not been documented to affect mRNA export (32, 33). These observations suggest that redundant proteins may exist to compensate for the absence of Gfd1 or, alternatively, that Gfd1 may catalyze a step in mRNA export that is not rate-limiting under normal conditions. One possibility that has been proposed is that Gfd1 facilitates Nab2 export through bridging the interaction between Gle1 and Nab2 (32).
Here we report molecular details of the interaction between Nab2 and Gfd1 and exploit this information to evaluate the functional significance of the Nab2-N/Gfd1 interaction. We describe the crystal structure of a fragment of Gfd1 complexed with Nab2-N. The crystal structure, together with complementary NMR data, indicated that residues 126–150 of Gfd1 form a single α-helix that binds primarily to helix 2 of Nab2-N. Expression in rat8-2 (dbp5) cells of an engineered Nab2 mutant that does not bind to Gfd1 caused a growth defect and nuclear accumulation of poly(A) RNA. Furthermore, Gfd1 mutants that show decreased binding to Nab2 only partially suppressed rat8-2 (dbp5) temperature sensitivity. These results are consistent with a model in which Gfd1 functions to coordinate Dbp5 and Gle1 to facilitate the removal of Nab2 from mRNPs at the cytoplasmic face of nuclear pores.
EXPERIMENTAL PROCEDURES
Plasmids, Strains, and Chemicals
All DNA manipulations were performed according to standard methods (36), and all media were prepared by standard procedures (37). S. cerevisiae strains and plasmids used are described in Table 1. All chemicals were obtained from Sigma-Aldrich, United States Biological (Swampscott, MA), or Fisher Scientific unless otherwise noted.
TABLE 1.
Yeast strains and plasmids
| Strain/plasmid | Description | Source |
|---|---|---|
| nab2Δ (ACY427) | MATa leu2Δ ura3Δ his3D nab2::HIS3 (pAC636) | (52) |
| nab2Δgfd1Δ (ACY1987) | MATa leu2Δ ura3Δ his3D nab2::HIS3 gfd1::KANMX4 (pAC636) | This study |
| nab2Δrat8-2 (dbp5) (SWY3597) | MATa rat8-2 (dbp5) nab2::HIS3 trp1Δ ura3Δ leu2Δ his3Δ (pAC636) | (14) |
| nab2Δrat8-2 (dbp5)gfd1Δ (ACY1989) | MATa rat8-2 (dbp5) nab2::HIS3 gfd1::KANMX4 trp1Δ ura3Δ leu2Δ his3Δ (pAC636) | This study |
| pGEX-4T-1 | GST, AMPR, bacterial expression vector | GE Healthcare |
| pGEX-TEV | GST-TEV, AMPR, bacterial expression vector | (38) |
| pGEX-TEV-Gfd1122–151 | GST-Gfd1 (residues 122–151) in pGEX-TEV, AMPR | This study |
| pGEX-TEV-Gfd1random | GST-Gfd1 (randomized residues 122–151) in pGEX-TEV, AMPR | This study |
| pAC2053 | GST-Nab2-N-WT (residues 1–97) in pGEX-TEV, AMPR | This study |
| pAC2766 | GST-Nab2-N-Y34A (residues 1–97) in pGEX-TEV, AMPR | This study |
| pAC2767 | GST-Nab2-N-L55W (residues 1–97) in pGEX-TEV, AMPR | This study |
| pAC2768 | GST-Nab2-N-F56D (residues 1–97) in pGEX-TEV, AMPR | This study |
| pET28a | His6, KANR, bacterial expression vector | Novagen |
| pET30a | His6-S-Tag, KANR, bacterial expression vector | Novagen |
| pET30a-TEV | His6-S-Tag-TEV, KANR, bacterial expression vector | (53) |
| pET28a-Nab2-N | His-Nab2-N (residues 1–105) in pET28a, KANR | This study |
| pET30a-Nab2-N | Nab2-N (residues 1–105) in pET30a, KANR | This study |
| pAC2801 | His6-S-Tag-Gfd1 in pET30a, KANR | This study |
| pAC2803 | His6-S-Tag-Gfd1-Δ130–143 in pET30a-TEV, KANR | This study |
| pAC2804 | His6-S-Tag-Gfd1-K135-A-K136 in pET30a-TEV, KANR | This study |
| pAC2805 | His6-S-Tag-Gfd1-K135-AA-K136 in pET30a-TEV, KANR | This study |
| pRS315 | CEN, LEU2, AMPR | (54) |
| pAC717 | NAB2, CEN, LEU2, AMPR | (26) |
| pAC1152 | nab2 ΔN, CEN, LEU2, AMPR | (26) |
| pAC2746 | nab2 Y34A, CEN, LEU2, AMPR | This study |
| pAC2747 | nab2 L55W, CEN, LEU2, AMPR | This study |
| pAC2748 | nab2 F56D, CEN, LEU2, AMPR | This study |
| pRS424 | 2μ, TRP1, AMPR | (39) |
| pAC2755 | GFD1, 2μ, TRP1, AMPR | This study |
| pAC2756 | gfd1-Δ122–143, 2μ, TRP1, AMPR | This study |
| pAC2757 | gfd1-Δ130–143, 2μ, TRP1, AMPR | This study |
| pAC2758 | gfd1-K135-A-K136, 2μ, TRP1, AMPR | This study |
| pAC2759 | gfd1-K135-AA-K136, 2μ, TRP1, AMPR | This study |
| pAC636 | NAB2, CEN, URA3, AMPR | (23) |
Generation of Nab2 and Gfd1 Mutants
GST-TEV-Gfd1122–151 (pGEX-TEV-Gfd1122–151) was constructed by cloning Gfd1122–151 in the pGEX-TEV vector (38), which contains an in-frame TEV protease site after the GST tag in pGEX-4T-1 (GE Healthcare). GST-TEV-Gfd1random (pGEX-TEV-Gfd1random) was constructed by generating a control randomized peptide sequence of Gfd1 122–151, Gfd1random (LKEKNRIQQLTKDKEHESKITHLKQMASKK), using the RandSeq website and cloning Gfd1random into the pGEX-TEV vector (38). His-Nab2-N (pET28a-Nab2-N) and untagged Nab2-N (pET30a-Nab2-N) were constructed by cloning Nab2-N (residues 1–105) into pET28a (Novagen) and pET30a (Novagen), respectively. Plasmids that expressed nab2-Y34A (pAC2746), nab2-L55W (pAC2747), and nab2-F56D (pAC2748) were generated by site-directed mutagenesis using oligonucleotides (Integrated DNA Technologies) encoding the Y34A, L55W, or F56D amino acid substitution, NAB2 (pAC717) plasmid template, and the QuikChange site-directed mutagenesis kit (Stratagene). Wild-type Nab2-N, GST-Nab2-N-WT (pAC2053), and Nab2-N mutants, GST-Nab2-N-Y34A (pAC2766), GST-Nab2-N-L55W (pAC2767), and GST-Nab2-N-F56D (pAC2768), were generated by PCR using oligonucleotides to Nab2-N (residues 1–97) and NAB2 plasmid templates pAC717, pAC2746, pAC2747, and pAC2748 and cloning into pGEX-TEV. Wild-type GFD1 (pAC2755) was generated by PCR using oligonucleotides including the 5′- and 3′-untranslated regions of GFD1 and yeast genomic DNA and cloning into pRS424 (39). Plasmids that expressed gfd1-Δ122–143 (pAC2756) and gfd1-Δ130–143 (pAC2757) were generated by PCR using pairs of oligonucleotides including the 5′-untranslated region and GFD1 residues 1–121 or 1–129 and GFD1 residues 144-3′-untranslated region and GFD1 (pAC2755) plasmid template and cloning into pRS424 (39). Plasmids that expressed gfd1-Lys135-A-Lys136 (pAC2758) and gfd1-Lys135-AA-Lys136 (pAC2759) were generated by site-directed mutagenesis using oligonucleotides encoding one or two alanine insertions between Lys135 and Lys136 and GFD1 (pAC2755) plasmid template. Wild-type Gfd1, His-Gfd1 (pAC2801), and Gfd1 mutants, His-Gfd1-Δ130–143 (pAC2803), His-gfd1-Lys135-A-Lys136 (pAC2804), and His-gfd1-Lys135-AA-Lys136 (pAC2805), were generated by PCR using oligonucleotides to GFD1 and gfd1 plasmid templates pAC2755, pAC2757, pAC2758, and pAC2759 and cloning into pET30a-TEV (50). All constructs were sequenced to ensure the presence of each desired mutation and the absence of any additional mutations.
Protein Expression and Purification
For expression of GST-TEV-Gfd1122–151 (pGEX-TEV-Gfd1122–151), GST-TEV-Gfd1random (pGEX-TEV-Gfd1random), His-Nab2-N (pET28a-Nab2-N), and untagged Nab2 (pET30a-Nab2-N), plasmids were transformed into Escherichia coli strain BL21(DE3) RIL (Stratagene), and 10 ml of overnight cultures derived from single colonies were added to 1 liter of 2× TY medium containing the appropriate antibiotic (25 μg/ml kanamycin for pET28a/pET30a-based constructs or 100 μg/ml ampicillin for pGEX-TEV-based constructs). Cultures were grown at 37 °C to an A600 of 0.6, induced with 1 mm isopropyl 1-thio-d-galactopyranoside, and grown overnight at 25 °C. Cells were harvested and stored in sucrose buffer (50 mm Tris-HCl, pH 8.0, 25% (w/v) sucrose, 1 mm EGTA) at −20 °C before use. Cell pellets were lysed using an Emulsiflex-C5 high pressure homogenizer (Avestin, Ottawa, ON, Canada). Lysate was centrifuged at 20,000 rpm at 4 °C for 20 min, and the supernatant was collected for affinity purification. To obtain the Nab2-N·Gfd1122–151 complex, cell pellets of GST-TEV-Gfd1122–151 and Nab2-N were mixed together and lysed. The complex was purified using glutathione-Sepharose resin followed by on-resin TEV protease digestion (50 μg/liter GST-TEV-Gfd1122–151 cell culture, with the addition of 20 mm dithiothreitol (DTT)) at room temperature overnight. The unbound fraction was collected and purified using a Superdex S75 column in 20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm EDTA, 1 mm DTT. Protein samples were concentrated to 10–20 mg/ml and stored at −20 °C.
For in vitro solution binding assays, recombinant GST fusion and His-tagged proteins were expressed in bacteria and purified. GST (pGEX-TEV), GST-Nab2-N-WT (pAC2053), GST-Nab2-Y34A (pAC2766), GST-Nab2-N-L55W (pAC2767), GST-Nab2-N-F56D (pAC2768), His-Gfd1 (pAC2801), His-Gfd1-Δ130–143 (pAC2803), His-gfd1-Lys135-A-Lys136 (pAC2804), and His-gfd1-Lys135-AA-Lys136 (pAC2805) were expressed in E. coli DE3 cells and purified by batch purification. Overnight cultures were used to inoculate 100 ml of LB medium. Cultures were grown at 37 °C to an A600 of 0.6–0.8, induced with 200 μm isopropyl 1-thio-d-galactopyranoside, and grown at 30 °C for 5 h. For batch purification of GST fusion proteins, cells were collected and lysed in 10 ml of phosphate-buffered saline (PBS) supplemented with protease inhibitor mixture (1 mm phenylmethylsulfonyl fluoride, 3 ng/ml pepstatin A, leupeptin, aprotinin, and chymostatin) by incubation with lysozyme (100 mg/ml) for 30 min on ice followed by sonication. Lysates were cleared by centrifugation and incubated with glutathione-Sepharose 4B (GE Healthcare) for 2 h at 4 °C with mixing. The beads were then washed once with 10 ml of PBS supplemented with 0.5% Triton-X-100 and twice with 10 ml of PBS. GST proteins were eluted from beads with 1 ml of 50 mm Tris-HCl, pH 8, containing 10 mm reduced glutathione and dialyzed into PBS supplemented with 2 mm DTT. For batch purification of His-tagged proteins, cells were collected and lysed in 10 ml of lysis buffer (50 mm NaH2PO4, pH 7.4, 300 mm NaCl, 10 mm imidazole) supplemented with protease inhibitor mixture by incubation with lysozyme and sonication. Lysates were cleared by centrifugation and incubated with nickel-nitrilotriacetic acid agarose (Qiagen) in lysis buffer for 2 h at 4 °C with mixing. The beads were then washed twice with 10 ml of wash buffer (50 mm NaH2PO4, pH 7.4, 300 mm NaCl, 20 mm imidazole). His-tagged proteins were eluted from agarose with 1 ml of elution buffer (50 mm NaH2PO4, pH 7.4, 300 mm NaCl, 250 mm imidazole) and dialyzed into PBS supplemented with 2 mm dithiothreitol.
Solution Binding Assays
For wild-type Gfd1 binding to Nab2-N wild-type and mutants, purified soluble GST, GST-Nab2-N-WT, GST-Nab2-N-Y34A, GST-Nab2-N-L55W, or GST-Nab2-N-F56D (6 μg) was incubated with 14 μg of purified soluble His-Gfd1 and 20 μl of glutathione-Sepharose 4B (GE Healthcare) in 1 ml of binding buffer (PBS, 2 mm DTT, 0.2 mm phenylmethylsulfonyl fluoride, 0.1% Tween 20, 10 mg/ml bovine serum albumin) for 2 h at 4 °C with mixing. Unbound fractions were collected, and the beads were washed three times with 1 ml of PBS for 10 s each. Bound fractions were eluted with loading buffer, and samples were analyzed by SDS-PAGE followed by Coomassie Blue staining. Unbound fractions (20 μl) were mixed with loading buffer and analyzed by SDS-PAGE followed by immunoblotting with an anti-His antibody coupled to horseradish peroxidase (Invitrogen). Binding of Gfd1 mutants, His-gfd1-Lys135-A-Lys136, His-gfd1-Lys135-AA-Lys136, and His-Gfd1-Δ130–143, to Nab2-N wild-type, GST-Nab2-N-WT, was performed identically. Input purified soluble Gfd1 wild-type and mutants (1 μg; 7.1%) were analyzed by SDS-PAGE followed by Coomassie Blue staining.
NMR Titration
NMR samples were dissolved in 50 mm potassium phosphate buffer, pH 6.0, and 10% (v/v) D2O. [1H, 15N]-HSQC spectra were acquired at 297 K using a Bruker DMX600 spectrometer. Spectra were processed using XWINNMR (Bruker Biospin Ltd., Milton, ON, Canada) and visualized in Sparky (40). The concentration of Gfd1122–151 was determined by the BCA assay (Pierce).
NMR spectra were recorded at 300 K on Bruker Av500 and DMX600 spectrometers equipped with cryo-probes. Chemical shift titrations were carried out using [15N]-labeled Nab2 N-terminal domain in 50 mm potassium phosphate buffer, pH 6.0, at a concentration of 100 μm. [15N]-HSQC spectra were recorded with the pure Nab2 sample and after the stepwise addition of Gfd1122–151 to 3.56, 7.1, 16.43, 32.95, 65.7, 131.4 μm. Chemical shift perturbation (CSP) data for Nab2-N binding to Gfd1122–151 were obtained for each residue by comparing the amide chemical shift values of [15N]-Nab2-N alone with those of [15N]-Nab2-N in the presence of a 2-fold molar excess Gfd1122–151. Values were calculated using the equation
In this equation, ΔN and ΔH represent the changes in the amide nitrogen and proton chemical shifts (in parts per million), respectively. The perturbation profiles were plotted against the sequence at a Gfd1 concentration of 131.4 μm. The assignments of the peaks in slow exchange were confirmed by reassigning the [13C, 15N]-labeled Nab2 N-terminal domain in the presence of a 2-fold excess of a Gfd1 peptide including residues 122–151 (AB Bioscience) on the basis of CBCAcoNH and CBCANH spectra (41, 42).
Crystallography
Crystals of Gfd1122–151 complexed with residues 1–105 of Nab2 (Nab2-N) were grown in Linbro plates. A 2-μl sample of the Gfd1122–151·Nab2-N complex (in 20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm DTT, 1 mm EDTA) and 2 μl of reservoir buffer (150 mm zinc acetate, 12% (w/v) polyethylene glycol 400) were mixed and allowed to equilibrate over 0.5 ml of reservoir buffer at 18 °C. Typically, crystals appeared within 1 day as blocks (with a generally cuboid shape) and reached maximum dimensions of ∼0.1 × 0.05 × 0.05 mm. Crystals were mounted in nylon cryo-loops before being flash-frozen in a nitrogen stream at 100 K. 30% (w/v) polyethylene glycol 400 was used as cryo-protectant. A 2.0 Å resolution native data set was collected at the European Synchrotron Radiation Facility (ESRF) (Grenoble, France) beamline ID23-1 using a wavelength of 0.9879 Å. The crystal belonged to the orthorhombic space group P21212 with unit cell dimensions, a = 55.83 Å, b = 121.76 Å, c = 37.38 Å. The data were processed and scaled using the packages MOSFLM (43) and SCALA (44). The data collection statistics are given in Table 2. The structure was solved by molecular replacement using PHASER (44) and a search model based on the crystal structure of Nab2-N (Protein Data Bank (PDB) accession code 2V75 (30)). A strong anomalous signal was detected and was subsequently shown to be due to the presence of three zinc atoms in the asymmetric unit. These phases were combined with those from molecular replacement and used to build an initial model using ArpWarp (44). After iterative cycles of rebuilding and refinement using REFMAC5 (44) and PHASER (45), a final model was obtained having an R-factor of 18.7% (Rfree 22.6%) and excellent geometry (see Table 2). The final model had a MolProbity (46) score of 1.15, corresponding to the 100th percentile of structures at this resolution.
TABLE 2.
Crystal data
| Crystals | |
| Symmetry | P21212 |
| a, b, c (Å) | 55.83, 121.76, 37.38 |
| Data collection | |
| Wavelength (Å) | 0.97980 |
| Resolution range (Å)a | 20–2.0 (2.11–2.0) |
| Total observationsa | 121838 (18152) |
| Unique observationsa | 17741 (2547) |
| Completeness (%)a | 98.9 (95.3) |
| Multiplicity | 6.9 (7.1) |
| Rmerge (%)a | 7.2 (33.5) |
| Mean I /σ(I)a | 13.5 (4.9) |
| Refinement | |
| Rcryst/Rfree (%) | 18.7/22.6 |
| Bond length r.m.s.d.b (Å) | 0.007 |
| Bond angle r.m.s.d.b (°) | 0.86 |
| MolProbity score/percentile | 1.15/100 |
| Ramachandran plot (%) | |
| Favored | 98.7 |
| Allowed | 1.3 |
| Forbidden | 0 |
a Parentheses refer to final resolution shell.
b r.m.s.d., root mean square deviation.
Circular Dichroism
CD measurements were carried out on an automated J-810 spectropolarimeter (Jasco UK, Ltd.). The spectra were measured over a wavelength range of 260 and 197 nm, using a 0.1-cm path length cell at 20 °C with Gfd1122–151 or control Gfd1 random samples at final concentrations of 0.5 mg/ml.
Nab2 Functional Assay
In vivo functional analysis of Nab2 was carried out through a standard plasmid shuffle assay combined with serial dilution and spotting or growth curves. nab2Δ cells (ACY427) containing a NAB2 URA3 maintenance plasmid (pAC636) were transformed with vector alone (pRS315), NAB2 (pAC717), nab2-ΔN (pAC1152), nab2-Y34A (pAC2746), nab2-L55W (pAC2747), or nab2-F56D (pAC2748) LEU2 plasmids and selected on Leu−Ura− minimal medium with 2% (w/v) glucose. Cells were grown overnight at 30 °C to saturation in Leu−Ura− minimal medium with 2% glucose. Cell concentrations were normalized by A600, and cultures were serially diluted in sterile H2O to obtain ∼10,000, 1000, 100, 10, or 1 cell(s) per 3-μl volume. These dilutions were spotted onto control Leu−Ura− minimal medium, where the NAB2 URA3 maintenance plasmid is maintained, or Leu− minimal medium containing 5-fluoroorotic acid, which selects for cells that have lost the NAB2 URA3 maintenance plasmid (47). Growth of nab2Δ cells harboring NAB2, nab2-ΔN, nab2-Y34A, nab2-L55W, or nab2-F56D as the sole copy of NAB2 was examined at 25, 30, and 37 °C. To generate double mutants of nab2 and rat8-2 (dbp5), nab2Δrat8-2 (dbp5) cells (SWY3597) containing a NAB2 URA3 maintenance plasmid (pAC636) were transformed with NAB2 (pAC717), nab2-Y34A (pAC2746), nab2-L55W (pAC2747), or nab2-F56D (pAC2748) LEU2 plasmid and selected on Leu−Ura− minimal medium with 2% glucose. Cells were streaked onto Leu− minimal medium plates containing 5-fluoroorotic acid to remove the NAB2 URA3 maintenance plasmid and restreaked onto Leu− minimal medium plates with growth at 25 °C. nab2Δ rat8-2 (dbp5) cells harboring NAB2, nab2-Y34A, nab2-L55W, or nab2-F56D as the sole copy of NAB2 were grown over 2 days at 25 °C to saturation in Leu− minimal medium. Cell concentrations were normalized by A600 and serially diluted, and dilutions were spotted onto Leu− minimal medium and examined at 25, 30, and 32 °C. For growth curve analysis, nab2Δ rat8-2 (dbp5) cells harboring NAB2, nab2-Y34A, nab2-L55W, or nab2-F56D as the sole copy of NAB2 were grown to saturation over 2 days at 25 °C in Leu− minimal medium. Cell concentrations were normalized by A600, diluted 100-fold in 100 μl of Leu− minimal medium with 2% glucose, and added to the wells of a MicroWell F96 microtiter plate (Nunc). Samples were loaded in quintuplicate. Cells in plate wells were grown at 30 °C with shaking, and absorbance at A600 was measured every 30 min for 25–48 h in an ELX808 Ultra microplate reader with KCjunior software (Bio-Tek Instruments, Inc.). Quintuplicate sample absorbances for time points were averaged and plotted using Microsoft® Excel® 2008 for Mac (Microsoft Corp.).
gfd1Δ Growth Assay
Growth of gfd1Δ or rat8-2 (dbp5) gfd1Δ cells was analyzed by serial dilution and spotting or growth curves. nab2Δ (ACY427), nab2Δ gfd1Δ (ACY1987), nab2Δ rat8-2 (dbp5) (SWY3597), and nab2Δ rat8-2 (dbp5) gfd1Δ (ACY1989) cells, all harboring a NAB2 URA3 maintenance plasmid (pAC636), were grown to saturation over 2 days at 25 °C, serially diluted, and spotted on Ura− minimal medium with 2% glucose and analyzed at 25, 30, and 32 °C. For growth curve analysis, cells were grown over 2 days at 25 °C to saturation in Ura− minimal medium. Cell concentrations were normalized by A600, and then samples were diluted 100-fold in 100 μl of Ura− minimal medium with 2% glucose and added to wells of a microtiter plate. Cell samples were loaded in quintuplicate, and growth at 30 °C was measured and analyzed as described under “Nab2 Functional Assay.”
GFD1 Suppression Assay
Suppression of rat8-2 (dbp5) strain temperature sensitivity by GFD1 wild-type or gfd1 mutants was performed by transformation of overexpression plasmids into rat8-2 (dbp5) cells and analysis of growth via serial dilution and spotting or growth curves. nab2Δrat8-2 (dbp5) cells (SWY3597) containing NAB2 URA3 maintenance plasmid (pAC636) were transformed with vector alone (pRS424), GFD1 (pAC2755), gfd1-Δ122–143 (pAC2756), gfd1-Δ130–143 (pAC2757), gfd1-Lys135-A-Lys136 (pAC2758), and gfd1-Lys135-AA-Lys136 (pAC2759) 2μ TRP1 plasmids and selected on Trp−Ura− minimal medium with 2% glucose at 25 °C. Cells were grown over 2 days at 25 °C to saturation in Trp−Ura− minimal medium with 2% glucose. Cell concentrations were normalized by A600, then cultures were serially diluted and spotted onto Trp−Ura− minimal medium, and growth of cells was analyzed at 25, 30, and 32 °C. For growth curve analysis, nab2Δ rat8-2 (dbp5) cells harboring GFD1 or gfd1 plasmids were grown over 2 days at 25 °C to saturation in Trp−Ura− minimal medium, and cell concentrations were normalized by A600, diluted 100-fold in 100 μl of Trp−Ura− minimal medium with 2% glucose, and added to the wells of a microtiter plate. Cell samples were loaded in quintuplicate, and growth at 32 °C was measured and analyzed as described under “Nab2 Functional Assay.”
Immunoblotting
Expression levels of Gfd1 or Nab2 variant proteins in rat8-2 (dbp5) cells were determined by immunoblotting (48) using anti-Gfd1 (32) or anti-Nab2 3F2 (24) antibodies. For detection of Gfd1 variant protein levels, nab2Δ rat8-2 (dbp5) (SWY3597) cells containing NAB2 URA3 maintenance plasmid (pAC636) and vector alone (pRS424), GFD1 (pAC2755), gfd1-Δ122–143 (pAC2756), gfd1-Δ130–143 (pAC2757), gfd1-Lys135-A-Lys136 (pAC2758), or gfd1-Lys135-AA-Lys136 (pAC2759) 2μ TRP1 plasmids were grown overnight at 25 °C to saturation in Ura−Trp− minimal medium. Cells were then normalized by A600, diluted into fresh medium, and grown at 32 °C for 6 h. Whole cell lysates were prepared from cells and analyzed by SDS-PAGE and immunoblotting with a rabbit anti-Gfd1 polyclonal antibody (1:1000) and peroxidase-conjugated anti-rabbit antibody (Jackson ImmunoResearch Laboratories, Inc.). For detection of Nab2 variant protein levels, nab2Δ rat8-2 (dbp5) (SWY3597) cells containing NAB2 (pAC717), nab2-Y34A (pAC2746), nab2-L55W (pAC2747), or nab2-F56D (pAC2748) LEU2 plasmids were grown overnight at 25 °C to saturation in Leu− minimal medium. Cells were then normalized by A600, diluted into fresh medium, and grown at 30 °C for 6 h. Whole cell lysates were prepared from cells and analyzed by SDS-PAGE and immunoblotting with an anti-Nab2 3F2 monoclonal antibody (1:2000) and peroxidase-conjugated anti-rabbit antibody. As a loading control, 3-phosphoglycerate kinase (Pgk1) protein levels in lysate samples were detected using an anti-Pgk1 monoclonal antibody (1:5000; Invitrogen) and peroxidase-conjugated anti-mouse antibody (Jackson ImmunoResearch Laboratories).
Fluorescence in Situ Hybridization (FISH)
The intracellular localization of poly(A) RNA was assayed by FISH (23). nab2Δrat8-2 (dbp5) cells containing NAB2 (pAC717), nab2-Y34A (pAC2746), nab2-L55W (pAC2747), or nab2-F56D (pAC2748) plasmid were grown in minimal medium with 2% glucose overnight at 25 °C, transferred to minimal medium with 2% glucose, and grown to log phase at 16 °C. Cells were prepared as described previously by Wong et al. (49). A digoxigenin-labeled oligo(dT) probe and fluorescein isothiocyanate-conjugated anti-digoxigenin antibody (1:200 dilution, Roche Applied Science) were used to localize poly(A) RNA. For all samples, cells were examined by fluorescence microscopy. Cells were also stained with 4′,6-diamidino-2-phenylindole-dihydro chloride to detect chromatin and hence the position of the nucleus.
To quantitate the percentage of cells processed for FISH that showed nuclear accumulation of poly(A) RNA, rat8-2 (dbp5) NAB2, rat8-2 (dbp5) nab2-Y34A, rat8-2 (dbp5) nab2-L55W, and rat8-2 (dbp5) nab2-F56D cells were probed with oligo(dT) to visualize poly(A) RNA, and five fields of cells for each strain were imaged. For each field of cells, cells were counted and scored using the ImageJ v1.4 software (National Institute of Health), and the percentage of cells with nuclear poly(A) RNA was calculated. Cells were scored as showing nuclear poly(A) RNA if the fluorescent signal in the nucleus was greater than the signal in the cytoplasm. The percentage of cells with nuclear poly(A) RNA was calculated from five fields, all of which contained at least 31 cells. All scoring of nuclear poly(A) RNA was performed blind to avoid biasing results. In each field of cells, at least 31 rat8-2 NAB2 and rat8-2 (dbp5) nab2 mutant cells were counted (∼200 cells in total). The -fold increase in nuclear poly(A) RNA accumulation in rat8-2 (dbp5) nab2 mutant cells relative to that in rat8-2 (dbp5) NAB2 cells was calculated by dividing the percentage of nuclear poly(A) RNA in rat8-2 (dbp5) nab2 mutant cells by the percentage in corresponding rat8-2 (dbp5) NAB2 wild-type cells.
Deposition of Coordinates
Coordinates for the structure of the Gfd1122–151·Nab2-N complex have been deposited with the PDB with accession number 3LCN.
RESULTS
Crystal Structure of the Nab2-N·Gfd1 Complex
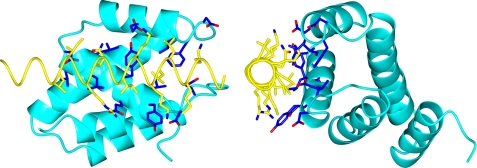
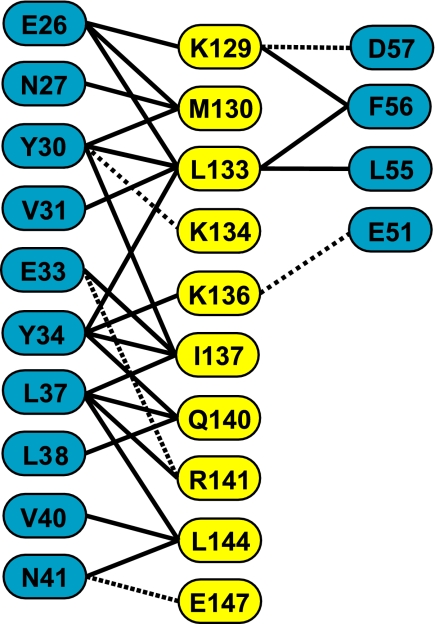
Preliminary experiments indicated that a fragment of Gfd1 containing residues 122–151 retained the capacity to bind to Nab2-N. Crystals of this fragment complexed with Nab2-N (the Nab2-N·Gfd1122–151 complex) that had P21212 symmetry were obtained by vapor diffusion in the presence of zinc ions. These crystals diffracted to 2.0 Å resolution and were solved using a combination of molecular replacement and zinc anomalous multiwavelength anomalous dispersion phasing (Table 2). The asymmetric unit contained two copies of the Nab2·Gfd1 complex and, after iterative cycles of refinement and rebuilding, the final model had excellent geometry (Table 2) and contained residues 4–100 of Nab2 and residues 126–150 of Gfd1 together with three zinc atoms and 111 water molecules. Both copies of Nab2-N showed the same five-helix bundle fold observed previously (30). Thus, the Cα root mean square deviation between chain A of the current structure and chain A of the previous Nab2-N structure (PDB 2V75) was 0.69 Å. Gfd1 residues 126–150 formed an α-helix that bound to the surface of Nab2-N, interacting primarily with Nab2 helix 2 (Fig. 1). The Gfd1/Nab2-N interface buried 1357 Å2 of surface area. Fig. 2 shows the principal interactions observed between the two chains in each complex. Both hydrophobic and polar contacts were present in the interface. Thus, Met130, Leu133, Ile137, and Leu144 of Gfd1, together with the aliphatic regions of the side chains of Lys129, Lys134, Lys136, Gln140, and Arg141, were inserted into a hydrophobic pocket on the surface of Nab2 that was formed by Tyr30, Val31, Tyr34, Leu37, Leu38, Val40, Leu55, and Phe56 together with the aliphatic regions of Glu26, Asn27, Glu33, and Asn41. Nab2 Tyr-34 was a central feature of the interface (Figs. 1 and 2) and was almost completely buried. These interactions were augmented by putative salt bridges formed between Glu33Nab2–Arg141Gfd1, Glu51Nab2–Lys136Gfd1, and Asp57Nab2–Lys129Gfd1 together with hydrogen bonds between Tyr30Nab2–Lys134Gfd1, Asn41Nab2–Gln140Gfd1, and Asn41Nab2 and the main-chain carbonyl of Gln140Gfd1. The Gfd1-binding site on Nab2-N is located on the opposite side of the molecule to the Mlp1-binding site, which is centered on Phe73 (see supplemental movie S1), consistent with Nab2 being able to bind Gfd1 and Mlp1 simultaneously (30).
FIGURE 1.
Crystal structure of the Nab2-N·Gfd1 complex. Two views of the complex rotated 90° about the vertical axis relative to each other are shown. Nab2-N is shown in blue, and Gfd1 is in yellow. Residues 122–151 of Gfd1 form a single α-helix that binds to Nab2-N through a series of hydrophobic interactions supplemented by H-bonds and salt bridges. The side chains of the residues that form the interaction interface are also shown. The Gfd1-binding interface of Nab2-N is located primarily on helix h2 and is centered on Tyr34 (Y34), which is substantially buried in the interface.
FIGURE 2.
Schematic showing key residues of the interaction interface between Gfd1 (yellow) and Nab2-N (blue). The Gfd1-binding interaction interface on Nab2 involves primarily helix h2, and Tyr34 is a central component that is buried between the side chains that protrude from the Gfd1 helix. The interface contains a number of hydrophobic interactions (full lines) supplemented by H-bonds and salt bridges (broken lines).
Three zinc atoms were clearly resolved in the asymmetric unit but were involved in interactions between complexes and not between the two chains within a complex. Thus, although the zinc ions were required to form the crystals themselves, they were not involved directly in the binding of Gfd1 to Nab2-N.
NMR Titrations Confirm the Nab2-N/Gfd1 Interaction Interface Observed by Crystallography
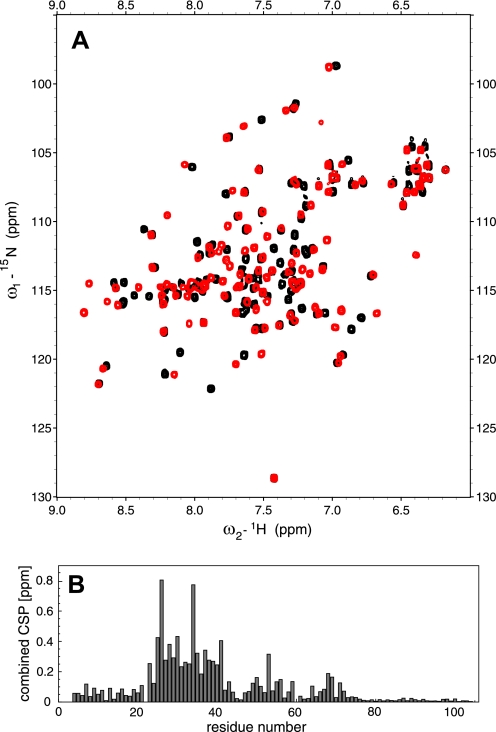
NMR titrations confirmed the general nature of the interaction interface observed in the crystal structure. Thus, the strongest chemical shift perturbations in the Nab2 15N-HSQC spectrum were observed for residues Glu26 and Tyr34 (Fig. 3A), and all of the perturbations greater than 0.2 ppm were clustered between residues Asn23 and Asn41 and so comprised all of helix 2 in Nab2 (Fig. 3B). When mapped onto the crystal structure of free Nab2 (PDB accession code 2V75), the chemical shift perturbation values clearly highlight a region around residue Tyr34 together with some perturbations in helix 3 and the core of the protein, whereas other areas of the structure showed virtually no change in chemical shift. When compared with the crystal structure of the Nab2·Gfd1 complex (supplemental Fig. S1), the perturbation data were in excellent agreement with the binding interface between the molecules in the crystal except for residues Asn25 and Glu26, for which the strong perturbations were more likely caused by a change in Chi-2 of Phe24 than by the direct interaction between Nab2 and Gfd1.
FIGURE 3.
Chemical shift perturbations in Nab2-N upon Gfd1 binding confirm the Gfd1-binding region within Nab2-N. A, 15N-HSQC spectra of Nab2-N in the absence (black) and presence of Gfd1 residues 122–151 (red). The direction of the movement of the most prominently altered signals is indicated by arrows. B, the combined chemical shift perturbation (combined CSP) plotted onto the sequence of Nab2. With the exception of residue Ala53, all perturbations larger than 0.2 ppm are confined to the region between residues Asn23 and Gly41.
Gfd1122–151 Takes Up Its Helical Conformation on Binding Nab2-N
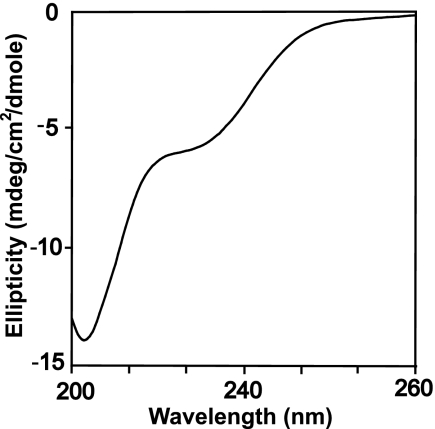
The crystal structure of the Gfd1122–151·Nab2-N complex showed that residues 126–150 of Gfd1 bound to Nab2-N as a single α-helix. This finding raised the question as to whether this region of Gfd1 forms a helix independently or only upon interaction with Nab2-N. To address this question, the structure of this fragment was assessed using CD. The CD spectrum of Gfd1122–151 (Fig. 4) showed a large negative ellipticity below 210 nm that is a characteristic feature of random coil structure (50), indicating that the region between residues 122 and 151 of Gfd1 lacks significant secondary structure in solution and adopts a helical conformation only in the complex. A similar CD spectrum was observed with a randomized peptide of Gfd1 residues 122–151, Gfd1random (data not shown). A lack of secondary structure in this region of Gfd1 when not bound to Nab2-N could, in principle, allow it to sweep out a larger volume and so facilitate its contacting a Nab2 chain.
FIGURE 4.
CD analysis of the Nab2-binding domain of Gfd1. Residues 122–151 of Gfd1 show a CD spectrum (ellipticity plotted versus wavelength) indicative of random coil with no evidence of significant secondary structure. mdeg, millidegrees.
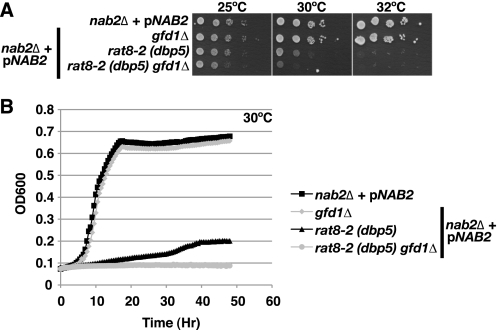
Deletion of GFD1 Exacerbates the Growth Defect of rat8-2 (dbp5) Cells
Although GFD1 is not essential for cell viability, overexpression of Gfd1 suppresses the temperature-sensitive growth of the rat8-2 (dbp5) mutant (33, 34). Amino acid substitutions in Nab2 that impair RNA binding, such as nab2-C437S, also suppress the temperature-sensitive growth of rat8-2 (dbp5) cells (14). To confirm that Gfd1 is not essential in cells with a wild-type genetic background, we compared the growth of gfd1Δ and wild-type cells both by spotting following serial dilution and by growth curve analysis (Fig. 5, A and B). As expected, we found that gfd1Δ cells grew identically to wild-type cells. We therefore explored how Gfd1 might influence cell growth in cells with a rat8-2 (dbp5) mutant genetic background in which an integrated rat8-2 (dbp5) allele serves as the only copy of the essential DBP5 gene (14, 15). As shown in Fig. 5, A and B, deletion of GFD1 exacerbated the growth defect seen in rat8-2 (dbp5) cells at the semipermissive temperature of 30 °C, consistent with Gfd1 contributing to the function of Dbp5 in mRNA export.
FIGURE 5.
GFD1 is important for proper cell growth in a rat8-2 (dbp5) genetic background. A, nab2Δ, nab2Δ gfd1Δ (gfd1Δ), nab2Δ rat8-2 (dbp5) (rat8-2 (dbp5)), and nab2Δ rat8-2 (dbp5) gfd1Δ (rat8-2 (dbp5) gfd1Δ) cells maintained by a NAB2 URA3 plasmid (pNAB2) were grown to saturation, serially diluted in 10-fold dilutions, and spotted on Ura− minimal medium plates. gfd1Δ cells show growth identical to wild-type cells, but rat8-2 (dbp5) gfd1Δ cells grow more slowly when compared with rat8-2 (dbp5) cells at 30 °C. Plates were grown at 25, 30, and 32 °C. B, growth curve analysis confirms that rat8-2 (dbp5) gfd1Δ cells exhibit slower growth than rat8-2 (dbp5) cells at 30 °C. nab2Δ, nab2Δ gfd1Δ, nab2Δ rat8-2 (dbp5), and nab2Δ rat8-2 (dbp5) gfd1Δ cells maintained by pNAB2 were grown to saturation and diluted, and their optical density (OD) was measured at A600 for 48 h as described under “Experimental Procedures.”
The Gfd1 α-Helix That Binds Nab2 Is Required for Efficient GFD1-mediated Suppression of rat8-2 (dbp5)
To understand how the interaction between Gfd1 and Nab2-N contributes to the functional link between Gfd1 and Dbp5, we determined whether the Nab2-binding region of Gfd1 is required for GFD1 overexpression suppression of the temperature-sensitive growth phenotype of rat8-2 (dbp5) cells. rat8-2 (dbp5) cells containing vector alone, GFD1, gfd-Δ122–143, or gfd1-Δ130–143 high copy plasmids were grown to saturation, serially diluted, spotted on selective medium plates, and grown at 25, 30, and 32 °C (Fig. 6A). Results indicated that the α-helix deletion mutants, gfd1-Δ122–143 and gfd1-Δ130–143, only partially suppress rat8-2 (dbp5) relative to suppression by full-length GFD1. Overall, these data suggest that the Gfd1 α-helix interaction with Nab2 is required for complete suppression of rat8-2 (dbp5) temperature sensitivity; however, because the gfd1 α-helix deletion mutants still retain the ability to partially suppress rat8-2 (dbp5), the data also suggest that Gfd1 interactions with other proteins, such as Gle1 or Nup42/Rip1, contribute to Gfd1 function. As a complementary approach to disrupt the Gfd1/Nab2 interaction, we inserted either a single alanine (gfd1-Lys135-A-Lys136) or two alanines (gfd1-Lys135-AA-Lys136) in the center of the Gfd1 helix (after residue Lys135), effectively altering the azimuthal register of the helix by rotating one-half by 100 or 200°, respectively, relative to the other. This approach has been used previously to disrupt an analogous interaction between Sus1 and Sgf11 (51). As shown in Fig. 6A, these alanine insertion mutants, like the deletion mutants, only partially suppressed the temperature-sensitive growth of rat8-2 (dbp5) cells. Further examination of the suppression of rat8-2 (dbp5) growth at 32 °C by gfd1 mutants using growth curve analysis confirmed that the gfd1 mutants did suppress rat8-2 (dbp5) temperature-sensitive growth, albeit with lower efficiency than wild-type GFD1 (Fig. 7B). Notably, although the exponential growth rate of the rat8-2 (dbp5) gfd1 mutant cells was slower than that of the rat8-2 (dbp5) GFD1 cells, the rat8-2 (dbp5) gfd1 mutant cells ultimately reached a similar cell density to the rat8-2 (dbp5) GFD1 cells. This observation suggests that impairing the Nab2/Gfd1 interaction may decrease the rate of mRNA export, resulting in a reduced exponential growth rate. Immunoblotting confirmed that each of the gfd1 mutant proteins was produced to the same extent as wild-type Gfd1 (Fig. 6C), confirming that the impaired suppression of rat8-2 (dbp5) by the gfd1 variants was not due to differential expression or protein stability.
FIGURE 6.
gfd1 mutants that are impaired for binding to Nab2 show only partial suppression of rat8-2 (dbp5) temperature-sensitive growth. A, gfd1 mutants with alanine insertions (gfd1-Lys135-A-Lys136; gfd1-Lys135-AA-Lys136) or deletions (gfd1-Δ122–143; gfd-Δ130–143) in the Nab2-binding α-helix only partially suppress the temperature-sensitive growth of rat8-2 (dbp5) cells. rat8-2 (dbp5) cells containing vector alone, GFD1, gfd1-Δ122–143, gfd1-Δ130–143, gfd1-Lys135-A-Lys136, or gfd1-Lys135-AA-Lys136 2μ TRP1 test plasmids were grown to saturation, serially diluted in 10-fold dilutions, and spotted on Trp− minimal medium plates. Plates were grown at 25, 30, and 37 °C. B, growth curve analysis of gfd1 mutants confirms that they only partially suppress rat8-2 (dbp5) temperature sensitivity. rat8-2 (dbp5) cells containing GFD1 or gfd1 mutant 2μ TRP1 test plasmids were grown to saturation and diluted, and their optical density (OD) was measured at A600 for 48 h as described under “Experimental Procedures.” C, gfd1 mutants are overproduced to the same level. To examine the expression levels, rat8-2 (dbp5) cells containing vector alone, GFD1, or gfd1 variant plasmids were grown at 32 °C, and whole cell lysates prepared from these cells were analyzed by immunoblotting with a polyclonal anti-Gfd1 antibody. As a loading control, Pgk1 protein levels in each lysate were detected with a monoclonal anti-Pgk1 antibody. D, gfd1 mutants with alanine insertions or deletions in the Nab2-binding α-helix do not bind to the Nab2-N. Recombinant GST-Nab2-N-wild-type was incubated with recombinant His-tagged Gfd1 wild-type (Gfd1), His-tagged gfd1 alanine insertion mutants (gfd1-Lys135-A-Lys136; gfd1-Lys135-AA-Lys136), or His-tagged gfd1 deletion mutant (gfd1-Δ130–143) and glutathione-Sepharose beads as described under “Experimental Procedures.” Bound fractions were analyzed by SDS-PAGE and Coomassie Blue staining. Unbound fractions were analyzed by immunoblotting with anti-His antibody. Input recombinant His-tagged Gfd1 proteins were analyzed by SDS-PAGE and Coomassie Blue staining.
FIGURE 7.
nab2-Y34A shows impaired binding to Gfd1 and interacts genetically with rat8-2 (dbp5). A, the Nab2-N mutant nab2-Y34A does not interact with Gfd1 in vitro, but Nab2-N mutants L55W and F56D retain their ability to bind Gfd1 in vitro. Recombinant GST, GST-Nab2-N-wild-type (WT), or GST-Nab2-N mutant (Y34A, L55W, or F56D) was incubated with recombinant His-tagged Gfd1 (His-Gfd1) and glutathione-Sepharose beads as described under “Experimental Procedures.” Bound fractions were analyzed by SDS-PAGE and Coomassie Blue staining. Unbound fractions were analyzed by immunoblotting with anti-His antibody. B, nab2-Y34A cells are viable and grow similarly to wild-type NAB2 cells. nab2Δ cells maintained by a NAB2 URA3 plasmid and containing vector alone, NAB2, nab2-ΔN, nab2-Y34A, nab2-L55W, or nab2-F56D LEU2 test plasmids were grown to saturation, serially diluted in 10-fold dilutions, and spotted on control and 5-fluoroorotic acid (5-FOA) plates. Cells were grown at 25, 30, and 37 °C. C, rat8-2 (dbp5) nab2-Y34A cells have a slow growth phenotype. nab2Δ rat8-2 (dbp5) cells containing NAB2, nab2-Y34A, nab2-L55W, or nab2-F56D LEU2 test plasmids as the sole copy of NAB2 were grown to saturation, serially diluted in 10-fold dilutions, and spotted on Leu− minimal medium plates. Cells were grown at 25, 30, and, 32 °C. D, growth curve analysis of rat8-2 (dbp5) nab2-Y34A cells confirms that they grow more slowly than rat8-2 (dbp5) NAB2 cells. rat8-2 (dbp5) nab2Δ cells carrying NAB2, nab2-Y34A, nab2-L55W, or nab2-F56D plasmids as the sole copy of Nab2 were grown to saturation and diluted, and their optical density (OD) was measured at A600 for 46 h as described under “Experimental Procedures.” E, nab2 mutants are expressed at similar levels. To examine the protein level of each nab2 variant, rat8-2 (dbp5) cells containing NAB2 or nab2 variant plasmids were grown at 30 °C, and whole cell lysates prepared from these cells were analyzed by immunoblotting with a polyclonal anti-Nab2 antibody. As a loading control, Pgk1 protein levels in each lysate were detected with a monoclonal anti-Pgk1 antibody. The nab2-Y34A protein level was comparable with that seen with wild-type and, although the levels of the two controls, nab2-L55W and nab2-F56D, were a little lower, this did not produce any phenotype. F, overexpression of GFD1 suppresses the slow growth phenotype of rat8-2 (dbp5) nab2-Y34A cells. nab2Δ rat8-2 (dbp5) cells containing NAB2 or nab2-Y34A LEU2 plasmids as the sole copy of NAB2 and vector alone or GFD1 TRP1 plasmid were grown to saturation, serially diluted in 10-fold dilutions, and spotted on Leu−Trp− minimal medium plates. Cells were grown at 25, 30, and, 32 °C.
We employed a direct in vitro binding assay to confirm that both the deletions and the alanine insertions in Gfd1 impair binding of Gfd1 to Nab2-N (Fig. 6D). Purified recombinant GST-Nab2-N was incubated with His-tagged Gfd1 mutants as well as control His-Gfd1 wild-type, as described under “Experimental Procedures.” These results indicate that binding was readily detected between wild-type Gfd1 and Nab2-N but that no binding was detected with any of the Gfd1 mutants that show impaired suppression of rat8-2 (dbp5). Taken together, these results confirm that the interaction between Gfd1 and Nab2 contributes to Gfd1-mediated suppression of the rat8-2 (dbp5) mutant.
Engineered nab2 Mutants That Disrupt the Nab2/Gfd1 Interaction
To further probe the functional role of the Nab2/Gfd1 interaction, we generated several amino acid substitutions in Nab2 that lie in the Nab2/Gfd1 interface. Tyr34 forms a central feature of the Gfd1-binding site on Nab2-N observed by crystallography (Figs. 1 and 2 and supplemental Fig. S2) and also shows a strong perturbation in NMR titrations (Fig. 3). We therefore changed Tyr34 to alanine, creating nab2-Y34A. As a control for the general effect of modifications in this region, we also mutated Leu55 and Phe56 (generating nab2-L55W and nab2-F56D), which were located at the periphery of the Gfd1-binding site on Nab2. To assess whether these amino acid substitutions impair binding between Nab2 and Gfd1, we used a direct in vitro binding assay that examined the interaction between purified, recombinant GST-Nab2-N mutants and His-tagged Gfd1 wild-type. As shown in Fig. 7A, whereas GST-Nab2-N wild-type, L55W, and F56D all bound to Gfd1 to a similar extent, no binding could be detected between GST-nab2-N-Y34A and Gfd1 in this assay. NMR spectra of these mutants (supplemental Fig. 3) indicated that they retained the same fold as wild-type Nab2-N. These results confirm that Tyr34 in Nab2 is important for the interaction with Gfd1.
To determine whether interaction with Gfd1 is critical for the essential function of Nab2, we employed a plasmid shuffle assay to test whether the nab2-Y34A mutant that perturbs Gfd1 binding can function in cells as the sole copy of NAB2. As shown by the serial dilution growth assay in Fig. 7B, cells expressing any of the nab2 mutants showed growth comparable with the control wild-type NAB2 cells at all temperatures examined. This plasmid shuffle assay is able to detect impaired Nab2 function because control cells that contain vector alone are not viable and nab2-ΔN cells, which express a variant of Nab2 lacking the N-terminal domain (26), show severely impaired growth. These findings indicate that interaction with Gfd1 is not critical for the function of Nab2 in a wild-type genetic background, consistent with the observation that Gfd1 is not essential for cell viability.
A nab2 Mutant That Does Not Bind to Gfd1 Shows Genetic Interaction with rat8-2 (dbp5)
To assess whether the nab2-Y34A mutant that showed impaired binding to Gfd1 impacted cell function in a genetic background where mRNA export is suboptimal, we examined the function of nab2 mutants in the rat8-2 (dbp5) mutant strain. nab2Δ rat8-2 (dbp5) cells expressing only NAB2 or nab2 mutants (nab2-Y34A, nab2-L55W, or nab2-F56D) were grown to saturation, serially diluted and spotted on selective medium plates, and grown at 25, 30, and 32 °C (Fig. 7C). At 30 °C, rat8-2 (dbp5) nab2-Y34A cells exhibited slower growth when compared with cells expressing wild-type Nab2. In contrast, rat8-2 (dbp5) cells expressing the nab2-L55W or nab2-F56D mutants, which retain the capacity to bind Gfd1 in vitro, showed growth similar to cells expressing wild-type NAB2. These data highlight the functional importance of Nab2 interaction with Gfd1. We confirmed the growth results obtained with the nab2 mutants in a rat8-2 (dbp5) background by growth curve analysis at 30 °C (Fig. 7D). As observed by spotting, rat8-2 (dbp5) nab2-Y34A cells grew more slowly than rat8-2 (dbp5) NAB2 cells, and rat8-2 (dbp5) nab2-F56D and rat8-2 (dbp5) nab2-L55W cells grew similarly to rat8-2 (dbp5) NAB2 cells. These synthetic growth defects were not due to altered expression levels of Nab2 because the mutant nab2-Y34A protein was present at approximately the same level as wild-type Nab2 (Fig. 7E). The expression levels of the control Nab2 mutants, nab2-L55W or nab2-F56D, were slightly lower than wild-type Nab2, but these mutants did not exhibit an impaired growth phenotype (Fig. 7E). In summary, the synthetic genetic interaction observed between nab2-Y34A and rat8-2 (dbp5) strongly suggests that the Gfd1/Nab2 interaction is required for efficient Dbp5 activity.
Because nab2-Y34A showed impaired binding to Gfd1 (Fig. 7A), we tested whether overexpression of GFD1 could suppress the growth defect of nab2-Y34A rat8-2 (dbp5) cells. As shown in Fig. 7F, GFD1 overexpression suppressed the growth defect of these cells at 32 °C (the non-permissive temperature), consistent with this phenotype arising because the interaction between Nab2-N and Gfd1 was impaired.
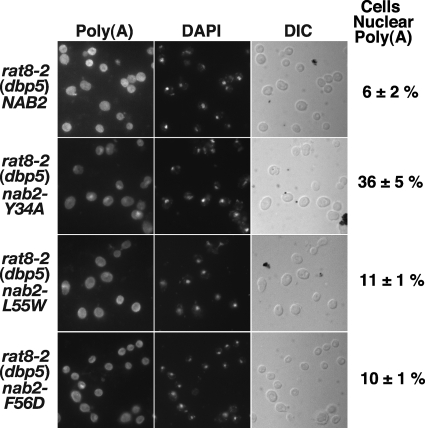
rat8-2 (dbp5) Cells Expressing a nab2 Mutant That Does Not Bind Gfd1 Show Nuclear Accumulation of poly(A) RNA
We used FISH with an oligo(dT) probe to visualize bulk poly(A) RNA to investigate whether rat8-2 (dbp5) cells expressing the nab2-Y34A mutant showed any defect in mRNA export that might account for the impaired growth phenotype observed at 30 °C. For this assay, we examined nab2Δ rat8-2 (dbp5) cells expressing NAB2, nab2-Y34A, nab2-L55W, or nab2-F56D at a temperature of 16 °C where rat8-2 (dbp5) cells show minimal accumulation of poly(A) RNA (35), and the percentage of cells with nuclear poly(A) RNA accumulation was quantitated as described under “Experimental Procedures.” rat8-2 (dbp5) nab2-Y34A cells showed a 6-fold increase in nuclear accumulation of poly(A) RNA when compared with rat8-2 (dbp5) NAB2 cells (Fig. 8). Control rat8-2 (dbp5) nab2-L55W, or rat8-2 (dbp5) nab2-F56D cells showed increases of less than 2-fold. This result is consistent with the interaction of Nab2 with Gfd1 contributing to efficient mRNA export.
FIGURE 8.
rat8-2 (dbp5) nab2-Y34A cells show accumulation of poly(A) RNA in the nucleus. nab2Δ rat8-2 (dbp5) cells carrying NAB2, nab2-Y34A, nab2-L55W, or nab2 F56D plasmid as the sole copy of NAB2 were grown in liquid culture at 16 °C, and poly(A) RNA (Poly(A)) was visualized by FISH using an oligo(dT) probe as described under “Experimental Procedures.” Cells were stained with 4′,6-diamidino-2-phenylindole-dihydro chloride (DAPI) to visualize the position of the nucleus. Corresponding differential interference contrast (DIC) images are shown. FISH results were quantified as described under “Experimental Procedures” to assess the percentage (means ± S.E.) of cells with nuclear accumulation of poly(A) RNA within five fields containing at least 31 cells (indicated to the right of the differential interference contrast images). Thus, rat8-2 (dbp5) nab2-Y34A cells exhibit a 6-fold increase in nuclear poly(A) RNA accumulation relative to rat8-2 (dbp5) NAB2 cells, whereas rat8-2 (dbp5) nab2-L55W and rat8-2 (dbp5) nab2-F56D cells show less than a 2-fold increase. The percentage of nuclear poly(A) RNA accumulation observed in rat8-2 (dbp5) nab2-Y34A cells was typical of multiple independent experiments.
DISCUSSION
Our results define the structural basis for the interaction between Nab2-N and Gfd1 and show that this interaction is important in coordinating the Dbp5-based cytoplasmic termination step of nuclear mRNA export. The crystal structure of the Nab2-N·Gfd1 complex shows that residues 126–150 of Gfd1 bind to the Nab2-N and identified the principal interactions in the interface, which also corresponded to those identified by NMR titrations as those showing chemical shift perturbations on binding. Although the Nab2-N-binding region of Gfd1 appears to have little secondary structure when isolated in solution, it takes up an α-helical conformation on binding.
The structural information obtained on the Nab2-N/Gfd1 interface was used to engineer Nab2 and Gfd1 mutants in which this interaction was impaired in vitro and which were then used to probe the functional significance of the interaction in vivo. Because GFD1 is not essential, we exploited the observation that overexpression of GFD1 suppresses the rat8-2 (dbp5) mutant (17) and, moreover, showed that deletion of GFD1 resulted in a severe growth defect in a rat8-2 (dbp5) background. Using these assays, we demonstrated that when overproduced, gfd1 mutants that lack Nab2 interaction are unable to suppress the rat8-2 (dbp5) phenotype as effectively as wild-type Gfd1. Moreover, when the nab2-Y34A mutant, in which the binding to Gfd1 is impaired, was the sole copy in rat8-2 (dbp5) cells, there was an enhanced growth defect and nuclear accumulation of poly(A) mRNA. Overall, these results demonstrate that the Nab2/Gfd1 interaction is directly involved in the mRNA export pathway and are consistent with this interaction, contributing to the step where the mRNP export complex is disassembled at the cytoplasmic face of NPCs.
Our results support the hypothesis (32) that Gfd1 may increase the efficiency of the mRNA nuclear export pathway by coordinating interactions between the poly(A) RNA-binding protein, Nab2, and the putative RNA DEAD box helicase, Dbp5, in the terminal mRNP disassembly step. Nab2 is important in modulating the length of the 3′-poly(A) tail of mRNAs and is thought to become attached to the mRNP at a late stage of nuclear processing (32). However, Nab2 remains incorporated into the mRNP during its passage through the NPC and is only removed at the cytoplasmic face through the action of Dbp5, consistent with the phenotype of the rat8-2 (dbp5) mutant being alleviated by nab2 variants in which the affinity for poly(A) RNA is reduced (14). Indeed, Nab2 is a physiological target for Dbp5 remodeling in vivo, and mRNPs are disassembled by Dbp5 in vitro (14). In yeast cells, under normal laboratory growth conditions, it is likely that although removal of Nab2 from the mRNP is crucial to the overall gene expression pathway, it is not rate-limiting. Thus, a decrease in transport rate associated with the absence of Gfd1 would only produce a dramatic growth defect if it changed the rate of Nab2 removal so radically that this step then became rate-limiting. However, under conditions where the efficiency of Dbp5 is impaired, such as in the rat8-2 (dbp5) background, the coordination effected by Gfd1 becomes critical, and so variants in which this interaction is impaired then show significant defects.
The N terminus of Nab2 interacts not only with Gfd1 but also with the Mlp1 protein (30), which localizes to the nuclear face of the NPCs (31). The Nab2-N/Mlp1 interaction requires a hydrophobic patch centered on residue Phe73 in Nab2-N (30), which is on the opposite face of Nab2-N from the Gfd1-binding site (supplemental Movie S1). This finding is consistent with previous work showing that a F73D mutant of Nab2-N, which is severely impaired for binding to Mlp1, still binds to Gfd1 (30). Thus, Nab2-N has the potential to bind simultaneously to Mlp1 and Gfd1 while bound to poly(A) RNA via its C-terminal zinc finger domain (19, 27). Simultaneous binding of Nab2 to Mlp1 and Gfd1 could help to coordinate targeting of the mRNA transcripts to the NPC, via interactions with Mlp1, and subsequent mRNP remodeling when the mRNP reaches the cytoplasmic face of the NPC for disassembly.
Our observation that overproduction of Gfd1 variants in which the formation of the Nab2·Gfd1 complex is impaired in vitro can still partially suppress the rat8-2 (dbp5) mutant phenotype indicates that Gfd1 may have additional functions in the mRNA nuclear export pathway. Although it might be possible that some of the single residue mutants employed could have retained sufficient binding capacity in vivo to account for this observation, this explanation would seem highly improbable for the variants in which the Gfd1 α-helix that binds to Nab2-N was deleted (gfd1-Δ122–143 and gfd1-Δ130–143, Fig. 6A). It is likely that in addition to the Gfd1/Nab2-N interaction, some interaction of Gfd1 with other components of the Dbp5-based mRNA export disassembly machinery, such as Gle1, Nup159, Nup42/Rip1, or Dbp5 itself, is also required for complete suppression of the rat8-2 (dbp5) phenotype. This hypothesis would also be consistent with the genetic interactions that have been observed between GFD1 and these components (17, 18, 25). Although further work will be required to define precisely the molecular mechanism by which complete suppression is achieved, these results are certainly consistent with Gfd1 functioning to coordinate the other components of the machinery that facilitates the disassembly of the mRNP export complex at the cytoplasmic face of the NPC.
Acknowledgments
We are most grateful to our many colleagues in Cambridge and in the Corbett laboratory in Atlanta for the many helpful comments, criticisms, and suggestions. We also thank David Neuhaus and Ji-Chun Yang for assistance with NMR and Divyang Jani and Ana Maria Goncalves for assistance with data collection at the ESRF.
This work was supported, in whole or in part, by National Institutes of Health Grants (to A. H. C. and S. R. W.). This work was also supported in part by a Federation of the Societies of Biochemistry and Molecular Biology Long Term Fellowship (to C. B.) and a Wellcome Trust Programme Grant (to M. S.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 3LCN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movie S1 and Figs. S1–S3.
- mRNP
- messenger ribonucleoprotein
- DTT
- dithiothreitol
- GST
- glutathione S-transferase
- Mlp1
- myosin-like protein 1
- Nab2-N
- N-terminal domain of Nab2
- NPC
- nuclear pore complex
- poly(A) RNA
- polyadenosine RNA
- PBS
- phosphate-buffered saline
- TEV
- tobacco etch virus
- WT
- wild type
- HSQC
- heteronuclear single quantum correlation
- FISH
- fluorescence in situ hybridization.
REFERENCES
- 1.Shatkin A. J., Manley J. L. (2000) Nat. Struct. Biol. 7, 838–842 [DOI] [PubMed] [Google Scholar]
- 2.Hastings M. L., Krainer A. R. (2001) Curr. Opin. Cell Biol. 13, 302–309 [DOI] [PubMed] [Google Scholar]
- 3.Akker S. A., Smith P. J., Chew S. L. (2001) J. Mol. Endocrinol. 27, 123–131 [DOI] [PubMed] [Google Scholar]
- 4.Kelly S. M., Corbett A. H. (2009) Traffic 10, 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart M. (2007) Mol. Cell 25, 327–330 [DOI] [PubMed] [Google Scholar]
- 6.Vinciguerra P., Stutz F. (2004) Curr. Opin. Cell Biol. 16, 285–292 [DOI] [PubMed] [Google Scholar]
- 7.Dower K., Rosbash M. (2002) RNA 8, 686–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen T. H., Rosbash M. (2003) Nat. Struct. Biol. 10, 10–12 [DOI] [PubMed] [Google Scholar]
- 9.Stutz F., Izaurralde E. (2003) Trends Cell Biol. 13, 319–327 [DOI] [PubMed] [Google Scholar]
- 10.Fasken M. B., Corbett A. H. (2005) Nat. Struct. Mol. Biol. 12, 482–488 [DOI] [PubMed] [Google Scholar]
- 11.Köhler A., Hurt E. (2007) Nat. Rev. Mol. Cell Biol. 8, 761–773 [DOI] [PubMed] [Google Scholar]
- 12.Carmody S. R., Wente S. R. (2009) J. Cell Sci. 122, 1933–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos-Rosa H., Moreno H., Simos G., Segref A., Fahrenkrog B., Panté N., Hurt E. (1998) Mol. Cell Biol. 18, 6826–6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran E. J., Zhou Y., Corbett A. H., Wente S. R. (2007) Mol. Cell 28, 850–859 [DOI] [PubMed] [Google Scholar]
- 15.Snay-Hodge C. A., Colot H. V., Goldstein A. L., Cole C. N. (1998) EMBO J. 17, 2663–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tseng S. S., Weaver P. L., Liu Y., Hitomi M., Tartakoff A. M., Chang T. H. (1998) EMBO J. 17, 2651–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alcázar-Román A. R., Tran E. J., Guo S., Wente S. R. (2006) Nat. Cell Biol. 8, 711–716 [DOI] [PubMed] [Google Scholar]
- 18.Weirich C. S., Erzberger J. P., Flick J. S., Berger J. M., Thorner J., Weis K. (2006) Nat. Cell Biol. 8, 668–676 [DOI] [PubMed] [Google Scholar]
- 19.Anderson J. T., Wilson S. M., Datar K. V., Swanson M. S. (1993) Mol. Cell Biol. 13, 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy R., Wente S. R. (1996) Nature 383, 357–360 [DOI] [PubMed] [Google Scholar]
- 21.Murphy R., Watkins J. L., Wente S. R. (1996) Mol. Biol. Cell 7, 1921–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarkar S., Hopper A. K. (1998) Mol. Biol. Cell 9, 3041–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green D. M., Marfatia K. A., Crafton E. B., Zhang X., Cheng X., Corbett A. H. (2002) J. Biol. Chem. 277, 7752–7760 [DOI] [PubMed] [Google Scholar]
- 24.Hector R. E., Nykamp K. R., Dheur S., Anderson J. T., Non P. J., Urbinati C. R., Wilson S. M., Minvielle-Sebastia L., Swanson M. S. (2002) EMBO J. 21, 1800–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fasken M. B., Stewart M., Corbett A. H. (2008) J. Biol. Chem. 283, 27130–27143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marfatia K. A., Crafton E. B., Green D. M., Corbett A. H. (2003) J. Biol. Chem. 278, 6731–6740 [DOI] [PubMed] [Google Scholar]
- 27.Kelly S. M., Pabit S. A., Kitchen C. M., Guo P., Marfatia K. A., Murphy T. J., Corbett A. H., Berland K. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12306–12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batisse J., Batisse C., Budd A., Böttcher B., Hurt E. (2009) J. Biol. Chem. 284, 34911–34917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Guisbert K., Duncan K., Li H., Guthrie C. (2005) RNA 11, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant R. P., Marshall N. J., Yang J. C., Fasken M. B., Kelly S. M., Harreman M. T., Neuhaus D., Corbett A. H., Stewart M. (2008) J. Mol. Biol. 376, 1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strambio-de-Castillia C., Blobel G., Rout M. P. (1999) J. Cell Biol. 144, 839–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suntharalingam M., Alcázar-Román A. R., Wente S. R. (2004) J. Biol. Chem. 279, 35384–35391 [DOI] [PubMed] [Google Scholar]
- 33.Hodge C. A., Colot H. V., Stafford P., Cole C. N. (1999) EMBO J. 18, 5778–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strahm Y., Fahrenkrog B., Zenklusen D., Rychner E., Kantor J., Rosbach M., Stutz F. (1999) EMBO J. 18, 5761–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Estruch F., Hodge C. A., Rodríguez-Navarro S., Cole C. N. (2005) J. Biol. Chem. 280, 9691–9697 [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 37.Adams A., Gottschling D. E., Kaiser C. A., Stearns T. (1997) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor [Google Scholar]
- 38.Matsuura Y., Stewart M. (2004) Nature 432, 872–877 [DOI] [PubMed] [Google Scholar]
- 39.Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992) Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]
- 40.Goddard T. D., Kneller D. G.SPARKY 3, University of California, San Francisco [Google Scholar]
- 41.Grzesiek S., Bax A. (1992) J. Magn. Reson. 99, 201–207 [Google Scholar]
- 42.Grzesiek S., Bax A. (1992) J. Am. Chem. Soc. 115, 11620–11621 [Google Scholar]
- 43.Leslie A. G. W. (1992) Newsletter on Protein Crystallography 26, 27–33 [Google Scholar]
- 44.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 45.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 46.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. (1987) Methods Enzymol. 154, 164–175 [DOI] [PubMed] [Google Scholar]
- 48.Towbin H., Staehelin T., Gordon J. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong D. H., Corbett A. H., Kent H. M., Stewart M., Silver P. A. (1997) Mol. Cell Biol. 17, 3755–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greenfield N., Fasman G. D. (1969) Biochemistry 8, 4108–4116 [DOI] [PubMed] [Google Scholar]
- 51.Ellisdon A. M., Jani D., Köhler A., Hurt E., Stewart M. (2010) J. Biol. Chem. 285, 3850–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lange A., Mills R. E., Devine S. E., Corbett A. H. (2008) J. Biol. Chem. 283, 12926–12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fairall L., Chapman L., Moss H., de Lange T., Rhodes D. (2001) Mol. Cell 8, 351–361 [DOI] [PubMed] [Google Scholar]
- 54.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]