Abstract
Variation in pathology of human prion disease is believed to be caused, in part, by distinct conformations of aggregated protein resulting in different prion strains. Several prions also exist in yeast and maintain different self-propagating structures, referred to as prion variants. Investigation of the yeast prion [PSI+] has been instrumental in deciphering properties of prion variants and modeling the physical basis of their formation. Here, we describe the generation of specific variants of the [RNQ+] prion in yeast transformed with fibers formed in vitro in different conditions. The fibers of the Rnq1p prion-forming domain (PFD) that induce different variants in vivo have distinct biochemical properties. The physical basis of propagation of prion variants has been previously correlated to rates of aggregation and disaggregation. With [RNQ+] prion variants, we found that the prion variant does not correlate with the rate of aggregation as anticipated but does correlate with stability. Interestingly, we found that there are differences in the ability of the [RNQ+] prion variants to faithfully propagate themselves and to template the aggregation of other proteins. Incorporating the mechanism of variant formation elucidated in this study with that previously proposed for [PSI+] variants has provided a framework to separate general characteristics of prion variant properties from those specific to individual prion proteins.
Keywords: Amyloid, Prions, Protein Conformation, Protein Folding, Protein Self-assembly, [RNQ+], Yeast Prion Variants
Introduction
Prion diseases are fatal neurodegenerative disorders that affect many mammals. Prions are responsible for diseases such as scrapie in sheep, bovine spongiform encephalopathy (BSE) in cows and Creutzfeldt-Jakob disease in humans (1). These diseases are linked to either mutations in the gene encoding the prion protein (PRNP) or aggregation of the prion protein (PrP) itself (2, 3). Self-propagating aggregates of PrP are implicated in the infectious transmission of prion disease among mammals (4). One interesting feature of prion diseases lies in the apparent heterogeneity of clinical symptoms and disease pathology (5–7). It has been hypothesized that this heterogeneity in pathology is partly due to the existence of different prion strains (8). The mechanism of prion strain generation is not entirely understood, but is an important aspect of prion disease. Elucidating mechanisms that dictate the generation of prion strains is crucial to understanding the biology underlying prion diseases, protein misfolding, and aggregation.
Naturally occurring prions that exist in fungi such as the budding yeast Saccharomyces cerevisiae may not be associated with disease, but can act as a mechanism to alter phenotype. Recent studies have uncovered several prion-like proteins in yeast (9–13). The three extensively characterized prions in yeast are the [PSI+], [RNQ+], and [URE3] prions (14–16). The [PSI+] prion manifests as a result of the aggregation of Sup35p (17, 18), which is the S. cerevisiae eukaryotic release factor 3 (eRF3) (19). In the [psi−] state, Sup35p binds Sup45p, and this complex is required for translation termination at stop codons (20). In the [PSI+] state, however, much of the Sup35p is in the aggregated state and not available to function in translation termination (17). A premature stop codon in a metabolic pathway, such as that in the ade1-14 allele, provides a convenient method of detecting the [PSI] state of yeast cells. Yeast grown on nutrient-rich medium that harbor the ade1-14 allele are white in color in the [PSI+] state, but red in color in the [psi−] state due to the accumulation of a metabolic intermediate in the adenine biosynthetic pathway (21). Yeast can maintain different variants of the [PSI+] prion and these cause different levels of read through of the ade1-14 premature stop codon that correlate to the amount of aggregated Sup35p (22–24). This results in different levels of pigment accumulation in cells and weaker strains of [PSI+] manifest as shades of pink (24). This phenotype has proven very useful in investigating the mechanism of prion variant generation.
The structure of different [PSI+] prion variants have been revealed by biophysical and nuclear magnetic resonance (NMR) studies on conformationally-distinct fibers of the prion-forming domain (PFD)2 of Sup35p (Sup35NM) formed in vitro at different temperatures (25–28). These studies suggest that the protected amyloid core of the strong variant is short relative to the amyloid core of the weak variant. A model designed to explain the propagation of prion variants related structural and biochemical properties of amyloid fibers to the prion variant phenotype (29). This model suggests that the propagation of prion variants can be explained by differences in the rate of fiber growth and sensitivity to shearing. Because the addition of new protein appears to occur on fiber ends (30, 31), it follows that the most efficient propagation requires more free ends to template the addition of new protein. Thus, more free ends may relate to a faster rate of aggregation. The mechanism for prion variant generation has been proposed primarily based on work conducted using Sup35NM and the [PSI+] prion.
Two other prions in yeast, [URE3] and [RNQ+], have also been reported to display variant phenotypes (32, 33). The [URE3] prion in S. cerevisiae is propagated by the Ure2 protein (16). Recombinant Ure2p can form fibers in vitro and when transformed into yeast can induce different variants of the [URE3] prion (33). The [RNQ+] prion is caused by the aggregation of the Rnq1 protein (34). In the [rnq−] state, the Rnq1 protein has no known function. However, in the [RNQ+] state, aggregated Rnq1p facilitates the induction of the [PSI+] prion (35–37). [RNQ+] also exists as different variants (38) that appear when recombinant Rnq1p fibers are transformed into yeast (39). However, the generation of specific [RNQ+] variants by altering the conditions of in vitro amyloid formation has not been investigated. As such, the physical basis for the generation of [URE3] and [RNQ+] variants has not been described and it remains to be determined if the properties described for [PSI+] variants can be generalized.
We have taken advantage of the [RNQ+] reporter protein (RRP) previously described by our laboratory to phenotypically assess the [RNQ+] prion status as well as [RNQ+] variants (40). The RRP molecule is a chimeric protein with the proposed PFD of Rnq1p (amino acids 153–405) fused to the translation termination domain of Sup35p (the MC region, amino acids 124–685) (41, 42). The RRP fusion presumably co-aggregates with Rnq1p in the [RNQ+] state, which results in prion-dependent nonsense suppression. As such, this provides a phenotypic assay for the [RNQ] state. This assay allowed us to detect both intragenic and extragenic effectors of [RNQ+] propagation (40, 43). We found that the level of nonsense suppression afforded by RRP changes with [RNQ+] prion variants, and this enables us to distinguish variants by phenotype.
In this study, we created and characterized variants of the [RNQ+] prion by controlling conditions of fiber formation and transforming the fibers into yeast strains with Rnq1p in the non-prion state, [rnq−]. We show that the proposed PFD of Rnq1p forms amyloid fibers with distinct characteristics when generated in different conditions. Additionally, infection of these fibers into yeast induces different [RNQ+] prion variants in vivo. Each [RNQ+] variant maintains and propagates a unique set of characteristics in vivo and in vitro. Finally, our data suggest the existence of both similarities and differences between the mechanisms that dictate [RNQ+] and [PSI+] variant propagation.
EXPERIMENTAL PROCEDURES
Protein Purification
The pHis10-Rnq1PD (amino acids 132–405) construct (39) was transformed into Escherichia coli BL21(DE3) cells. Resultant transformants were scraped to inoculate large cultures in CircleGrow medium, and the cells were grown to an A600 ∼0.6. The expression of Rnq1PFD was induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside for 6 h. The cells were harvested and incubated with agitation at room temperature for 1 h in 20 mm Tris-HCl, 8 m urea (Buffer A, pH 8). The cells were then further lysed by sonication. The lysate was cleared by spinning at 10,000 × g for 30 min. The supernatant was incubated with Ni2+-Sepharose beads for 2 h. Subsequently, the beads were packed in a column and washed with 25 column volumes (CV) Buffer A, pH 8. The beads were then washed successively with four CV each of Buffer A, pH 6.3 and Buffer A, pH 5.9, before eluting with Buffer A, pH 4.5. Rnq1p in the eluted fractions was verified for purity by SDS-PAGE and Coomassie staining. The fractions containing the protein were filtered through a 100-kD molecular mass cut off column (Amicon) and stored in methanol at −80 °C.
Amyloid Fiber Formation
Kinetic assays of fiber formation were done in a SpectraMax M2e fluorimeter microplate reader. Rnq1PFD fibers were diluted 75-fold from 7 m guanidine hydrochloride to a concentration of 4 μm in FFB buffer (150 mm NaCl, 5 mm KPO4, 2 m urea, pH 7.4) with 50-fold molar excess Thioflavin-T to initiate fiber formation in the presence of glass beads for agitation. The change in Thioflavin-T fluorescence over time was measured using an excitation wavelength of 450 nm and emission wavelength of 481 nm. The plate was agitated each minute prior to reading for 10 s. Fibers were also formed in a rotator (end-over-end) at a monomer concentration of 4 μm for thermostability assays and to obtain seeds for seeding experiments. For the seeded experiments, the fibers were seeded using 5% (w/w) seed.
Thermostability Assay
Pre-formed fibers were treated with 2% SDS for 5 min at different temperatures (gradient from 55 to 95 °C, with 5–7 °C increments). The treated samples were analyzed by SDS-PAGE and Western blot using an anti-Rnq1p antibody. The bands were quantified using ImageJ software and values were normalized to the 95 °C band. Results were plotted using Origin 6.1 statistical software.
Electron Microscopy
Samples of fibrillar Rnq1PFD were allowed to settle onto freshly glow-discharged 200 mesh carbon-formvar coated copper grids for 10 min. Grids were then rinsed twice with water and stained with 1% uranyl acetate (Ted Pella) for 1 min. Samples were viewed on a JEOL 1200EX transmission electron microscope (JEOL USA).
Protein Transformation
Transformation of Rnq1PFD fibers into a [rnq−] 74-D694 (ade1-14, ura3-52, leu2-3,112, trp1-289, his3-200, sup35::RRP) yeast strain was conducted as described (44). The resulting colonies were replica plated onto rich medium (YPD) plates to assay for colony color. Colonies that appeared to have acquired the prion state by nonsense suppression were picked and spotted on YPD, YPD containing 3 mm GdnHCl and SD-ade for phenotypic analyses.
Mitotic Stability of [RNQ+]
Three independent protein transformants for each of the [RNQ+] prion variants were grown in liquid YPD medium overnight (∼16 h) to an A600 of ∼2.0. The cultures were diluted 8,000-fold in water and 250 μl were plated on 13-cm (diameter) YPD plates. Approximately 750 colonies for each transformant and a total of >2,000 colonies were counted for each variant. The average and standard deviation is reported for each variant. The percentage of cells that lost the prion was calculated by dividing the number of red cells by the total number of cells. The loss of the [RNQ+] prion was confirmed by Rnq1p solubility assays for 10 representative colonies of each variant.
[PSI+] Induction Assay
Yeast strains containing the [RNQ+] prion were transformed with pEMBL-SUP2 (41) and plated on SD-ura. Transformants were grown in selective medium to an A600 of ∼1.6 and plated on YPD. After 5 days of growth, [PSI+] cells were counted as any cell with a light pink sector. Representative colonies were checked for curing by transient growth on medium containing 3 mm GdnHCl.
[RNQ+] Solubility Assay
The protocol was adapted from previously published work (34). The yeast cells were lysed in buffer containing 100 mm Tris-HCl, pH 7, 200 mm NaCl, 1 mm EDTA, 5% glycerol, 0.5% dithiothreitol, 50 mm N-ethylmaleimide (NEM), 3 mm phenylmethanesulfonyl fluoride, and EDTA-free complete Mini protease inhibitor mixture (Roche). Lysis was performed by vortexing with glass beads. Following lysis, an equal volume of radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7, 200 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS) was added to the lysate, and cell debris was cleared by centrifugation at 3,300 × g for 15 s. A portion of this cleared lysate was saved for the total protein sample. Aggregated protein was pelleted by centrifugation at 80,000 rpm in a Beckman TLA-100 rotor for 30 min. The supernatant containing the soluble protein was removed, and the pellet was resuspended in a 1:1 mix of lysis buffer and RIPA buffer. The total, supernatant, and pellet fractions were subjected to SDS-PAGE, transferred to polyvinylidene difluoride membrane, and probed with an anti-Rnq1p antibody.
RESULTS
Temperature Has a Profound Effect on the Kinetics of Rnq1PFD Fiber Formation
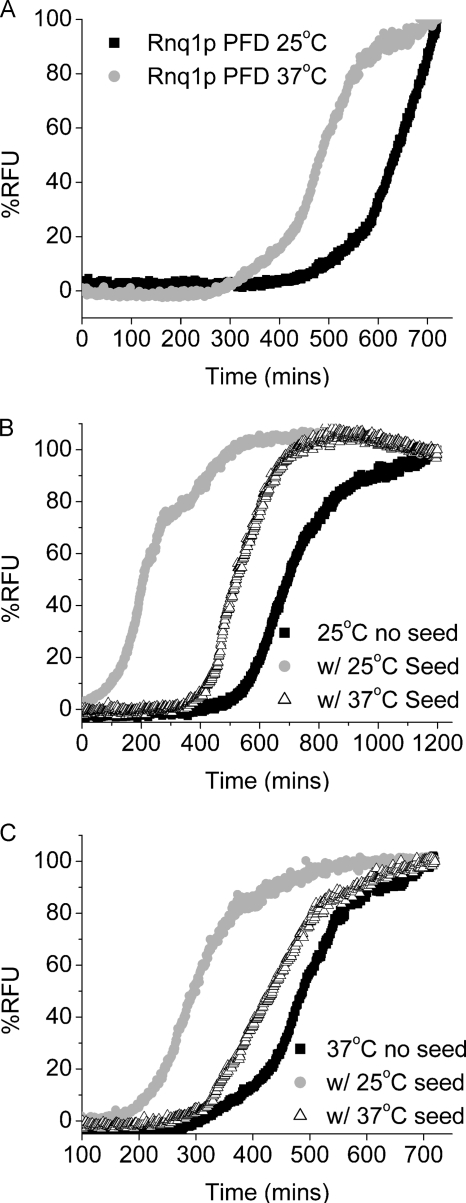
We purified recombinant Rnq1PFD in 8 m urea as described under “Experimental Procedures.” The purified Rnq1PFD protein formed amyloid fibers when diluted out of denaturant. The length of the lag phase of fiber formation is a parameter that can be used to measure the ability of the protein to aggregate and is one determinant for prion variant formation; a longer lag phase indicates a reduced ability to aggregate. In order to create [RNQ+] prion variants, we used different temperatures to generate structural variation in the fibers of Rnq1PFD. We followed fiber formation of Rnq1PFD at 25 and 37 °C using a Thioflavin-T fluorescence assay. At 25 °C, Rnq1PFD fibers had a lag phase of ∼470 min followed by a logarithmic growth phase (Fig. 1A). In contrast, fiber formation of Rnq1PFD at 37 °C had a shorter lag phase of ∼300 min.
FIGURE 1.
Fiber formation of Rnq1p is faster at higher temperatures and enhanced by the addition of preformed seeds. Purified Rnq1PFD was diluted from denaturant (75-fold) to 4 μm, and fiber formation was followed using Th-T fluorescence. Graphs are plotted as % RFU versus time. A, Rnq1PFD unseeded at 25 °C (black squares) and 37 °C (gray circles). B, Rnq1PFD fiber formation at 25 °C unseeded (black squares), with 5% seeds formed at 25 °C (gray circles), and with 5% seeds formed at 37 °C (open triangles). C, Rnq1PFD fiber formation at 37 °C unseeded (black squares), with 5% seeds formed at 25 °C (gray circles) and with 5% seeds formed at 37 °C (open triangles). Graphs are representative of at least three independent trials.
The ability of seeds to enhance the conversion of monomeric protein into fibers is an important feature of nucleated conformational conversion for amyloid fiber formation (45). We formed Rnq1PFD fibers at 25 and 37 °C and tested the ability of the fibers to act as seeds and template the addition of freshly diluted monomeric Rnq1PFD. Interestingly, even though Rnq1PFD fiber formation occurs with a shorter lag phase at 37 °C, the seeds obtained from fibers formed at 25 °C were more efficient at templating new fiber formation at both 25 and 37 °C (Fig. 1, B and C). Seeds obtained from fibers formed at 37 °C also enhanced fiber formation reactions but were less efficient at seeding Rnq1PFD monomer at either temperature (Fig. 1, B and C).
Rnq1PFD Fibers Formed at Different Temperatures Induce Distinct Distributions of Prion Variants
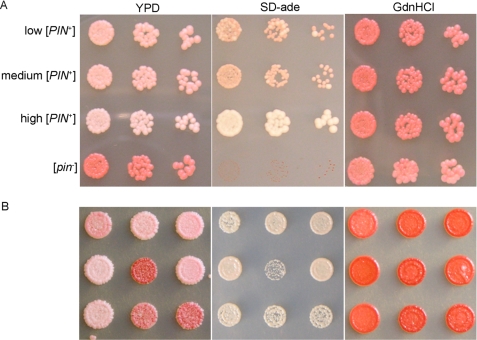
Prion proteins in yeast can maintain different variants of the prion state that are a consequence of different self-propagating structures (46). The variants differ from each other based on their phenotype, the ratio of soluble to aggregated protein, and their ability to maintain and propagate the prion (23, 24, 38). Previous work has extensively characterized variants of the [PSI+] prion generated in vitro (25–27, 29, 47, 48). Variants of the [RNQ+] prion have also been characterized in vivo and display different abilities to induce the [PSI+] prion (38, 49). However, these variants arose spontaneously and were originally referred to as [PIN+] strains for [PSI+] inducibility (37, 50). Additionally, some variants of [PIN+] were acquired when fibers of Rnq1PFD were transformed into [rnq−] yeast, but these were not characterized (39). Characterization of [RNQ+] variants has been limited since the only methods to assay the [RNQ+]/[PIN+] status of yeast cells involved either [PSI+] induction or Rnq1p solubility assays. In our laboratory, we have been able to circumvent some of these tedious procedures by taking advantage of the previously described RRP ([RNQ+] Reporter Protein), which enables the use of a colony color assay to determine the presence of different prion variants of [RNQ+]. We found that RRP not only reported on the presence of the [RNQ+] prion, but also showed changes in phenotype depending on the [RNQ+]/[PIN+] variant ((40) and Fig. 2A). High [PIN+] and [pin−] are readily discernible from low [PIN+] and medium [PIN+] when RRP-expressing strains are grown on nutrient-rich medium. Low [PIN+] and medium [PIN+] strains are phenotypically similar to each other in this assay.
FIGURE 2.
RRP differentiates [RNQ+]/[PIN+] variants. A, yeast strains expressing the RRP reporter display different levels of nonsense suppression in [PIN+] variants. B, transformation of Rnq1PFD amyloid fibers into [rnq−] yeast induces variants of the [RNQ+] prion. The [RNQ+] phenotype was assayed using the RRP reporter to show color on YPD, growth on SD-ade, and curability by growth on medium containing GdnHCl. Colony color on YPD is indicative of different levels of suppression of the ade1-14 premature stop codon to produce Ade1p. [RNQ+] prion variants co-aggregate with RRP and cause different phenotypes (color on YPD). The strength of the variants is also reflected by their ability to grow on SD-ade medium.
To create [RNQ+] variants for characterization, we first asked whether amyloid fibers of Rnq1PFD formed at different temperatures corresponded to the generation of different prion variants, as previously observed with Sup35NM and [PSI+] (48, 51). We transformed [rnq−] 74-D694 cells expressing both wild-type RNQ1 and the RRP chimeric protein with amyloid fibers of recombinant Rnq1PFD formed at 18, 25, and 37 °C. Transformants were picked, diluted, and spotted on plates to assess phenotype. We scored the transformants that turned white, light pink, or dark pink as strong, medium, or weak [RNQ+] variants, respectively (Fig. 2B and Table 1). We also correlated the colony color phenotype of the variants to their growth on medium lacking adenine (SD-ade). Transformants were also grown on medium containing guanidine hydrochloride (GdnHCl), which inactivates Hsp104 (52) and cures all variants of the [RNQ+] prion. [RNQ+] variants were verified for curability on medium containing GdnHCl using color as an indicator of change in nonsense suppression. [RNQ+] RRP strains converted from either white or pink to red, and the cured strains remained red upon removal from medium containing GdnHCl. Rnq1PFD fibers formed at 18 °C only induced strong and medium variants of [RNQ+] when transformed into [rnq−] cells (Table 1). Fibers formed at 25 °C induced weak, medium, and strong [RNQ+] variants when transformed into [rnq−] cells. In contrast, cells infected with fibers formed at 37 °C yielded primarily medium and weak [RNQ+] variants and very few strong [RNQ+] variants. These data suggest that the temperature at which fibers were formed in vitro influenced the distribution of [RNQ+] variants propagated in cells. Specifically, increasing the temperature decreased the fraction of strong [RNQ+] variants and increased the weak variants of [RNQ+].
TABLE 1.
Distribution of [RNQ+] prion variants
Yeast colonies that resulted from infection of Rnq1PFD fibers formed at 18, 25, and 37 ○C were grouped into weak, medium, and strong [RNQ+] strains based on RRP-mediated colony color (on YPD) and growth on medium lacking adenine (SD-ade). The numbers in the table reflect the percentage of variants obtained by infectivity assays after counting over 300 transformed colonies from more than three experiments for each temperature at which fibers were formed.
| Distribution of [RNQ+] variants |
|||
|---|---|---|---|
| Weak | Medium | Strong | |
| 18 °C | 0 | 25 | 75 |
| 25 °C | 15 | 46 | 39 |
| 37 °C | 29 | 68 | 3 |
Rnq1PFD Fibers Formed at Different Temperatures Are Differentially Stable
In the case of Sup35NM and the [PSI+] prion, weaker variants have a longer amyloid core and stronger variants have a shorter amyloid core (25, 26). The length of the core positively correlates with the stability of the amyloid fibers. Additionally, the more stable amyloid fibers with a longer core propagate weaker prion variants while less stable fibers with a shorter core propagate stronger prion variants.
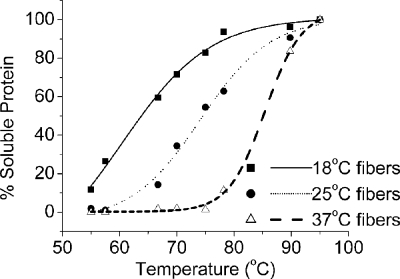
We next asked if the stability of the amyloid fibers correlated to the [RNQ+] variants induced in a manner similar to the observations with [PSI+] and Sup35NM. Therefore, we used thermostability to determine the relative stability of Rnq1PFD fibers formed at different temperatures. We formed fibers at 18, 25, and 37 °C and subjected them to increasing temperatures in 2% SDS. We then analyzed the soluble protein by SDS-PAGE and Western blot. We found that fibers formed at 18 °C were labile and showed ∼50% disruption at 65 °C (Fig. 3). Fibers formed at 25 °C were slightly more stable and showed ∼50% dissociation at 75 °C. Rnq1PFD fibers formed at 37 °C were the most stable and required 85 °C to dissociate. Thus, stronger amyloid fibers yield weaker prion variants, and this trend recapitulates the data observed previously for Sup35NM.
FIGURE 3.
Rnq1PFD fibers formed at higher temperatures are more stable. Preformed fibers were treated across a temperature gradient in the presence of 2% SDS. Following SDS-PAGE and Western blot, the amount of soluble Rnq1PFD was quantified, and the results were graphed as a percentage of total protein at 95 °C using OriginPro 6.1 with a sigmoidal fit function.
TEM Reveals Dramatic Morphological Differences in Rnq1PFD Fibers
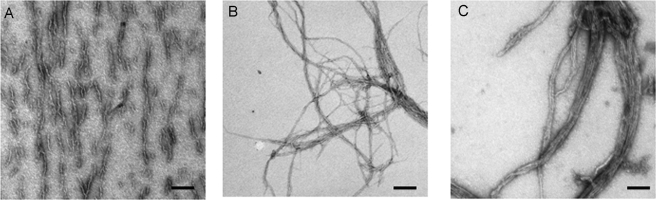
The observed dramatic differences in kinetics and stability of amyloid fibers of Rnq1PFD formed at different temperatures and their ability to induce different variants of [RNQ+] led us to hypothesize that the morphology of the Rnq1PFD fibers formed at these temperatures may be distinct. To determine if there are gross morphological differences between the Rnq1PFD fibers formed at 18, 25, and 37 °C we obtained TEM images of these amyloid fibers. In fact, we did observe significant differences in the morphology of the fibers formed at the different temperatures. We noted that the Rnq1PFD fibers formed at 18 °C were short and curly (Fig. 4A) compared with the long fibers formed at 25 °C (Fig. 4B). Lastly, the fibers formed at 37 °C were also long and appeared to be more heavily bundled with one another as compared with the fibers formed at 25 °C (Fig. 4C). These results indicate that temperature has a profound impact on the properties of the amyloid fibers formed by Rnq1PFD.
FIGURE 4.
The morphology of amyloid fibers formed from Rnq1PFD at different temperatures is distinct. Amyloid fibers of Rnq1PFD formed in vitro were imaged by TEM. The scale bar represents 200 nm. A, 18 °C fibers are short and curly; B, 25 °C fibers are long and curvy; and C, 37 °C fibers are long and bundled.
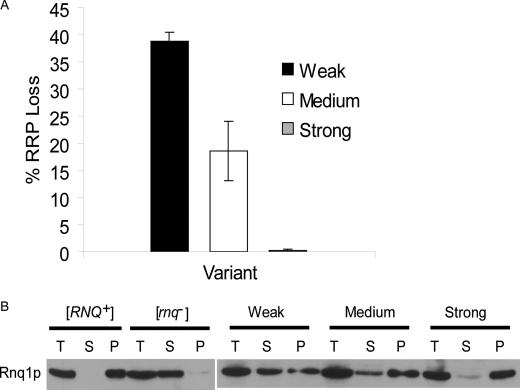
[RNQ+] prion variants have distinct protein aggregation and propagation properties in vivo. Variants of the [PSI+] prion have differential abilities to faithfully propagate the prion through mitosis (24). We utilized the RRP marker to determine the relative mitotic stability of the [RNQ+] prion variants. We grew the three different variants in liquid medium overnight and plated them on rich medium to assess [RNQ+] stability by color. The colonies that did not maintain the prion through mitotic division turned red and were counted and plotted as a fraction of the total number of colonies (Fig. 5A). The strong [RNQ+] prion variant propagated the prion with nearly 100% fidelity and rarely lost the prion phenotype. In comparison, the medium [RNQ+] variant lost the prion in ∼19% of the progeny and the weak [RNQ+] variant lost the prion in almost 40% of the progeny. The loss of the [RNQ+] prion was verified by conducting Rnq1p solubility assay on a subset of candidate [rnq−] cells for each variant. Thus, the stability of prion propagation is correlated to the strength of the prion variant and is similar to what has been observed with the [PSI+] prion.
FIGURE 5.
Variants of the [RNQ+] prion show distinct propagation and aggregation properties in vivo. A, mitotic stability of the [RNQ+] prion was dependent on the strength of the variant. The weak [RNQ+] variant lost the prion in ∼40% of the colonies (black bar), while medium [RNQ+] lost the prion in ∼19% of the colonies (white bar), and strong [RNQ+] lost the prion in <1% of the colonies (gray bar). Three independent experiments were performed for each variant to yield the resulting means and standard deviations. B, full-length endogenous Rnq1p in lysates from different [RNQ+] variants was fractionated by high speed centrifugation into total (T), supernatant (S), and pellet (P) fractions. The amount of protein in the supernatant fraction decreased as the strength of the variant increased. A representative blot from four independent experiments is shown.
Even though there are differences in mitotic stability, all prions should be transmitted in a non-Mendelian fashion via mating and meiosis (53). To verify that the cells infected with in vitro formed fibers of Rnq1PFD truly harbor self-propagating prion elements, we tested the transmission of the prion. We mated the transformed cells harboring different variants of the [RNQ+] prion to [psi−] [rnq−] cells. The diploids that we obtained were [psi−] [RNQ+] and red in color because wild-type Sup35p does not aggregate with RRP and allows for faithful termination of translation (data not shown). We sporulated the diploids to determine if the [RNQ+] prion variants were faithfully inherited in meiotic progeny as expected for the prion state. We then performed Rnq1p solubility assays on at least three complete tetrads from each strain. We noted that in each case, the [RNQ+] prion was inherited in a non-Mendelian fashion with all four progeny being [RNQ+] (data not shown).
Prion variants of both [PSI+] and [RNQ+] have also been shown to display differences in the ratio of soluble to aggregated protein (23, 38). In order to determine whether the different [RNQ+] variants that we had classified based on phenotype had corresponding biochemical differences, we performed Rnq1p solubility assays on the variants (Fig. 5B). In cells containing the weak [RNQ+] variants, most of the protein was in the soluble fraction and a small amount of Rnq1p was in the pellet fraction. In the medium [RNQ+] variants, a small portion of the protein was soluble, while most was found in the aggregated pellet fraction. However, in the strong [RNQ+] variant, almost all of Rnq1p was aggregated and very little protein was found in the soluble fraction. Interestingly, this variant-dependent change in solubility was reproducible in fresh transformants but was not as apparent in new meiotic progeny (data not shown). However, the [RNQ+] solubility assay has not always provided a robust measure of differences in prion variants (38, 40).
Variants of [RNQ+] Induce [PSI+] with Different Efficiencies
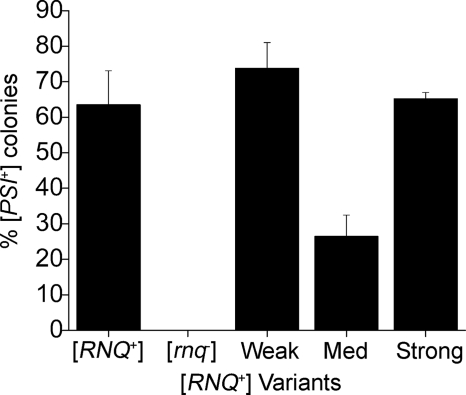
The only phenotype associated with the [RNQ+] prion state is its ability to induce the [PSI+] prion by acting as [PIN+]. While other protein aggregates can act as [PIN+], to date only [RNQ+] has been found commonly in lab and wild strains (34, 54, 55) and as such, is likely the most common, naturally occurring [PIN+] element. Because we observed that [RNQ+] induced by protein transformation resulted in distinct variants based on nonsense suppression, we next wanted to test the ability of the variants to act as [PIN+] and induce [PSI+] (24, 37). Red [psi−] [RNQ+] cells that were propagating different variants of the [RNQ+] prion were obtained by mating and sporulation as described in the previous section. The progeny that expressed wild-type SUP35 and did not have the RRP reporter were transformed with a plasmid expressing a second copy of SUP35 from its own promoter to induce [PSI+]. Transformants were grown in selective medium overnight to allow for Sup35p overexpression and then plated on YPD. The efficiency of [PSI+] induction was determined by counting the number of colonies with light pink sectors and dividing that by the total number of colonies. In the [rnq−] controls, we did not observe any induction of the [PSI+] prion when SUP35 was overexpressed in the cell, while in [RNQ+] cells we obtained efficient induction of [PSI+] (Fig. 6). As expected, the strong [RNQ+] variants showed a high level of induction of [PSI+], with >60% of the colonies sectoring (Fig. 6). The medium [RNQ+] variants had lower induction of [PSI+] when compared with strong [RNQ+]. Surprisingly, the weak [RNQ+] variants demonstrated high levels of [PSI+] induction (Fig. 6) that were comparable to the strong variants of [RNQ+]. Interestingly, the induction of [PSI+] we observed with our strong and weak [RNQ+] variants were reminiscent of the previously characterized high and very high [PIN+] variants, respectively, that were previously published (38) and reconfirmed (data not shown).
FIGURE 6.
The [PSI+] induction capacity of the [RNQ+] variants is distinct. The overexpression of Sup35p in the control [RNQ+] strain induced [PSI+] in ∼62% of the colonies. [PSI+] was not induced in [rnq−] cells. Overexpression of Sup35p in weak [RNQ+] variants induced [PSI+] in ∼72% of the colonies, medium [RNQ+] variants induced [PSI+] in ∼25% of the colonies, and strong [RNQ+] variants induced [PSI+] in ∼65% of the colonies.
DISCUSSION
The mechanism underlying the generation of prion variants was extensively investigated for only one yeast prion protein previously. Both the physical and structural basis for variant formation of the [PSI+] prion from Sup35NM has been elucidated (25, 26, 29). Here, we created and characterized different variants of the [RNQ+] prion. We assayed for prion variants in vivo using the chimeric protein RRP (40) that displayed distinct nonsense suppression phenotypes. We used changes in temperature as a means to generate different amyloid structures and correlated those to prion variant formation in vitro and in vivo. Rnq1PFD fibers formed at 18, 25, and 37 °C have distinct biochemical and morphological properties. Interestingly, fibers formed at 37 °C have a shorter lag phase when compared with the 25 °C fibers. As expected, the stability of the fibers increased with the temperature at which the fibers were formed, which was also reflected by the morphology of the fibers observed by TEM. We speculate that the fibers formed at 18 °C are the least stable because they are short. Fibers formed at 37 °C appear to be the most stable and this may be due, in part, to the interactions between fibers. The more stable 37 °C fibers also show reduced seeding capacity as compared with fibers formed at 25 °C. The ability of 37 °C Rnq1PFD fibers to seed new reactions was not enhanced by increased shearing, however (data not shown). Finally, fibers formed at 18, 25, or 37 °C were transformed into [rnq−] yeast cells and each resulted in distinct distributions of [RNQ+] prion variants.
The correlation we observed between the length of the lag phase and the resulting [RNQ+] prion variants, however, was not in accordance with previously observed properties of Sup35NM fibers and [PSI+] variants. In the case of Sup35NM, fibers formed at 25 °C have a longer lag phase when compared with 4 °C fibers (56). When Sup35NM fibers are transformed into cells, strong [PSI+] variants are generated from fibers with a short lag phase and weak [PSI+] variants result from fibers that formed with a longer lag phase. In stark contrast, although Rnq1PFD fibers formed faster at 37 °C, they induced weaker [RNQ+] variants. Rnq1PFD fibers formed slower at 25 °C and induced strong [RNQ+] variants. We would like to note that unlike Sup35NM, Rnq1PFD was unable to form discernable amyloid fibers at 4 °C (data not shown).
The correlation between the stability of Rnq1PFD amyloid fibers and the resulting prion variant distribution is similar to what was observed with Sup35NM and the [PSI+] prion (48). We observed that an increase in the stability of the Rnq1PFD fibers correlated to an increase in weak [RNQ+] variants and a decrease in strong [RNQ+] variants. From this, we suggest that the stability of the fibers is a critical property in determining the previously proposed parameters that dictate prion variants. Since the rate of aggregation does not always correlate to the generation of specific prion variants, we suggest that this in vitro property cannot be generalized to determine the physical basis of variants in vivo. As the addition of new protein occurs at fiber ends (30), it follows that the most efficient propagation requires more free ends for templating. Faster aggregation, but slower disaggregation, would be unlikely to create strong variants, as is the case with Rnq1PFD fibers formed at 37 °C. However, fast “disaggregation” or shearing could compensate for some reduction in the rate of aggregation, as is the case with Rnq1PFD fibers formed at 25 °C.
We were also able to determine the differences in the mitotic stability of the different [RNQ+] variants using the convenient RRP marker. The ability to maintain the [RNQ+] prion state was intimately linked to the strength of the prion, with strong [RNQ+] being more stable than medium [RNQ+], and medium [RNQ+] being more stable than weak [RNQ+]. The stability of the different variants could be related to the ratio of aggregated to soluble protein, the size of the aggregates, and the ability of Hsp104p and other chaperones to generate seeds that can be passed on to daughter cells.
We further characterized the [RNQ+] variants by assaying their ability to act as [PIN+] and induce [PSI+]. Surprisingly, both weak [RNQ+] and strong [RNQ+] induced [PSI+] at high efficiencies (>60%) when compared with the medium [RNQ+] variant. Interestingly, the weak [RNQ+] variant was denoted as weak based on solubility of Rnq1p and low levels of nonsense suppression using RRP, but it maintained the greatest capacity to induce [PSI+]. We compared our variant to the previously described variants of the [RNQ+] prion that were obtained spontaneously (38). They were classified based on their ability to induce [PSI+] into low, medium, high, and very high [PIN+] variants. The extent of aggregation of Rnq1p correlated to an increase in [PSI+] induction efficiency with the low, medium, and high variants. However, the very high [PIN+] variant did not fit the trend as it showed the largest amount of soluble Rnq1p, but had the greatest capacity to induce [PSI+]. Therefore, we propose that the weak [RNQ+] variants we obtained by protein transformation are most similar to the previously obtained very high [PIN+] variant (38). Because the RRP phenotype likely reflects co-aggregation with wild-type Rnq1p in the [RNQ+] state and the induction of [PSI+] likely reflects some transient physical interaction with Sup35p (40, 57), we suggest that very high [PIN+]/weak [RNQ+] forms a structure that affords less efficient self-templating but more efficient interaction with Sup35p than other variants. While the medium and strong [RNQ+] variants correspond to medium and high [PIN+], we note one important caveat in the interpretation of the differences in the ability to induce [PSI+]. The reduced [PSI+] induction we observed with medium [RNQ+] is probably due in part to the decreased mitotic stability of medium [RNQ+] when compared with high [RNQ+]. This is not likely to explain the differences entirely, however, as different structures of Rnq1p in [RNQ+] are likely to interact differently with Sup35p to induce [PSI+].
Here, we have defined conditions for the aggregation of Rnq1PFD in vitro that result in the induction and propagation of specific [RNQ+] variants in vivo. These data are consistent with a direct relationship between the biochemical properties of amyloid fibers and prion variants. The information gleaned from the generation of [RNQ+] prion variants suggests that there may be some general properties dictating prion strain propagation, while others are unique to individual prion proteins. Using the [RNQ+] variants and the ability to investigate their in vitro properties, we may be able to delineate the mechanisms involved in the self-seeding of Rnq1p to maintain variants of [RNQ+] from those involved in cross-seeding Sup35p to induce [PSI+]. Hence, this provides a tractable system to investigate mechanisms that impact protein aggregation and the onset of protein conformational disorders.
Acknowledgments
We thank Dr. J. Patrick Bardill for creating the RRP strain and plasmids used in this study. We thank Susan Liebman (University of Illinois, Chicago) for the pHis10-Rnq1PD plasmid. We thank Wandy Beatty (EM core facility, Molecular Microbiology department, WA University School of Medicine) for help with the TEM images and Dr. John Cooper for allowing us to use equipment and providing helpful advice. We thank Dr. J. Phillip Miller, Dr. Joanna Masel, and Dr. Jingqin (Rosy) Luo for advice on data analysis. We thank members of the True Laboratory for helpful discussions and critical comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM072228 (to H. L. T.) and the National Science Foundation.
- PFD
- prion-forming domain
- TEM
- transmission electron microscope
- RRP
- [RNQ+] reporter protein
- GdnHCl
- guanidine hydrochloride
- Th-T
- thioflavin-T
- RFU
- relative fluorescence units
- YPD
- richmedia.
REFERENCES
- 1.Prusiner S. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chesebro B. (2003) Br. Med. Bull. 66, 1–20 [DOI] [PubMed] [Google Scholar]
- 3.Wadsworth J. D., Hill A. F., Beck J. A., Collinge J. (2003) Br. Med. Bull. 66, 241–254 [DOI] [PubMed] [Google Scholar]
- 4.Prusiner S. B. (1982) Science 216, 136–144 [DOI] [PubMed] [Google Scholar]
- 5.Croes E. A., Theuns J., Houwing-Duistermaat J. J., Dermaut B., Sleegers K., Roks G., Van den Broeck M., van Harten B., van Swieten J. C., Cruts M., Van Broeckhoven C., van Duijn C. M. (2004) J. Neurol. Neurosurg Psychiatry 75, 1166–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King A., Doey L., Rossor M., Mead S., Collinge J., Lantos P. (2003) Neuropathol. Appl. Neurobiol. 29, 98–105 [DOI] [PubMed] [Google Scholar]
- 7.Kovács G. G., Trabattoni G., Hainfellner J. A., Ironside J. W., Knight R. S., Budka H. (2002) J. Neurol. 249, 1567–1582 [DOI] [PubMed] [Google Scholar]
- 8.Collinge J. (2001) Annu. Rev. Neurosci. 24, 519–550 [DOI] [PubMed] [Google Scholar]
- 9.Du Z., Park K. W., Yu H., Fan Q., Li L. (2008) Nat. Genet. 40, 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel B. K., Gavin-Smyth J., Liebman S. W. (2009) Nat. Cell Biol. 11, 344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alberti S., Halfmann R., King O., Kapila A., Lindquist S. (2009) Cell 137, 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemecek J., Nakayashiki T., Wickner R. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1892–1896 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Brown J. C., Lindquist S. (2009) Genes Dev. 23, 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uptain S. M., Lindquist S. (2002) Annu. Rev. Microbiol. 56, 703–741 [DOI] [PubMed] [Google Scholar]
- 15.Wickner R. B., Masison D. C., Edskes H. K. (1995) Yeast 11, 1671–1685 [DOI] [PubMed] [Google Scholar]
- 16.Wickner R. B. (1994) Science 264, 566–569 [DOI] [PubMed] [Google Scholar]
- 17.Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. (1996) EMBO J. 15, 3127–3134 [PMC free article] [PubMed] [Google Scholar]
- 18.Patino M. M., Liu J. J., Glover J. R., Lindquist S. (1996) Science 273, 622–626 [DOI] [PubMed] [Google Scholar]
- 19.Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M. (1995) EMBO J. 14, 4065–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stansfield I., Jones K. M., Kushnirov V. V., Dagkesamanskaya A. R., Poznyakovski A. I., Paushkin S. V., Nierras C. R., Cox B. S., Ter-Avanesyan M. D., Tuite M. F. (1995) EMBO J. 14, 4365–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serio T. R., Lindquist S. L. (1999) Annu. Rev. Cell Dev. Biol. 15, 661–703 [DOI] [PubMed] [Google Scholar]
- 22.Kochneva-Pervukhova N. V., Chechenova M. B., Valouev I. A., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. (2001) Yeast 18, 489–497 [DOI] [PubMed] [Google Scholar]
- 23.Uptain S. M., Sawicki G. J., Caughey B., Lindquist S. (2001) EMBO J. 20, 6236–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., Liebman S. W. (1996) Genetics 144, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan R., Lindquist S. L. (2005) Nature 435, 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toyama B. H., Kelly M. J., Gross J. D., Weissman J. S. (2007) Nature 449, 233–237 [DOI] [PubMed] [Google Scholar]
- 27.Tessier P. M., Lindquist S. (2007) Nature 447, 556–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka M., Chien P., Yonekura K., Weissman J. S. (2005) Cell 121, 49–62 [DOI] [PubMed] [Google Scholar]
- 29.Tanaka M., Collins S. R., Toyama B. H., Weissman J. S. (2006) Nature 442, 585–589 [DOI] [PubMed] [Google Scholar]
- 30.Collins S. R., Douglass A., Vale R. D., Weissman J. S. (2004) PLoS Biol. 2, e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheibel T., Kowal A. S., Bloom J. D., Lindquist S. L. (2001) Curr. Biol. 11, 366–369 [DOI] [PubMed] [Google Scholar]
- 32.Schlumpberger M., Prusiner S. B., Herskowitz I. (2001) Mol. Cell. Biol. 21, 7035–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edskes H. K., McCann L. M., Hebert A. M., Wickner R. B. (2009) Genetics 181, 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sondheimer N., Lindquist S. (2000) Mol. Cell 5, 163–172 [DOI] [PubMed] [Google Scholar]
- 35.Derkatch I. L., Bradley M. E., Hong J. Y., Liebman S. W. (2001) Cell 106, 171–182 [DOI] [PubMed] [Google Scholar]
- 36.Osherovich L. Z., Weissman J. S. (2001) Cell 106, 183–194 [DOI] [PubMed] [Google Scholar]
- 37.Derkatch I. L., Bradley M. E., Zhou P., Chernoff Y. O., Liebman S. W. (1997) Genetics 147, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradley M. E., Edskes H. K., Hong J. Y., Wickner R. B., Liebman S. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, Suppl. 4, 16392–16399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel B. K., Liebman S. W. (2007) J. Mol. Biol. 365, 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bardill J. P., True H. L. (2009) J. Mol. Biol. 388, 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ter-Avanesyan M. D., Kushnirov V. V., Dagkesamanskaya A. R., Didichenko S. A., Chernoff Y. O., Inge-Vechtomov S. G., Smirnov V. N. (1993) Mol. Microbiol. 7, 683–692 [DOI] [PubMed] [Google Scholar]
- 42.Vitrenko Y. A., Pavon M. E., Stone S. I., Liebman S. W. (2007) Curr. Genet 51, 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bardill J. P., Dulle J. E., Fisher J. R., True H. L. (2009) Prion 3, 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka M., Weissman J. S. (2006) Methods Enzymol. 412, 185–200 [DOI] [PubMed] [Google Scholar]
- 45.Harper J. D., Lansbury P. T., Jr. (1997) Annu. Rev. Biochem. 66, 385–407 [DOI] [PubMed] [Google Scholar]
- 46.Perrett S., Jones G. W. (2008) Curr. Opin. Struct Biol. 18, 52–59 [DOI] [PubMed] [Google Scholar]
- 47.Ohhashi Y., Ito K., Toyama B. H., Weissman J. S., Tanaka M. (2010) Nat. Chem. Biol. 6, 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S. (2004) Nature 428, 323–328 [DOI] [PubMed] [Google Scholar]
- 49.Bradley M. E., Liebman S. W. (2003) Genetics 165, 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Derkatch I. L., Bradley M. E., Masse S. V., Zadorsky S. P., Polozkov G. V., Inge-Vechtomov S. G., Liebman S. W. (2000) EMBO J. 19, 1942–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chien P., DePace A. H., Collins S. R., Weissman J. S. (2003) Nature 424, 948–951 [DOI] [PubMed] [Google Scholar]
- 52.Ferreira P. C., Ness F., Edwards S. R., Cox B. S., Tuite M. F. (2001) Mol. Microbiol. 40, 1357–1369 [DOI] [PubMed] [Google Scholar]
- 53.Tuite M. F., Cox B. S. (2003) Nat. Rev. Mol. Cell Biol. 4, 878–890 [DOI] [PubMed] [Google Scholar]
- 54.Resende C. G., Outeiro T. F., Sands L., Lindquist S., Tuite M. F. (2003) Mol. Microbiol. 49, 1005–1017 [DOI] [PubMed] [Google Scholar]
- 55.Nakayashiki T., Kurtzman C. P., Edskes H. K., Wickner R. B. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10575–10580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serio T. R., Cashikar A. G., Kowal A. S., Sawicki G. J., Moslehi J. J., Serpell L., Arnsdorf M. F., Lindquist S. L. (2000) Science 289, 1317–1321 [DOI] [PubMed] [Google Scholar]
- 57.Vitrenko Y. A., Gracheva E. O., Richmond J. E., Liebman S. W. (2007) J. Biol. Chem. 282, 1779–1787 [DOI] [PubMed] [Google Scholar]