Abstract
IL-10 is a potent anti-inflammatory cytokine that is crucial for down-regulating pro-inflammatory genes, which are induced by Toll-like receptor (TLR) signaling. In this study, we have examined whether modulation of microRNAs plays a role in the inhibitory effect of IL-10 on TLR4 signaling. Analyzing microRNAs known to be induced by TLR4, we found that IL-10 could inhibit the expression of miR-155 in response to lipopolysaccharide but had no effect on miR-21 or miR-146a. IL-10 inhibited miR-155 transcription from the BIC gene in a STAT3-dependent manner. This inhibitory effect of IL-10 on miR-155 led to an increase in the expression of the miR-155 target, SHIP1. This is the first example of IL-10 playing a role in microRNA function and suggests that through its inhibitory effect on miR-155, IL-10 has the ability to promote anti-inflammatory gene expression.
Keywords: Immunology, Interleukin, MicroRNA, Signal Transduction, Toll-like Receptors (TLR), IL-10, miR-155
Introduction
IL-10 is a potent anti-inflammatory cytokine that is crucial for dampening the inflammatory response after pathogen invasion and acts to protect the host from excessive inflammation (1). For example, mice deficient in IL-10 have been shown to die from excessive inflammatory responses when exposed to bacterial pathogens (1). In addition, many inflammatory diseases in humans can be associated with poor IL-10 expression such as ulcerative colitis, Crohn disease, and asthma (2, 3). One mechanism whereby IL-10 mediates its anti-inflammatory effect is through the down-regulation of pro-inflammatory genes induced downstream of Toll-like receptor (TLR)2 signaling such as those encoding IL-1, IL-12, tumor necrosis factor α, and IL-6. This is accomplished when IL-10 signals through the JAK1-STAT3 pathway, resulting in the induction of as yet unknown STAT3-responsive genes, which are thought to be responsible for the inhibition of these pro-inflammatory proteins (1, 4). In this study, we set out to examine whether modulation of microRNAs (miRNAs) might play a role in the inhibitory effect of IL-10 on signaling by TLR4, the receptor responsible for sensing the Gram-negative bacterial product, lipopolysaccharide (LPS).
The discovery of miRNAs has revealed an entirely new mechanism of negative regulation within the cell (5–8). miRNAs are small endogenous RNA molecules (∼22 nucleotides) that have the ability to base pair to mRNA sequences from protein-coding genes, leading to partial or full degradation of the mRNA transcript (5–8). With the identification of over 500 miRNAs to date and the prediction that each miRNA may recognize several hundred target sequences, the current challenge is to identify these targets and understand how miRNAs are regulated within the cell. This is particularly important considering the mounting evidence demonstrating their contribution to disease and their roles in cellular mechanisms such as differentiation, metabolism, and immunity (5–8).
A role for miRNAs in the innate immune response was demonstrated when miRNAs such as miR-146a, miR-155, and miR-21 were shown to become induced in response to TLR4 signaling in monocytes (9–11). miR-146a and miR-21 are both induced by LPS, where the former has been shown to target TRAF6 and IRAK1, two upstream signaling components within the TLR4 pathway, whereas miR-21 was shown to negatively regulate programmed cell death 4, a pro-inflammatory protein that promotes NF-κB activation and suppresses IL-10 (9, 11). miR-155 is also induced by LPS, as well as other TLR ligands and pro-inflammatory cytokines (10). Numerous targets have been identified for miR-155 such as c-Maf, Bach1, PU.1, C/ebpβ, and SHIP1; however, their role in TLR signaling has never been extensively explored before (12–17). Mice deficient in miR-155 have defects in B cell differentiation, as well as possessing severe deficiencies in immune responses when exposed to pathogens, thus highlighting the important role miR-155 plays in the immune system as a whole (12, 18, 19).
In this study, we demonstrated that IL-10 could inhibit the expression of miR-155 in response to LPS but had no effect on miR-21 or miR-146a. This inhibition of miR-155 by IL-10 led to an increase in the expression of the miR-155 target, SHIP1. Because SHIP1 has been shown to limit TLR signaling (20), the ability of IL-10 to increase its expression via inhibition of miR-155 provides new insights into the complex signaling mechanism of IL-10. This finding also identifies a novel mechanism of control on miR-155, an miRNA that has been implicated in the innate immune response and cancer progression.
EXPERIMENTAL PROCEDURES
Reagents
LPS from Escherichia coli, Serotype 0111:B4, was from Alexis. Recombinant mouse and human IL-10 were from R&D Biosystems. Precursor-miR-155 (pre-155) oligonucleotide was obtained from Ambion.
Cell Culture
Immortalized bone marrow-derived macrophages (BMDM), a kind gift from Douglas Golenbock (University of Massachusetts), and Raw264.7 cell lines, obtained from the European Cell Culture Collection, were maintained in Dulbecco's modified Eagle's medium. Wild-type and IL-10-deficient bone marrow obtained from Peter Murray (University of Memphis) were isolated from the tibias and femurs of C57/Bl6 mice, and primary BMDM were generated as described previously (11). Human peripheral blood mononuclear cells (hPBMC) were isolated from whole blood using a Ficoll gradient (21). Splenocytes from wild-type and Eμ-miR-155 transgenic mice, obtained from Carlo Croce (Ohio State University), were maintained in RPMI and 50 μm β-mercaptoethanol. In all cases, Dulbecco's modified Eagle's medium and RPMI medium were supplemented with 10% fetal calf serum, 2 mm l-glutamine, 1% penicillin/streptomycin solution (v/v).
RT-PCR
Immortalized BMDM (I-BMDM) and differentiated primary BMDM or hPBMC were set up at 4 × 105 or 1 × 106, respectively, in 24-well plates 1 day prior to stimulation. Cells were stimulated with LPS and/or IL-10 as indicated in the figure legends. Total RNA was extracted using the RNeasy kit (Qiagen), modified to obtain small RNA species. For miRNA analysis, miRNA TaqMan assays for miR-21, miR-146a, miR-155, miR-191, and RNU6B (Applied Biosystems) were used according to the manufacturer's instructions where 5 ng/ml total RNA was used as starting material. For mRNA expression analysis, cDNA was prepared from 20 to 100 ng/ml total RNA using the High-Capacity cDNA archive kit (Applied Biosystems) according to the manufacturer's instructions. mRNA expression was then monitored using SYBR Green-based chemistry (Invitrogen) using the following primers: glyceraldehyde-3-phosphate dehydrogenase, 5′-gaa cgg gaa gct tgt cat caa-3′, forward, 5′-cta agc agt tgg tgg tgc ag-3′, reverse; Pri-mmu-155, 5′-gac aca agg cct gtt act agc ac-3′, forward, 5′-gtc tga cat cta cgt tca tcc agc-3′, reverse; Pre-mmu-155, 5′-gct aat tgt gat agg ggt ttt gg-3′, forward, 5′-gtt aat gct aac agg tag gag tc-3′, reverse; SHIP1, 5′-ggt ggt acg gtt tgg aga ga-3′, forward, 5′-atg ctg agc ctc tgt ggt ct-3′, reverse. miRNA and mRNA expression were measured on the 7900 RT-PCR system (Applied Biosystems), and -fold changes in expression were calculated by the Delta Delta CT method using miR-191 (BMDM/hPBMC) (22) or RNU6B (splenocytes) as an endogenous control for miRNA analysis and glyceraldehyde-3-phosphate dehydrogenase as an endogenous control for mRNA expression. All -fold changes are expressed normalized to non-stimulated control for each cell type.
Enzyme-linked Immunosorbent Assay
Murine IL-10 expression was measured from the supernatants of stimulated cells using an enzyme-linked immunosorbent assay DuoSet kit (R&D Biosystems) according to the manufacturer's instructions.
Luciferase Assays
BIC luciferase plasmid along with the NF-κB, AP1, and Ets1 mutants were a kind gift from Eric Flemington (Tulane University, New Orleans, LA). Raw264.7 cells seeded at 2 × 105/ml in 24-well plates were transfected using 6% GeneJuice with each plasmid and TK-Renilla and 25 nm murine si-control and si-STAT3 (Santa Cruz Biotechnology, sc-29494) for small interfering RNA experiments. Cells were rested for 24–48 h prior to stimulation with LPS or LPS + IL-10 for 18 h. pMir-SHIP1 and SHIP1 mutant 3′-UTR luciferase plasmids (kind gifts from David Baltimore, California Institute of Technology) were co-transfected with TK-Renilla and increasing concentrations of pre-155 oligonucleotide into Raw264.7 or stimulated with LPS or LPS + IL-10 for 8 h. In all cases, cells were lysed in passive lysis buffer before being analyzed for both luciferase and TK-Renilla activity as described previously (23). Data were normalized to TK-Renilla activity and represented as mean ± S.D. for triplicate determinations where -fold changes are expressed normalized to non-stimulated control.
Protein Expression
Differentiated IL-10-deficient BMDM cells seeded at 4 × 105/ml in 6-well plates were stimulated with LPS and/or IL-10 as indicated in the figure legends. Cells were lysed in low stringency lysis buffer complete with protease inhibitors, and protein concentration was determined using the Coomassie Bradford reagent (Pierce). Lysates were resolved on 10% SDS-PAGE gels and transferred onto polyvinylidene difluoride membrane before being immunoblotted with anti-SHIP1 (P1C1, Santa Cruz Biotechnology) or anti-β-actin (AC-15, Sigma). Blots were developed by enhanced chemiluminescence (ECL) (Cell Signaling Technology Inc.).
RESULTS
IL-10 Inhibits miR-155 Expression in Response to TLR4 Stimulation
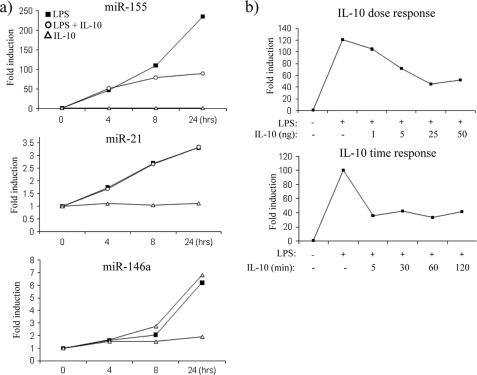
In an effort to determine whether IL-10 could modulate any miRNA downstream of TLR4 signaling, we first decided to investigate the effect of IL-10 on miR-155, miR-146a, and miR-21, miRNAs that are known to be induced downstream of TLR4 (9–11). As shown in Fig. 1a, in I-BMDM, LPS gradually induced the expression of miR-155 (upper graph), miR-21 (middle graph), and miR-146a (lower graph) over time where the expression of each miRNA was greatest at 24 h. The effect of LPS was particularly evident on miR-155, which was induced over 200-fold when compared with non-stimulated control. When cells were treated with LPS in the presence of IL-10, the expression of miR-155 was inhibited, the effect being most evident at 24 h (upper graph). In contrast, IL-10 had no effect on miR-21 or miR-146a (middle and lower graph). This indicated a specific effect for IL-10 on the sole expression of one miRNA, although all three were induced by LPS. IL-10 alone appeared to have no effect on miR-155, miR-21, or miR-146a, suggesting that IL-10 only works to inhibit miR-155 after TLR4 stimulation.
FIGURE 1.
IL-10 inhibits miR-155 expression in response to TLR4 stimulation. a, I-BMDM were stimulated with LPS (100 ng/ml), with LPS + IL-10 (20 ng/ml), or with IL-10 alone for the times indicated. Expression of miR-155, miR-21 and miR-146a was measured by RT-PCR. b, I-BMDM were pretreated for 5 min with increasing doses of IL-10 or pretreated for various times with IL-10 (20 ng/ml) prior to the addition of LPS (100 ng/ml) for 24 h. miR-155 expression was measured by RT-PCR. In both cases, results were normalized and represented as -fold stimulation over the non-stimulated control and are representative of at least three separate experiments.
We next investigated the expression of miR-155 in response to LPS when pretreated with varying concentrations of IL-10 or when pretreated with IL-10 for various times (Fig. 1b). In response to LPS stimulation alone, miR-155 expression was induced 120-fold; however, pretreatment with increasing concentrations of IL-10 gradually decreased miR-155 expression, where the optimal inhibition occurred when IL-10 was used at 25 ng/ml (Fig. 1b, upper graph). miR-155 expression was then analyzed in I-BMDM, which were pretreated with IL-10 for various times prior to the addition of LPS for 24 h (Fig. 1b, lower graph). miR-155 expression was induced 100-fold in response to LPS alone, and interestingly, irrespective of the length of time cells were pretreated with IL-10, miR-155 expression was reduced. For these reasons, cells were pretreated for the minimum time of 5 min with IL-10 at a concentration of 20 ng/ml prior to the addition of LPS for all future experiments unless otherwise indicated.
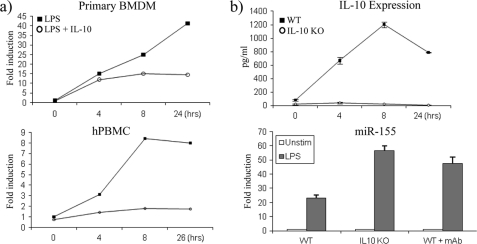
We then set out to examine whether IL-10 had the same effect on miR-155 in primary BMDM and hPBMC (Fig. 2a). LPS induced the expression of miR-155 40-fold in primary BMDM (upper graph) and 8-fold in hPBMC (lower graph) after 24 h of stimulation. In both these cell types, IL-10 inhibited the expression of miR-155 from as early as 4 h, although the effect of IL-10 appeared to occur earlier in hPBMC (lower graph).
FIGURE 2.
IL-10 inhibits miR-155 in primary BMDM and hPBMC. a, primary BMDM and hPBMC were stimulated with LPS (100 ng/ml) or LPS + IL-10 (20 ng/ml) and measured for miR-155 expression by RT-PCR. b, WT and IL-10-deficient BMDM (IL-10 KO) were untreated or pretreated with monoclonal (mAb) IL-10 antibody (5 μg/ml) for 1 h prior to stimulation with LPS (100 ng/ml) for 24 h. Cells were analyzed for IL-10 expression by enzyme-linked immunosorbent assay and for miR-155 expression by RT-PCR. In all cases, graphs are representative of at least three separate experiments. Error bars indicate S.D. Unstim, unstimulated.
We next compared the effect of LPS on miR-155 expression in primary wild-type and IL-10-deficient BMDM. It is well known that in addition to LPS inducing pro-inflammatory cytokines, LPS also induces IL-10 to negatively feed back on the pathway and switch off this pro-inflammatory response. As shown in Fig. 2b, upper graph, wild-type BMDM cells treated with LPS induced IL-10 protein expression, where the maximal amount produced occurred at 8 h. As expected, LPS could not induce IL-10 in IL-10-deficient BMDM (Fig. 2b, upper graph). The fact that LPS cannot induce IL-10 in IL-10-deficient BMDM predicts that miR-155 expression should be higher in these cells. LPS induced the expression of miR-155 ∼25-fold over non-stimulated control in wild-type BMDM, and as predicted, miR-155 expression was greater in IL-10-deficient BMDM where miR-155 expression had more than doubled to ∼55-fold over non-stimulated control (Fig. 2b, lower graph). As a control, a monoclonal IL-10 antibody was used to block the action of endogenous IL-10 in response to LPS in wild-type BMDM. Blocking IL-10 induction in wild-type BMDM relieved the inhibition of miR-155 and demonstrated that its expression could be increased to a level similar to that found in IL-10-deficient BMDM (Fig. 2b, lower graph). This demonstrated that the induction of endogenous IL-10 by LPS in wild-type cells can feed back on the expression of miR-155 by LPS, thus keeping its expression in check.
IL-10 Inhibits the Transcription of miR-155 in a STAT3-dependent Manner
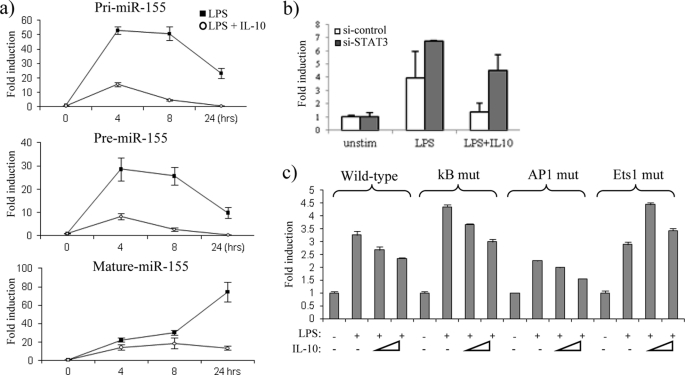
We next addressed whether IL-10 could inhibit transcription of miR-155. miR-155 is transcribed as a primary (pri-) transcript from the third exon of the B cell integration cluster gene referred to as BIC (24), after which it is sequentially processed by the enzymes Drosha and Dicer to form a precursor (pre-) and mature miR-155, respectively. We designed primers for the pri- and pre-miR-155 transcripts and stimulated IL-10-deficient BMDM with LPS alone or LPS in the presence of IL-10 (Fig. 3a). In response to LPS, pri-miR-155 was rapidly induced, where the highest expression was induced 50-fold at 4 h, after which its expression began to decline (Fig. 3a, upper graph), most likely due to the processing of pri-miR-155 into the pre- and mature form. In contrast, in the presence of IL-10, generation of pri-miR-155 was inhibited, where pri-miR-155 expression was reduced to 15-fold at 4 h (Fig. 3a, upper graph). A similar trend was observed when the pre-miR-155 transcript was measured, where its highest expression in response to LPS was also observed at 4 h, and IL-10 inhibited this expression from 27-fold down to 7-fold over non-stimulated control (Fig. 3b, middle graph). It could also be noted that the expression of pre-miR-155 in response to LPS appeared more gradual, indicating that pre-miR-155 generation occurs after pri-miR-155. Mature miR-155 was also measured, demonstrating that IL-10 had the same inhibitory effect on miR-155 in IL-10-deficient BMDM as that observed in I-BMDM, primary wild-type BMDM, and hPBMC (Fig. 3a, lowest graph). The observation that IL-10 inhibits the expression of both the pri-transcripts and the pre-transcripts suggests that IL-10 acts upstream to inhibit the actual transcription of the BIC gene.
FIGURE 3.
IL-10 inhibits the transcription of miR-155. a, IL-10-deficient BMDM were stimulated with LPS (100 ng/ml) or LPS + IL-10 (20 ng/ml). pri-miR-155, pre-miR-155, and mature miR-155 were measured by RT-PCR. b, Raw264.7 cells were co-transfected with wild-type BIC promoter luciferase plasmid and either si-control or si-STAT3. Cells were stimulated with LPS (100 ng/ml) or with LPS + IL-10 (25 ng/ml) for 18 h. Unstim, unstimulated. c, Raw264.7 cells transfected with wild-type BIC, NF-κB mutant (kB mut), AP1 mutant (AP1 mut), or Ets1 mutant (Ets1 mut) promoter luciferase plasmids were stimulated with LPS (100 ng/ml) or with LPS + IL-10 at two different doses (25 and 50 ng/ml). Luciferase activity was measured where results were normalized for TK-Renilla activity and represented as -fold stimulation over the non-stimulated control. In all cases, results are expressed as mean ± S.D. for triplicate determinations and are representative of three separate experiments.
We further analyzed the effect of IL-10 on transcriptional regulation of the BIC gene through the use of a luciferase reporter plasmid containing 1200 bp of the BIC promoter region (25). In addition, we investigated whether any of the IL-10 effects were mediated through STAT3 by using small interfering RNA (siRNA) targeted against STAT3. In the presence of control siRNA, LPS induced luciferase expression 3-fold from the BIC promoter, and this expression was returned to near basal levels in the presence of IL-10 (Fig. 3b), validating that IL-10 appears to inhibit miR-155 at the transcriptional level. In the presence of si-STAT3, IL-10 could no longer suppress luciferase expression, demonstrating that the effect of IL-10 on miR-155 transcription is dependent on STAT3.
The Ets1 Binding Site Is Required for the IL-10-mediated Suppression of the BIC Gene
To investigate how IL-10 acts to suppress BIC transcription, we analyzed the effect of IL-10 when consensus transcription factor binding sites for NF-κB, AP1, and Ets1 found within the BIC promoter were mutated. In response to LPS, luciferase expression was induced 3-fold from the wild-type BIC promoter. A similar effect was observed when the NF-κB motif and Ets1 site were mutated (Fig. 3c). Luciferase activity was, however reduced in a promoter with a mutated AP-1 motif, suggesting that this transcription factor plays a role in the LPS induction of miR-155 (Fig. 3c), supporting previous studies, which show that AP1 is required for BIC gene induction in response to B cell stimulation (25).
In concordance with our previous results, IL-10 inhibited luciferase activity in the wild-type BIC promoter (Fig. 3c, bars 3 and 4). IL-10 was also able to reduce luciferase activity in both the NF-κB and the AP1 promoter mutants. In contrast, IL-10 could no longer inhibit luciferase activity when the Ets1 site was mutated, suggesting that this site is important in mediating the IL-10-driven suppression of miR-155.
IL-10 Increases the Expression of the miR-155 Target, SHIP1
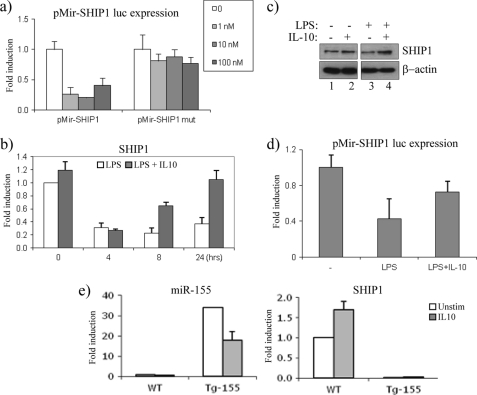
We next wanted to investigate a functional outcome for the IL-10-mediated suppression of miR-155. We therefore set out to examine the effects of LPS and IL-10 on the miR-155 target, SHIP1 (15, 16). We first verified that the 3′-UTR of SHIP1 is targeted by miR-155. The 3′-UTR for SHIP1 was cloned into the pMir luciferase reporter system. Under basal conditions, luciferase expression was present (Fig. 4a, white bars). However, in the presence of increasing concentrations of pre-miR-155 (1, 10, and 100 nm), luciferase expression was lost. In contrast, increasing concentrations of pre-miR-155 had no effect when the miR-155 seed sequence within the 3′-UTR of SHIP1 was mutated.
FIGURE 4.
IL-10 increases the expression of the miR-155 target, SHIP1. a, pMir-SHIP1 and pMir-SHIP1 mutant 3′-UTR luciferase plasmids were co-transfected with increasing amounts of pre-155 (1, 10, and 100 nm) in Raw264.7 cells. Luciferase activity was measured, and results were normalized for TK-Renilla activity. b, IL-10-deficient BMDM were stimulated with LPS (100 ng/ml, white bars) or LPS + IL-10 (20 ng/ml, gray bars) for the times indicated. mRNA expression for SHIP1 was measured by RT-PCR. c, IL-10-deficient BMDM were stimulated with IL-10 (20 ng/ml), LPS (100 ng/ml), or LPS + IL-10 for 24 h. SHIP1 and β-actin protein expression were measured. d, pMir-SHIP1 3′-UTR luciferase activity was measured in Raw264.7 cells after stimulation with LPS (100 ng/ml) or LPS + IL-10 (20 ng/ml) for 8 h. Luciferase activity was measured, and results were normalized for TK-Renilla activity. e, WT and Tg miR-155 splenocytes were stimulated with IL-10 (50 ng/ml) for 16 h. miR-155 and SHIP1 expression was measured by RT-PCR. In all cases, results were represented as -fold stimulation over non-stimulated control and expressed as mean ± S.D. for triplicate determinations where each experiment is representative of three separate experiments. Unstim, unstimulated.
The fact that LPS potently induces mature miR-155 suggests that as miR-155 expression increases, its ability to bind to the 3′-UTR of SHIP1 should result in a decrease of SHIP1 mRNA expression. As shown in Fig. 4b, the expression of SHIP1 decreased over time upon LPS stimulation, reciprocal to the increase in miR-155 expression. However, in the presence of IL-10, when miR-155 expression is inhibited, the expression of SHIP1 increased (Fig. 4b), particularly at 8 and 24 h, when the effect of IL-10 on miR-155 suppression was greatest (Fig. 1a). This effect of LPS and IL-10 was also apparent on SHIP1 protein expression, where in response to LPS alone, SHIP1 expression was reduced (Fig. 4c, lane 3), whereas in the presence of IL-10, SHIP1 protein expression increased (Fig. 4c, lane 4).
We next wanted to examine whether the increase in SHIP1 expression in response to IL-10 is dependent on miR-155. We first used the pMir-SHIP1 luciferase reporter system, where luciferase expression will only decrease in the presence of miR-155 (Fig. 4a). As LPS can potently drive miR-155, this would suggest that LPS has also the ability to decrease luciferase expression. As shown in Fig. 4d, LPS decreased SHIP1 luciferase activity, whereas in the presence of IL-10, this inhibition was relieved, implicating that the suppression of miR-155 by IL-10 is mediating this effect.
This was further demonstrated by using transgenic (Tg) splenocytes, which overexpress miR-155 under the control of the Eμ-B cell promoter (26). We postulated that in cells where miR-155 is overexpressed, IL-10 would no longer be able to maintain its effect on miR-155 targets. As shown in Fig. 4e, left graph, miR-155 was overexpressed 35-fold in Tg splenocytes when compared with wild-type (WT) cells. IL-10 was able to decrease this expression somewhat, although substantial levels of miR-155 still remained in Tg cells. As expected, IL-10 could increase the expression of SHIP1 in WT cells, whereas in miR-155 Tg cells, SHIP1 expression was no longer detected, decreasing well below basal levels, and as predicted, IL-10 could not rescue this effect (Fig. 4e, right graph).
DISCUSSION
This study highlights a novel mechanism of miR-155 regulation. We showed that IL-10 can inhibit the expression of miR-155, a miRNA induced downstream of TLR4 signaling, but had no effect on miR-21 and miR-146a, two other miRNAs also induced by TLR4. This not only demonstrated a very specific effect for IL-10 but was also the very first example of IL-10 playing a role in miRNA function. miR-155 expression was doubled in response to LPS in IL-10-deficient cells, demonstrating that endogenous IL-10 can feed back on the system to keep miR-155 expression in check, highlighting an additional mechanism of IL-10 control on the pro-inflammatory response.
miR-155 was the first oncogenic miRNA to be discovered. It has been shown to be highly expressed in several types of B cell lymphoma, in particular Hodgkin lymphoma and diffuse large B cell lymphoma (27, 28). In addition, transgenic mice overexpressing miR-155 succumb to B cell malignancies (26). miR-155 was also found overexpressed in patients with acute myeloid leukemia and rheumatoid arthritis (14, 29). Together, this information demonstrates how miR-155 may provide a potential link between inflammatory diseases and cancer. It is therefore essential that miR-155 is tightly regulated in the cell, and we propose that IL-10 is a likely candidate for this regulation.
We sought to investigate how IL-10 inhibits miR-155 expression. Measuring pri-mir-155 and pre-miR-155 expression, we demonstrated that IL-10 could potently inhibit generation of both transcripts, illustrating that IL-10 acts to inhibit miR-155 upstream of primary transcript generation. In addition, IL-10 reduced BIC promoter activity in a STAT3-dependent manner, indicating that the IL-10 suppression of the BIC gene is mediated through the canonical IL-10-STAT3 signaling pathway.
In an effort to further investigate the effect of IL-10 on miR-155 transcription, we analyzed the effect of IL-10 when various transcription factor sites within the BIC promoter were mutated. IL-10 was able to suppress wild-type BIC promoter luciferase expression as well as suppressing luciferase activity when the NF-κB and AP1 sites were mutated. In contrast, IL-10 could no longer suppress luciferase activity when the Ets1 site was mutated, demonstrating that the Ets1 site is required for mediating IL-10 suppression. To date, over 30 Ets family members exist that each have the potential to bind to the canonical Ets consensus sequence found in the BIC promoter (30). Etv3, an Ets family transcriptional repressor, was recently identified as a novel IL-10-induced gene (31). It is possible that the IL-10 suppression of miR-155 could involve recruitment of Etv3, and we are currently investigating this mechanism.
We also explored the functional outcomes for IL-10 suppression of miR-155. Taking SHIP1 as a well characterized miR-155 target, we were able to illustrate that overexpressing miR-155 decreased the expression of a SHIP1 3′-UTR reporter. We went on to show that IL-10 could rescue the LPS-driven down-regulation of SHIP1 at the mRNA and protein level, and through the use of the SHIP1 3′-UTR reporter and miR-155 Tg splenocytes, we demonstrated that this effect was mediated by miR-155.
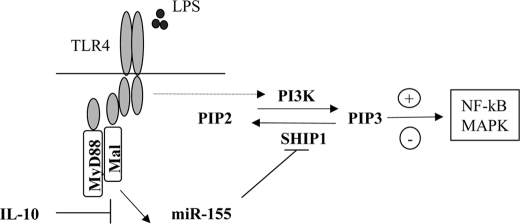
SHIP1 is an inositol phosphatase that is known to convert the signaling molecule PIP3 back to PIP2, whereas PI3K is responsible for the opposite reaction. TLR signaling can promote the pro-inflammatory response through the activation of PI3K, resulting in generation of PIP3 and activation of MAPK and NF-κB (20). The induction of miR-155 by LPS supports this model, whereby expression of miR-155 can target SHIP1, decreasing its expression and promoting the conversion of PIP2 to PIP3 by PI3K. In this setting, IL-10 increases SHIP1, and in this way, it acts to switch the pro-inflammatory response off by decreasing the levels of PIP3 (Fig. 5).
FIGURE 5.
Schematic illustrating a possible role for miR-155 in TLR4 and SHIP1 signaling. In response to LPS, miR-155 expression is induced, resulting in a decrease in SHIP1 expression, thus allowing PI3K activation of NF-κB and MAPK to proceed and promote the pro-inflammatory response. However, in the presence of IL-10, miR-155 expression is inhibited, allowing SHIP1 expression to recover and promote the conversion of PIP3 back to its inactive PIP2 state, switching off the pro-inflammatory response. Mal, MyD88-like adapter protein.
This study therefore sheds light on a novel role for IL-10 in miR-155 regulation. miR-155 has been shown to be directly involved in the regulation of more than 30 innate immune genes (32). Taking SHIP1 as an example, our data suggest that IL-10 could impact on these genes via its inhibitory effect on miR-155. It may therefore be possible that through the inhibition of miR-155, we may elicit some of the properties mediated by IL-10. With increasing studies performed on how to inhibit or increase miRNAs for therapeutic use in vivo (33), our study could provide new approaches in the effort to develop anti-inflammatory therapeutics.
Acknowledgments
We are grateful to Carlo Croce and Stefan Costinean for providing us with transgenic splenocytes as well as Nigel Stevenson for advice and reagents regarding STAT3. We also acknowledge the Monash Institute of Medical Research, in particular Bryan Williams, Michael Gantier, Tony Sadler, and Dakang Xu, for facilitating the revision of this manuscript.
This work was supported by grants from the Science Foundation Ireland, Health Research Board Ireland, and Marie Curie Actions.
- TLR
- Toll-like receptor
- miRNA
- microRNA
- pri
- primary
- pre
- precursor
- miR
- miRNA
- pMir
- plasmid-Mir REPORT
- LPS
- lipopolysaccharide
- BMDM
- bone marrow-derived macrophages
- I-BMDM
- immortalized BMDM
- hPBMC
- human peripheral blood mononuclear cells
- RT-PCR
- real-time PCR
- si
- small interfering
- UTR
- untranslated region
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PIP3
- phosphatidylinositol 3,4,5-trisphosphate
- PI3k
- phosphatidylinositol 3-kinase
- MAPK
- mitogen-activated protein kinase
- Tg
- transgenic.
REFERENCES
- 1.Murray P. J. (2006) Curr. Opin. Pharmacol. 6, 379–386 [DOI] [PubMed] [Google Scholar]
- 2.O'Garra A., Barrat F. J., Castro A. G., Vicari A., Hawrylowicz C. (2008) Immunol. Rev. 223, 114–131 [DOI] [PubMed] [Google Scholar]
- 3.Asadullah K., Sterry W., Volk H. D. (2003) Pharmacol. Rev. 55, 241–269 [DOI] [PubMed] [Google Scholar]
- 4.Murray P. J. (2006) Biochem. Soc. Trans. 34, 1028–1031 [DOI] [PubMed] [Google Scholar]
- 5.Taganov K. D., Boldin M. P., Baltimore D. (2007) Immunity 26, 133–137 [DOI] [PubMed] [Google Scholar]
- 6.Gantier M. P., Sadler A. J., Williams B. R. (2007) Immunol. Cell Biol. 85, 458–462 [DOI] [PubMed] [Google Scholar]
- 7.Lindsay M. A. (2008) Trends Immunol. 29, 343–351 [DOI] [PubMed] [Google Scholar]
- 8.Kanellopoulou C., Monticelli S. (2008) Semin. Cancer Biol. 18, 79–88 [DOI] [PubMed] [Google Scholar]
- 9.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheedy F. J., Palsson-McDermott E., Hennessy E. J., Martin C., O'Leary J. J., Ruan Q., Johnson D. S., Chen Y., O'Neill L. A. (2010) Nat. Immunol. 11, 141–147 [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez A., Vigorito E., Clare S., Warren M. V., Couttet P., Soond D. R., van Dongen S., Grocock R. J., Das P. P., Miska E. A., Vetrie D., Okkenhaug K., Enright A. J., Dougan G., Turner M., Bradley A. (2007) Science 316, 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin Q., McBride J., Fewell C., Lacey M., Wang X., Lin Z., Cameron J., Flemington E. K. (2008) J. Virol. 82, 5295–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Connell R. M., Rao D. S., Chaudhuri A. A., Boldin M. P., Taganov K. D., Nicoll J., Paquette R. L., Baltimore D. (2008) J. Exp. Med. 205, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connell R. M., Chaudhuri A. A., Rao D. S., Baltimore D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7113–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedersen I. M., Otero D., Kao E., Miletic A. V., Hother C., Ralfkiaer E., Rickert R. C., Gronbaek K., David M. (2009) EMBO. Mol. Med. 1, 288–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costinean S., Sandhu S. K., Pedersen I. M., Tili E., Trotta R., Perrotti D., Ciarlariello D., Neviani P., Harb J., Kauffman L. R., Shidham A., Croce C. M. (2009) Blood 114, 1374–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thai T. H., Calado D. P., Casola S., Ansel K. M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J. L., Schmidt-Supprian M., Rajewsky N., Yancopoulos G., Rao A., Rajewsky K. (2007) Science 316, 604–608 [DOI] [PubMed] [Google Scholar]
- 19.Faraoni I., Antonetti F. R., Cardone J., Bonmassar E. (2009) Biochim. Biophys. Acta 1792, 497–505 [DOI] [PubMed] [Google Scholar]
- 20.An H., Xu H., Zhang M., Zhou J., Feng T., Qian C., Qi R., Cao X. (2005) Blood 105, 4685–4692 [DOI] [PubMed] [Google Scholar]
- 21.Palsson-McDermott E. M., Doyle S. L., McGettrick A. F., Hardy M., Husebye H., Banahan K., Gong M., Golenbock D., Espevik T., O'Neill L. A. (2009) Nat. Immunol. 10, 579–586 [DOI] [PubMed] [Google Scholar]
- 22.Peltier H. J., Latham G. J. (2008) RNA 14, 844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCoy C. E., Carpenter S., Pålsson-McDermott E. M., Gearing L. J., O'Neill L. A. (2008) J. Biol. Chem. 283, 14277–14285 [DOI] [PubMed] [Google Scholar]
- 24.Eis P. S., Tam W., Sun L., Chadburn A., Li Z., Gomez M. F., Lund E., Dahlberg J. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin Q., Wang X., McBride J., Fewell C., Flemington E. (2008) J. Biol. Chem. 283, 2654–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., Croce C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7024–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kluiver J., van den Berg A., de Jong D., Blokzijl T., Harms G., Bouwman E., Jacobs S., Poppema S., Kroesen B. J. (2007) Oncogene 26, 3769–3776 [DOI] [PubMed] [Google Scholar]
- 28.Rai D., Karanti S., Jung I., Dahia P. L., Aguiar R. C. (2008) Cancer Genet. Cytogenet. 181, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanczyk J., Pedrioli D. M., Brentano F., Sanchez-Pernaute O., Kolling C., Gay R. E., Detmar M., Gay S., Kyburz D. (2008) Arthritis Rheum. 58, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 30.Oikawa T., Yamada T. (2003) Gene 303, 11–34 [DOI] [PubMed] [Google Scholar]
- 31.El Kasmi. K. C., Smith A. M., Williams L., Neale G., Panopolous A., Watowich S. S., Häcker H., Foxwell B. M., Murray P. J. (2007) J. Immunol. 179, 7215–7219 [DOI] [PubMed] [Google Scholar]
- 32.Gantier M. P. (2010) J. Interferon. Cytokine Res. 30, 283–289 [DOI] [PubMed] [Google Scholar]
- 33.Liu Z., Sall A., Yang D. (2008) Int. J. Mol. Sci. 9, 978–999 [DOI] [PMC free article] [PubMed] [Google Scholar]