Abstract
The Trypanosoma brucei genome has four highly similar genes encoding sphingolipid synthases (TbSLS1–4). TbSLSs are polytopic membrane proteins that are essential for viability of the pathogenic bloodstream stage of this human protozoan parasite and, consequently, can be considered as potential drug targets. TbSLS4 was shown previously to be a bifunctional sphingomyelin/ethanolamine phosphorylceramide synthase, whereas functions of the others were not characterized. Using a recently described liposome-supplemented cell-free synthesis system, which eliminates complications from background cellular activities, we now unambiguously define the enzymatic specificity of the entire gene family. TbSLS1 produces inositol phosphorylceramide, TbSLS2 produces ethanolamine phosphorylceramide, and TbSLS3 is bifunctional, like TbSLS4. These findings indicate that TbSLS1 is uniquely responsible for synthesis of inositol phosphorylceramide in insect stage parasites, in agreement with published expression array data (17). This approach also revealed that the Trypanosoma cruzi ortholog (TcSLS1) is a dedicated inositol phosphorylceramide synthase. The cell-free synthesis system allowed rapid optimization of the reaction conditions for these enzymes and site-specific mutagenesis to alter end product specificity. A single residue at position 252 (TbSLS1, Ser252; TbSLS3, Phe252) strongly influences enzymatic specificity. We also have used this system to demonstrate that aureobasidin A, a potent inhibitor of fungal inositol phosphorylceramide synthases, does not significantly affect any of the TbSLS activities, consistent with the phylogenetic distance of these two clades of sphingolipid synthases. These results represent the first application of cell-free synthesis for the rapid preparation and functional annotation of integral membrane proteins and thus illustrate its utility in studying otherwise intractable enzyme systems.
Keywords: Lipid Synthesis, Membrane Enzymes, Parasitology, Sphingolipid, Trypanosome
Introduction
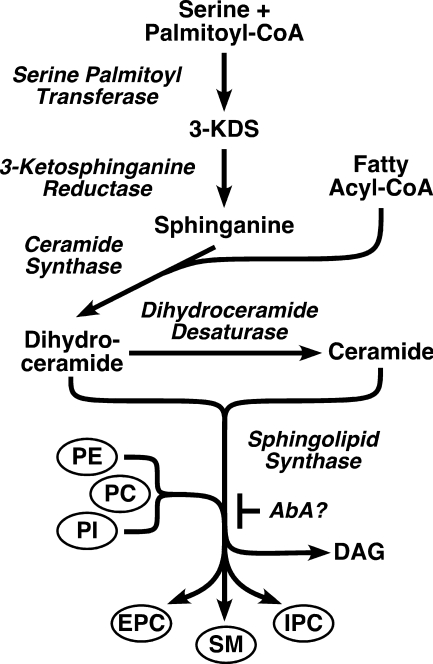
Sphingolipids (SLs)5 are ubiquitous in eukaryotic membranes, where they perform multiple functions. Besides acting as structural components of biological membranes, they participate in protein sorting and cell signaling via membrane rafts (1) and serve as critical apoptotic and anti-apoptotic second messengers. As such, sphingolipids have important influence in human disease (2). SLs also act as precursors of other essential lipid biosynthetic pathways, e.g. generation of ethanolamine phosphate for use in the Kennedy pathway (3) and subsequent synthesis of phospholipids and glycosylphosphatidyl inositols. The common SL synthesis pathway (Fig. 1) begins with the condensation of serine and palmitate by serine palmitoyl transferase to make the sphingoid base 3-ketodihydrosphinganine, which is then reduced to make sphinganine. Sphinganine is the acceptor for N-fatty acylation by ceramide synthase to make dihydroceramide, which is then desaturated to make ceramide. Ceramide synthase also utilizes sphingosine generated by ceramide turnover. Specific sphingolipid synthases (SLS) then transfer the polar head groups from phosphoglycerolipid donors to generate the phosphosphingolipids: sphingomyelin (SM), ethanolamine phosphorylceramide (EPC), and inositol phosphorylceramide (IPC).
FIGURE 1.
Sphingolipid synthesis pathway. 3-KDS, 3-keto-dihydrosphingosine; PL, phospholipid; DAG, diacylglycerol. Homologs of all indicated enzymes (italics) are found in the T. brucei and L. major genomes: serine palmitoyl transferase, Tb927.4.1020 (subunit 1) and Tb10.70.3220 (subunit 2); 3-ketosphinganine reductase, Tb927.10.4040; ceramide synthase, Tb927.8.7730; dihydroceramide desaturase, Tb927.6.3000; sphingolipid synthases, Tb09.211.1000-1030. The proposed site of action of aureobasidin A (AbA?) is indicated.
The major phosphosphingolipid in mammals is SM, although EPC is present, whereas in fungi, the major phosphosphingolipid is IPC. Among the parasitic trypanosomatid protozoa, Leishmania synthesizes IPC exclusively (4, 5). The South American trypanosome Trypanosoma cruzi synthesizes IPC (6–8) but has also been reported to contain SM (9, 10). The African trypanosome Trypanosoma brucei synthesizes SM, EPC, and IPC (11). Interestingly, SL synthesis is developmentally regulated in that IPC is only synthesized in the procyclic insect stage (PCF), and EPC synthesis is restricted to the mammalian bloodstream form (BSF), whereas SM synthesis is constitutive throughout the life cycle (11).
SL synthesis in mammals is carried out by a family of related sphingomyelin synthases (SMSs) with distinct specificity and function (12–15). SMS1 is responsible for de novo synthesis of SM in the Golgi, SMS2 regenerates SM from ceramide derived from turnover in the plasma membrane, and SMSr synthesizes EPC in the ER. The trypanosomatids have an orthologous family of SLSs. In Leishmania major, the single SLS present has been shown to be an IPC synthase (LmjIPCS) (11, 16). T. brucei has a family of four paralogs (TbSLS1–4), one of which (TbSLS4) has been shown by transgene expression in L. major to be a bifunctional SM/EPC synthase (11). The activities of TbSLS1–3 are unknown, although TbSLS1 mRNA levels are specifically up-regulated in PCF trypanosomes, suggesting that this enzyme could be the agent of stage-specific IPC synthesis (17). T. cruzi has a single SLS locus with minor polymorphisms in the two allelic genes. All of these SLS enzymes are modeled to have a six-transmembrane topology with putative catalytic residues located on the lumenal side of the organellar membrane (11, 12). Furthermore, simple reciprocal homology searches (BlastP) are consistently positive between the mammalian and trypanosomatid orthologs but fail to detect any of the fungal IPC synthases, suggesting a more distant relationship for this clade of SLS enzymes.
In this work, we use a cell-free synthesis system designed for in vitro expression of active polytopic membrane proteins (18) to define the enzymatic specificity of the trypanosomatid SLS enzymes. Our results reveal unexpected diversity in the biosynthetic specificities of these enzymes and explain the basis for stage-specific content of sphingolipids in African trypanosomes. Additional data are presented raising doubt about the utility of aureobasidin A, a potent inhibitor of fungal IPC synthases, as a chemotherapeutic agent targeting trypanosomatid SLSs. Finally, this work illustrates and validates the utility of this novel expression system for structure-function investigation of complex polytopic membrane proteins.
EXPERIMENTAL PROCEDURES
In Vitro Expression Constructs
Complete SLS open reading frames (ORFs) were amplified by PCR using specific primers containing 5′ SgfI and 3′ PmeI sites for cloning into the 5′ SgfI and 3′ EcoICRI sites of the pEU-C-His FlexiVector plasmid (19, 20). Pfu high fidelity DNA polymerase (Agilent Technologies, Stratagene, San Diego, CA) was used for all cloning reactions. The plasmid contains (in order, 5′–3′: Sp6 RNA polymerase promoter, 5′ internal ribosome entry site, restriction sites for insertion of cloned ORFs, in-frame C-terminal His6 tag, 3′ homology region for cloning efficiency, and 3′-UTR for translational efficiency). Specific trypanosomatid SLS ORFs (TryTrp database accession numbers) were: TbSLS1, nt 1–1011, Tb09.211.1030; TbSLS2, nt 1–969, Tb09.211.1020; TbSLS3, nt 1–987, Tb09.211.1010; TbSLS4, 1–1041, Tb09.211.1000; LmjIPCS, nt 1–1014, LmjF35.4990; and TcSLS1.1, nt 1–1005, Tc00.1047053506885.124. The start codons for the T. brucei paralogs were as defined by reverse transcription PCR in Ref. 11. The numbering of TcSLS alleles is according to (16). T. brucei SLS genes were amplified from Lister 427 strain genomic DNA, T. cruzi SLS (TcSLS1.1) was amplified from CL-Brenner strain genomic DNA, and L. major IPCS was amplified from a previous PCR clone of the FV39-clone 5 gene (11). T. cruzi SLS has a second allelic copy (TcSLS1.2, Tc00.1047053510729.290) with minor polymorphisms relative to TcSLS1.1; this gene product was not characterized. All PCR products were verified by sequencing using primers flanking the FlexiVector insertion site. The deduced amino acid sequences are compared with the homologous ORFs from their respective annotated genome strains in supplemental Fig. S1. Each TbSLS product was cloned and sequenced from at least two independent PCR reactions, and the deduced amino acid sequences reported here for TbSLS1, TbSLS3, and TbSLS4 were identical for each independent clone. Two distinct but highly related PCR products were consistently obtained from TbSLS2 reactions (data not shown). These are likely to be polymorphic alleles. TbSLS2.1 was expressed in cell-free translation but was inactive in catalytic assays (data not shown) and is not considered further in this work. TbSLS2.2 was active; its sequence is reported in supplemental Fig. S1 and is characterized herein. TbSLS2.2 is referred to as TbSLS2 throughout the remainder of text. Overall, these analyses reveal that the Lister 427 strain TbSLS sequences display minor polymorphisms relative to their homologs from the TREU 927 genome strain.
Site-specific Mutagenesis
Mutagenesis was performed using the PCR-based QuikChange® Multi Site-directed Mutagenesis kit (Stratagene). The cloned TbSLS1 and TbSLS3 genes in the FlexiVector and were used as templates. TbSLS1 was altered by an S252F mutation, and TbSLS3 was altered by the equivalent F252S mutation. Mutagenic primer sequences are presented in supplemental Table S1. Mutated DNA was heat-shocked into XL10-Ultracompetent cells (Stratagene), and mutant TbSLS sequences were confirmed by plasmid sequencing.
Preparation of Liposomes
Liposomes were prepared as described previously (18, 21) using an Avanti mini-extruder and soybean lecithin (Soy PC, 20%, category no. 541601G, Avanti Polar Lipids, Alabaster, AL). These liposomes contain soy phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI) in an approximate molar ratio of 2:2:1. To make liposomes of defined phospholipid composition, PC (16:0/18:2, Avanti product no. 850458P), PE (16:0/18:2, Avanti product no. 850756P), and PI (16:0/18:2, Avanti product no. 840044P) were mixed in equimolar ratios and processed as above.
In Vitro Transcription and Cell-free Synthesis
In vitro transcription of cloned SLS ORFs with bacteriophage Sp6 RNA polymerase (Promega, Madison, WI) was carried out as described previously (18, 21). Standard cell-free translation (18, 21) was carried out in 50-μl reactions in 12 MWCO dialysis cups (Cosmo Bio, Tokyo, Japan) with wheat germ extract (Cell Free Sciences, Yokohama, Japan) and 60 μg of liposomes. Reactions were initiated with mRNA from a transcription reaction programmed with 4 μg of plasmid DNA and allowed to proceed overnight at 26 °C. Proteoliposomes (70 μl) were recovered from two pooled translation reactions by floatation, and a sample (5 μl) was reserved for estimation of translation product by SDS-PAGE and Coomassie Blue staining relative to known amounts of creatine kinase. Typical recovery in floated proteoliposomes was 18–28 μg of specific translation product. Total phosphate analysis of Folch extracted lipids by the phosphorus ash method (22) indicates a typical total phospholipid content of 2.1 mm in the final recovered proteoliposomes. Proteoliposomes were stored at −80 °C until use. Pilot experiments indicated that SLS activities in proteoliposomes were stable through four cycles of freeze-thaw (−80 °C, room temperature).
Liposome Sphingolipid Synthesis Assay
Standard reaction buffer was MES-buffered saline (MBS: 50 mm MES, pH 6.5, 50 mm NaCl, 5 mm KCl, 1 mm EDTA). For pH titrations, HEPES was substituted for MES in some cases (HBS). Reactions contained proteoliposomes (typically 25–40 μl containing ∼10 μg of translation product) in a 100-μl volume and were initiated by addition of 6-((N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoyl)sphingosine (NBD C6-ceramide, Invitrogen, Molecular Probes) to final concentrations ranging from 0.1–5 μm as indicated in the text. Reactions were terminated by addition of 200 μl distilled H2O, followed by 2.0 ml of chloroform:methanol (1:1 v/v). Samples were mixed, insoluble material was removed by centrifugation, and the organic extract was dried under N2. Dried lipids were subjected to n-butanol/water partitioning, and lipids recovered from the butanol phase were fractionated by thin layer chromatography in chloroform:methanol:acetic acid:water (25:15:4:2, v/v/v) on TLC Silica Gel 60 plates (Merck, Gibbsburg, NJ). Fluorescent lipids were visualized on a Storm 860 imaging system with ImageQuant software. NBD-C6-SM (Avanti Polar Lipids) and NBD-C6-IPC (preparation described in Ref. 11) were run as mobility standards. The identity of NBD-C6-EPC products were previously inferred by relative mobility (11).
Maintenance and Generation of Transgenic Trypanosomes
Standard propagation of BSF Lister 427 strain Trypanosoma brucei brucei is described elsewhere (11). The TbSLS1 ORF was PCR-amplified from genomic DNA with primers designed to introduce an in-frame C-terminal HA epitope tag (YPYDVPDYA), and flanking 5′ HindIII and 3′ XmaI sites for insertion into the multicloning region of our constitutive pXS5 expression vector (23). The construct was linearized with XhoI, electroporated into BSF trypanosomes, and clonal neor cell lines derived as described recently (24). Likewise, metabolic radiolabeling and immunoprecipitation have been described previously.
Preparation of Trypanosome Membranes
Semi-intact trypanosome membranes were prepared by a modification of Masterson et al. (25). Cultured BSF trypanosomes constitutively expressing TbSLS1HA were harvested at late log phase (1–2 × 106/ml) and washed in HBS, pH 7.4, containing 70 mm d-glucose. Cells were pelleted and resuspended (1.1 × 108/ml) on ice in distilled H2O containing 0.1 mm tosyl-lysyl-choromethylketone and 2 μg/ml each leupeptin, antipain, pepstatin, and chymostatin. After 5 min, 0.1 volume of 10× MBS was added, and the membranes were washed twice in MBS. The semi-intact membranes were resuspended in MBS at 108 cell equivalents/ml and stored at −80 °C until use (not longer than 3 days). NBD-SL synthesis was assayed exactly as described above for liposomes using 107 cell equivalents of membranes per reaction.
Preparation of Perforated Yeast Spheroplasts
A modified protocol of Rexach et al. (26) was followed. The GPY60 Saccharomyces cerevisiae strain (MATαleu2-3, 112 ura3-52 his4-579trp1-289 prb1 gal2pep4::URA3) was grown to early log phase (2–4 A600/ml) at 30 °C in YP medium. An equivalent of 1,500 A600 cells (1 A unit is defined as 107 cells) were pelleted (3,000 rpm for 5 min) and resuspended to 50 A600/ml in 0.1 mm Tris, pH 9.4, 10 mm dithiothreitol (5 min, room temperature). Cells were centrifuged (3,000 rpm for 2 min at room temperature) and resuspended to 50 A600/ml in spheroplasting medium (0.75× YP, 10 mm Tris, pH 7.5, 0.7 m sorbitol, 0.5% glucose) with yatalase (2 mg/ml, 10 lytic units/mg, Takara Bio, Inc., Otsu, Japan) and lyticase (3 mg/ml, 2000 units/mg, Sigma Aldrich). Cells were incubated at 30 °C, and samples were periodically diluted 1:100 in H2O for A600 determination. When measured A600 was <30% of the initial value, spheroplasts were harvested (3,000 rpm, 2 min, room temperature) and resuspended (50 A600/ml) in regeneration medium (0.75× YP, 0.7 m sorbitol, 1% glucose). After 30 min at 30 °C, spheroplasts were again harvested (3,000 rpm, 5 min, 4 °C) and resuspended in lysis buffer (20 mm HEPES, pH 6.8, 400 mm sorbitol, 150 mm KOAc, 2 mm MgOAc, 0.5 mm EGTA) and pelleted (5,000 rpm, 5 min, 4 °C). This step was repeated. Perforated spheroplasts were washed and resuspended to 300 A600 in MBS, aliquoted, and frozen over liquid nitrogen vapors for 45 min. Perforated spheroplasts were then stored at −80 °C.
Data Analyses
Statistical analyses and curve fitting were performed in Prizm4 (GraphPad Software, La Jolla, CA).
RESULTS
Production of Active TbSLS4 in a Wheat Germ Cell-free System
We previously characterized TbSLS4 as a bifunctional SM/EPC synthase by the laborious process of generating a transgenic L. major promastigote cell line constitutively expressing the TbSLS4 gene (11). Analyses by ESI mass spectrometry and by in vivo incorporation of NBD-ceramide substrate unequivocally demonstrated the presence and synthesis, respectively, of both SM and EPC, which are absent in wild type L. major (27). To continue our characterization of the TbSLS gene family and the orthologous trypanosomatid SLSs in a more expeditious manner, we have exploited a recently developed system for in vitro synthesis of active polytopic membrane proteins (18, 21). In this system, mRNA for the gene of interest is generated by in vitro transcription and is in turn used to program cell-free translation in a wheat germ extract. The translation reaction is supplemented with unilamellar liposomes prepared from soy phospholipids composed predominantly of PC, PE, and PI (∼2:2:1 ratio). The liposomes presumably capture and stabilize nascent transmembrane domains as they emerge from the ribosome thereby facilitating protein folding within the membrane bilayers. Although the precise biophysics and biochemistry of this process are not fully understood, proper folding can be monitored by enzymatic assay (18, 21).
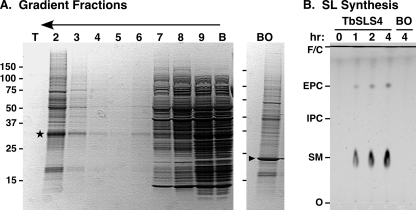
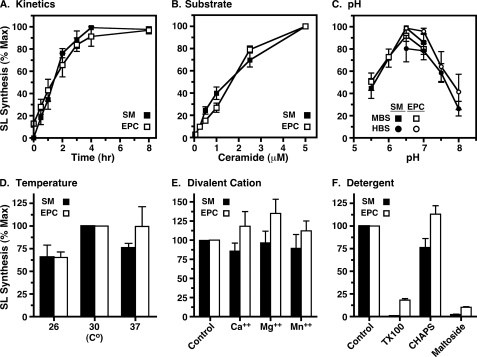
To validate this system for assaying TbSLS function, we programmed cell-free synthesis with TbSLS4 mRNA and recovered proteoliposomes by floatation (Fig. 2A). A band of expected size (Fig. 2A, star) was recovered. This band is not observed in cell-free reactions programmed with other mRNAs such as bacteriorhodopsin. Variable amounts of wheat germ proteins were present in the proteoliposomes. These contaminants are typical and have been partially characterized (18). The recovered proteoliposomes were assayed for SL synthesis using exogenous fluorescent NBD-ceramide substrate and endogenous liposome phospholipids as head group donors (Fig. 2B). Consistent with our previous characterization of this enzyme, NBD-SM, and to a lesser extent NBD-EPC, were produced in a time- and transcript-dependent manner. The wheat germ extract and proteoliposomes containing bacteriorhodopsin (Fig. 2B) had no SL synthesis activity. This system was used to further investigate and optimize enzymatic activity (Fig. 3). Synthesis of both NBD-SM and NBD-EPC were found to be linear for ∼3 h with maximal production of each at 2.5–5 μm NBD-ceramide substrate. The kinetic profiles of product formation were essentially identical at 0.25 (Fig. 3A) and 2.5 μm (data not shown) NBD-ceramide, indicating that the extent of the reaction was not controlled by substrate depletion. The pH optimum was fairly sharp at 6.5–7.0, and reactions were modestly superior at 30 °C. Divalent cations had no effect on activity. Exogenous detergents had differential effects on TbSLS4 activity (Fig. 3F). Both Triton X-100 and dodecyl-maltoside dramatically reduced (≥90%) both SM and EPC synthesis, but CHAPS had minimal effect on either activity. Based on these results, final reaction conditions for subsequent assays were set as 2.5 h at 30 °C in MBS buffer, pH 6.5, with 2.5 μm NBD-ceramide.
FIGURE 2.
Cell-free synthesis of active TbSLS4. TbSLS4 was synthesized in the cell-free system, and proteoliposomes were recovered by floatation and assayed for conversion of NBD-ceramide substrate. A, samples from the gradient were fractionated by SDS-PAGE and stained with Coomassie Blue. Gradient fractions are indicated from top (T) to bottom (B), and the direction of floatation is indicated (arrow). A star indicates a TbSLS4-specific band recovered in the proteoliposomes. This band is absent in floated fraction 2 from reactions programmed for synthesis of bacteriorhodopsin (BO, indicated by arrowhead). B, recovered TbSLS4 or bacteriorhodopsin proteoliposomes were used for NBD-SL synthesis over a 4-h time course in MBS (pH 6.5, 0.25 μm NBD-ceramide, 30 °C); products were visualized by TLC and phosphorimaging. Mobilities of NBD standards, solvent front (F), NBD-ceramide (C), and origin (O) are indicated.
FIGURE 3.
Optimization of cell-free enzymatic parameters. Formation of NBD-SM (black symbols and bars) and NBD-EPC (open symbols and bars) were used to optimize enzyme assay conditions for TbSLS4 synthesized in the cell-free system. Unless specifically varied, reaction conditions in all cases were 4 h at 30 °C in MBS, pH 6.5, with 0.25 μm NBD-ceramide. Fluorescent products were extracted and analyzed by TLC as in Fig. 2. SM and EPC products were individually quantified by phosphorimaging and normalized to maximum production (A–C) or to standard control conditions (D–F). All assays were performed in triplicate, and results are presented as mean ± S.E. A, kinetics of product formation. B, NBD-ceramide substrate dependence. C, pH dependence using MES (MBS, pKa of 6.15) or HEPES (HBS, pKa of 7.55) buffering agent. D, temperature dependence. E, effect of 10 mm CaCl2, MgCl2, or MnCl2. Note that under standard buffer conditions all reactions contain 1 mm EDTA. F, effect of detergents: Triton X-100, 0.1% (v/v); CHAPS, 0.1% (w/v); n-dodecyl-maltoside, 0.1% (w/v). The detergent:phospholipid molar ratios for these assays were Triton X-100 (TX100), 2.8; CHAPS, 2.7; and n-dodecyl-maltoside, 3.3.
Enzymatic Specificity of Trypanosomatid Sphingolipid Synthases
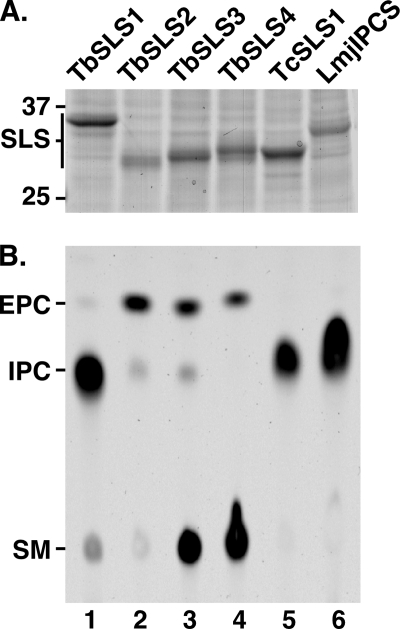
The trypanosomatid SL synthases were produced individually in the cell-free system. For these experiments, liposomes were prepared with equimolar pure preparations of PC, PE, and PI to further normalize reactions for substrate concentration. Consequently, these assays represent a fair comparison of the relative activity and specificity of each enzyme. SDS-PAGE of recovered proteoliposomes indicated that each SLS was synthesized to similar levels (Fig. 4A). The enzymatic activities of each were determined in parallel in standard NBD-ceramide incorporation assays programmed with normalized amounts of TbSLS protein (Fig. 4B). As demonstrated above, TbSLS4 synthesized both NBD-SM and NBD-EPC (Fig. 4B, lane 4). Although not apparent in this image, TbSLS4 also made a trace amount of NBD-IPC (see also Fig. 6, lanes 5 and 6). TbSLS3 has identical activity to TbSLS4 (Fig. 4B, lane 3), along with the production of a minor amount of IPC. Interestingly, TbSLS2 made predominantly NBD-EPC (Fig. 4B, lane 2), with minor amounts of NBD-SM and NBD-IPC detected. More surprisingly, TbSLS1 predominantly synthesized NBD-IPC with only minor amounts of NBD-SM and NBD-EPC detected (Fig. 4B, lane 1). As expected (11, 16), LmjIPCS synthesized NBD-IPC exclusively (Fig. 4B, lane 6). Finally, TcSLS1.1 also made NBD-IPC exclusively (Fig. 4B, lane 5). These results indicate that the TbSLSs have overlapping enzymatic profiles, but each has a distinct dominant specificity.
FIGURE 4.
Enzymatic activities of trypanosomatid sphingolipid synthases. The trypanosomatid SLSs were individually synthesized in the cell-free system using equimolar PC/PE/PI liposomes, and proteoliposomes were recovered by floatation. A, samples of proteoliposomes containing the individual trypanosomatid SLS translation products were analyzed by SDS-PAGE and Coomassie Blue staining. The positions of molecular mass standards (kDa) and TbSLS translation products are indicated. This gel was used to normalize the amount of TbSLS protein used in subsequent synthesis reactions. B, proteoliposomes containing equivalent amounts of TbSLS translation products (∼10 μg) were assayed for activity in a standard 2.5-h reaction at 30 °C with 2.5 μm NBD-ceramide. NBD-SL product analysis was performed in parallel on a single TLC plate. A phosphorimage is presented with mobility standards indicated.
FIGURE 6.
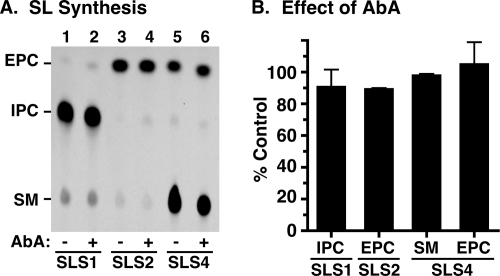
Effect of AbA on SLS activities in the cell-free system. TbSLSs were produced in the cell-free system using standard equimolar donor liposomes. A, proteoliposomes containing TbSLS1 (lanes 1 and 2), TbSLS2 (lanes 3 and 4), or TbSLS4 (lanes 5 and 6) were assayed in standard reactions for synthesis of NBD-SLs as in Fig. 4. Reactions were performed in the absence (−) or presence (+) of 500 nm AbA. B, products on TLC plates were quantified and are presented as percent of the AbA minus control (mean ± S.D., n = 3) for each TbSLS.
Modulation of TbSLS Specificity by Site-specific Mutagenesis
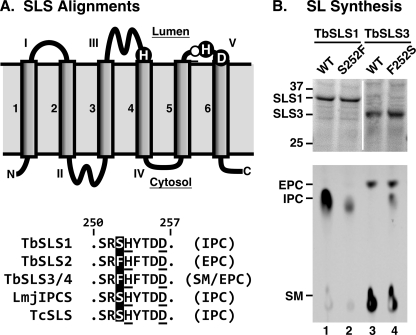
The differences in TbSLS specificities are striking given that the primary sequences of the four TbSLSs are >90% identical (excluding the C-terminal cytoplasmic domain), and, when conservative substitutions in transmembrane helices are discounted, e.g. Leu for Ile, identity exceeds 95% (see supplemental Fig. S1 in Ref. 11 for a comprehensive alignment). This leaves few residues to account for the differences in substrate specificity. The chemistry of SL synthase reactions is thought to proceed via formation of a covalent phosphoenzyme intermediate (28). Thus, specificity is likely to be defined by residues in the lumenal catalytic domain that interact with the polar head groups of the phospholipid donors. The aligned trypanosomatid SLS sequences (Fig. 5A) reveal a single residue immediately adjacent to a catalytic histidine in lumenal Loop V that is invariably associated with either IPC (Ser252) or SM/EPC (Phe252) synthesis by the trypanosomatid SLSs. Conversion of this residue in TbSLS1 (S252F) dramatically reduced IPC synthesis (Fig. 5B, compare lanes 1 and 2). In contrast, conversion of the same residue in TbSLS3 (F252S) enhanced IPC synthesis without affecting innate SM or EPC synthetic activity, essentially creating a trifunctional enzyme (Fig. 5B, compare lanes 3 and 4). These results provide the first functional mapping of substrate binding determinants in the sphingolipid synthase family and suggest that differences in amino acid sequence are the primary determinants of substrate/product specificity, as opposed to substrate availability, in the native organisms.
FIGURE 5.
Site-specific mutagenesis of TbSLSs. A, alignment of trypanosomatid SLS Loop V residues. Top, diagram of the modeled topology of trypanosomatid SLSs (adapted from Refs. 28 and 11). Transmembrane domains and solvent exposed loops are numbered in Arabic and Roman numerals, respectively. Key catalytic residues are silhouetted. Residue 252 targeted for mutagenesis is indicated by an open circle. Bottom, alignment of the Loop V region from trypanosomatid SLSs as indicated. Key catalytic residues are underlined, and residue 252 is silhouetted. Proven enzymatic activities are in parentheses. Numbering is according to the T. brucei orthologs. B, TbSLS1 and TbSLS3 mutants were produced in the cell-free system, and normalized amounts of translation product (∼10 μg) were assayed for enzymatic activity as in Fig. 4. Top, SDS-PAGE and Coomassie Blue staining of purified proteoliposomes. Equivalent synthesis of matched wild type and mutant TbSLSs is evident. The vertical white stripe indicates intervening lanes that were digitally excised from the gel. Equivalent amounts of each cell-free translation product were used in the SLS assays. Bottom, NBD-SL product analysis was performed in parallel on a single plate. A phosphorimage is presented with mobility standards indicated. WT, wild type.
Aureobasidin A Is Not a TbSLS Inhibitor
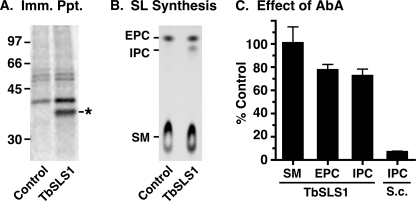
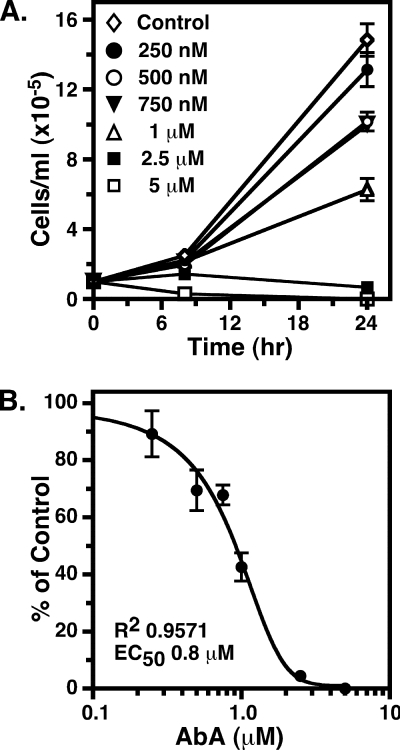
Fungal IPC synthases are potently inhibited (subnanomolar) by aureobasidin A (AbA) (29), and fungal growth is blocked in the low nm range (30). It has recently been reported (31) that AbA is a potent inhibitor of TbSLS4 (IC50 of 0.42 nm) when assayed in membranes prepared from transgenic yeast and likewise that AbA is an inhibitor of growth of cultured BSF trypanosomes (EC50 < 250 nm). Given the reported lack of inhibition by AbA with T. cruzi IPC synthase in native membranes (tested at 7.3 μm) (8) or LmjIPCS in membranes from transgenic yeast (IC50 > 100 μm) (31), we decided to re-examine the inhibitory activity of AbA against the T. brucei orthologues using three distinct assays. First, 500 nm AbA had little effect on the formation of any of the NBD-SL products of TbSLS1 (Fig. 6A, lanes 1 and 2), TbSLS2 (Fig. 6A, lanes 3 and 4), or TbSLS4 (Fig. 6A, lanes 5 and 6) produced using the cell-free system. These data indicate that AbA is not an inhibitor of the TbSLSs when assayed in isolation. Second, to rule out that insensitivity of recombinant TbSLSs to AbA was an artifact of the cell-free system, we assayed native TbSLS activities in semi-intact membranes prepared from a transgenic BSF cell line constitutively expressing HA-tagged TbSLS1. This system allows simultaneous monitoring of synthesis of SM, EPC, and IPC in a single assay. Immunoprecipitation of metabolically radiolabeled control versus TbSLS1HA cells confirmed expression of a polypeptide of the expected size (Fig. 7A, star), and SL synthesis assays revealed specific production of NBD-IPC in addition to the normal production of NBD-SM and NBD-EPC (Fig. 7B). ESI-MS analyses of total lipid extracts also confirmed elevated steady state levels of d18:1/16:0-IPC in this cell line (supplemental Fig. S2). By using membranes from this cell line, the effect of 500 nm AbA on synthesis of all three SLs was determined (Fig. 7C). AbA gave no inhibition of NBD-SM synthesis and only minor inhibition of NBD-EPC and NBD-IPC synthesis (20–25%). This is in contrast to NBD-IPC synthesis by yeast membranes containing yeast IPC synthase, which was inhibited by >90% (a typical reaction with yeast membranes is presented in supplemental Fig. S3). Collectively, these data indicate that the TbSLS enzymes, in both native and recombinant forms, are relatively refractory (IC50 > 500 μm) to the inhibitory action of AbA. Finally, we determined the effect of AbA on growth of cultured BSF trypanosomes (Fig. 8). Proliferation was increasingly impaired at concentrations of >250 nm and was completely ablated at 2.5 μm. Similar results were obtained with multiple lots of commercially available AbA. The EC50 for growth inhibition was determined to be 0.8 μm, consistent with the relative lack of effect on SL synthesis by AbA in both the cell-free and semi-intact membrane systems.
FIGURE 7.
Effect of AbA on SLS activities in semi-intact trypanosome membranes. Permeabilized whole cell membranes were prepared from transgenic BSF cells ectopically expressing TbSLS1:HA (indicated as TbSLS1). A, immunoprecipitation of metabolically radiolabeled control and transgenic cells extracts with anti-HA antibody. A band of the expected size for TbSLS:HA is specifically detected in the TbSLS1:HA membranes (star). B, NBD-SL synthesis assays were performed under the standard conditions defined in the cell-free system. Reactions were programmed with 2 × 107 cell equivalents of semi-intact membranes. Reaction products were analyzed by TLC and phosphorimaging. In addition to NBD-SM and NBD-EPC synthesized in control membranes, TbSLS1:HA-containing membranes also synthesize NBD-IPC. C, the effect of 500 nm AbA on NBD-SL synthesis in TbSLS:HA semi-intact membranes was determined. As a positive control for inhibition, assays were also programmed with 5 μg of perforated membranes prepared from S. cerevisiae. Products on TLC plates were quantified by phosphorimaging, and data are presented as percent of the AbA minus control (mean ± S.D., n = 5).
FIGURE 8.
Effect of AbA on growth of BSF trypanosomes. A, BSF trypanosome cultures were initiated in the presence of increasing concentrations of AbA, and cell density was monitored for 24 h by microscopic cell counting. B, cell densities at 24 h were normalized as a percent of control AbA minus cultures. All data are presented as mean ± S.D. for triplicate independent cultures.
DISCUSSION
T. brucei has a unique linear array of four tandemly linked genes encoding sphingolipid synthases. Previously, we demonstrated that TbSLS4 is a bifunctional SM/EPC synthase, but the activities of the remaining paralogs were uncertain (11). Using a recently developed cell-free system for synthesis of active polytopic membrane proteins, we now have determined unambiguously the dominant activities of the entire enzyme family. TbSLS1 is an IPC synthase, TbSLS2 is an EPC synthase, and TbSLS3 and TbSLS4 are bifunctional SM/EPC synthases. The assignment of primary IPC synthesis activity to TbSLS1 confirms a recent prediction to this effect made on the basis of stage-specific patterns of TbSLS gene expression (17). Our conclusion is supported by the specific synthesis of IPC in BSF cells ectopically expressing TbSLS1. Thus, TbSLS1 is responsible for the stage-specific production of IPC in PCF trypanosomes. We have also used the cell-free system to confirm that LmjIPCS is an exclusive IPC synthase and to establish for the first time that the T. cruzi ortholog (TcSLS1.1) is an IPC synthase. T. cruzi has a second allelic copy (TcSLS1.2, Tc00.1047053510729.290) that has minor polymorphisms, and it is possible that this also is an IPC synthase. However, T. cruzi has also been reported to contain SM (9, 10, 32), so it may be that TcSLS1.2 has SM synthase activity.
Our determinations of TbSLS activities were made in liposomes containing equimolar concentrations of all three potential head group donor phospholipids (PC, PE, PI), and thus provide new information on the specificity of each enzyme. Sequence alignments and site-specific mutagenesis indicate that these specificities are exquisitely controlled by subtle differences in active site residues. For instance, Ser252 in Loop V is permissive for synthesis of all three SL products, but Phe252 is restrictive for synthesis of the cationic SLs, i.e. SM and EPC.
Recently, the enzymatic activity of TbSLS4 was investigated by Mina et al. (31) using transgene expression in a conditional aur1 (IPC synthase) mutant of S. cerevisiae. Their conclusions differ in several key aspects from our current and previously published findings. Mina et al. (31) found that TbSLS4 is a bifunctional IPC/SM synthase. Other than leaky expression of endogenous IPC synthase in the yeast cell line, we can offer no certain explanation for this result. However, multiple lines of evidence support our conclusion that TbSLS4 is a bifunctional SM/EPC synthase with minimal IPC synthase activity. First, transgene expression of TbSLS4 in L. major, which only synthesizes IPC, resulted in synthesis and steady state accumulation of both SM and EPC (11). Second, unique production of TbSLS4 in the cell-free system, which is devoid of other SLS activities, results in robust synthesis of SM and EPC but little IPC. Finally, TbSLS4 is constitutively expressed in both BSF and PCF trypanosomes (17), yet significant IPC is only found in PCF stage parasites (11). The later two results in particular would be quite unlikely if native TbSLS4 had significant IPC synthase activity. The fact that TbSLS1, a confirmed IPC synthase, is active when expressed ectopically in BSF trypanosomes also rules out any inherent problems with IPC synthesis in this stage of the life cycle.
Another discrepancy with the findings of Mina et al. (31) is the sensitivity of TbSLS4 activity to inhibition by AbA. By again using the transgenic yeast system, they found an IC50 of 0.42 nm for putative synthesis of IPC, a value that is remarkably similar to the published sensitivity of yeast IPC synthase (∼0.2 nm) (29). This result is quite surprising because the trypanosomatid SLSs are phylogenetically distant from fungal IPC synthases (16), and neither of the closely related T. cruzi (8) nor L. major (31) orthologs are sensitive to AbA. Furthermore, the bifunctional TbSLS4-mediated SM synthesis assigned in the yeast system by Mina et al. (31) should have also been sensitive to AbA, but these data were not reported (31).
We independently investigated the AbA sensitivity of the TbSLSs in three complementary ways. First, we find that recombinant TbSLS1, TbSLS2, and TbSLS4 in the cell-free system are not significantly inhibited by AbA. Likewise, we find that native SLS activities in semi-intact trypanosome membranes containing all four TbSLSs are relatively insensitive to AbA. It is worth noting that in both these systems, TbSLS activities were assayed at 500 nm AbA, a concentration >1000-fold higher than the IC50 reported by Mina et al. (31). Again, one possible explanation of this discrepancy is leaky expression of endogenous IPC synthase in the yeast system. Alternatively, a critical protein interaction partner for yeast IPC synthase has recently been identified (Kei1p) (33). Kei1p is essential for both yeast IPC synthase activity and for its sensitivity to AbA. Perhaps this protein, assuming it is present in aur1 cells, affected the activity of TbSLS4 when expressed in yeast. Finally, we do find that AbA inhibits growth of cultured BSF trypanosomes, but at a higher concentration (EC50 of 0.8 μm) than reported by Mina et al. (31) (EC50 < 0.25 μm). In summary, the present results indicate that AbA is not a potent inhibitor of TbSLS activities. Instead, inhibition of parasite growth is likely due to off-target effects. Alternatively, even weak inhibition of TbSLS activities may subtly alter ratios of ceramide substrate and diacylglycerol product, both of which are potent intracellular signaling molecules and can have apototic effects (2, 34).
In closing, it is worth commenting on the advantages of the cell-free system used in this work. The speed and simplicity with which we were able to generate and assay the individual TbSLS enzymes (days to weeks) greatly exceeds the time commitment and complexity of our prior strategy of transgene expression in Leishmania (months) (11). This method was particularly advantageous in pursuing mutagenesis studies of the determinants of enzymatic specificity. In addition, the ability to synthesize and assay a single enzyme in isolation eliminates any confounding issues of background activities associated with endogenous expression in living systems. For instance, it would not have been possible to unambiguously determine the IPC synthase activity of TbSLS1 in transgenic Leishmania because of the background activity of endogenous LmjIPCS. The wheat germ system can also be used for large scale protein production (in mg quantities) for subsequent structure/function studies (21), which provides another advantage relative to the presently known methods to produce SLS enzymes. All of these qualities are potentially applicable to the study of other polytopic membrane proteins. Consequently, this system should become an important tool for investigation of a wide variety of previously inaccessible biological processes.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Beverley (Washington University, St. Louis, MO) for thoughtful discussions. We are grateful to Dr. Rick Tarleton (University of Georgia) for T. cruzi genomic DNA.
This work was supported by United States Public Health Service, NIAID, National Institutes of Health Grant R01 AI35739 (to J. D. B.), NIGMS, National Institutes of Health Grant RO1 GM50853 (to B. G. F.), and NIGMS, National Institutes of Health Protein Structure Initiative Grant 1 U54 GM074901 (to J. L. Markley, G. N. Phillips, Jr., and B. G. F.). The Washington University Department of Medicine Mass Spectroscopy Facility is supported by United States Public Health Service Grants P41-RR00954, P60-DK20579, and P30-DK56341.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- SL
- sphingolipid
- BSF
- bloodstream form
- PCF
- procyclic form
- SLS
- sphingolipid synthase
- SM
- sphingomyelin
- EPC
- ethanolamine phosphorylceramide
- ORF
- open reading frame
- IPC
- inositol phosphorylceramide
- nt
- nucleotide(s)
- HA
- hemagglutinin
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol
- AbA
- aureobasidin A
- NBD
- 6-((N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)
- MES
- 4-morpholineethanesulfonic acid
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Schuck S., Simons K. (2004) J. Cell Sci. 117, 5955–5964 [DOI] [PubMed] [Google Scholar]
- 2.Lahiri S., Futerman A. H. (2007) Cell. Mol. Life Sci. 64, 2270–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang K., Pompey J. M., Hsu F. F., Key P., Bandhuvula P., Saba J. D., Turk J., Beverley S. M. (2007) EMBO J. 26, 1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaneshiro E. S., Jayasimhulu K., Lester R. L. (1986) J. Lipid Res. 27, 1294–1303 [PubMed] [Google Scholar]
- 5.Zhang K., Hsu F. F., Scott D. A., Docampo R., Turk J., Beverley S. M. (2005) Mol. Microbiol. 55, 1566–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uhrig M. L., Couto A. S., Colli W., de Lederkremer R. M. (1996) Biochim. Biophys. Acta 1300, 233–239 [DOI] [PubMed] [Google Scholar]
- 7.Bertello L. E., Gonçalvez M. F., Colli W., de Lederkremer R. M. (1995) Biochem. J. 310, 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueiredo J. M., Dias W. B., Mendonça-Previato L., Previato J. O., Heise N. (2005) Biochem. J. 387, 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franco, da Silveira J., Colli W. (1981) Biochim. Biophys. Acta 644, 341–350 [DOI] [PubMed] [Google Scholar]
- 10.Oliveira M. M., Timm S. L., Costa S. C. (1977) Comp. Biochem. Physiol. 58B, 195–199 [DOI] [PubMed] [Google Scholar]
- 11.Sutterwala S. S., Hsu F. F., Sevova E. S., Schwartz K. J., Zhang K., Key P., Turk J., Beverley S. M., Bangs J. D. (2008) Mol. Microbiol. 70, 281–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huitema K., van den Dikkenberg J., Brouwers J. F., Holthuis J. C. (2004) EMBO J. 23, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaoka S., Miyaji M., Kitano T., Umehara H., Okazaki T. (2004) J. Biol. Chem. 279, 18688–18693 [DOI] [PubMed] [Google Scholar]
- 14.Vacaru A. M., Tafesse F. G., Ternes P., Kondylis V., Hermansson M., Brouwers J. F., Somerharju P., Rabouille C., Holthuis J. C. (2009) J. Cell Biol. 185, 1013–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ternes P., Brouwers J. F., van den Dikkenberg J., Holthuis J. C. (2009) J. Lipid Res. 50, 2270–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denny P. W., Shams-Eldin H., Price H. P., Smith D. F., Schwarz R. T. (2006) J. Biol. Chem. 281, 28200–28209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabani S., Fenn K., Ross A., Ivens A., Smith T. K., Ghazal P., Matthews K. R. (2009) BMC Genomics 11, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goren M. A., Fox B. G. (2008) Prot. Express. Purif. 62, 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawasaki T., Hasegawa Y., Tsuchimochi M., Kasahara Y., Endo Y. (2000) Nucleic Acids Symp. Ser., 44, 9–10 [DOI] [PubMed] [Google Scholar]
- 20.Blommel P. G., Martin P. A., Wrobel R. L., Steffen E., Fox B. G. (2006) Prot. Express. Purif. 47, 562–570 [DOI] [PubMed] [Google Scholar]
- 21.Goren M. A., Nozawa A., Makino S., Wrobel R. L., Fox B. G. (2009) Methods Enzymol. 463, 647–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson R. L., Davis S. (1982) Clinica. Chimica. Acta 121, 111–116 [DOI] [PubMed] [Google Scholar]
- 23.Alexander D. L., Schwartz K. J., Balber A. E., Bangs J. D. (2002) J. Cell Sci. 115, 3255–3263 [DOI] [PubMed] [Google Scholar]
- 24.Sevova E. S., Bangs J. D. (2009) Mol. Biol. Cell 20, 4739–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masterson W. J., Doering T. L., Hart G. W., Englund P. T. (1989) Cell 56, 793–800 [DOI] [PubMed] [Google Scholar]
- 26.Rexach M. F., Latterich M., Schekman R. W. (1994) J. Cell Biol. 126, 1133–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K., Showalter M., Revollo J., Hsu F. F., Turk J., Beverley S. M. (2003) EMBO J. 22, 6016–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tafesse F. G., Ternes P., Holthuis J. C. (2006) J. Biol. Chem. 281, 29421–29425 [DOI] [PubMed] [Google Scholar]
- 29.Nagiec M. M., Nagiec E. E., Baltisberger J. A., Wells G. B., Lester R. L., Dickson R. C. (1997) J. Biol. Chem. 272, 9809–9817 [DOI] [PubMed] [Google Scholar]
- 30.Takesako K., Ikai K., Haruna F., Endo M., Shimanaka K., Sono E., Nakamura T., Kato I., Yamaguchi H. (1991) J. Antibiot. 44, 919–924 [DOI] [PubMed] [Google Scholar]
- 31.Mina J. G., Pan S. Y., Wansadhipathi N. K., Bruce C. R., Shams-Eldin H., Schwarz R. T., Steel P. G., Denny P. W. (2009) Mol. Biochem. Parasitol. 168, 16–23 [DOI] [PubMed] [Google Scholar]
- 32.Quiñones W., Urbina J. A., Dubourdieu M., Luis, Concepción J. (2004) Exp. Parasitol. 106, 135–149 [DOI] [PubMed] [Google Scholar]
- 33.Sato K., Noda Y., Yoda K. (2009) Mol. Biol. Cell 20, 4444–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shea J., Del Poeta M. (2006) Cur. Opin. Microbiol. 9, 352–358 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.