Abstract
Ectodomain shedding of the amyloid precursor protein (APP) by the two proteases α- and β-secretase is a key regulatory event in the generation of the Alzheimer disease amyloid β peptide (Aβ). At present, little is known about the cellular mechanisms that control APP shedding and Aβ generation. Here, we identified a novel protein, transmembrane protein 59 (TMEM59), as a new modulator of APP shedding. TMEM59 was found to be a ubiquitously expressed, Golgi-localized protein. TMEM59 transfection inhibited complex N- and O-glycosylation of APP in cultured cells. Additionally, TMEM59 induced APP retention in the Golgi and inhibited Aβ generation as well as APP cleavage by α- and β-secretase cleavage, which occur at the plasma membrane and in the endosomes, respectively. Moreover, TMEM59 inhibited the complex N-glycosylation of the prion protein, suggesting a more general modulation of Golgi glycosylation reactions. Importantly, TMEM59 did not affect the secretion of soluble proteins or the α-secretase like shedding of tumor necrosis factor α, demonstrating that TMEM59 did not disturb the general Golgi function. The phenotype of TMEM59 transfection on APP glycosylation and shedding was similar to the one observed in cells lacking conserved oligomeric Golgi (COG) proteins COG1 and COG2. Both proteins are required for normal localization and activity of Golgi glycosylation enzymes. In summary, this study shows that TMEM59 expression modulates complex N- and O-glycosylation and suggests that TMEM59 affects APP shedding by reducing access of APP to the cellular compartments, where it is normally cleaved by α- and β-secretase.
Keywords: Diseases/Alzheimer Disease, Diseases/Amyloid, Enzymes/Proteolytic, Glycoproteins/Biosynthesis, Membrane/Proteins, Proteases/Secretases
Introduction
Processing of the amyloid precursor protein (APP)3 by two different proteases, called α- and β-secretase, is a central regulatory event in the generation of the amyloid β peptide (Aβ), which has a key role in Alzheimer disease (AD) pathogenesis (1). Both α- and β-secretase cleave the type I membrane protein APP within its ectodomain close to its transmembrane domain (2). This leads to the secretion of soluble forms of APP (APPs) and is referred to as APP shedding. The β-secretase is the aspartyl protease BACE1 and cleaves APP at the N terminus of the Aβ peptide domain, thus catalyzing the first step in Aβ peptide generation (3, 4). After the initial cleavage of APP by BACE1, the remaining C-terminal APP fragment is cleaved by γ-secretase within its transmembrane domain at the C terminus of the Aβ domain, leading to the secretion of the Aβ-peptide (5). In contrast to β-secretase, α-secretase cleaves within the Aβ-sequence of APP, and thereby precludes Aβ peptide generation. Additionally, α- but not β-secretase cleavage generates a secreted form of APP (APPsα), which has neurotrophic and neuroprotective properties (6–8). The α-secretase is a member of the ADAM family (A disintegrin and metalloprotease) of proteases (9–12). At present little is known about how the cell controls access of APP to its secretases and the amount of α- and β-secretase cleavage (reviewed in Refs. 13 and 14). Recent studies increasingly imply intracellular APP trafficking as a mechanism to regulate access of APP to its secretases and thus, the amount of APP processing (15). For example, the neuronal sorting receptor SorL1 (sorLA, LR11) influences how much APP is present in the endosomes and is available for cleavage by β-secretase (16, 17). Likewise, changes in endocytic trafficking control APP α- versus β-secretase cleavage and are associated with early neuropathological changes observed in AD brains (18). Additionally, previous reports indicated that changes in Golgi glycosylation may affect APP trafficking and α- and β-secretase cleavage, but the underlying molecular mechanisms remain unclear. For example, increased sialylation of APP increases α- and β-secretase cleavage, whereas inhibition of the Golgi-localized mannosidase II reduces APP shedding (19, 20). APP has two N-glycosylation sites at amino acids Asn467 and Asn496 (with regard to the 695-amino acid long APP isoform). Depending on the cell line only one or both of them may be used (21, 22). N-Glycosylation begins in the endoplasmic reticulum, where a core of sugars of the high-mannose type is co-translationally added to APP and then gradually trimmed. As APP transits the Golgi, further post-translational modifications occur (23, 24). The N-glycans are processed to complex glycans containing galactose and sialic acid. The generation of complex glycans can be monitored by their sensitivity to endoglycosidase H, which only removes high-mannose sugars, but not sugars of the complex type. APP also undergoes O-glycosylation in the Golgi, leading to a marked increase in its molecular mass. As additional post-translational modification, APP may be phosphorylated and sulfated.
In general, O-glycosylation and complex N-glycosylation, which occur in the Golgi, are multistep processes, which involve a number of different enzymes. New proteins controlling the correct cellular localization and the activity of glycosylation enzymes continue to be identified. Among them are the subunits of the conserved oligomeric Golgi (COG) complex, which is a hetero-oligomeric complex localized to the cytoplasmic face of the Golgi (25, 26). The eight subunits of the COG complex are soluble proteins named COG1 through COG8. The COG complex is assumed to act as a retrograde vesicle tethering factor in intra-Golgi trafficking and is particularly required for the correct localization and activity of Golgi glycosylation enzymes. Mutations or deletions of COG proteins lead to multiple defects in glycoprotein processing and are linked to human disorders, called congenital disorders of glycosylation (27). It is not yet known whether COG proteins influence APP processing.
Here, we report that expression of the novel type I transmembrane protein 59 (TMEM59) inhibits the Golgi glycosylation of APP, reduces APP cell surface levels, and blocks APP cleavage by both α- and β-secretase. The protein was identified from a human cDNA library, which we screened for modulators of APP shedding (28, 29). TMEM59 has a distant homolog called TMEM59L or BSMAP (brain-specific membrane-associated protein), which is expressed in brain, but also in peripheral tissues and in human embryonic kidney 293 cells (30) (sequence alignment shown in Fig. 1). TMEM59 and TMEM59L do not have any known functional domains apart from the signal peptide and the transmembrane domain. Moreover, both proteins have not yet been functionally described. Interestingly, a similar inhibition of APP glycosylation and shedding as for TMEM59 expression was observed in COG1- or COG2-deficient cells, suggesting that TMEM59 affects the Golgi glycosylation machinery, which also requires the COG proteins. These results underline that changes in Golgi glycosylation have a profound effect on APP shedding.
FIGURE 1.
Sequence alignment of TMEM59 and its homolog TMEM59L. Alignment of the protein sequences of human TMEM59 (323 amino acids) and its human homolog TMEM59L (342 amino acids, 32% identity between TMEM59 and TMEM59L). Both proteins are type I transmembrane proteins. The signal peptide is underlined, the transmembrane domain is marked by a box, the putative N-glycosylation site is marked with an asterisk, and the antibody epitopes are underlined (dashed). Identical amino acids are on a dark background, similar amino acids are on a light gray background. Between humans and mice, the amino acid sequence of TMEM59 is highly conserved (93.8% identity, 97.2% similarity to mouse). Apart from the transmembrane domain, no other known functional domains are found in TMEM59. TMEM59 is mostly found in vertebrates, but orthologs are also found in some insects (28.5% identity, 44% similarity to Ixodes scapularis), but not in other invertebrates, such as Caenorhabditis elegans or Drosophila melanogaster.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
The following antibodies were used: anti-HA HA.11 (Covance) and 12CA5 (Roche), anti-FLAG (Sigma), anti-GFP (Clontech), anti-β-actin, (Sigma), anti-calnexin (Stressgene), horseradish peroxidase-coupled goat anti-mouse and anti-rabbit (Promega), Alexa 555/Alexa 488-coupled secondary anti-mouse (Molecular Probes), Alexa 555-coupled secondary anti-rat antibody (Molecular Probes), Alexa 488-coupled anti-rabbit secondary antibody (Molecular Probes), anti-giantin (Alexis) (31), 6E10 (against Aβ1–17, Senetek Inc.), 6687 (against APP C terminus) (32), 22C11 (against APP ectodomain, provided by Konrad Beyreuther), 192wt (specific for the C terminus of APPsβ, provided by Dale Schenk), W02 (against amino acids 5–8 of Aβ, provided by Konrad Beyreuther) (33), 3552 (against Aβ1–40) (34), EE-17 (against N terminus of BACE1, Sigma), Nicastrin (N1660, Sigma), and the antibodies GM130, GS15, GS27, GS28, p230, Syntaxin 6, Vti1a, and Vti1b of the Golgi Sampler Kit, as well as the antibody against TGN38 (both BD Transduction Laboratories). Polyclonal TMEM59-antiserum 93 was generated against a synthetic peptide (H2N-309–323-CONH2) from the C terminus of TMEM59 (Eurogentec Seraing, Belgium), 3F4 (anti-mouse PrP) (35). Rat monoclonal antibody 4E5 (IgG2b), binding to the C terminus of TMEM59 was generated by immunization with a GST fusion protein of amino acids 261–323 of TMEM59 (GST-TMEM59-CT). GST-TMEM59-CT was expressed in Escherichia coli BL21 cells and purified according to the manufacturer's protocol (Amersham Biosciences). Control rat antibody 4G8 (IgG2b) was generated against the mouse Fcγ-receptor. Rat monoclonal antibody BAWT (IgG2a), specific for APPsβ, was generated against the peptide ISEVKM derived from the APP β-secretase cleavage site.
Plasmid Construction
TMEM59 in the peak8-vector was obtained from a human brain cDNA library (Edgebio). The TMEM59 nucleotide sequence corresponds to accession number AF047439. Plasmids p12/APP695 and peak12/BACE1 have been described (36–38). Soluble APPsβ was cloned into peak12-vector (ending with amino acids KM at the β-secretase cleavage site). cDNAs of TMEM59 (without UTRs, with C-terminal fusions to GFP and HA tag or N-terminal fusions to HA tag), TMEM59L (C-terminal fusion to HA tag), and HA-TMEM59-KKXX with an endoplasmic reticulum-retention signal added to the C terminus of TMEM59 (…SEIKKTN) were cloned into peak12-vector. GST-TMEM59-CT was cloned in pGEX5.1. Peak12/FLAG-TNFα-HA, peak12/HA-SEAP, peak12/GFP (transfection control), and peak12/luciferase (negative control) have been described (29, 36, 39, 40). The pcDNA3.1/wtPrP construct was cloned as described (41). The pShuttle/CMV-YFP-APP was obtained from Kai Simons (42). The pCAG-IRES2-golgiVENUS was obtained from Timm Schroeder (43).
Cell Culture, Transfection, and Western Blot
Human embryonic kidney 293 EBNA (HEK293) cells, COS cells, H4 cells, and U373 cells were cultured in Dulbecco's modified Eagle's medium (Cambrex) supplemented with 10% fetal bovine serum (HyClone), 50 units/ml of penicillin and 50 μg/ml of streptomycin (Invitrogen) (44). Chinese hamster ovary (CHO) wt, ldlB, ldlC knockout, and [ldlB] rescue CHO cells were obtained from Monty Krieger and cultured as above. Transfections were done using Lipofectamine 2000 (Invitrogen). One day after transfection the medium was changed. After an additional overnight incubation cell lysates (in 50 mm Tris, pH 7.5, 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40) were prepared and medium was collected and analyzed. To detect secreted and cellular APP or other cellular proteins, the protein concentration in the cell lysate was measured, and corresponding aliquots of lysate or medium were separated by SDS-PAGE and analyzed by Western blot.
Knockdown of TMEM59
TMEM59 and TMEM59L double knockdown was achieved using siRNAs (Dharmacon): siRNA-pool against TMEM59 and a combination of the single siRNAs number 1 and 3 against TMEM59L. A non-targeting siRNA-pool (composed of non-targeting siRNA numbers 3 and 4) was used to assess unspecific effects of siRNA delivery. HEK293 cells were transfected with 5 nm siRNAs using Lipofectamine RNAiMAX (Invitrogen). Fresh medium was added after 24 h, completely changed 48 h post-transfection, and cells were analyzed 72 h after transfection. Knockdown of TMEM59 protein was visualized by immunofluorescence using antibody 93. Knockdown efficiency was determined using quantitative real-time PCR, because the antibodies generated against TMEM59 were not sensitive enough to detect endogenous TMEM59 in the Western blot. Total RNA was isolated (using the RNeasy mini kit from Qiagen), checked for quality by agarose gel electrophoresis, and reverse transcribed into cDNA (using the high capacity cDNA reverse transcription kit from Applied Biosystems.) real-time PCR was performed with a 7500 Fast Real-Time PCR System (Applied Biosystems) using the TaqMan Universal PCR Master Mix (ROX) and TMEM59- and TMEM59L-specific probes as well as the corresponding primers (Applied Biosystems). The mRNA level was normalized against the amount of actin gene transcript.
Immunoprecipitations
For HA-soluble enzyme secretory alkaline phosphatase (SEAP) immunoprecipitation, cell lysates were incubated with antibody (anti-HA, 12CA5, Roche) overnight (4 °C) using protein G-Sepharose (Amersham Biosciences). After washing with STEN-NaCl (STEN buffer + 0.35 m NaCl) and twice with STEN (0.05 m Tris-HCl, pH 7.5, 0.15 m NaCl, 2 mm EDTA, and 0.2% Nonidet P-40), bound proteins were resolved by SDS-PAGE and analyzed by Western blot. For Aβ detection in the conditioned medium immunoprecipitation with antibody 3552 was performed prior to SDS-PAGE. Precipitates were transferred to nitrocellulose membranes. Aβ was analyzed with antibody 6E10 using a Tris-Tricine gel according to Ref. 45. For APPsβ detection in the conditioned medium, immunoprecipitation with antibody BAWT was performed prior to Western blot using 192wt. Interaction between TMEM59 and APP was tested by coimmunoprecipitation. HEK293 cells were transfected with HA-TMEM59 and lysed as described above. Lysates were incubated with HA tag antibody overnight (4 °C) using protein G-Sepharose (Amersham Biosciences). After three washing steps (as above), bound proteins were resolved by SDS-PAGE and analyzed by Western blot using antibody 6687 against the APP C terminus.
Northern Blot Analysis
Multiple tissue Northern blot analysis was performed according to the supplier's protocol (Clontech, BD Biosciences). Briefly, a [α-32P]dCTP (Amersham Biosciences)-labeled TMEM59 or actin probe (Random Primer DNA Labeling System, Invitrogen) was added to a human multiple tissue Northern blot (BD Biosciences). Hybridization was performed overnight at 65 °C. Excess probe was removed by washing with 2× SSC, 0.1% SDS at room temperature followed by washing with 0.1× SSC, 0.1% SDS at 65 °C. The blot was exposed to Super RX film (Fuji).
Immunocytochemistry
COS cells plated on coated coverslips were left untreated or were transfected with GFP-tagged TMEM59, respectively. After 24 h the cells were washed in ice-cold phosphate-buffered saline supplemented with 1 mm CaCl2, 0.5 mm MgCl2, transferred to 4% paraformaldehyde, 4% sucrose in phosphate-buffered saline, and fixed for 20 min and stained with 1:200 diluted anti-TMEM59 93 antibody/peptide blocked antibody or undiluted anti-TMEM59 4E5-antibody/IgG2b antibody and 1:200 diluted giantin antibody. As secondary antibodies, 1:500 diluted Alexa 555-/Alexa 488-coupled secondary anti-mouse, Alexa 555-coupled secondary anti-rat antibody, and Alexa 555-/Alexa 488-coupled anti-rabbit secondary antibody were used. Cells were analyzed using a Zeiss 510Meta confocal system equipped with a 40/1.3 objective.
Live Cell Imaging
HEK293 cells plated in poly-l-lysine-coated 15-μm slide 8-well plates (Ibidi) were cotransfected with yellow fluorescent protein (YFP)-APP and either control plasmid or HA-TMEM59. HEK293 cells transfected with pCAG-IRES2-golgiVENUS were also incubated in this formate. After 24 h the cells were analyzed using a Zeiss 510Meta confocal system equipped with a 100/1.3 objective.
N-Linked Glycosylation Analysis
HEK293 cells were transfected with TMEM59-HA. In additional experiments BACE1 was cotransfected with either control plasmid or TMEM59. Cell lysates were divided into four aliquots and treated with or without 1 unit of N-glycosidase F or with and without 1 milliunit of endoglycosidase H for 17 h at 37 °C in the presence of the buffers recommended by the supplier (Roche). Samples were loaded onto a SDS-PAGE and analyzed by Western blot using the APP-antibody 22C11, anti-HA HA.11, or anti-BACE1 EE-17.
ADAM and BACE Fluorimetric Assays
ADAM activity was measured in intact cells as described previously (46, 47). HEK293 cells were transfected with either control plasmid or TMEM59. The metalloprotease inhibitor TAPI-1 (50 μm; Peptides International) and the fluorogenic substrate (10 μm; Mca-PLAQAV(Dpa)RSSSR-NH2; R & D Systems) were used. ADAM activity was measured as fluorescence and recorded every 30 min. At the end of the incubation the obtained fluorescence signals were normalized to protein content and samples were checked for TMEM59 effects on APP shedding and maturation (data not shown).
To investigate BACE activity in TMEM59 expressing cells a fluorimetric assay (48, 49) was used. HEK293 cells were co-transfected with BACE1 and either control plasmid or TMEM59. Samples were prepared by harvesting membranes (50), solving them in STET buffer (0.05 m Tris-HCl, pH 7.5, 0.15 m NaCl, 2 mm EDTA, and 1% Triton X-100), and diluting to 3.3 mg/ml of protein in 10 mm sodium acetate buffer, pH 4.5. Hydrolysis of a fluorogenic BACE substrate (10 μm; Mca-SEVNLDAEFRK(Dnp)RR-NH2; R & D Systems) was recorded in the absence or presence of BACE inhibitor C3 (1 μm; Calbiochem) in 100 μl of the described samples. Additionally samples were checked for TMEM59 effects on APP shedding and maturation (data not shown).
RESULTS
TMEM59 Modulates Maturation and Shedding of APP
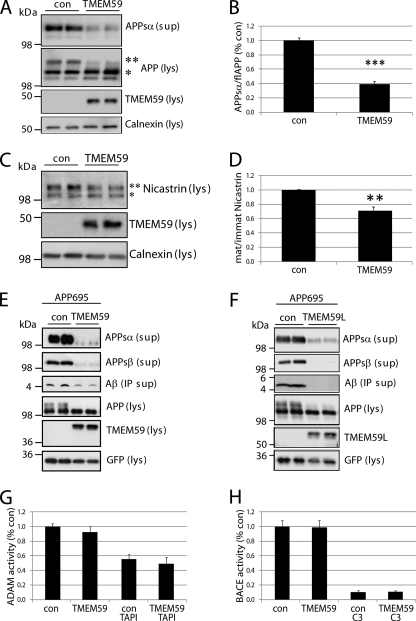
To analyze the effect of TMEM59 on APP processing, TMEM59 was transiently transfected into HEK293 cells expressing endogenous APP. Compared with control cells TMEM59 expression reduced the level of α-secretase cleaved, secreted APP (APPsα) in the conditioned medium by about 60% (Fig. 2A and for quantification, see B). In the cell lysate of control cells APP was present in lower (∼100 kDa) and higher (∼115 kDa) molecular mass forms (Fig. 2A). The lower molecular mass form corresponds to immature APP, which has obtained the core N-glycosylation in the endoplasmic reticulum (high-mannose form) but does not carry complex sugars and is not yet O-glycosylated. The higher molecular mass form (Fig. 2A) corresponds to mature APP, which is O-glycosylated and carries complex N-linked sugars (24). Both modifications are added as APP transits the Golgi complex. Surprisingly, in the TMEM59 expressing cells the mature form of APP was strongly diminished and showed a reduced apparent molecular mass. The immature form of APP carrying the core N-glycosylation was slightly increased (Fig. 2A), suggesting a reduction of complex APP glycosylation. Additionally, we observed that TMEM59 mildly affected the complex glycosylated form of the γ-secretase subunit nicastrin (Fig. 2C), but this effect was less pronounced than for APP (Fig. 2, B and D).
FIGURE 2.
TMEM59 effect on APP shedding and APP maturation. A, control plasmid (con) or TMEM59 were transfected into HEK293 cells. The lysate (lys) was blotted against cellular APP (22C11, **, mature and *, immature form of the APP751 splice variant; the band below the asterisk corresponds to the immature form of the shorter APP695 variant) and TMEM59 (HA.11), the supernatant (sup) was blotted against secreted APPsα (W02). B, TMEM59 expression reduced the amount of secreted APP (APPsα) by about 60% compared with control cells. Quantification of blots shown in A. Given are mean ± S.E. of 4 independent experiments. ***, p < 0.001, determined with t test. C, control plasmid (con) or TMEM59 were transfected into HEK293 cells. The lysate (lys) was blotted against nicastrin (N1660, **, mature and *, immature form of nicastrin) and TMEM59 (HA.11). D, TMEM59 expression reduced the amount of mature nicastrin by about 30% compared with control cells. Total amounts of nicastrin were not changed. Quantification of blots shown in C. Given are mean ± S.E. of 8 independent experiments. **, p < 0.01, determined with t test. E, APP695 and either control plasmid (con) or HA-TMEM59 with GFP as transfection control were transfected into HEK293 cells. The lysate was blotted against cellular APP (22C11), TMEM59 (HA.11), and GFP, the supernatant was blotted against secreted APPsα (W02), APPsβ (192wt), and Aβ (6E10, after immunoprecipitation). F, HEK293 cells were transfected with APP695 and either control plasmid (con) or TMEM59L-HA. The experiment was carried out as described in E. G, control plasmid (con) or TMEM59 were transfected into HEK293 cells. ADAM activity was measured in intact cells for 6 h in the presence or absence of TAPI-1 (50 μm). Shown is the relative ADAM activity in % of control cells. Given are mean ± S.E. of 5 (con/TMEM59) or 3 (con TAPI-1/TMEM59 TAPI-1) independent experiments. H, BACE1 and either control plasmid (con) or TMEM59 were transfected into HEK293 cells. BACE activity was recorded for 90 min in membranes in the presence or absence of BACE inhibitor C3 (1 μm). Shown is the relative BACE activity in % of control cells. Given are mean ± S.E. of 6 (con/TMEM59) or 3 (con C3/TMEM59 C3) independent experiments.
Because the amount of Aβ derived from endogenous APP is below the detection limit of the commonly used antibodies, we next coexpressed APP with either control vector or TMEM59 in HEK293 cells. Similar to endogenous APP, TMEM59 strongly inhibited APPsα secretion and APP maturation (Fig. 2E). The β-secretase cleaved, soluble APP (APPsβ) was reduced to a similar extent as APPsα. Consistent with the reduction in APPsβ levels, expression of TMEM59 reduced Aβ generation (Fig. 2E). The low amounts of APPsα and APPsβ that were still secreted upon TMEM59 expression showed a slightly enhanced electrophoretic mobility. This may be due to a reduced complex glycosylation similar to the observed reduction of the complex glycosylated, mature APP in the cell lysate. The apparent molecular mass of the immature APP (only core N-glycosylation) was not altered, demonstrating that TMEM59 did not affect the core N-glycosylation of APP (compare also Fig. 7A), but specifically blocked the O-glycosylation and the complex N-glycosylation of APP. Similar results on APP maturation and shedding as in HEK293 cells were observed in COS7 (data not shown) and CHO cells (see Fig. 8). The TMEM59 homolog TMEM59L had a similar inhibitory effect on APP maturation and shedding as TMEM59 (Fig. 2F), revealing that both proteins have similar functions, at least with regard to APP. An N-terminal or C-terminal HA-epitope tag or fusion to GFP did not alter the effect of TMEM59 on APP glycosylation or shedding (data not shown).
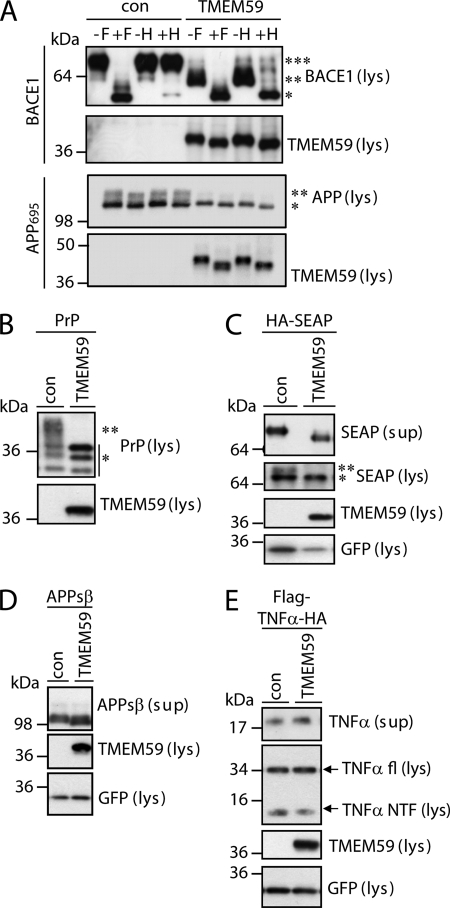
FIGURE 7.
TMEM59 inhibits glycosylation of different proteins. HEK293 cells were transiently transfected with the indicated constructs and either control vector (con) or TMEM59. GFP served as transfection control, where indicated. Lysates (lys) and supernatants (sup) were blotted against TMEM59 (93) and the indicated proteins. A, lysates of BACE1 transfected cells were treated with N-glycosidase F (+F) or endoglycosidase H (+H), controls remained untreated (−F, −H). The samples were blotted against BACE1 (EE-17, ***, mature, **, immature, and *, de-glycosylated BACE1) and TMEM59 (93). As a control, APP was deglycosylated in the same manner, revealing that TMEM59 does not affect the core N-glycosylation, but only the complex glycosylation (mature form) of APP. B, transfected PrP was detected in the lysate (3F4, **, complex glycosylated and *, di-/mono-/unglycosylated forms of PrP). C, secreted alkaline phosphatase (HA-SEAP) was detected in the supernatant (after immunoprecipitation) or in lysate (**, mature and *, immature SEAP) and detected with anti-HA antibody. D, transfected APPsβ was detected in the supernantant (192wt) and TMEM59 in the lysate (93). E, FLAG-TNFα-HA was transfected. fl-TNFα and TNFα-NTF were detected in the lysate (FLAG), secreted TNFα (HA) in the supernatant.
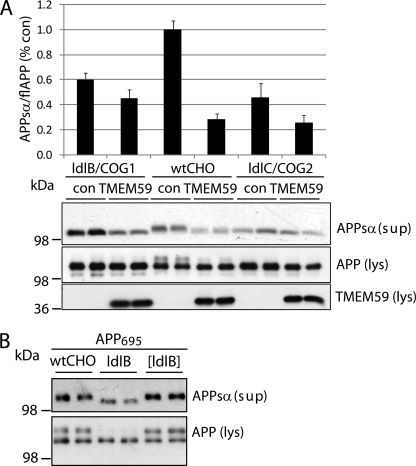
FIGURE 8.
A knockout of COG proteins shows similar defects of APP glycosylation as TMEM59 expression. A, wtCHO, ldlB-CHO, and ldlC-CHO cells were cotransfected with APP695 and either control plasmid (con) or TMEM59. The lysates (lys) were blotted against cellular APP (22C11) and TMEM59 (93), the supernatant (sup) was blotted against secreted APPsα (W02). The quantification shows the secreted APPsα normalized to the cellular APP and is represented as mean ± S.E. of three to four independent experiments. B, wtCHO, ldlB-CHO, and [ldlB]-CHO cells were transfected with APP695. [ldlB]-CHO cells are COG1-deficient ldlB cells retransfected with COG1. The lysates (lys) were blotted against cellular APP (22C11) and the supernatants (sup) against secreted APPsα (W02).
TMEM59 expression did not affect the activity of α- and β-secretase, as measured by established assays using fluorogenic substrates (46, 48) (Fig. 2, G and H). As a control for the specificity of the assay, cleavage of the fluorogenic α-secretase substrate could be inhibited to about 50% by the metalloprotease inhibitor TAPI-1 (Fig. 2G), in agreement with a previous study (46). Likewise, cleavage of the fluorogenic β-secretase substrate could be inhibited by about 90% using the specific BACE1 inhibitor C3 (51) (Fig. 2H). This experiment demonstrates that the TMEM59-induced reduction of APPsα and APPsβ levels is not due to a reduction of secretase activities, but may be caused by altered APP glycosylation and trafficking (see below).
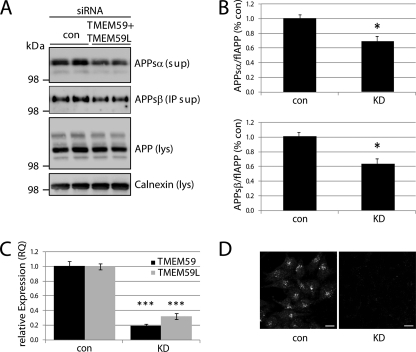
Next, we analyzed how knockdown of TMEM59 affects APP maturation and shedding. HEK293 cells were transiently transfected with control siRNAs or siRNAs against TMEM59 or TMEM59L or against both. The single knockdown of TMEM59 or TMEM59L did not significantly affect APP shedding or maturation (data not shown). This may be due to the fact that even in the absence of one or both proteins, the other homolog is still present, because both proteins are expressed in HEK293 cells (30). In fact, the combined knockdown of TMEM59 and TMEM59L reduced shedding of the endogenous APPsα and APPsβ by 30–40%, respectively (Fig. 3, A and B). APP maturation showed a mild reduction, which, however, did not reach statistical significance. This may be due to the fact that despite an efficient knockdown, ∼20% of TMEM59 and ∼30% of TMEM59L RNA were still remaining, as measured by quantitative reverse transcription-PCR (Fig. 3C). The successful knockdown of TMEM59 was also verified by immunofluorescence (Fig. 3D). For TMEM59L no antibody is available that detects the endogenous protein, either by immunoblot or immunofluorescence. The finding that both knockdown and overexpression of TMEM59 and TMEM59L affected APP shedding in a similar manner is reminiscent of what has been reported for other proteins, such as endocytic endophilins (29, 52, 53), adaptor proteins FE65 (54), and c-Jun N-terminal kinase-interacting proteins (55). Typically, these proteins form hetero-oligomeric complexes. A protein knockdown leads to a loss of fully assembled, functional complexes, and protein overexpression is assumed to lead to too many complexes that are not fully assembled, thus again resulting in a loss of functional complexes. Potentially, TMEM59 and TMEM59L also form complexes with as yet unidentified proteins, such that overexpression and knockdown affect APP maturation and shedding in a similar manner.
FIGURE 3.
Knockdown of TMEM59 and TMEM59L affects APP shedding. A, HEK293 cells were transfected with control siRNA or siRNAs against TMEM59 and TMEM59L. Endogenous APP and its cleavage products were detected as described in the legend to Fig. 2A. B, knockdown (KD) of TMEM59 and its homolog reduced the amount of secreted APP (APPsα and APPsβ) by about 30–40% compared with control cells. Quantification of blots shown in A. Given are mean ± S.E. of 4 independent experiments. *, p < 0.05, determined with t test. C, siRNA treatment reduced mRNA levels of TMEM59 and TMEM59L by about 70–80%, as determined by quantitative reverse transcription-PCR. Given are mean ± S.E. of 4 independent experiments. ***, p < 0.001, determined with t test. D, endogenous TMEM59 (antibody 93) was detected in permeabilized HEK293 cells treated with control siRNAs (con) but not in cells treated with siRNAs against TMEM59 and TMEM59L (KD). Scale bars = 10 μm. For a more detailed immunofluorescence analysis, see Fig. 5.
TMEM59 Reduces APP Cell Surface Levels
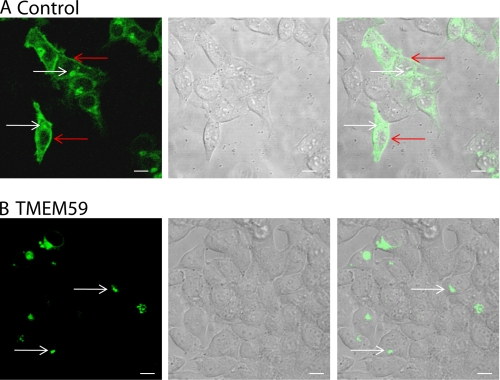
In addition to the reduction of APP maturation and shedding, the expression of TMEM59 strongly reduced APP levels at the cell surface, as observed by live cell imaging using a fusion protein (YFP-APP) of YFP and APP. In control transfected HEK293 cells YFP-APP was found at the plasma membrane and in the Golgi (Fig. 4A). In contrast, in cells transfected with TMEM59, YFP-APP fluorescence was confined to the Golgi and perinuclear sites, but no plasma membrane staining was observed (Fig. 4B). This analysis reveals that TMEM59 strongly reduced the APP levels in the later cellular compartments where APP normally undergoes shedding and is consistent with the reduced levels of APPsα and APPsβ upon TMEM59 expression. Taken together, TMEM59 strongly reduced the amount of mature APP (complex N- and O-glycosylated), APP staining at the cell surface, as well as APP cleavage by α- and β-secretase.
FIGURE 4.
TMEM59 leads to retention of APP in the Golgi. HEK293 cells were transiently cotransfected with YFP-APP and either control plasmid (con)(A) or TMEM59 plasmid (B). The living cells were analyzed using a Zeiss 510Meta confocal system. The arrows point to the plasma membrane (red) and the Golgi (white), respectively. Scale bars = 10 μm.
TMEM59 Is a Glycosylated, Golgi-localized Protein
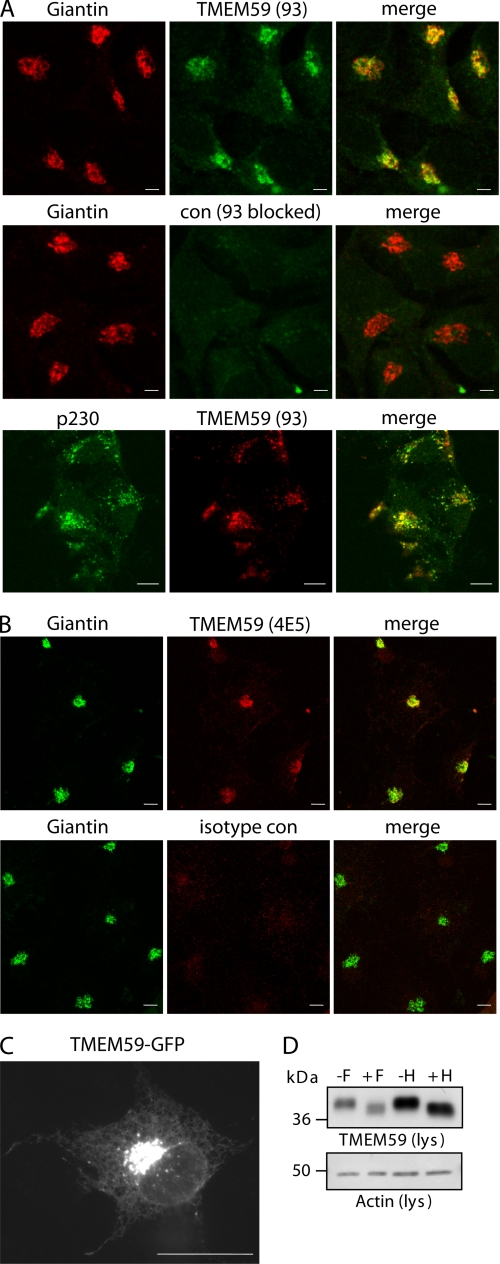
Given that TMEM59 reduced the Golgi glycosylation of APP and confines APP staining to the Golgi, we next tested whether TMEM59 itself is a Golgi-localized protein. Indeed, immunofluorescence analysis of COS cells demonstrated Golgi staining of the endogenous TMEM59, which overlapped with the well characterized Golgi markers giantin (cis-medial Golgi) (31, 56) and p230 (trans-Golgi and TGN) (57) (Fig. 5, A and B). With both markers a partial colocalization was observed, indicating localization of TMEM59 throughout all Golgi cisternae. A similar partial colocalization was observed with several other markers for the Golgi cisternae, such as GM130, GS15, and GS28 (data not shown).
FIGURE 5.
TMEM59 localizes to the Golgi. A, endogenous TMEM59 was detected in permeabilized COS cells with polyclonal antibody 93 (green) and showed costaining with the endogenous Golgi marker giantin (red). As a control the TMEM59 antibody did not detect TMEM59 when blocked by the antigenic peptide. TMEM59 was detected in permeabilized COS cells with the polyclonal antibody 93 (red) and showed costaining with the endogenous Golgi marker p230 (green). B, endogenous TMEM59 was detected in permeabilized COS cells with the monoclonal antibody 4E5 (red) and showed a costaining with the endogenous Golgi marker giantin (green). An isotype antibody served as control for the specificity of the 4E5 antibody. C, GFP-tagged TMEM59 was transiently transfected into COS cells. Scale bars = 10 μm. D, HEK293 cells were transiently transfected with HA-TMEM59. Aliquots of the cell lysate were treated (+) or not (−) with endoglycosidase F or H as indicated. The actin blot shows equal loading in all lanes.
The costaining with giantin was observed using two different antibodies against TMEM59, a polyclonal (Fig. 5A) and a monoclonal one (Fig. 5B). As a specificity control, TMEM59 was not detected, when the polyclonal antibody was blocked by the antigenic peptide (Fig. 5A) or when the isotype control antibody for the monoclonal antibody was used (Fig. 5B). Endogenous TMEM59 also showed Golgi staining in human neuroglioma H4 and human astroglioma U373 cells (data not shown), which is in agreement with its ubiquitous expression, as determined by Northern blot analysis (data not shown). GFP-tagged TMEM59 showed the same Golgi localization as the endogenous protein and to a lower extent additional reticular staining, presumably of the endoplasmic reticulum (Fig. 5C). These experiments show that TMEM59 is a Golgi-localized protein and may be used as a novel Golgi marker for immunofluorescence studies.
The Golgi localization of TMEM59 is in line with its glycosylation pattern. In the immunoblot the transfected TMEM59 showed an apparent molecular mass of about 40 kDa (Fig. 3D). Endogenous TMEM59 was not detected, presumably because the endogenous expression level was below the detection limit of the antibodies in the immunoblot. The apparent molecular mass of 40 kDa is slightly higher than its calculated molecular mass of 36 kDa. This difference is likely due to N-glycosylation at amino acid 90, where a typical N-glycosylation amino acid motif is located (Asn-Arg-Thr, Fig. 1). In fact, treatment with N-glycosidase F reduced the apparent molecular mass to about 38 kDa (Fig. 5D). The sugar moiety could also be removed with endoglycosidase H (Fig. 5D), which only removes sugars that are not of the complex type. This suggests that TMEM59 is localized in an early compartment of the secretory pathway and is in good agreement with Golgi localization observed by fluorescence microscopy.
TMEM59 Inhibits Complex Glycosylation in the Golgi
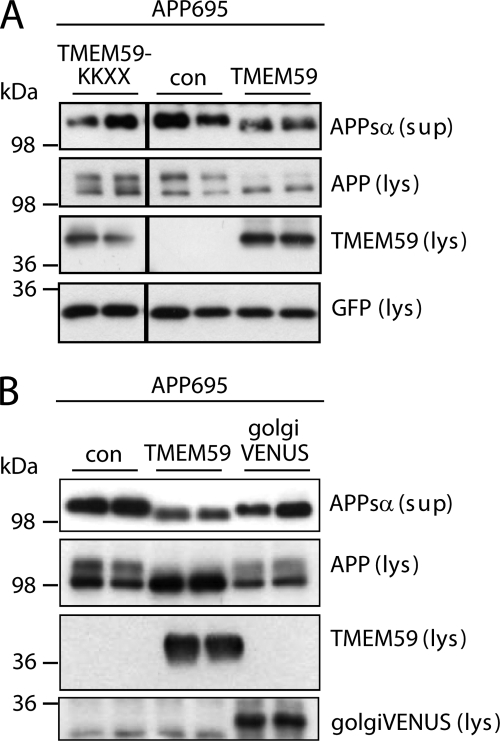
Next, we investigated in more detail the mechanism by which TMEM59 inhibits the maturation of APP. Given that TMEM59 localizes to the Golgi and blocks the Golgi glycosylation of APP, we generated a TMEM59 mutant with an added endoplasmic reticulum retention signal (TMEM59-KKXX). This mutant did not inhibit APP glycosylation and shedding compared with wild-type TMEM59 (Fig. 6A), revealing that TMEM59 must be able to leave the endoplasmic reticulum to inhibit APP shedding and maturation. Next, we tested whether an unrelated Golgi membrane protein could induce a similar effect on APP maturation and shedding as observed for TMEM59. To this aim we used golgiVENUS, which consists of the cytoplasmic and transmembrane domains of β-1,4-galactosyltransferase fused to the fluorescent protein VENUS. This protein showed Golgi staining (not shown), in agreement with a previous study (43). In contrast to TMEM59, golgiVENUS did not affect APP maturation and shedding (Fig. 6B), revealing that the inhibitory effect on APP glycosylation and shedding was specific to TMEM59.
FIGURE 6.
Control proteins do not affect APP shedding and maturation. A, control plasmid (con), HA-TMEM59 or HA-TMEM59-KKXX were transfected into HEK293 cells. The lysate (lys) was blotted against cellular APP (22C11), TMEM59 (HA.11), and GFP as a transfection control, the supernatant (sup) was blotted against secreted APPsα (W02). The vertical black lines in the gels indicate that the samples were run on the same gel, but not directly next to each other. B, control plasmid (con), HA-TMEM59, or golgiVENUS were transfected into HEK293 cells. The experiment was carried out as described in A, golgiVENUS was detected by anti-GFP.
By coimmunoprecipitation, no significant interaction was observed between TMEM59 and APP in HEK293 cells (data not shown), suggesting that TMEM59 does not directly act on APP. Instead, we considered the possibility that TMEM59 leads to a more general modulation of Golgi glycosylation reactions. To test this possibility, we analyzed whether TMEM59 also affects the complex N-glycosylation of two other proteins unrelated to APP, the β-secretase BACE1 and the prion protein (PrP). Both proteins are N-, but not O-glycosylated. Additionally, the immature (core N-glycosylation) and the mature (complex N-glycosylation) forms of both proteins can be well separated by gel electrophoresis. BACE1 was cotransfected with either control vector or TMEM59 into HEK293 cells. In control cells BACE1 was mainly present in a mature and to a low extent in an immature form, in agreement with previous publications (58, 59). The mature form is complex N-glycosylated and resistant to treatment with endoglycosidase H, which only removes N-linked sugars that are not complex glycosylated (Fig. 7A). In contrast, the immature form is sensitive to endoglycosidase H and lacks complex glycosylation (Fig. 7A) (58, 60). Expression of TMEM59 strongly suppressed the mature form of BACE1, but increased the amount of immature BACE1, which lacks complex glycosylation (Fig. 7A). This reveals that TMEM59 inhibits the complex N-glycosylation of BACE1.
PrP is a glycosylphosphatidylinositol-anchored membrane protein that is involved in the prion diseases. PrP expressed in HEK293 cells was present in differentially glycosylated forms (Fig. 7B), in agreement with previous publications (61, 62). The lower three bands correspond to the unglycosylated, the monoglycosylated, and the diglycosylated forms, which carry the core N-glycosylation in a high-mannose form. The upper broad band represents the complex N-glycosylated PrP (Fig. 7B). Similar to BACE1, expression of TMEM59 inhibited the complex N-glycosylation of PrP, but did not affect core glycosylation with the high-mannose sugars. Together with the data on APP these results suggest that TMEM59 induces a more general block of Golgi glycosylation reactions.
TMEM59 Does Not Affect the Secretion of Soluble Proteins
Because TMEM59 not only affects the Golgi glycosylation, but also the shedding of APP, we next tested, whether TMEM59 blocked general post-Golgi traffic. To this aim, the effect of TMEM59 on the secretion of the SEAP was investigated. SEAP is a complex N-glycosylated protein. Upon TMEM59 expression, the mature band of SEAP was absent in the cell lysate (Fig. 7C). In the conditioned medium the apparent molecular mass was decreased, which is in line with a lack of Golgi glycosylation. Despite the lack of SEAP maturation the total amount of SEAP secretion was not significantly reduced upon TMEM59 expression (Fig. 7C), demonstrating that TMEM59 does not affect the general protein secretion. Likewise, when the APP ectodomain (APPsβ) was expressed as a soluble protein lacking transmembrane and cytoplasmic domains, the amount of secretion was not altered upon TMEM59 expression (Fig. 7D). Similar to SEAP, the secreted APPsβ had a lower apparent molecular mass, indicating a loss of Golgi glycosylation. As a further control we tested whether TMEM59 affects shedding of the cytokine tumor necrosis factor α (TNFα), which is a non-glycosylated single-span membrane protein. TNFα undergoes shedding in an α-secretase like fashion similar to APP (63). In contrast to APP, TMEM59 did not alter TNFα shedding (Fig. 7E), demonstrating that it does not act on all proteins, which undergo shedding. Together, these results demonstrate that despite the more general inhibitory effect on Golgi glycosylation reactions and the reduction of APP shedding, TMEM59 does not inhibit all functions of the Golgi apparatus. Secretion of soluble proteins (SEAP, APPsβ) as well as shedding of an unglycosylated protein (TNFα) occurred normally.
TMEM59 Expression Resembles a Loss-of-function of the COG Complex
Next, we tested whether other proteins involved in Golgi N- and O-glycan processing would have a similar effect on APP maturation and shedding as TMEM59. Defects in Golgi glycan processing can also be caused by the loss of function of the COG complex. The COG complex is a hetero-octameric protein complex consisting of COG1 through COG8 and is required for correct localization and activity of Golgi-localized proteins, in particular enzymes involved in the glycosylation process (25, 26). For example, CHO cells lacking COG1 or COG2 show defects in the glycosylation and cell surface levels of the low density lipoprotein receptor and other protein glycoconjugates (64). This raises the possibility that the COG-deficient CHO cells show a similar defect of APP glycosylation and shedding as the TMEM59 expressing cells. To test this possibility, wild-type CHO cells as well as ldlB (COG1-deficient) and ldlC (COG2-deficient) CHO cells were cotransfected with APP and either control vector or TMEM59. In wild-type CHO cells, APP was detected in its mature and immature forms. Transfection of TMEM59 reduced APP maturation (Fig. 8A), consistent with the results in HEK293 cells (Fig. 8A). Likewise, APP shedding was reduced by about 70% as measured by the amount of APPsα detected with the W02 antibody (Fig. 8A). The remaining APP showed a slightly enhanced electrophoretic mobility, consistent with the results in HEK293 cells (Fig. 8A). Compared with the wild-type CHO cells, ldlB and ldlC cells showed no mature APP, indicating an inhibition of APP maturation (Fig. 8A), similar to the effect of TMEM59 observed in the wild-type cells. Additionally, when the amount of secreted APPsα was normalized to the total amount of APP in the cell lysate, a reduction of APP shedding by 40 and 55% was observed for ldlB and ldlC cells, respectively (Fig. 8A). Importantly, when COG1 was retransfected into ldlB (Cog1-deficient) cells ([ldlB]), APP maturation and shedding were restored to wild-type levels (Fig. 8B). This demonstrates that COG1 and COG2 are required for normal APP maturation and shedding. TMEM59 expression did not further reduce APP shedding in the ldlB and ldlC cells, suggesting that TMEM59 inhibits the same cellular Golgi glycosylation machinery, which requires the COG proteins.
DISCUSSION
APP shedding is a central event in Aβ generation, because shedding by β-secretase directly contributes to Aβ generation, whereas shedding by α-secretase prevents Aβ formation. The molecular mechanisms that regulate APP shedding are not yet well defined, but include cellular signaling pathways, such as the mitogen-activated protein kinase pathway, and a diverse group of compounds, including lipid derivatives, calcium, and cholinesterase inhibitors (14, 65). Additionally, recent studies increasingly imply intracellular APP trafficking as a mechanism to regulate access of APP to its secretases and thus, the amount of APP processing (15). This study identifies TMEM59 as a novel protein modulating cell surface APP levels, APP shedding by α- and β-secretase, and Aβ generation. Moreover, this study identifies three additional proteins: TMEM59L, COG1, and COG2 as novel modulators of APP shedding.
Our study shows that TMEM59 is a novel Golgi-localized protein, which affects glycosylation processes in the Golgi, but not in the endoplasmic reticulum. TMEM59 expression blocked both O-glycosylation and complex N-glycosylation, which occur in the Golgi, but did not affect the cotranslational addition of the core N-glycosylation in the endoplasmic reticulum. A function of TMEM59 as a modulator of Golgi glycosylation was further supported by the finding that TMEM59 induced a similar phenotype on APP glycosylation and shedding as the loss of COG1 and COG2. The COG complex is a hetero-octameric protein complex consisting of subunits COG1 through COG8. The COG complex is assumed to act as a retrograde vesicle tethering factor in intra-Golgi trafficking and is particularly required for the correct localization and activity of Golgi glycosylation enzymes (25, 26). Deficiencies in individual COG complex proteins are linked to human congenital disorders of glycosylation and result in different defects in protein glycosylation, presumably due to mislocalization or degradation of enzymes involved in the glycosylation process (26, 27). In particular, CHO cells lacking COG1 (ldlB cells) or COG2 (ldlC cells) show defects in glycosylation and cell surface levels of the low density lipoprotein receptor and other protein glycoconjugates (64) without apparent defects in overall secretion or endocytosis (66).
In contrast to the COG proteins, which are soluble proteins binding to the cytoplasmic face of the Golgi, TMEM59 and its homolog TMEM59L are membrane proteins. Potentially, both proteins affect the activity of one or several Golgi glycosylation enzymes, which are also transmembrane proteins. Alternatively, TMEM59 may indirectly modulate Golgi glycosylation by affecting the correct cellular localization of the corresponding glycosylation enzymes.
The reduction of APP Golgi glycosylation was accompanied by reduced levels of mature APP and slightly increased levels of immature APP in TMEM59 expressing cells. Additionally, TMEM59 induced a predominant Golgi staining of APP and a reduction of cell surface APP staining. These results suggest that TMEM59 prevents access of APP to the plasma membrane and the endosomes, where it is normally cleaved by α- and β-secretase. The activity of both enzymes was found to be not affected by TMEM59 expression. Additionally, the shedding of TNFα as well as the secretion of soluble proteins, such as SEAP, were not affected, demonstrating that TMEM59 did not block general protein transport to the cell surface. Currently, it is unclear why TMEM59 induces the inhibition of cell surface transport of APP but not TNFα or soluble proteins. We consider the following possibilities. Potentially, there is a cellular control mechanism ensuring that Golgi glycosylation of APP, but not the other proteins, is complete, before APP can exit the Golgi or the TGN for its transport to the plasma membrane. However, this scenario seems unlikely, because a previous study found that APP shedding was normal in CHO cells expressing an APP mutant lacking N-glycosylation sites or when APP lacked normal O-glycosylation (67).
Given that TMEM59 not only inhibits the glycosylation of APP, an alternative explanation could be that an as yet unknown glycoprotein is required for APP transport to the cell surface, regardless of whether APP is fully glycosylated or not. This conclusion has also been drawn in a previous study that analyzed APP shedding in CHO cells with reduced N- or O-glycosylation (67). We would expect that upon TMEM59 expression such a glycoprotein would no longer be complex glycosylated and may have lost its function of allowing APP transport to the cell surface. Although such a protein is not yet known, it is interesting to note that APP does associate with glycosylated membrane proteins, such as SorL1 and LRP1, and that this interaction occurs in the secretory pathway (16, 68). Whether these or other proteins require their glycosylation for correct transport of APP remains to be tested in future studies.
In an alternative scenario, the plasma membrane transport of APP may occur in a manner different from those proteins, which were secreted normally, even in the absence of complex glycosylation. In fact, different membrane proteins use different transport routes from the TGN to the plasma membrane. For example, distinct TGN exit sites have been reported for the transport of TNFα and E-cadherin to the plasma membrane. Although TNFα leaves the TGN through p230/golgin-245-coated tubules, E-cadherin exits through golgin-97-coated tubules (69). If APP exits the TGN through other sites than TNFα and the soluble proteins, we would expect that TMEM59 blocks the exit site for APP but not for the other proteins.
Our finding that changes in Golgi glycan processing not only affect APP glycosylation, but also APP cell surface levels and APP shedding by α- and β-secretase is in agreement with previous studies. One study blocked complex N-glycosylation in the Golgi by use of the drug swainsonine, which inhibits Golgi-localized mannosidase II (19). Under these conditions APP showed increased Golgi localization, which supports the idea that changes in the Golgi glycosylation process can impair the intracellular transport of APP. Additionally, APP shedding was reduced, presumably both by α- and β-secretase. Conversely, transfection of the α2,6-sialyltransferase ST6Gal-I increased APP sialylation and enhanced APP shedding by both α- and β-secretase, as well as Aβ secretion (19, 20). In this case, the increased APP shedding appeared to result specifically from the enhanced sialylation of APP and not other cellular proteins (20).
Interestingly, changes in the activity of glycan processing enzymes have been observed in AD brains compared with age-matched controls, such as reduction in the activity of sialyltransferases and a corresponding change in general protein glycosylation (70, 71). Because sialylation increases APP secretion, a reduced sialyltransferase activity may contribute to the reduced levels of the neuroprotective and neurotrophic APPsα, as it is found in Alzheimer disease (72). However, it remains to be determined whether the altered sialyltransferase activity directly contributes to the disease process or occurs as a consequence of the disease process.
Acknowledgment
We thank Monty Krieger for providing COG-deficient and wild-type CHO cells.
This work was supported by grants from Deutsche Forschungsgemeinschaft SFB596 Projects B12 (to S. F. L.), B4 (to J. T.), and Z2 (to E. K.), the competence network degenerative dementias (Bundesministerium für Bildung und Forschung) (to S. F. L.), the European Commission (NeuroNE), and the Friedrich-Baur-Stiftung (to S. F. L.).
- APP
- amyloid precursor protein
- AD
- Alzheimer disease
- ADAM
- a disintegrin and metalloprotease
- APPs
- soluble form of APP
- COG
- conserved oligomeric golgi
- HEK
- human embryonic kidney
- YFP
- yellow fluorescent protein
- TGN
- trans-Golgi network
- SEAP
- soluble enzyme secretory alkaline phosphatase
- TNFα
- tumor necrosis factor α
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- CHO
- Chinese hamster ovary
- siRNA
- small interfering RNA
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- PrP
- prion protein
- TMEM59
- transmembrane protein 59
- wt
- wild-type.
REFERENCES
- 1.Selkoe D. J., Schenk D. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 545–584 [DOI] [PubMed] [Google Scholar]
- 2.Haass C. (2004) EMBO J. 23, 483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole S. L., Vassar R. (2008) Curr. Alzheimer Res. 5, 100–120 [DOI] [PubMed] [Google Scholar]
- 4.Rossner S., Sastre M., Bourne K., Lichtenthaler S. F. (2006) Prog. Neurobiol. 79, 95–111 [DOI] [PubMed] [Google Scholar]
- 5.Steiner H., Fluhrer R., Haass C. (2008) J. Biol. Chem. 283, 29627–29631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa K., Sopher B. L., Rydel R. E., Begley J. G., Pham D. G., Martin G. M., Fox M., Mattson M. P. (1996) J. Neurochem. 67, 1882–1896 [DOI] [PubMed] [Google Scholar]
- 7.Meziane H., Dodart J. C., Mathis C., Little S., Clemens J., Paul S. M., Ungerer A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12683–12688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stein T. D., Anders N. J., DeCarli C., Chan S. L., Mattson M. P., Johnson J. A. (2004) J. Neurosci. 24, 7707–7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allinson T. M., Parkin E. T., Condon T. P., Schwager S. L., Sturrock E. D., Turner A. J., Hooper N. M. (2004) Eur. J. Biochem. 271, 2539–2547 [DOI] [PubMed] [Google Scholar]
- 10.Koike H., Tomioka S., Sorimachi H., Saido T. C., Maruyama K., Okuyama A., Fujisawa-Sehara A., Ohno S., Suzuki K., Ishiura S. (1999) Biochem. J. 343, 371–375 [PMC free article] [PubMed] [Google Scholar]
- 11.Lammich S., Kojro E., Postina R., Gilbert S., Pfeiffer R., Jasionowski M., Haass C., Fahrenholz F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3922–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slack B. E., Ma L. K., Seah C. C. (2001) Biochem. J. 357, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allinson T. M., Parkin E. T., Turner A. J., Hooper N. M. (2003) J. Neurosci. Res. 74, 342–352 [DOI] [PubMed] [Google Scholar]
- 14.Lichtenthaler S. F. (2006) Neurodegener. Dis. 3, 262–269 [DOI] [PubMed] [Google Scholar]
- 15.Sannerud R., Annaert W. (2009) Semin. Cell Dev. Biol. 20, 183–190 [DOI] [PubMed] [Google Scholar]
- 16.Andersen O. M., Reiche J., Schmidt V., Gotthardt M., Spoelgen R., Behlke J., von Arnim C. A., Breiderhoff T., Jansen P., Wu X., Bales K. R., Cappai R., Masters C. L., Gliemann J., Mufson E. J., Hyman B. T., Paul S. M., Nykjaer A., Willnow T. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13461–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogaeva E., Meng Y., Lee J. H., Gu Y., Kawarai T., Zou F., Katayama T., Baldwin C. T., Cheng R., Hasegawa H., Chen F., Shibata N., Lunetta K. L., Pardossi-Piquard R., Bohm C., Wakutani Y., Cupples L. A., Cuenco K. T., Green R. C., Pinessi L., Rainero I., Sorbi S., Bruni A., Duara R., Friedland R. P., Inzelberg R., Hampe W., Bujo H., Song Y. Q., Andersen O. M., Willnow T. E., Graff-Radford N., Petersen R. C., Dickson D., Der S. D., Fraser P. E., Schmitt-Ulms G., Younkin S., Mayeux R., Farrer L. A., St. George-Hyslop P. (2007) Nat. Genet. 39, 168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nixon R. A. (2005) Neurobiol. Aging 26, 373–382 [DOI] [PubMed] [Google Scholar]
- 19.McFarlane I., Georgopoulou N., Coughlan C. M., Gillian A. M., Breen K. C. (1999) Neuroscience 90, 15–25 [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa K., Kitazume S., Oka R., Maruyama K., Saido T. C., Sato Y., Endo T., Hashimoto Y. (2006) J. Neurochem. 96, 924–933 [DOI] [PubMed] [Google Scholar]
- 21.Yazaki M., Tagawa K., Maruyama K., Sorimachi H., Tsuchiya T., Ishiura S., Suzuki K. (1996) Neurosci. Lett. 221, 57–60 [DOI] [PubMed] [Google Scholar]
- 22.Påhlsson P., Shakin-Eshleman S. H., Spitalnik S. L. (1992) Biochem. Biophys. Res. Commun. 189, 1667–1673 [DOI] [PubMed] [Google Scholar]
- 23.Oltersdorf T., Ward P. J., Henriksson T., Beattie E. C., Neve R., Lieberburg I., Fritz L. C. (1990) J. Biol. Chem. 265, 4492–4497 [PubMed] [Google Scholar]
- 24.Weidemann A., König G., Bunke D., Fischer P., Salbaum J. M., Masters C. L., Beyreuther K. (1989) Cell 57, 115–126 [DOI] [PubMed] [Google Scholar]
- 25.Smith R. D., Lupashin V. V. (2008) Carbohydr. Res. 343, 2024–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ungar D., Oka T., Krieger M., Hughson F. M. (2006) Trends Cell Biol. 16, 113–120 [DOI] [PubMed] [Google Scholar]
- 27.Zeevaert R., Foulquier F., Jaeken J., Matthijs G. (2008) Mol. Genet. Metab. 93, 15–21 [DOI] [PubMed] [Google Scholar]
- 28.Schöbel S., Neumann S., Hertweck M., Dislich B., Kuhn P. H., Kremmer E., Seed B., Baumeister R., Haass C., Lichtenthaler S. F. (2008) J. Biol. Chem. 283, 14257–14268 [DOI] [PubMed] [Google Scholar]
- 29.Schöbel S., Neumann S., Seed B., Lichtenthaler S. F. (2006) Int. J. Dev. Neurosci. 24, 141–148 [DOI] [PubMed] [Google Scholar]
- 30.Elson G. C., de Coignac A. B., Aubry J. P., Delneste Y., Magistrelli G., Holzwarth J., Bonnefoy J. Y., Gauchat J. F. (1999) Biochem. Biophys. Res. Commun. 264, 55–62 [DOI] [PubMed] [Google Scholar]
- 31.Linstedt A. D., Hauri H. P. (1993) Mol. Biol. Cell 4, 679–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steiner H., Kostka M., Romig H., Basset G., Pesold B., Hardy J., Capell A., Meyn L., Grim M. L., Baumeister R., Fechteler K., Haass C. (2000) Nat. Cell Biol. 2, 848–851 [DOI] [PubMed] [Google Scholar]
- 33.Ida N., Hartmann T., Pantel J., Schröder J., Zerfass R., Förstl H., Sandbrink R., Masters C. L., Beyreuther K. (1996) J. Biol. Chem. 271, 22908–22914 [DOI] [PubMed] [Google Scholar]
- 34.Yamasaki A., Eimer S., Okochi M., Smialowska A., Kaether C., Baumeister R., Haass C., Steiner H. (2006) J. Neurosci. 26, 3821–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kascsak R. J., Rubenstein R., Merz P. A., Tonna-DeMasi M., Fersko R., Carp R. I., Wisniewski H. M., Diringer H. (1987) J. Virol. 61, 3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lichtenthaler S. F., Dominguez D. I., Westmeyer G. G., Reiss K., Haass C., Saftig P., De Strooper B., Seed B. (2003) J. Biol. Chem. 278, 48713–48719 [DOI] [PubMed] [Google Scholar]
- 37.Lichtenthaler S. F., Ida N., Multhaup G., Masters C. L., Beyreuther K. (1997) Biochemistry 36, 15396–15403 [DOI] [PubMed] [Google Scholar]
- 38.Lammich S., Schöbel S., Zimmer A. K., Lichtenthaler S. F., Haass C. (2004) EMBO Rep. 5, 620–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhn P. H., Marjaux E., Imhof A., De Strooper B., Haass C., Lichtenthaler S. F. (2007) J. Biol. Chem. 282, 11982–11995 [DOI] [PubMed] [Google Scholar]
- 40.Neumann S., Schöbel S., Jäger S., Trautwein A., Haass C., Pietrzik C. U., Lichtenthaler S. F. (2006) J. Biol. Chem. 281, 7583–7594 [DOI] [PubMed] [Google Scholar]
- 41.Gilch S., Winklhofer K. F., Groschup M. H., Nunziante M., Lucassen R., Spielhaupter C., Muranyi W., Riesner D., Tatzelt J., Schätzl H. M. (2001) EMBO J. 20, 3957–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ehehalt R., Keller P., Haass C., Thiele C., Simons K. (2003) J. Cell Biol. 160, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okita C., Sato M., Schroeder T. (2004) BioTechniques 36, 418–422, 424 [DOI] [PubMed] [Google Scholar]
- 44.Chatterton J. E., Hirsch D., Schwartz J. J., Bickel P. E., Rosenberg R. D., Lodish H. F., Krieger M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schägger H., von Jagow G. (1987) Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 46.Cissé M. A., Gandreuil C., Hernandez J. F., Martinez J., Checler F., Vincent B. (2006) Biochem. Biophys. Res. Commun. 347, 254–260 [DOI] [PubMed] [Google Scholar]
- 47.Alfa Cissé M., Sunyach C., Slack B. E., Fisher A., Vincent B., Checler F. (2007) J. Neurosci. 27, 4083–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrau D., Dumanchin-Njock C., Ayral E., Vizzavona J., Farzan M., Boisbrun M., Fulcrand P., Hernandez J. F., Martinez J., Lefranc-Jullien S., Checler F. (2003) J. Biol. Chem. 278, 25859–25866 [DOI] [PubMed] [Google Scholar]
- 49.Lefranc-Jullien S., Lisowski V., Hernandez J. F., Martinez J., Checler F. (2005) Br. J. Pharmacol. 145, 228–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sastre M., Steiner H., Fuchs K., Capell A., Multhaup G., Condron M. M., Teplow D. B., Haass C. (2001) EMBO Rep. 2, 835–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stachel S. J., Coburn C. A., Steele T. G., Jones K. G., Loutzenhiser E. F., Gregro A. R., Rajapakse H. A., Lai M. T., Crouthamel M. C., Xu M., Tugusheva K., Lineberger J. E., Pietrak B. L., Espeseth A. S., Shi X. P., Chen-Dodson E., Holloway M. K., Munshi S., Simon A. J., Kuo L., Vacca J. P. (2004) J Med. Chem. 47, 6447–6450 [DOI] [PubMed] [Google Scholar]
- 52.Verstreken P., Kjaerulff O., Lloyd T. E., Atkinson R., Zhou Y., Meinertzhagen I. A., Bellen H. J. (2002) Cell 109, 101–112 [DOI] [PubMed] [Google Scholar]
- 53.Sugiura H., Iwata K., Matsuoka M., Hayashi H., Takemiya T., Yasuda S., Ichikawa M., Yamauchi T., Mehlen P., Haga T., Yamagata K. (2004) J. Biol. Chem. 279, 23343–23348 [DOI] [PubMed] [Google Scholar]
- 54.Pietrzik C. U., Yoon I. S., Jaeger S., Busse T., Weggen S., Koo E. H. (2004) J. Neurosci. 24, 4259–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitmarsh A. J. (2006) Biochem. Soc. Trans. 34, 828–832 [DOI] [PubMed] [Google Scholar]
- 56.Linstedt A. D., Foguet M., Renz M., Seelig H. P., Glick B. S., Hauri H. P. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5102–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gleeson P. A., Anderson T. J., Stow J. L., Griffiths G., Toh B. H., Matheson F. (1996) J. Cell Sci. 109, 2811–2821 [DOI] [PubMed] [Google Scholar]
- 58.Huse J. T., Pijak D. S., Leslie G. J., Lee V. M., Doms R. W. (2000) J. Biol. Chem. 275, 33729–33737 [DOI] [PubMed] [Google Scholar]
- 59.Westmeyer G. G., Willem M., Lichtenthaler S. F., Lurman G., Multhaup G., Assfalg-Machleidt I., Reiss K., Saftig P., Haass C. (2004) J. Biol. Chem. 279, 53205–53212 [DOI] [PubMed] [Google Scholar]
- 60.Capell A., Steiner H., Willem M., Kaiser H., Meyer C., Walter J., Lammich S., Multhaup G., Haass C. (2000) J. Biol. Chem. 275, 30849–30854 [DOI] [PubMed] [Google Scholar]
- 61.Endo T., Groth D., Prusiner S. B., Kobata A. (1989) Biochemistry 28, 8380–8388 [DOI] [PubMed] [Google Scholar]
- 62.Winklhofer K. F., Heller U., Reintjes A., Tatzelt J. (2003) Traffic 4, 313–322 [DOI] [PubMed] [Google Scholar]
- 63.Hooper N. M., Karran E. H., Turner A. J. (1997) Biochem. J. 321, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kingsley D. M., Kozarsky K. F., Segal M., Krieger M. (1986) J. Cell Biol. 102, 1576–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vardy E. R., Catto A. J., Hooper N. M. (2005) Trends Mol. Med. 11, 464–472 [DOI] [PubMed] [Google Scholar]
- 66.Oka T., Krieger M. (2005) J. Biochem. 137, 109–114 [DOI] [PubMed] [Google Scholar]
- 67.Påhlsson P., Spitalnik S. L. (1996) Arch. Biochem. Biophys. 331, 177–186 [DOI] [PubMed] [Google Scholar]
- 68.Waldron E., Heilig C., Schweitzer A., Nadella N., Jaeger S., Martin A. M., Weggen S., Brix K., Pietrzik C. U. (2008) Neurobiol. Dis. 31, 188–197 [DOI] [PubMed] [Google Scholar]
- 69.Lieu Z. Z., Lock J. G., Hammond L. A., La Gruta N. L., Stow J. L., Gleeson P. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3351–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Breen K. C., Coughlan C. M., Hayes F. D. (1998) Mol. Neurobiol. 16, 163–220 [DOI] [PubMed] [Google Scholar]
- 71.Maguire T. M., Breen K. C. (1995) Dementia 6, 185–190 [DOI] [PubMed] [Google Scholar]
- 72.Fellgiebel A., Kojro E., Müller M. J., Scheurich A., Schmidt L. G., Fahrenholz F. (2009) J. Geriatr. Psychiatry Neurol. 22, 3–9 [DOI] [PubMed] [Google Scholar]