Abstract
Hsp90 requires cochaperone Cdc37 to load its clients to the Hsp90 superchaperone complex. The purpose of this study was to utilize split Renilla luciferase protein fragment-assisted complementation (SRL-PFAC) bioluminescence to study the full-length human Hsp90-Cdc37 complex and to identity critical residues and their contributions for Hsp90/Cdc37 interaction in living cells. SRL-PFAC showed that full-length human Hsp90/Cdc37 interaction restored dramatically high luciferase activity through Hsp90-Cdc37-assisted complementation of the N and C termini of luciferase (compared with the set of controls). Immunoprecipitation confirmed that the expressed fusion proteins (NRL-Hsp90 and Cdc37-CRL) preserved their ability to interact with each other and also with native Hsp90 or Cdc37. Molecular dynamic simulation revealed several critical residues in the two interaction patches (hydrophobic and polar) at the interface of Hsp90/Cdc37. Mutagenesis confirmed the critical residues for Hsp90-Cdc37 complex formation. SRL-PFAC bioluminescence evaluated the contributions of these critical residues in Hsp90/Cdc37 interaction. The results showed that mutations in Hsp90 (Q133A, F134A, and A121N) and mutations in Cdc37 (M164A, R167A, L205A, and Q208A) reduced the Hsp90/Cdc37 interaction by 70–95% as measured by the resorted luciferase activity through Hsp90-Cdc37-assisted complementation. In comparison, mutations in Hsp90 (E47A and S113A) and a mutation in Cdc37 (A204E) decreased the Hsp90/Cdc37 interaction by 50%. In contrast, mutations of Hsp90 (R46A, S50A, C481A, and C598A) and mutations in Cdc37 (C54S, C57S, and C64S) did not change Hsp90/Cdc37 interactions. The data suggest that single amino acid mutation in the interface of Hsp90/Cdc37 is sufficient to disrupt its interaction, although Hsp90/Cdc37 interactions are through large regions of hydrophobic and polar interactions. These findings provides a rationale to develop inhibitors for disruption of the Hsp90/Cdc37 interaction.
Keywords: Computer Modeling, Mutant, Protein Structure, Protein/Protein Interactions, Tumor Therapy, Cdc37, Critical Residues, Hsp90, SRL-PFAC
Introduction
The 90-kDa heat shock protein (Hsp90)4 is a ubiquitous and essential molecular chaperone with multiple functions in eukaryotic cells under both stressed and nonstressed conditions (1, 2). It plays a central role in post-translational folding and stability of over 100 signaling proteins, including steroid hormone receptors, the dioxin receptor, growth factor receptors, transcription factors, protein kinases, and enzymes (3). Many of these Hsp90 clients are crucial in tumorigenesis, and when these proteins are dysregulated, they contribute to the hallmark traits of cancer. It has also been reported that the expression of Hsp90 in cancer cells is 2–10-fold higher compared with their normal counterparts (4). Therefore, tumor cells show much more sensitivity when subjected to Hsp90 inhibition than nontransformed cells (5), which suggests the important function of Hsp90 in tumor progression.
Hsp90 consists of the following three highly conserved domains: a 25-kDa N-terminal domain, a 35-kDa middle domain, and a 10-kDa C-terminal domain (6). A nucleotide binding pocket is located on the N terminus, by which ATP binds Hsp90 and is subsequently hydrolyzed to induce a conformational change of Hsp90. These conformational changes help Hsp90 to interact with various cochaperones to form different complexes in the chaperoning cycles (7–9). The middle segment of Hsp90 is considered to be a major site for binding client proteins (10), and the C terminus is essential for Hsp90 dimerization (8).
Hsp90 interacts with multiple cochaperones to form a superchaperone complex, including Cdc37, Aha1, p23/Sba1, Hop, Hsp70, and Hsp40 (8, 11, 12). Each component in the complex has its own function associated with different types of clients. Cdc37, originally named as p50, was reported as an accessory factor to load the subset of protein kinases to Hsp90 in the intermediate Hsp90 superchaperone complex (13–15). Silencing Cdc37 reduces expression of the Hsp90 clients ERBB2, CRAF, CDK4, CDK6, and phosphorylated AKT, which are highly relevant to cancer progression (16). The involvement of the Hsp90-Cdc37 complex in the maturation and activity of oncogenic protein kinases makes the complex a potential therapeutic target for cancer chemotherapy. Recently, a quinone methide triterpene compound celastrol was shown to disrupt Hsp90/Cdc37 interaction and exhibited anticancer activity (17, 18), supporting the potential application of disrupting the protein/protein interaction of the Hsp90-Cdc37 complex in cancer therapy.
Cdc37 is composed of the following three domains: a 15.5-kDa N-terminal domain (residues 1–127), a 16-kDa middle domain (residues 147–276), and a 10.5-kDa C-terminal domain (residues 283–378) (19, 20). Cdc37 forms a complex with the N terminus of Hsp90 through its middle and C-terminal portions (21, 22). Crystal structures of yeast Hsp90 and human Cdc37 revealed that the interaction between the two proteins is through a flat hydrophobic patch and is reinforced by a network of polar interactions, burying ≈1056 Å2 of the molecular surface (21). NMR mapping using human N-terminal Hsp90 (residues 18–223) and middle terminal Cdc37 (residues 147–276) fragments further showed a series of residues in the interaction patch, including Ser-113, Lys-116, Ala-117, Glu-120, Ala-121, Ala-124, Ala-126, Met-130, Gln-133, and Phe-134 of Hsp90 and His-161, Met-164, Leu-165, Arg-166, Arg-167, Asp-170, Trp-193, Ala-204, Leu-205, and Gln-208 of Cdc37 (23).
However, the crystallographic study of yeast Hsp90/human Cdc37 interaction and NMR of human Hsp90/human Cdc37 interaction only used fragments of Hsp90 and Cdc37, due to the large size of full-length Hsp90 and Cdc37. In addition, the crystal structure of Hsp90/Cdc37 using yeast Hsp90, and the amino acids involved in Hsp90/Cdc37 interactions revealed by NMR and crystallography have not been fully validated (21, 23). Furthermore, the study of Hsp90/Cdc37 interactions has been limited to the use of purified protein fragments or immunoprecipitation using cell lines (24–26). Because these methods are sensitive to the ionic strength of the detergents/buffers, they may not truly reflect the nature of interactions between Hsp90/Cdc37 in intact living cells.
Therefore, in this study, we have utilized a bioluminescence imaging, SRL-PFAC, to study full-length human Hsp90/human Cdc37 interaction in living cells. SRL-PFAC imaging system was developed by Paulmurugan and Gambhir (27). It is a complementation-based bioluminescence assay to quantitatively measure real time protein/protein interactions in living cells. It is based on division of the full-length Renilla luciferase into two separate inactive halves that can reconstitute function upon complementation. When fused to two interacting proteins, the luciferase reporter fragments are complemented upon association of the interacting proteins, thus showing different degrees of bioluminescence due to different levels of protein interaction (27–29). In our study, we applied SRL-PFAC to identify critical amino acid residues for the formation of the full-length human Hsp90-Cdc37 complex and evaluated the contributions of the critical amino acid residues in the interaction of Hsp90/Cdc37 in living cells. In addition, we used computational modeling and molecular dynamics simulations to evaluate the details of the interaction interface of Hsp90-Cdc37 complex. These critical interacting amino acid residues were confirmed by mutagenesis, and their contributions in Hsp90/Cdc37 interaction were evaluated using the SRL-PFAC system in living cells. We found that although Hsp90/Cdc37 interactions are through both hydrophobic and polar interactions, mutation in a single amino acid residue in the hydrophobic patch or the polar interaction patch of either Hsp90 or Cdc37, including Ala-121, Gln-133, and Phe-134 in Hsp90 and Met-164, Arg-167, Leu-205, and Gln-208 in Cdc37, is sufficient to disrupt the Hsp90/Cdc37 interaction. These finding provides a rationale to develop inhibitors for disruption of the Hsp90/Cdc37 interaction.
EXPERIMENTAL PROCEDURES
Chemicals
The pCR-BluntII-TOPO vector encoding cDNA for human Hsp90α and pINCY vector encoding human Cdc37 were purchased from Open Biosystems (Huntsville, AL). The pG5Luc vector encoding the full-length firefly luciferase (FL) and pGL4.75 [hRluc/CMV] vector encoding Renilla luciferase (RL), the Dual-Luciferase reporter assay system kit, and the EnduRen Live Cell Substrate were purchased from Promega (Madison, WI). Endonuclease enzymes were purchased from New England Biolabs (Ipswich, MA). pcDNA3.1(+) vector, TaqDNA polymerase, T4 DNA ligase, and Lipofectamine2000 transfection reagents, bacteria culture media, and ampicillin were purchased from Invitrogen. Plasmid purification, DNA gel extraction, and PCR purification kits were purchased from Qiagen (Valencia, CA). All site-directed mutants were performed using the Stratagene multisite-directed mutagenesis kit (La Jolla, CA). Native coelenterazine was purchased from Nanolight Technology (Pinetop, AZ). Cell culture media, fetal bovine serum, penicillin, streptomycin, and trypsin were purchased from Invitrogen. The cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 1% penicillin/streptomycin in 5% CO2, 37 °C.
Construction of Plasmids
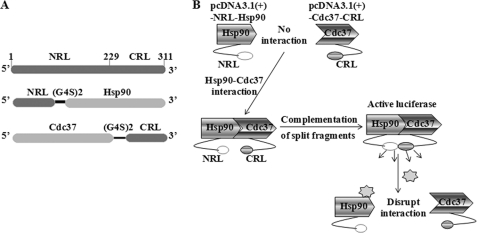
The full-length firefly luciferase was directly subcloned into pcDNA3.1(+) vector through BamHI and XhoI restriction enzyme sites. The N-terminal (NRL, amino acid 1–229) and the C-terminal (CRL, amino acid 230–311) Renilla luciferases were PCR-amplified using the forward primers designed with NheI or BamHI with a start codon and the corresponding reverse primers designed with BamHI with a stop codon or XhoI. The NRL and CRL were also PCR-amplified with linker DNA sequences GGTGGCGGAGGGAGCGGTGGCGGAGGGAGC (corresponding to peptide GGGGSGGGGS) designed to the NRL reverse primers or the CRL forward primers (27). The full-length human Hsp90α was amplified and subcloned downstream of NRL with linker using the corresponding restriction enzymes. The full-length human Cdc37 was PCR-amplified and subcloned upstream of linker with CRL. The N terminus of human Hsp90α (amino acids 1–223 corresponding to the cDNA base pairs) was PCR-amplified using the forward primers designed with a BamHI site and reverse primers with an XhoI site and a stop codon. The middle and C termini of human Hsp90α (amino acid 224–732) was also PCR-amplified with the forward and reverse primers with BamHI and XhoI sites, respectively. The constructed Hsp90N and Hsp90MC were subcloned into the downstream of the NRL with linker using corresponding restriction enzymes, respectively. All constructs (Fig. 1A) were verified by restriction enzyme digestion and DNA sequencing.
FIGURE 1.
A, schematic diagram for the plasmid constructs. The two interacting proteins Hsp90 and Cdc37 are fused to the NRL (amino acids 1–229) and CRL (amino acids 230–311) portion of the RL through an (G4S)2 peptide linker. B, schematic diagram for monitoring Hsp90/Cdc37 interaction using SRL-PFAC system. The interactions between Hsp90 and Cdc37 bring NRL and CRL in close proximity and lead to complementation of RL enzyme activity and photon production in the presence of the substrate coelenterazine.
Cell Transfection, FL, and RL Assay
Cells were plated in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and no antibiotics 1 day before transfection and cultured 24 h to reach a 90% confluence. Dulbecco's modified Eagle's medium were then replaced with serum-free Opti-MEM prior to transfection. For transfection, 0.25 μg of pcDNA3.1(+) molar equivalent of each constructed plasmid was used in each well of 24-well plates, and 17.5 ng/well pcDNA3.1(+)-FL plasmids were cotransfected to normalize the transfection efficiency. The Lipofectamine2000 from Invitrogen was used as described in the manual. After 6 h, the culture medium was replaced with Dulbecco's modified Eagle's medium, 10% fetal bovine serum, and no antibiotics.
The cells were cultured at 37 °C with 5% CO2 for 48 h. After 48 h, the cells were washed twice with cold phosphate-buffered saline and lysed with Passive Lysis Buffer. The luminometer assays for FL and RL activity were performed using Promega Dual-Luciferase reporter assay system according to instructions. The luciferase activity was measured using Bio-Tek Synergy 2 microplate reader. RL activities were normalized for transfection efficiency using FL activity.
Computational Modeling and Molecular Dynamics Simulations
The complex of human Hsp90 with human Cdc37 was obtained from the Protein Data Bank data base, code 2K5B. This model was created by utilizing the crystal structure of Cdc37 and NMR-based constrained fitting of Hsp90 (23). Molecular dynamic (MD) simulations were performed in Amber for 3 ns using explicit TIP3P solvent. After initial minimization of the solvent, the system was further relaxed with constraints on the backbone before final minimization. MD simulations involved a gradual increase in temperature to 300 K over 30 ps, while holding the solute constrained, followed by another 30 ps of simulation with the constraint only on the backbone. Further equilibration was performed for 40 ps before the production run. Shaking was applied to all bonds involving hydrogen to permit a time step of 0.002 ps. The B-factors for the residues were computed to identify the relative stability of the residues at the interface during the simulation and to identify the residues that might be involved in strong interactions. The enthalpic contributions of the residues at the interface were obtained by utilizing the MM/GBSA (30) method over the structures saved every 1 ps. This analysis involved the in silico mutation of the interfacial residues to Ala (Gly for Ala residues). A set of residues at the interface was subsequently selected for mutagenesis study.
Mutageneses of Hsp90 and Cdc37 Critical Residues
Mutagenesis of NRL-Hsp90 and Cdc37-CRL was generated using the Stratagene mutagenesis kit and confirmed by direct sequencing. Briefly, 100 ng of pcDNA3.1(+)-NRL-Hsp90 or pcDNA3.1(+)-Cdc37-CRL template and 100 ng of each mutagenic primer were used in the reaction with a total reaction volume of 25 μl. After 30 cycles of PCR amplification of DNA template, 1 μl of DpnI restriction enzyme was added to 10 μl of each amplification reaction and incubated at 37 °C for 3 h. 1.5 μl of the DpnI-treated DNA from each mutagenesis reaction was transferred to XL10-Gold ultracompetent cells. The DNA was purified using Qiagen purification kit (Qiagen, Valencia, CA). The mutations were confirmed by DNA sequencing.
Western Blotting and Immunoprecipitation
To assess protein/protein interactions, we performed Western blotting of endogenous or vector-expressed proteins. Cells were plated in 100-mm tissue culture dishes and transiently transfected with different combinations of constructed plasmids. After culturing for 48 h, cells were washed twice with ice-cold phosphate-buffered saline and collected in lysis buffer (20 mmol/liter Tris (pH 7.5), 1% Nonidet P-40, 150 mmol/liter NaCl, 5 mmol/liter EDTA, 1 mmol/liter Na3VO4) supplemented with a protease inhibitor mixture (Sigma; added at a 1:100 dilution) as described previously (17). Equal amounts of protein were subjected to SDS-PAGE. The protein was transferred to polyvinylidene difluoride membrane using semidry transfer apparatus. The membrane was blocked with nonfat milk and incubated with primary antibody against Hsp90 (1:1000, Cayman Chemical Inc., Ann Arbor, MI) and Cdc37 (1:1000, Santa Cruz Biotechnology, Santa Cruz, CA). The protein levels were detected using ECL detection system (Thermo Fisher Scientific Inc., Waltham, MA).
Immunoprecipitation was performed as described previously (17, 18). Briefly, cells were lysed and centrifuged. Supernatant was recovered, and protein concentrations were determined with BCA protein assay reagents (Pierce). A total of 500 μg of each protein in 500 μl of lysis buffer was first incubated with H9010 antibody (Axxora, San Diego) at 4 °C for 1 h, followed by addition of 20 μl of protein A/G-agarose (Santa Cruz Biotechnology) and incubation at 4 °C overnight. Beads were collected the next day and washed with phosphate-buffered saline. Precipitates were resuspended with lysis buffer and electrophoresis sample buffer. 20 μl of each sample were subjected to SDS-PAGE for Western blot analysis.
Optical Charge-coupled Device Imaging in Living Cells
To visually assess the Hsp90/Cdc37 interactions using the SRL-PFAC system in live cells, HEK293 cells were transfected with different combinations of constructed plasmids on 12-well plates and cultured for 24 h. Cells were then washed with phosphate-buffered saline buffer and trypsinized. Then 1 × 105 transfected cells were plated in each well on 96-well plates and allowed to attach for 24 h. EnduRen live cell substrate (10 μg/ml; diluted in 50 μl of medium) was added to each well for 1.5 h, and RL activities were determined by bioluminescence imaging with an acquisition time of 1 min.
RESULTS
SRL-PFAC to Detect Hsp90/Cdc37 Interactions
SPL-PFAC system has been established as an imaging method for studying protein/protein interactions in mammalian cells (27–29), yet it is uncertain whether it could be used for studying full-length Hsp90/Cdc37 interactions. We hypothesized that the N terminus of Hsp90 would interact with the C terminus of Cdc37, thus bringing two inactive halves of full-length RL (NRL and CRL) together and lead to complementation of RL activity, which could be imaged using coelenterazine. In contrast, when the interactions between Hsp90 and Cdc37 are disrupted, the luminescence would subsequently disappear (Fig. 1B).
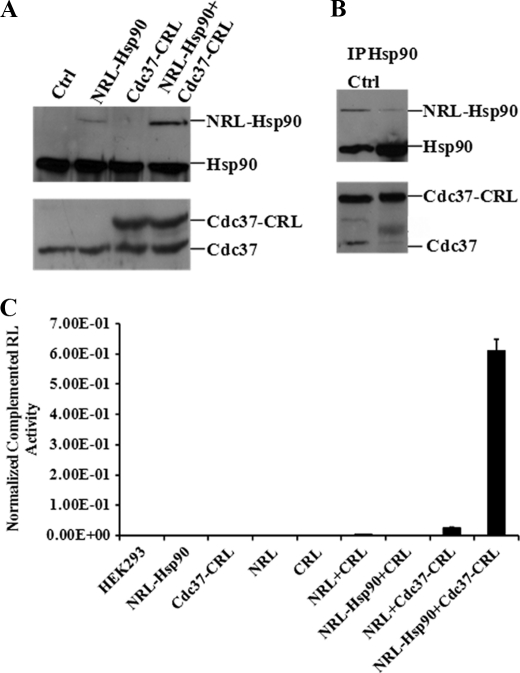
To test our hypothesis, we created constructs to express the N and C termini of RL, NRL-Hsp90 (full-length) fusion protein, and Cdc37 (full-length)-CRL fusion proteins in pcDNA3.1(+) vector (Fig. 1A). Constructed plasmids were transfected into human HEK293 cells. pcDNA3.1(+) vector-expressed FL was cotransfected with each plasmid or combination for transfection efficiency normalization. After 48 h, the cells were collected to detect the expressed protein using Western blotting. The results showed that the specific bands of recombined NRL-Hsp90 (117 kDa) and Cdc37-CRL (60 kDa) fusion proteins could be clearly detected in the transfected cells (Fig. 2A). These bands are distinct from native Hsp90 (90 kDa) and Cdc37 (50 kDa) bands in the cells.
FIGURE 2.
SRL-PFAC system is sensitive and specific for monitoring Hsp90/Cdc37 interactions. A, Western blotting shows specific band of NRL-Hsp90 and Cdc37-CRL expression using monoclonal antibody against Hsp90 and Cdc37, respectively. HEK293 cells were transiently transfected with NRL-Hsp90, Cdc37-CRL, and NRL-Hsp90 + Cdc37-CRL, respectively. HEK293 cells without transfection were used as control (Ctrl) (1st lane). B, NRL-Hsp90 and Cdc37-CRL forms complex in cells. HEK293 cells were transiently cotransfected with NRL-Hsp90 and Cdc37-CRL. Cell lysate was immunoprecipitated with Hsp90 antibody. Western blot was performed for detection of Hsp90, Cdc37, NRL-Hsp90, and Cdc37-CRL. Cell lysate without immunoprecipitation was used as control to mark the band position (1st lane). C, SPL-PFAC system shows highly complemented RL activity and low background. HEK293 cells were transiently transfected with NRL, CRL, NRL-Hsp90, Cdc37-CRL, NRL + CRL, NRL-Hsp90 + CRL, Cdc37-CRL + NRL, and NRL-Hsp90 + Cdc37-CRL, FL was used to normalize the transfection efficiency. Luciferase assay was performed as described under “Experimental Procedures.” Data are presented as mean ± S.D. (n = 3).
To test if the expressed fusion proteins (NRL-Hsp90 and Cdc37-CRL) could interact with each other in cells, we performed immunoprecipitation (IP) study after transfection. Fig. 2B showed that Hsp90 antibody is able to precipitate both native Hsp90 and fusion protein NRL-Hsp90. The immunoprecipitated complex was also probed with anti-Cdc37 antibody. The results showed that both native Cdc37 and fusion proteins Cdc37-CRL were present in IP samples. These data suggest that both the expressed fusion proteins (NRL-Hsp90 and Cdc37-CRL) are able to interact with each other and also able to interact with native Hsp90 and Cdc37. Thus, the fusion proteins preserve the function of Hsp90 and Cdc37.
To test the complementation of two split RL fragments by the interaction of full-length Hsp90 and Cdc37, we transfected HEK293 cells with various vectors (Fig. 2C). The cell lysates were used to measure the luciferase activity after transfection. When the cells were transfected with each fragment alone (NRL, CRL, NRL-Hsp90, and Cdc37-CRL), the luciferase activity was low. When cotransfected with different pairs of fragments, NRL and CRL, NRL-Hsp90 and CRL, and NRL and Cdc37-CRL to HEK293 cells, the two fragments all showed low levels of complementation compared with NRL or CRL alone. However, when the two fragments NRL-Hsp90 and Cdc37-CRL were cotransfected into HEK293 cells, the complementation of the fragments was enhanced by 170-, 220-, and 35-fold, compared with the transfection of NRL + CRL, NRL-Hsp90 + CRL, and Cdc37-CRL + NRL, respectively. These data suggest that the interaction of full-length Hsp90 and Cdc37 was able to complement split luciferase fragments, and the SRL-PFAC method is sensitive and specific to monitor the interaction of full-length human Hsp90 and Cdc37.
MD Simulation and Selection of Hsp90 and Cdc37 Residues for Mutation
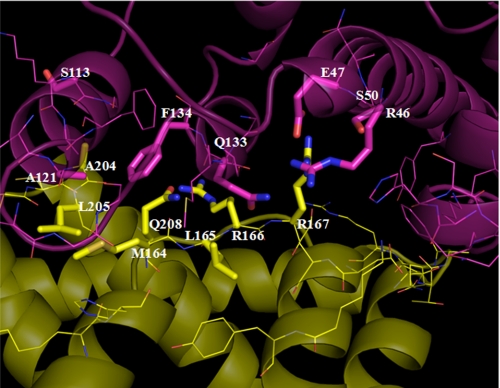
To study in detail the interaction of Hsp90/Cdc37 and to identify the residues at the interface of Hsp90/Cdc37, we performed a 3-ns MD simulation of the Hsp90-Cdc37 complex and calculated the B-factors of residues in the hydrophobic core and the polar network (Fig. 3). The residues with low B-factors suggest less fluctuation during the course of simulation, which suggest that these residues are either structurally restricted or are involved in stable interactions at the interface (Table 1). The residues of one protein within 5 Å of the other were examined more closely to select residues that are at the interface and are involved in complex formation. A larger number of Hsp90 residues were found to be within this cutoff indicating the nature of the complex where Hsp90 clamps over the Cdc37 protein. However, this also means that the Hsp90 residues at the interface were more labile than those of Cdc37 with larger B-factors.
FIGURE 3.
Interface residues of Hsp90-Cdc37 complex. Hsp90 is shown in magenta with Cdc37 in yellow. Mutated residues are shown in stick representation, and others are shown with lines.
TABLE 1.
B-factors for Hsp90 residues within 5 Å of Cdc37 and Cdc37 residues within 5 Å of Hsp90, calculated from 3 ns of MD simulations of a complex of Hsp90/Cdc37
Differences in the calculated interaction energy (enthalpic component) on mutating the residues to Ala or Gly (in italics) were obtained using the MM/GBSA method in Amber. Residues selected for mutation are shown in boldface. HIE is a histidine residue with a proton at the Nϵ position.
| Hsp90 |
Cdc37 |
||||||
|---|---|---|---|---|---|---|---|
| Residue | Residue no. | Residue B-factor from MD | (MM/GBSA) Ala/Gly mutation | Residue | Residue no. | Residue B-factor from MD | (MM/GBSA) Ala/Gly mutation |
| Arg | 46 | 18 | −1.28 | His | 161 | 59 | −1.81 |
| Glu | 47 | 20 | −9.26 | Met | 164 | 23 | −4.76 |
| Ser | 50 | 12 | −0.36 | Leu | 165 | 13 | −1.77 |
| Arg | 60 | 97 | −2.39 | Arg | 166 | 21 | −0.99 |
| Tyr | 61 | 101 | −1.80 | Arg | 167 | 14 | −10.97 |
| Leu | 64 | 115 | −1.62 | Asp | 169 | 23 | −0.46 |
| Ser | 113 | 84 | 0.03 | Asp | 170 | 20 | −3.96 |
| Gly | 114 | 23 | Lys | 173 | 63 | −0.35 | |
| Lys | 116 | 61 | −0.10 | Trp | 193 | 23 | −0.84 |
| Ala | 117 | 24 | −0.99 | Leu | 197 | 29 | −0.21 |
| Glu | 120 | 42 | −2.67 | Lys | 202 | 47 | −2.11 |
| Ala | 121 | 22 | −1.00 | Ala | 204 | 30 | −1.28 |
| Ala | 124 | 35 | −1.01 | Leu | 205 | 23 | −3.73 |
| Gly | 125 | 25 | Gln | 208 | 24 | −4.31 | |
| Ala | 126 | 18 | −0.53 | Lys | 242 | 51 | −0.5 |
| Met | 130 | 38 | −3.27 | Thr | 243 | 93 | −0.12 |
| Gly | 132 | 14 | Ala | 244 | 123 | −1.21 | |
| Gln | 133 | 13 | −7.74 | Asp | 245 | 122 | 0.03 |
| Phe | 134 | 22 | −5.00 | Arg | 246 | 86 | −0.79 |
| Gly | 137 | 11 | |||||
| Lys | 209 | 90 | 0.21 | ||||
| His | 210 | 34 | −0.20 | ||||
| Gln | 212 | 32 | −0.09 | ||||
| Phe | 213 | 31 | −2.53 | ||||
Further evaluation of the residues at the interface was performed by the calculations of the change in the enthalpic component of the free energy of binding of the two proteins by mutating each residue individually to Ala (Gly for Ala residues at the interface) and utilizing the MM/GBSA method to calculate the interaction energy. In general, residues found to impact the interaction energy significantly by this analysis also had lower B-factors as obtained from the simulation.
Several polar and hydrophobic residues were then selected from each protein for mutation analysis, based on their B-factor, impact on interaction energy, and potential interactions with residues of the other protein. Hsp90 residues selected for mutation included Arg-46, Glu-47, Ser-50, Ser-113, Ala-121, Gln-133, and Phe-134, whereas those selected from Cdc37 included Met-164, Leu-165, Arg-166, Arg-167, Ala-204, Leu-205, and Gln-208. Several of these residues were involved in interactions with each other such as Hsp90 Glu-47 with Cdc37 Arg-167 among the polar interactions, and Hsp90 Phe-134 with Cdc37 Met-164, Ala-204, and Leu-205 in the hydrophobic patch as described below.
SRL-PFAC Confirmed Hsp90 Critical Amino Acid Residues for Hsp90/Cdc37 Interactions
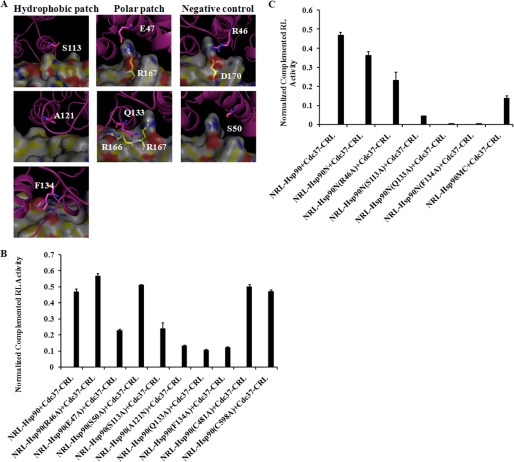
To validate the results from the MD simulation, we selected seven residues with low B-factor scores (Table 1) in Hsp90 for mutagenesis on constructed NRL-Hsp90. Three residues in Hsp90 hydrophobic interaction patch were mutated as follows: S113A, A121N, and F134A. Two residues in Hsp90 polar interaction patch were mutated as follows: E47A and Q133A. In addition, two residues unlikely to disrupt the interaction were also mutated for negative controls in Hsp90 as follow: R46A and S50A. Two Cys residues (Cys-481 and Cys-598), which are located in the middle and C-terminal domain of Hsp90 and distal from the interaction patch, were also chosen as controls. These mutants in NRL-Hsp90 were used for SRL-PFAC activity to confirm their importance for Hsp90/Cdc37 interaction.
As shown in Fig. 4, in the hydrophobic interaction region of Hsp90, Phe-134 in the α-helix of Hsp90 is found to be accommodated in a shallow hydrophobic pocket. The relatively smaller and shorter Ala substituent is unlikely to be able to reach the pocket and fill it, leaving a gap in the interaction surface and making it unfavorable for complex formation, and the computational analysis of the mutant predicted a significant decrease in affinity. Indeed, SRL-PFAC showed that mutation of F134A in NRL-Hsp90 reduced Hsp90/Cdc37 interaction by 75% because cotransfection of NRL-Hsp90 (F134A) and Cdc37-CRL only showed 25% restored luciferase activity through Hsp90-Cdc37-assisted complementation, compared with wild type of NRL-Hsp90 and Cdc37-CRL cotransfection. Another critical residue Ser-113 is located at the terminus of an α-helix in Hsp90 and interacts with Cdc37 Ala-204. Mutation of this residue was predicted to have a smaller impact compared with the F134A mutation, and SRL-PFAC showed that mutation of S113A in Hsp90 only reduced Hsp90/Cdc37 interaction by 50%. In comparison, Ala-121 is located in the pocket formed by several surrounding hydrophobic residues of Hsp90. Although it does not show direct interaction with Cdc37, it is located at a hydrophobic region of the interface. Thus, the mutation Ala-121 to a polar glutamine residue (Asn) causes 70% disruption of Hsp90/Cdc37 interaction. SRL-PFAC showed that NRL-Hsp90 (A121N) and Cdc37-CRL only restored 30% luciferase activity compared with wild type of NRL-Hsp90 and Cdc37-CRL cotransfection.
FIGURE 4.
Computational modeling and effect of mutagenesis on Hsp90 residues. A, interface residues of Hsp90-Cdc37 complex. Hsp90 is shown as schematic in pink with Cdc37 in surface representation. Hsp90 residues selected for mutation and polar Cdc37 residues that interact directly are shown in stick representation. B, luciferase assay for NRL-Hsp90 mutations. C, luciferase assay for NRL-Hsp90N mutations. FL was used to normalize the transfection efficiency. Luciferase assay was performed as described under “Experimental Procedures.” Data are presented as mean ± S.D. (n = 3).
In the polar interaction region of Hsp90, Hsp90 Glu-47 forms a salt bridge with Arg-167 in Cdc37. This interaction is quite stable during the course of MD simulation, suggesting a significant contribution to Hsp90-Cdc37 complex formation, and was also calculated to contribute the most to the interaction energy. However, mutation of E47A only reduced Hsp90/Cdc37 interaction by 50%, because SRL-PFAC showed that NRL-Hsp90 (E47A) and Cdc37-CRL restored 50% luciferase activity through Hsp90-Cdc37-assisted complementation, compared with wild type of NRL-Hsp90 and Cdc37-CRL cotransfection. Although Glu-47 in Hsp90 is a critical residue that interacts with Arg-167 of Cdc37, Cdc37 Arg-167 also interacts with Gln-133 in Hsp90. Therefore, mutation of Glu-47 is expected to only partially disrupt Hsp90/Cdc37 interaction, and thus the SRL-PFAC restored 50% luciferase activity in the cotransfection of this mutant (NRL-Hsp90 E47A) and Cdc37-CRL. In comparison, MD simulation showed that Gln-133 in Hsp90 interacts with multiple residues (Arg-166, Arg-167, and Asn-170) in Cdc37 forming an important set of polar interactions. The side chain carbonyl of Gln-133 in Hsp90 formed an H-bond with the backbone NH2 of Arg-166 and Arg-167 in Cdc37. In addition, the side chain NH2 of Gln-133 was placed in a location to form hydrogen bonds with Asp-170 of Cdc37. The multiple interactions of Gln-133 in Hsp90 with multiple residues in Cdc37 by MD simulation, and the large change in energy predicted by mutating this residue suggest that Gln-133 may play a critical role in the Hsp90-Cdc37 complex. To confirm its importance, we mutated Q133A in NRL-Hsp90 and transfected it with Cdc37-CRL. Indeed, SRL-PFAC showed that mutation Q133A in Hsp90 reduced Hsp90/Cdc37 interaction by 80%, because cotransfection of NRL-Hsp90 (Q133A) and Cdc37-CRL only restored 20% luciferase activity compared with wild type of NRL-Hsp90 and Cdc37-CRL cotransfection. These studies showed the contributions of these critical residues for Hsp90/Cdc37 interactions are ranked as follows: Gln-133 = Phe-134 > Ser-113 = Glu-47.
To validate if SRL-PFAC is specific to identify critical residues for Hsp90/Cdc37 interaction, we made four negative control mutants in two sets. The first set includes Ser-50 and Arg-46, both within proximity to the interaction patch. Ser-50 is located in an α-helix in Hsp90 with the possibility of forming an intramolecular hydrogen bond with the backbone carbonyl of Arg-46 in Hsp90. In addition, Arg-46 of Hsp90 is located at the interface close to Asp-170 in Cdc37. However, it does not show any direct interaction with the residues in Cdc37. Therefore, these two residues were not expected to influence Hsp90/Cdc37 interaction as much as other residues located on the interaction patch. We thus made two mutants (R46A and S50A) in NRL-Hsp90. As expected, SRL-PFAC showed that cotransfection of NRL-Hsp90 (R46A or S50A) and Cdc37-CRL restored 100% luciferase activity compared with wild type of NRL-Hsp90 and Cdc37-CRL cotransfection. The other set includes Cys-481 and Cys-598, which are distal from the interaction patch and thus not expected to affect the Hsp90/Cdc37 interaction. We mutated them to Ala in NRL-Hsp90 and found no disruption in Hsp90/Cdc37 interaction by either C481A or C598A, because the cotransfection with Cdc37-CRL restored 100% luciferase activity compared with the wild types.
Hsp90 has been reported to form a dimer through its C terminus. Therefore, NRL-Hsp90 may form a homodimer (Hsp90-Hsp90) or a heterodimer (NRL-Hsp90-Hsp90). To test whether the constructed NRL-Hsp90 would form a dimer with the endogenous Hsp90, thus leading to the interference to the RL complementation, we constructed an NRL-Hsp90N fragment, which cannot form a dimer because of the absence of the C-terminal dimerization domain, to complement Cdc37-CRL. In addition, we also performed mutagenesis on NRL-Hsp90N fragments. As shown in Fig. 4C, SRL-PFAC showed NRL-Hsp90N and Cdc37-CRL restored luciferase activity, which is slightly lower (25%) than that of NRL-Hsp90 and Cdc37-CRL complementation. Mutation of R46A or S113A in NRL-Hsp90N reduced Hsp90/Cdc37 interaction by 30 and 80%, respectively, compared with the wild type of NRL-Hsp90N and Cdc37-CRL cotransfection. The other two mutations NRL-Hsp90N(Q133A) and NRL-Hsp90N(F134A) completely disrupted the complementation with Cdc37-CRL. These reductions of luciferase complementation by these mutations were much more significant compared with the same mutations in NRL-Hsp90 (full length). The most significant reduction of complementation by mutation (Q133A) in NRL-Hsp90 with Cdc37-CRL still showed 20% residual complementation. These data suggest that the Hsp90 heterodimer may contribute the complementation to Cdc37-CRL.
To further test whether Cdc37-CRL may form complementation with the trans-subunit of heterodimer Hsp90 (NRL-Hsp90-Hsp90), we also constructed the NRL-Hsp90MC fragment, which has the C-terminal dimerization domain but lacks the N-terminal interaction domain with Cdc37. SRL-PFAC showed that cotransfection of NRL-Hsp90 MC and Cdc37-CRL still showed ∼25% restored luciferase activity compared with full-length NRL-Hsp90. Taken together, these data clearly show that the SRL-PFAC system can specifically identify critical residues and evaluate their contributions for Hsp90/Cdc37 interactions.
SRL-PFAC Confirmed the Cdc37 Critical Amino Acid Residues for Hsp90/Cdc37 Interactions
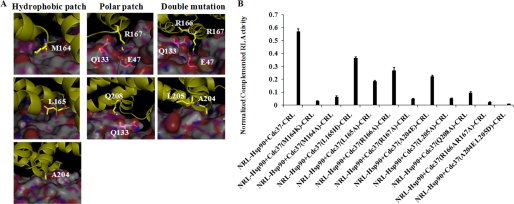
Based on the MD simulations and interaction energy analysis, we mutated Arg-166, Arg-167, and Gln-208 of Cdc37 in the polar interaction region, and we mutated Met-164, Leu-165, Ala-204, and Leu-205 of Cdc37 in the hydrophobic interaction region in Cdc37-CRL for SRL-PFAC study to validate its importance and evaluate their contributions for Hsp90/Cdc37 interaction.
As shown in Fig. 5, Cdc37 Arg-167 is involved in the most significant polar interactions among the Hsp90/Cdc37 residues at the interface. It interacts with multiple residues in Hsp90. The backbone carbonyl of the Arg-167 residue forms a hydrogen bond with the side chain of Gln-133 of Hsp90, whereas the guanidinium moiety is involved in salt bridge formation with the acidic side chain of Glu-47 of Hsp90 (Fig. 4A). The MM/GBSA analysis also indicated that this residue had the largest contribution to the interaction of the two proteins. Indeed, mutation of Arg-167 of Cdc3 reduced Hsp90/Cdc37 interaction by 90%, and SRL-PFAC showed that cotransfection of Cdc37-CRL(R167A) and NRL-Hsp90 only restored 10% luciferase activity when compared with cotransfection of two wide types Cdc37-CRL and NRL-Hsp90 (Fig. 4B).
FIGURE 5.
Computational modeling and effect of mutagenesis on Cdc37 residues. A, interface residues of Hsp90-Cdc37 complex. Cdc37 is shown as schematic in yellow with Hsp90 in surface representation. Cdc37 residues selected for mutation and polar Hsp90 residues that interact directly are shown in stick representation. B, luciferase assay for Cdc37-CRL mutations. FL was used to normalize the transfection efficiency. Luciferase assay was performed as described under “Experimental Procedures.” Data are presented as means ± S.D. (n = 3).
Cdc37 Arg-166 is reported to form a polar interaction with Hsp90 Gln-133 together with Cdc37 Arg-167. MD simulation showed that the backbone NH of Arg-166 of Cdc37 may be involved in hydrogen bonding with Gln-133 of Hsp90, but there is no direct interaction involving the side chain. Indeed, SRL-PFAC showed that mutation of Arg-166 of Cdc37 to Ala only reduced Hsp90/Cdc37 interaction by about 50%, compared with cotransfection of two wild types. We also made a double mutant R166A/R167A of Cdc37 to test if two mutations further decrease binding to Hsp90 as compared with single mutation R167A. SRL-PFAC showed that double mutant Cdc37-CRL (R166A/R167A) reduced Hsp90/Cdc37 interaction by 95%.
The amide in Gln-208 forms a hydrogen bond with the backbone carbonyl of Hsp90 Gln-133 and was predicted to have a significant contribution to the Hsp90-Cdc37 complex formation. As expected, the Q208A mutant had an 85% decrease of RL complementation.
In the hydrophobic interaction region of Cdc37, Met-164 is located at the central part of hydrophobic patch in Cdc37 at the interface formed with Hsp90. SRL-PFAC showed that mutation of Met-164 to a hydrophobic residue Ala or hydrophilic residue Lys reduced Hsp90/Cdc37 interaction by 90 and 95%, respectively, compared with wide type Cdc37-CRL and NRL-Hsp90. These data indicate that the hydrophobic interactions between Hsp90 and Cdc37 are critical, and even a single amino acid alteration is able to disrupt this interaction.
In comparison, Leu-165 is adjacent to Met-164 but not involved in significant direct interactions with Hsp90, although it is part of a general hydrophobic region. Mutation of this residue was predicted to impact the complex formation to a smaller extent, and the Ala mutation resulted in a 60% decrease in RL activity. Mutation of Cdc37 L165H was also tried and found to reduce Hsp90/Cdc37 interaction by 35% in RL activity.
Ala-204 of Cdc37 interacts with the hydrophobic regions of Ser-113 and Ala-117 of Hsp90. SRL-PFAC showed that mutation of Cdc37 (A204E) reduced Hsp90/Cdc37 by 60%. The adjacent Leu-205 was predicted to be more important for complex formation; mutating this residue to Ala resulted in the interaction decreasing by about 90%, significantly more than predicted. The double mutant A204E/L205D in Cdc37 almost abolished Hsp90/Cdc37 interaction by 97% as measured by the restored luciferase activity.
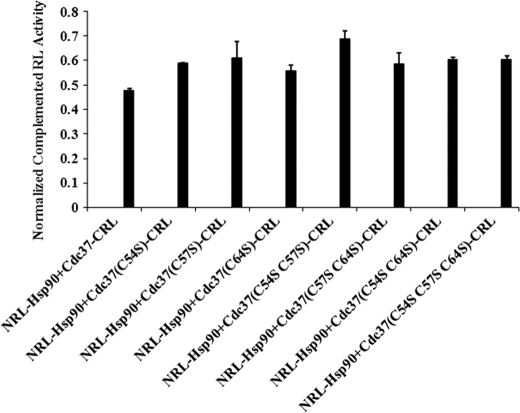
SRL-PFAC Showed That Cysteines at N Terminus of Cdc37 Do Not Directly Contribute to Hsp90-Cdc37 Complex Formation
N terminus of Cdc37 was reported to function as a kinase binding domain that interacts with a subset of client protein kinases and loads them to Hsp90 (31, 32). In a recent study, three Cys residues (Cys-54, Cys-57, and Cys-64) in N terminus of Cdc37 were reported to react with small molecules, which caused oxidation and oligomerization of Cdc37/ and thus disrupted Hsp90/Cdc37 interaction (33). To test whether direct mutations of these cysteine residues in the N terminus of Cdc37 could interfere with Hsp90 interaction, we mutated these three cysteine residues in Cdc37-CRL individually (C54S, C57S, and C64S), in pairs (C54S/C57S and C57S/C64S), and together (C54S/C57S/C64S). These mutants were cotransfected with NRL-Hsp90 in HEK293 cells. SRL-PFAC showed that these cysteine mutants did not impact the Hsp90/Cdc37 interaction, and they still complemented NRL-Hsp90 to restore 100% luciferase activity, which is similar to cotransfection of wild type Cdc37-CRL and NRL-Hsp90 (Fig. 6). These data suggested that these cystines at the N terminus of Cdc37 are not directly involved in the interaction with Hsp90. The small molecules that interact with these cysteines to disrupt Hsp90/Cdc37 interaction may be through its oxidation, oligomerization, or conformational change of Cdc37. Furthermore, these small molecules may also interact with Hsp90 in addition to Cdc37 to disrupt Hsp90/Cdc37 interactions.
FIGURE 6.
Effect of mutagenesis on Cdc37 N-terminal Cys residues. HEK293 cells were transfected with NRL-Hsp90 with Cdc37 (C54S, C57S, C64S, C54S/C57S, C57S/C64S, and C54S/C57S/C64S)-CRL. FL was used to normalize the transfection efficiency. Luciferase assay was performed as described under “Experimental Procedures.” Data are presented as mean ± S.D. (n = 3).
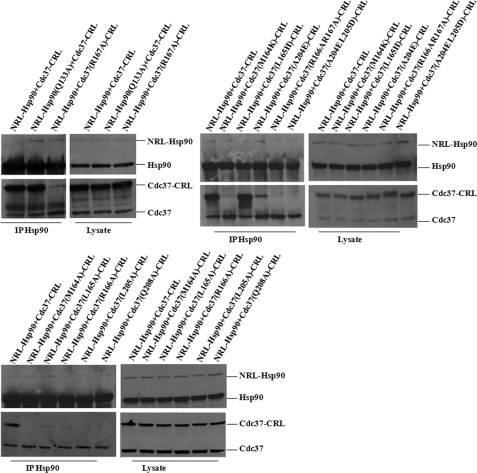
Immunoprecipitation Confirmed SRL-PFAC Results for Hsp90/Cdc37 Interaction and Critical Residues for Hsp90-Cdc37 Complex
To further confirm the SRL-PFAC results that mutations of critical residues in either NRL-Hsp90 or Cdc37-CRL can disrupt the complementation of NRL-Hsp90 and Cdc37-CRL, we performed immunoprecipitation assay in the HEK293 cells with transfection of various vectors. When the HEK293 cells were cotransfected with wild type NRL-Hsp90 and Cdc37-CRL, the antibody against Hsp90 was able to immunoprecipitate native Hsp90 or expressed fusion proteins (NRL-Hsp90). Therefore, Western blotting detected both native Hsp90 and NRL-Hsp90 fusion protein (using Hsp90 antibody) in the precipitated complex. In this case, either Hsp90 or NRL-Hsp90 was able to pull down native Cdc37 and Cdc37-CRL fusion protein in the precipitated complex as detected by Western blotting using Cdc37 antibody (Fig. 7).
FIGURE 7.
Immunoprecipitation of NRL-Hsp90(Q133A) and Cdc37-CRL different mutations. HEK293 cells were cotransfected with NRL-Hsp90 + Cdc37-CRL, NRL-Hsp90(Q133A) + Cdc37-CRL, and NRL-Hsp90 + Cdc37-CRL mutations. Cell lysate was immunoprecipitated with Hsp90 antibody. Western blotting was performed for detection of Hsp90, Cdc37, NRL-Hsp90, Cdc37-CRL, NRL-Hsp90(Q133A), and Cdc37-CRL mutations.
In comparison, when the HEK293 cells were cotransfected with wild type NRL-Hsp90 and different mutants of Cdc37-CRL, antibody against Hsp90 was able to immunoprecipitate Hsp90 and fusion NRL-Hsp90. In such cases, both Hsp90 and NRL-Hsp90 interacted with native Cdc37. Depending on the contributions of mutations in Cdc37-CRL for the RL complementation, different levels of the Cdc37-CRL mutants were pulled down by Hsp90 and NRL-Hsp90. As shown in Fig. 7, the L165H mutation showed almost the same pulldown level in the IP Hsp90 complex compared with the wide type Cdc37-CRL. The pulldown levels of mutants (L165A, R166A, and A204E) was significantly attenuated in IP Hsp90 complex. In contrast, the pulldown band of mutants (M164K, M164A, R167A, L205A, and Q208A and two double mutations) could hardly be detected in the IP Hsp90 complex by Cdc37 antibody indicating the disruption of the Hsp90-Cdc37 complex. All these IP results are consistent with the complementation of luciferase as described above (Fig. 7).
Interestingly, when the HEK293 cells were cotransfected with mutant NRL-Hsp90 (Q133A) and wild type Cdc37-CRL, Hsp90 antibody still immunoprecipitated both native Hsp90 and fusion NRL-Hsp90 (Q133A). In such cases, although NRL-Hsp90 (Q133A) does not interact with either native Cdc37 or Cdc37-CRL, native Hsp90 was still able to interact with both native Cdc37 and Cdc37-CRL. Therefore, Western blotting using Cdc37 antibody still detected both Cdc37 and Cdc37-CRL in the precipitated complex (Fig. 7). The immunoprecipitation results clearly confirmed the SRL-PFAC data for Hsp90/Cdc37 interaction and critical residues involved in Hsp90-Cdc37 complex formation. Furthermore, these data also demonstrate that immunoprecipitation is rather qualitative and cannot accurately quantify the contribution of these residues in protein/protein interaction. SRL-PFAC is also superior to immunoprecipitation to study protein/protein interactions and to identify critical residues in the complex to avoid the interference of native protein/protein interactions.
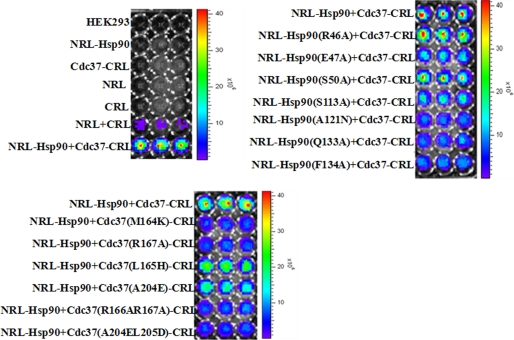
Bioluminescence Imaging to Visualize Hsp90/Cdc37 Interaction and Disruption of Hsp90-Cdc37 Complex by Mutagenesis in Living Cells
The SRL-PFAC and immunoprecipitation confirmed the Hsp90/Cdc37 interactions and identified the critical residues for formation of the complex. To visualize the interaction of Hsp90/Cdc37 and disruption of the complex by mutagenesis in living cells, we used bioluminescence imaging to monitor the process. Human HEK293 cells were transfected with different combinations of the constructed plasmids for 48 h. EnduRen live cell substrate was directly added to the cell culture medium. Bioluminescence images were taken with an acquisition time of 1 min. As shown in Fig. 8, the HEK293 cells without transfection or transfected with control plasmids (NRL alone, CRL alone, NRL-Hsp90 alone, and Cdc37-CRL alone) showed no visual bioluminescence, whereas cotransfection of NRL and CRL showed minimal background of bioluminescence. Cells transfected with NRL-Hsp90 and Cdc37-CRL showed extremely high luminescence. These data demonstrate the interaction of Hsp90 and Cdc37 for the complementation of the two fragments of Renilla luciferase restoring activity.
FIGURE 8.
Molecule imaging in living cells. Human HEK293 cells were transfected with different combinations of the constructed plasmids for 48 h. EnduRen live cell substrate was directly added to the cell culture medium. Bioluminescence images were taken with an acquisition time of 1 min.
In the mutagenesis study, cotransfection of wild type Cdc37-CRL with mutants of NRL-Hsp90 (A121N, Q133A, and F134A) showed significantly decreased bioluminescence, whereas NRL-Hsp90 (E47A and S113A) showed medium level reduction of bioluminescence, compared with cotransfection of the two wild types NRL-Hsp90 and Cdc37-CRL. In comparison, two negative control mutants of NRL-Hsp90 (R46A and S50A) did not change the interaction with Cdc37-CRL and thus did not significantly change the bioluminescence intensity.
Similarly, cotransfection of wild type NRL-Hsp90 with various mutants of Cdc37-CRL (M164K, R167A, R166A/R167A, and A204E/L205D) showed significant decrease, whereas Cdc37-CRL (L165H, A204E) exhibited medium level reduction of bioluminescence, compared with cotransfection of two wild types NRL-Hsp90 and Cdc37-CRL. The bioluminescence imaging in living cells is consistent with the luciferase activity using cell lysates. These data demonstrate that SRL-PFAC live cell imaging is able to also visualize protein/protein interaction in living cells, and the imaging is able to identify critical amino acid residues for protein/protein interactions.
DISCUSSION
The inhibition of molecular chaperone Hsp90 has become an attractive therapeutic target for cancer therapeutics. Currently, most Hsp90 inhibition strategies are to block the Hsp90 N-terminal ATP-binding site. The function of Hsp90 chaperoning activity requires cochaperones to form the superchaperone complex. The binding and release of cochaperones at various stages of Hsp90 superchaperone complexes regulate the folding, assembly, and maturation of Hsp90 client proteins in cancer cells (9). Therefore, investigating protein/protein interactions in Hsp90 superchaperone complex may offer a new strategy to inhibit Hsp90 activity.
Our preliminary data and recent literature demonstrate that Cdc37 plays a central role in loading kinase client proteins in the intermediate Hsp90 superchaperone complex (21, 34). A client protein first binds the Hsp70-Hsp40 chaperone complex and then interacts with Cdc37. Cdc37 helps load the client proteins to the Hsp90 complex. The N terminus of Cdc37 binds to the client protein, whereas the C-terminal side chain of Cdc37 inserts into the N-terminal ATP binding pocket of Hsp90. This holds Hsp90 in an “open” conformation in the Hsp90 intermediate complex (21). Upon ATP binding and hydrolysis, Cdc37 is ejected from the Hsp90 intermediate complex (21), and other cochaperones, including p23 and immunophilins, subsequently bind to Hsp90 to form the mature complex to regulate the maturation of client proteins (9).
In addition, Cdc37 is up-regulated in cancer cells and highly expressed in all prostate tumors and is absent from normal prostate epithelium. Transgenic mice expressing Cdc37 in the prostate epithelium have displayed dramatic proliferative disorders in the prostate, including epithelial hyperplasia and dysplasia (35, 36). Hepatocellular carcinomas also overexpress Cdc37 and Hsp90 compared with normal and surrounding tissues (37). Therefore, protein/protein interactions of the Hsp90-CDc37 complex may play a critical role in tumorigenesis. Study of the mechanisms and critical residues of Hsp90/Cdc37 interaction may offer a new strategy to develop a new class of Hsp90 inhibitors for cancer therapeutics.
However, the methods to study protein/protein interactions, such as immunoprecipitation, NMR, and crystallography, are rather tedious to purify various proteins. These methods can only qualitatively describe the amino acid residues in the interaction. In this study, we adapted an SRL-PFAC system to study full-length human Hsp90/human Cdc37 interaction in living cells. We also applied bioluminescence imaging to identify critical amino acid residues for the formation of full-length human Hsp90-Cdc37 complex in living cells. Finally, we evaluated the contribution of the critical amino acid residues in the interaction of Hsp90/Cdc37 in the living cells.
SRL-PFAC was first developed by Paulmurugan and Gambhir (27). It is achieved through a complementation of two fragments of split Renilla luciferase driven by two interacting proteins. The 36-kDa Renilla luciferase is a monomeric protein whose activity is independent of ATP or post-translational modification (38), making it an ideal reporter in mammalian cells. The N- and C-terminal fragments of Renilla luciferase, which is split between amino acids 229 and 230, was shown to have the most efficient complementation of luciferase activity (27). The SRL-PFAC system is advantageous in studying protein/protein interaction as follows. (a) Renilla luciferase has low molecular weight compared with other reporter genes, and it is easily expressed in mammalian cells. (b) The Renilla luciferase activity does not depend on ATP. (c) There is no cross-reaction between Renilla luciferase substrate coelenterazine and FL substrate d-luciferin (39), and thus two reporter genes can be cotransfected for normalization of transfection efficiency. (í) The small N- or C-terminal fragment of Renilla luciferase itself is inactive, which means the SRL-PFAC has low background and high sensitivity. (e) SRL-PFAC can also be used to study the kinetics of protein/protein interaction (40), thus providing a real time monitoring of Hsp90/Cdc37 interaction in living cells.
SRL-PFAC imaging has been validated to monitor protein/protein interactions, including the heterodimerization between MyoD/Id (27), the interacting location of Tyr-941/Src homology 2 domain (41), the interaction of each Hsp90 isoform (α/β) with p23 (29), homodimeric formation of herpes simplex virus type 1-thymidine kinase (42), and dimerization of ERK2 (43). Here, we optimized the SRL-PFAC system to study full-length human Hsp90/human Cdc37 interaction, and we applied this system to identify critical residues for human Hsp90/Cdc37 interactions in living cells. The contribution of mutated amino acids in human Hsp90 and Cdc37 was evaluated by the restoration of luciferase activity through Hsp90-Cdc37-assisted complementation of two luciferase fragments. The restored luciferase activities through Hsp90/Cdc37 interaction were also confirmed by immunoprecipitation.
Hsp90 and Cdc37 form a complex as dimers with 1:1 molar stoichiometry. The Hsp90 dimer has a shape like a “molecular clamp” with the Cdc37 dimer inside (21, 44). Therefore, this Hsp90-Cdc37 complex is distinct from other interacting proteins that were used for complementation of luciferase fragments as described above. The dimer “clamp”-like structure has the risk of blocking the complementation of two luciferase fragments. However, our results showed that the interaction of full-length of Hsp90 and Cdc37 still helped the complementation of the N terminus of Renilla luciferase and C terminus of Renilla luciferase with 170-, 220-, and 35-fold higher than the transfection of NRL + CRL, NRL-Hsp90 + CRL, and Cdc37-CRL + NRL, respectively (Fig. 2). The imaging results from living cells are consistent with the cell lysate results (Fig. 8). These data suggest that the SRL-PFAC system is sensitive and specific for monitoring the full-length human Hsp90 and Cdc37 interaction in living cells.
Once we established the SRL-PFAC imaging system, we applied this method to identify the critical residues in full-length human Hsp90/Cdc37 interactions. The crystal structure of yeast Hsp90 N terminus and human Cdc37 C terminus (residues 138–378) revealed a large interaction patch between the two proteins, consisting of the lid segment of Hsp90 nucleotide binding pocket and the large helix domain of Cdc37. In these interactions, two regions are present as follows: (a) the hydrophobic interaction patch formed by the C terminus of human Cdc37 (Met-164, Leu-165, Ala-204, and Leu-205) and the N terminus of yeast Hsp90 (Ala-103, Ala-107, Ala-110, Gly-111, Ala-112, Met-116, and Phe-120); (b) the polar/polar interaction patch formed by the main chain of yeast Hsp90 Gln-119 with the side chain of human Cdc37 Gln-208, whereas the side chain of Hsp90 Gln-119 forms hydrogen bonds with Asp-120, Arg-166, and Arg-167 of Cdc37. The yeast Hsp90 Glu-33, which may be involved in Hsp90 ATPase activity, also interacts with human Cdc37 Arg-167 through a hydrogen bond (21).
A few differences were observed by NMR mapping using N terminus of human Hsp90 and a fragment of Cdc37. The NMR showed that Hsp90 Glu-120 (corresponding to yeast Hsp90 Glu-106) forms a hydrogen bond with human Cdc37 Lys-202. Human Hsp90 Glu-47 (corresponding to yeast Glu-33) forms a hydrogen bond with Cdc37 Arg-167. However, the NMR study did not detect that human Hsp90 Gln-133 (corresponding to yeast Gln-119) interacts with human Cdc37 Gln-208, but Hsp90 Gln-133 may form an intramolecular hydrogen bond with Hsp90 Ser-113 (corresponding to yeast Hsp90 Ser-99) (23).
Our molecular dynamic simulation results showed, in the polar interaction patch, that Hsp90 Glu-47 (corresponding to yeast Hsp90 Glu-33) forms a salt bridge with Cdc37 Arg-167 (Figs. 4A and 5A), and this interaction is quite stable during the course of the simulation. The salt bridge between these two charged residues may or may not be critical in Hsp90/Cdc37 interaction. When human Hsp90 E47A was mutated, it only decreased 50% of Hsp90 (E47A)/Cdc37 interaction as measured by its contribution to complement two fragments of luciferase. In comparison, our MD simulation showed that Hsp90 Gln-133 formed a set of hydrogen bonds with Cdc37 Arg-166, Arg-167, and Asp-170 simultaneously. These interactions seem to be more critical in the Hsp90-Cdc37 complex. Indeed, mutating Hsp90 Q133A significantly reduced the Hsp90 (Q133A)/Cdc37 interaction by 80% as measured by restored luciferase activity through Hsp90/Cdc37-assisted complementation of two fragments of luciferase and diminished bioluminescence signal in living cells.
In contrast, MD simulation showed that even though Arg-46 is close to CDC37 Asp-170, there may not be any persistent interaction between these two residues. Arg-46 forms an intramolecular hydrogen bond with the backbone carbonyl of Ser-50 (Fig. 4A). Therefore, the side chains of Hsp90 Arg-46 and Ser-50 may not contribute significantly to the interaction with Cdc37, which can serve as negative controls. The SRL-PFAC showed that mutation of Hsp90 R46A or S50A did not change Hsp90/Cdc37 interactions and did not alter the Hsp90/Cdc37-assisted complementation of the two luciferase fragments leading to no reduction of bioluminescence in living cells.
Hsp90 Phe-134 is in the hydrophobic interaction patch of Hsp90 and makes hydrophobic contacts with Met-164, Ala-204, and Leu-205 residues of Cdc37 (Fig. 4A). Therefore, mutations of F134A diminished Hsp90/Cdc37 interaction by 75% as measured by restored luciferase activity through Hsp90/Cdc37-assisted complementation of two luciferase fragments in cell lysates and living cells (Fig. 4B and Fig. 8B). Hsp90 Phe-134 and Gln-133 are adjacent but involved in different type of interactions in the Hsp90-Cdc37 complex; Phe-134 serves as the hydrophobic core and Gln-133 forms the polar networks. Mutations of either of these residues led to a sharp decrease of Hsp90/Cdc37 interaction and diminished SRL-PFAC bioluminescence signal in living cells.
In comparison, the Hsp90 Ser-113 in the hydrophobic interaction patch is located near Cdc37 Glu-207 and Glu-208, but it has no direct interactions with either of these two residues; on the other hand, we observed that it has hydrophobic interactions with Ala-204 in Cdc37 (Fig. 4A and Fig. 5A). Mutation of Hsp90 S113A decreased Hsp90 (S113A)/Cdc37 interaction by only 50% (Fig. 4B).
It was reported that Hsp90 Ala-121 is located in the hydrophobic interaction patch and interacts with Cdc37 Leu-205 (23). A previous study also showed that the A121N mutation activates the inherent Hsp90 ATPase activity probably by favoring lid closure and helping ATP binding to the N terminus of Hsp90, thus blocking Cdc37 interaction with Hsp90 (21, 44). Our MD simulation showed that Hsp90 Ala-121 was located in a pocket formed by several surrounding hydrophobic residues (Fig. 4A), which may or may not be critical. However, the mutation of Hsp90 A121N significantly decreased Hsp90 (A121N)/Cdc37 interaction by 70% as measured by SRL-PFAC (Fig. 4B). Different explanations are possible for this effect as follows. (a) A121N may help ATP binding and activate Hsp90 inherent ATPase activity as reported previously and thus block Cdc37 interaction because Cdc37 only binds to ADP-bound form of Hsp90. (b) The mutation of the small hydrophobic Ala to a polar Glu residue may cause significant Hsp90 conformational change and thus block Cdc37 interaction.
Very interestingly, even though Gln-133 and Phe-134 of Hsp90 are shown to be important from our MD simulation and MM/GBSA analysis in the Hsp90-Cdc37 complex formation, the mutations Q133A and F134A still restored about 20–25% complemented luciferase activity. We speculated that Cdc37-CRL may still form complementation with the unique heterodimer (NRL-Hsp90-Hsp90), because the NRL-Hsp90 mutations may still form heterodimers with endogenous Hsp90 through the C-terminal dimerization domain and lead to a complementation with Cdc37-CRL binding to the endogenous Hsp90. We thus performed selective mutations (Ser-113 and Phe-134 in the hydrophobic patch, Gln-133 in the polar patch, and Arg-46 for negative control) on N-terminal Hsp90 fragments lacking the C-terminal dimerization domain (NRL-Hsp90N). The R46A and S113A mutations in NRL-Hsp90N further decrease luciferase complementation by 30% (Fig. 4C) compared with the same mutations (R46A and S113A) in full-length Hsp90. In addition, SRL-PFAC also showed decreased complementation in luciferase activity by both NRL-Hsp90N(Q133A) and NRL-Hsp90N(F134A) mutation compared with the same mutations in full-length of NRL-Hsp90. Furthermore, we fused NRL with a middle and C-terminal domain Hsp90 fragment (NRL-Hsp90MC). Although NRL-Hsp90MC does not directly interact with Cdc37-CRL, it restored 25% luciferase activity when cotransfected with Cdc37-CRL compared with the cotransfection of Cdc37-CRL and full-length NRL-Hsp90 (Fig. 4C). Taken together, these data suggest that Cdc37-CRL may be able to complement with the trans-subunit NRL-Hsp90 of heterodimer (NRL-Hsp90-Hsp90).
Similarly two interaction regions (polar interaction and hydrophobic interaction) are also observed on Cdc37. Cdc37 Met-164 is located in the center of the hydrophobic interaction region of Cdc37, which interacts with Gly-125 and Ala-126 of Hsp90. The mutation of this residue to a polar residue in the center of hydrophobic interaction region can block Hsp90/Cdc37 interaction. Therefore, mutation of Cdc37 M164K reduced its interaction with Hsp90 by more than 95% as measured by SRL-PFAC (Fig. 5B) and diminished bioluminescence signal in living cells (Fig. 8). The adjacent residue, Leu-165, is reported to be located in the hydrophobic interaction patch and form a hydrogen bond to Hsp90 Gln-133. However, we did not observe this interaction in our MD simulation (Fig. 5A); SRL-PFAC showed that mutation of Cdc37 L165A and L165H reduces Hsp90/Cdc37 interaction by 60 and 35% as measured by Hsp90-Cdc37-assisted complementation of two luciferase fragments (Fig. 5B), which suggests that Leu-165 may not contribute as significantly to Hsp90/Cdc37 interaction. In addition, Ala-204 is also involved in the hydrophobic interaction region of Cdc37. MD simulation revealed that it interacts with Hsp90 Ser-113 (Figs. 4A and 5A). The mutation of Cdc37, A204E, decreased Hsp90/Cdc37 interaction by 60% as measured by SRL-PFAC (Fig. 5B), which is similar to the reduction of Hsp90/Cdc37 interaction by mutation of Hsp90 S113A (50% reduction), indicating that the interaction in this area is less important in the Hsp90-Cdc37 complex formation. The L205A mutation on the other hand affects the hydrophobic interaction surface of the proteins and thus causes a significant decrease in complex formation by 90%.
Cdc37 Arg-167 is the main interaction residue of Cdc37 for the polar interaction with Hsp90. This residue was also predicted to have the most significant contribution to the interaction for these proteins by MM/GBSA calculations. Cdc37 Arg-167 inserts into the ATP-binding pocket to interact with Hsp90 Glu-47. On the other hand, Cdc37 Arg-167 backbone carbonyl forms hydrogen bonds with Hsp90 Gln-133 on the lid segment of the pocket of Hsp90. Therefore, mutation R167A in Cdc37 reduced Hsp90/Cdc37 interaction by 90% as measured by SRL-PFAC in the cell lysate (Fig. 5B) and diminished bioluminescence signal in living cells (Fig. 8). Similarly, Gln-208 in CDC37 is involved in forming a hydrogen bond with Gln-133 of Hsp90, and thus mutation of this residue resulted in a significant decrease in luciferase activity by 85%. Overall interaction between Hsp90 and Cdc37 is seen to be mediated by the residues at the interface that form a network of interactions where the one residue from the first protein interacts with a residue of the second that is in turn involved in interactions with other residues of the first. Thus, disrupting a single interaction could result in a significant decrease in complex formation as the effect is propagated along the network of interactions at the interface.
The N terminus of human Cdc37 is known as a kinase client binding domain (31, 32). Recently, three Cdc37 N-terminal Cys residues (Cys-54, Cys-57, and Cys-64) have been reported to be the binding sites for low molecular weight compounds, through which the small molecules bind and disrupt Hsp90/Cdc37 interaction (33). However, single (C54S, C57S, C64S), double (C54S/C57S and C57S/C64S), and triple mutations (C54S/C57S/C64S) did not reduce Hsp90/Cdc37 interaction as measured by SRL-PFAC, indicating that the N-terminal Cdc37 may not directly contribute to complex formation (Fig. 6). The molecules, which bind to the cysteines to disrupt Hsp90/Cdc37 interactions, may cause other conformational changes of Cdc37 to block Hsp90/Cdc37 interactions. It is also possible that the small molecules also interact with Hsp90 to disrupt the complex.
This work was partially supported by the National Institutes of Health (RO1 CA120023 and R21 CA143474); University of Michigan Cancer Center Research Grant (Munn); and University of Michigan Cancer Center Core Grant (to D. S.). This study was also partially supported by Chinese Scholarship Council (to Y. J.) (2009).
- Hsp90
- 90-kDa heat shock protein
- SRL-PFAC
- split Renilla luciferase protein fragment-assisted complementation
- MD
- molecular dynamic
- IP
- immunoprecipitation
- FL
- firefly luciferase
- RL
- Renilla luciferase
- NRL
- N-terminal luciferase
- CRL
- C-terminal luciferase
- MM-GBSA
- molecular mechanics-generalized born surface area.
REFERENCES
- 1.Finkelstein D. B., Strausberg S. (1983) J. Biol. Chem. 258, 1908–1913 [PubMed] [Google Scholar]
- 2.Lai B. T., Chin N. W., Stanek A. E., Keh W., Lanks K. W. (1984) Mol. Cell. Biol. 4, 2802–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamal A., Boehm M. F., Burrows F. J. (2004) Trends Mol. Med. 10, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrarini M., Heltai S., Zocchi M. R., Rugarli C. (1992) Int. J. Cancer 51, 613–619 [DOI] [PubMed] [Google Scholar]
- 5.Solit D. B., Chiosis G. (2008) Drug Discov. Today 13, 38–43 [DOI] [PubMed] [Google Scholar]
- 6.Stebbins C. E., Russo A. A., Schneider C., Rosen N., Hartl F. U., Pavletich N. P. (1997) Cell 89, 239–250 [DOI] [PubMed] [Google Scholar]
- 7.Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1997) Cell 90, 65–75 [DOI] [PubMed] [Google Scholar]
- 8.Terasawa K., Minami M., Minami Y. (2005) J. Biochem. 137, 443–447 [DOI] [PubMed] [Google Scholar]
- 9.Neckers L. (2003) Curr. Med. Chem. 10, 733–739 [DOI] [PubMed] [Google Scholar]
- 10.Hawle P., Siepmann M., Harst A., Siderius M., Reusch H. P., Obermann W. M. (2006) Mol. Cell. Biol. 26, 8385–8395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Picard D. (2002) Cell. Mol. Life Sci. 59, 1640–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wegele H., Müller L., Buchner J. (2004) Rev. Physiol. Biochem. Pharmacol. 151, 1–44 [DOI] [PubMed] [Google Scholar]
- 13.Hunter T., Poon R. Y. (1997) Trends Cell Biol. 7, 157–161 [DOI] [PubMed] [Google Scholar]
- 14.Pearl L. H. (2005) Curr. Opin. Genet. Dev. 15, 55–61 [DOI] [PubMed] [Google Scholar]
- 15.Reed S. I. (1980) Genetics 95, 561–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith J. R., Clarke P. A., de Billy E., Workman P. (2009) Oncogene 28, 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang T., Hamza A., Cao X., Wang B., Yu S., Zhan C. G., Sun D. (2008) Mol. Cancer Ther. 7, 162–170 [DOI] [PubMed] [Google Scholar]
- 18.Zhang T., Li Y., Yu Y., Zou P., Jiang Y., Sun D. (2009) J. Biol. Chem. 284, 35381–35399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacLean M., Picard D. (2003) Cell Stress Chaperones 8, 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao J., Irwin A., Hartson S. D., Matts R. L. (2003) Biochemistry 42, 12577–12588 [DOI] [PubMed] [Google Scholar]
- 21.Roe S. M., Ali M. M., Meyer P., Vaughan C. K., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. (2004) Cell 116, 87–98 [DOI] [PubMed] [Google Scholar]
- 22.Zhang W., Hirshberg M., McLaughlin S. H., Lazar G. A., Grossmann J. G., Nielsen P. R., Sobott F., Robinson C. V., Jackson S. E., Laue E. D. (2004) J. Mol. Biol. 340, 891–907 [DOI] [PubMed] [Google Scholar]
- 23.Sreeramulu S., Jonker H. R., Langer T., Richter C., Lancaster C. R., Schwalbe H. (2009) J. Biol. Chem. 284, 3885–3896 [DOI] [PubMed] [Google Scholar]
- 24.Rowlands M. G., Newbatt Y. M., Prodromou C., Pearl L. H., Workman P., Aherne W. (2004) Anal. Biochem. 327, 176–183 [DOI] [PubMed] [Google Scholar]
- 25.Soti C., Vermes A., Haystead T. A., Csermely P. (2003) Eur. J. Biochem. 270, 2421–2428 [DOI] [PubMed] [Google Scholar]
- 26.He H., Zatorska D., Kim J., Aguirre J., Llauger L., She Y., Wu N., Immormino R. M., Gewirth D. T., Chiosis G. (2006) J. Med. Chem. 49, 381–390 [DOI] [PubMed] [Google Scholar]
- 27.Paulmurugan R., Gambhir S. S. (2003) Anal. Chem. 75, 1584–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan C. T., Paulmurugan R., Gheysens O. S., Kim J., Chiosis G., Gambhir S. S. (2008) Cancer Res. 68, 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulmurugan R., Massoud T. F., Huang J., Gambhir S. S. (2004) Cancer Res. 64, 2113–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsui V., Case D. A. (2000) Biopolymers 56, 275–291 [DOI] [PubMed] [Google Scholar]
- 31.Grammatikakis N., Lin J. H., Grammatikakis A., Tsichlis P. N., Cochran B. H. (1999) Mol. Cell. Biol. 19, 1661–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao J., Prince T., Hartson S. D., Matts R. L. (2003) J. Biol. Chem. 278, 38117–38120 [DOI] [PubMed] [Google Scholar]
- 33.Sreeramulu S., Gande S. L., Göbel M., Schwalbe H. (2009) Angew. Chem. Inter. Ed. Engl. 48, 5853–5855 [DOI] [PubMed] [Google Scholar]
- 34.Siligardi G., Hu B., Panaretou B., Piper P. W., Pearl L. H., Prodromou C. (2004) J. Biol. Chem. 279, 51989–51998 [DOI] [PubMed] [Google Scholar]
- 35.Stepanova L., Finegold M., DeMayo F., Schmidt E. V., Harper J. W. (2000) Mol. Cell. Biol. 20, 4462–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stepanova L., Yang G., DeMayo F., Wheeler T. M., Finegold M., Thompson T. C., Harper J. W. (2000) Oncogene 19, 2186–2193 [DOI] [PubMed] [Google Scholar]
- 37.Pascale R. M., Simile M. M., Calvisi D. F., Frau M., Muroni M. R., Seddaiu M. A., Daino L., Muntoni M. D., De Miglio M. R., Thorgeirsson S. S., Feo F. (2005) Hepatology 42, 1310–1319 [DOI] [PubMed] [Google Scholar]
- 38.Matthews J. C., Hori K., Cormier M. J. (1977) Biochemistry 16, 85–91 [DOI] [PubMed] [Google Scholar]
- 39.Bhaumik S., Gambhir S. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loening A. M., Fenn T. D., Wu A. M., Gambhir S. S. (2006) Protein Eng. Des. Sel. 19, 391–400 [DOI] [PubMed] [Google Scholar]
- 41.Kaihara A., Kawai Y., Sato M., Ozawa T., Umezawa Y. (2003) Anal. Chem. 75, 4176–4181 [DOI] [PubMed] [Google Scholar]
- 42.Massoud T. F., Paulmurugan R., Gambhir S. S. (2004) FASEB J. 18, 1105–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaihara A., Umezawa Y. (2008) Chem. Asian J. 3, 38–45 [DOI] [PubMed] [Google Scholar]
- 44.Prodromou C., Panaretou B., Chohan S., Siligardi G., O'Brien R., Ladbury J. E., Roe S. M., Piper P. W., Pearl L. H. (2000) EMBO J. 19, 4383–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]