Abstract
Clustered protocadherins (Pcdhs) are a family of cadherin-like molecules arranged in gene clusters (α, β, and γ). γ-Protocadherins (Pcdh-γs) are involved in cell-cell interactions, but their prominent intracellular distribution in vivo and different knock-out phenotypes suggest that these molecules participate in still unidentified processes. We found using correlative light and electron microscopy that Pcdh-γA3 and -γB2, but not -γC4, -α1, or N-cadherin, generate intracellular juxtanuclear membrane tubules when expressed in cells. These tubules recruit the autophagy marker MAP1A/1B LC3 (LC3) but are not associated with autophagic vesicles. Lipidation of LC3 is required for its coclustering with Pcdh-γ tubules, suggesting the involvement of an autophagic-like molecular cascade. Expression of wild-type LC3 with Pcdh-γA3 increased tubule length whereas expression of lipidation-defective LC3 decreased tubule length relative to Pcdh-γA3 expressed alone. The tubules were found to emanate from lysosomes. Deletion of the luminal/extracellular domain of Pcdh-γA3 preserved lysosomal targeting but eliminated tubule formation whereas cytoplasmic deletion eliminated both lysosomal targeting and tubule formation. Deletion of the membrane-proximal three cadherin repeats resulted in tubes that were narrower than those produced by full-length molecules. These results suggest that Pcdh-γA and -γB families can influence the shape of intracellular membranes by mediating intraluminal interactions within organelles.
Keywords: Autophagy, Cell Adhesion, Electron Microscopy, Intracellular Trafficking, Lysosomes, Membrane Structure, Subcellular Organelles, Vesicles
Introduction
Clustered protocadherins (Pcdhs)3 comprise ∼50 different cadherin-like transmembrane proteins arranged into three genomic clusters, termed α, β, and γ (1, 2). The Pcdh genes within the α and γ clusters, numbering 14 and 22, respectively, share constant cytoplasmic domains within their respective clusters by alternative splicing (3). The Pcdh-γs participate in cell-cell interactions (4, 5) with homophilic properties (6) and have been localized at synapses (6–8), but also have a prominent intracellular distribution in cultured cells and in vivo (6, 8). For Pcdh-αs, conflicting evidence has been presented as to whether they mediate cell-cell interactions (9–11), but a synaptic localization has been reported (1).
Complete genetic deletion of Pcdh-γs reduced the number of synaptic specializations in spinal cord but also caused substantial apoptosis of spinal cord interneurons (7, 12). Conditional knock out in retina also increased apoptosis of some retinal cell types but, unlike in spinal cord, did not affect synaptic connectivity (13). Conditional knock out in astrocytes did not cause apoptosis but delayed synaptic development (14). The multiple effects of Pcdh-γ deletion may indicate still unidentified cellular roles for these molecules.
We show here that Pcdh-γA3 and -γB2 generate intracellular membrane tubules that recruit the autophagic vesicle protein LC3, the mammalian homologue of yeast Atg8. LC3 is lipidated via a reaction similar to the ubiquitin ligase cascade and targets to nascent autophagosomes where it is thought to stabilize the phagophore (15, 16) or promote its closure (17). How LC3 carries out this function(s) is completely unknown. LC3 is a member of a family of proteins including GABARAP, GEC-1, and GATE-16 (18), all of which are implicated in protein or vesicle trafficking. We found that LC3 recruitment to Pcdh-γ-induced tubules was dependent on LC3 lipidation. Coexpression of Pcdh-γA3 with lipidation-defective LC3 reduced tubule length whereas expression of wild-type LC3 increased tubule length compared with Pcdh-γA3 expressed alone. The tubules emanated from lysosomes. The luminal/extracellular domain of Pcdh-γA3 was found to be required for tubule formation, and shortening its length resulted in narrower tubules. Thus, Pcdh-γs, in concert with LC3 family members, could participate in the biogenesis of their own trafficking organelles via intraluminal homophilic interactions. The effect of LC3 perturbation on tubule length is consistent with a role for LC3 in promoting or stabilizing membrane expansion.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Pcdh-γA3-GFP has been described (8). Pcdh-γB2-YFP and Pcdh-γC4-YFP were provided by Dr. Joshua Weiner (University of Iowa). Luminal/extracellular deleted Pcdh-γA3-GFP was provided by Drs. Marcus Frank and Ingrid Haas (Max Planck Institute). Pcdh-α1-GFP was provided by Dr. Qiang Wu (Shanghai Jiao Tong University). N-cadherin (N-cad)-Venus was provided by Dr. Hidekazu Tanaka (Osaka University School of Medicine). RFP-LC3 was provided by Dr. Zhenyue Yue (Mount Sinai School of Medicine). hcRed-LC3wt and hcRed-LC3ΔG were provided by Dr. Isei Tanida (National Institute of Infectious Diseases, Tokyo, Japan). Cytoplasmic domain-deleted Pcdh-γA3-GFP (ΔCCD-GFP) has been described (6). Pcdh-γA3-GFP with the three membrane-proximal cadherin repeats deleted was generated by PCR amplification of the distal three cadherin repeats and ligating to the cytoplasmic, transmembrane, and extracellular membrane-proximal segment just after the end of the sixth cadherin repeat (corresponding to amino acid 667) using an introduced Sal1 site to fuse the segments.
Cell Cultures, Transfection, and Immunostaining
Human embryonic kidney (HEK293) cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Cells were plated on 35-mm live imaging dishes with gridded glass bottoms (Mattek) for analysis by correlative light and electron microscopy (CLEM). Cells were plated on 25-mm coverslips in 6-well plates for light microscopy alone. Cells were transfected with 4 μg of each plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For lysosomal inhibition, after transfection cells were treated overnight with 20 mm NH4Cl. Primary neurons were grown and transfected as described (6). Cells were fixed and immunostained exactly as described (6) except that the concentration of Triton X-100 was 0.2%.
Antibodies
Antibodies to the constant cytoplasmic domain of mouse Pcdh-γs and to the cytoplasmic domain of N-cad have been described (8, 19, 20). Antibodies to the constant cytoplasmic domain of mouse Pcdh-αs were generated using GST fused to the entire Pcdh-α constant cytoplasmic domain. Polyclonal anti-GFP was from Clontech. Chicken antibodies to MAP2 were from Covance. Monoclonal anti-KDEL was from Stressgen. Monoclonal anti-LAMP-2 (H4B4, developed by J. T. August and J. E. K. Hildreth) was from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology.
Microscopy
Detailed methods for CLEM after confocal microscopy have been described (21–23). For Pcdhs or N-cad transfected alone, cells were fixed 24 h after transfection with 4% glutaraldehyde in 0.1 m sodium cacodylate buffer with 1 mm CaCl2. For cotransfection with hcRedLC3wt or ΔG constructs, cells were imaged live prior to fixation with glutaraldehyde because we found hcRed fluorescence to be incompatible with glutaraldehyde fixation. Confocal stacks of transfected cells were acquired on a Zeiss LSM 510 META microscope. Differential interference contrast and fluorescent images were acquired. The location of the cell was documented with respect to the coverslip grid using brightfield illumination. The cells were then fixed if imaged live and processed for transmission electron microscopy. The material was washed in sodium cacodylate buffer and treated with 1% osmium tetroxide, 1.5% potassium ferracyanide in 0.1 m cacodylate buffer for 1 h at 4 °C. The cells were then dehydrated in solutions of ethanol at increasing concentrations of 50%, 60%, and 70%, kept in 2 ml of 3% uranyl acetate in 70% ethanol for 12 h at 4 °C, washed in 70% ethanol, and further dehydrated with increasing concentrations of 80%, 90%, and 100% ethanol. After dehydration, cells were infiltrated with a 1:1 solution of resin (Embed 812 kit; Electron Microscopy Sciences) and 100% ethanol for 24 h at room temperature. After infiltration, the resin/ethanol was replaced with a 1-ml layer of pure resin, and an open ended embedding capsule was placed on the dish surrounding the cell of interest. The resin was then hardened in a vacuum oven at 65 °C for 8–12 h. After the first layer was solidified, the capsule was topped off with more resin and put back in the oven for another 8–12 h. To separate the block from the dish, a hot plate was heated to 60 °C, and the dish was placed on a preheated hot plate for exactly 3 min. The dish was removed from the hot plate and the capsule carefully peeled free from the dish.
The imaged cell was relocated in the block face and sectioned through. Sections were contrasted with lead citrate and uranyl acetate, and serial sections of the cell of interest were documented at magnifications of ×10,000, ×15,000, and ×30,000. Confocal, differential interference contrast, and transmission electron microscopy images were realigned and oriented using nuclear and other morphological landmarks. Immunoelectron microscopy was performed as described (8).
Quantification
Induction of LC3 clustering by Pcdh-γs, -α1, or N-cad was quantified as follows. Confocal sections of random cells doubly transfected with RFP-LC3 and Pcdh or N-cad GFP/YFP/Venus constructs were acquired on a Zeiss LSM 410 microscope. It was then determined whether RFP-LC3 in each cell was punctate or diffuse. At least 75 cells were scored for each condition over three independent experiments, and the number of punctate LC3 cells was averaged. All conditions were counted blindly. ImageJ was used on imported transmission electron microscopy images to determine tubule length and diameter and averaged from 20–30 tubules from two cells/condition. A t test was used to determine significance. For immunoelectron microscopic quantification of anti-Pcdh-γ-labeled organelles, the overall number of gold particles/organelle was determined, and for each organelle, the orientation of particles was designated as “paired,” representing an adhesive-like conformation, or “unpaired.” The ratio of unpaired to paired labeled organelles was determined from 51 labeled organelles.
RESULTS
Pcdh-γs Generate Electron-dense Membrane Appositions
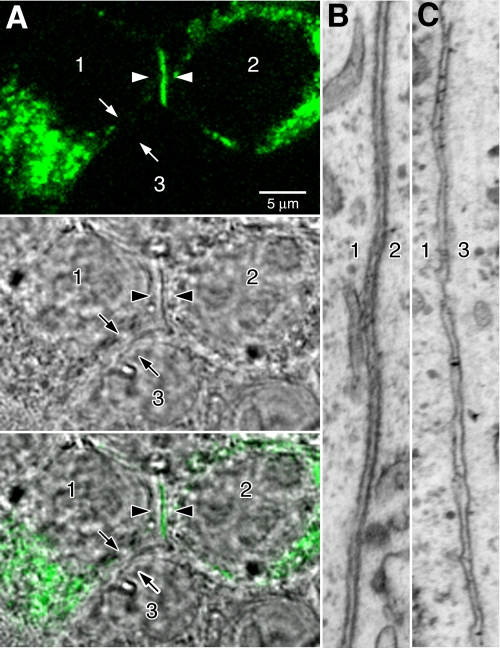
Pcdh-γ-GFP fusions are functional in vivo (7, 12–14, 24). When expressed in cultured hippocampal neurons and cell lines, Pcdh-γ-GFPs exhibited an intracellular distribution that shifted to the cell surface at cell-cell contacts upon deletion of the cytoplasmic region (6), indicating specific retention signals within the cytoplasmic domain. To determine the identity of the compartments that harbor full-length intracellular Pcdh-γs, we performed CLEM (21, 23) of transiently transfected cells. This allowed the precise relocalization of GFP-tagged Pcdh-γs under the electron microscope. Although largely intracellular, in some cases γB2-YFP (Fig. 1A, arrowheads) was found at the interface between two transfected cells (Fig. 1A, cells 1 and 2). Pcdh-γA3-GFP was previously observed to target to cell-cell interfaces (5, 6). γB2-YFP never accumulated at the interface between a transfected cell and a nontransfected cell (Fig. 1A, cells 1 and 3). Both Pcdh-γ-positive and -negative cell-cell interfaces were relocated under the electron microscope by CLEM. The γB2-YFP positive interface between cell 1 and cell 2 (Fig. 1B) was more electron-dense with rigid alignment of plasma membranes compared with the γB2-YFP-negative interface between cell 1 and the nonexpressing cell 3 (Fig. 1C).
FIGURE 1.
Pcdh-γs generate electron-dense membrane profiles when expressed in cells. A, three cells are labeled in the images. Cells 1 and 2 have been transfected with Pcdh-γB2-YFP, whereas cell 3 remains untransfected. Pcdh-γB2-YFP accumulates in a line at the junction between cells 1 and 2 (arrowheads) but does not accumulate at the junction between cells 1 and 3 (arrows), consistent with its participation in a transcellular interaction between cells 1 and 2. B and C, interfaces between cells 1 and 2 and cells 1 and 3 were examined by CLEM. The membranes at the junction between cells 1 and 2 were significantly more electron-dense (B) relative to the membranes at the junction between cells 1 and 3 (C).
Pcdh-γA3 and -γB2 Induce Intracellular Tubules
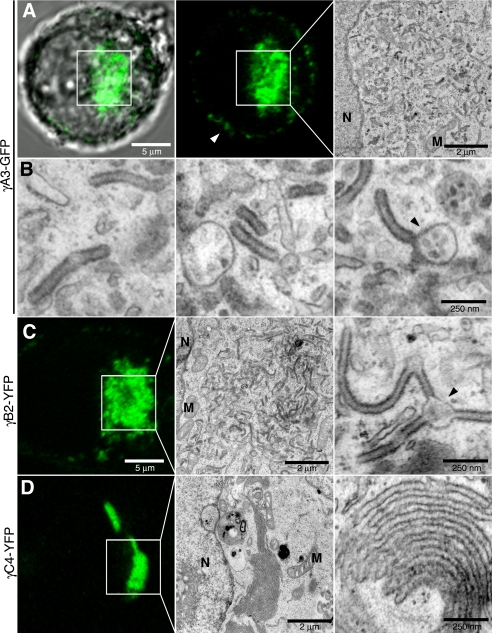
We took advantage of the property that Pcdh-γs render membranes electron-dense to identify the intracellular organelles that harbor the A, B, and C subclasses of Pcdh-γs and compare these with a Pcdh-α family member (Pcdh-α1) and the classical cadherin, N-cad. Three representatives of the Pcdh-γ cluster, γA3-GFP, γB2-YFP, and γC4-YFP, were compared with each other and with a member of the α cluster, Pcdh-α1-GFP (α1-GFP), as well as the classical N-cad-Venus. Intracellular juxtanuclear accumulations of γA3-GFP were present in most transfected cells (Fig. 2A, box). The area corresponding to γA3-GFP fluorescence (Fig. 2A, boxes) was analyzed by CLEM in individual cells (Fig. 2, A, right panel, and B). In all cells, precisely at the area of γA3-GFP fluorescence, electron-dense membrane tubules were observed that were completely absent in untransfected cells. High magnification of the γA3-GFP induced tubules (Fig. 2B) revealed a diameter of ∼60–70 nm. These tubules were distinct from rough and smooth ER-like profiles that were oftentimes found in the vicinity of the tubules but were also observed in untransfected cells. In some cases the tubules were observed to emanate from 100–250 nm vesicular organelles that also contained smaller dot-like profiles (Fig. 2B, right panel, arrowheads).
FIGURE 2.
Pcdh-γA3 and -γB2 but not -γC4 generate intracellular tubules. A and B, γA3-GFP accumulates mostly intracellularly in HEK293 cells (6). A, cell expressing γA3-GFP was identified by confocal microscopy (left and center) and processed for CLEM. Boxed region in confocal micrographs was visualized by electron microscopy (right). This region contained part of the nucleus, several mitochondria, and other tubulovesicular organelles resembling ER, which were also found in nontransfected cells. Darker, electron-dense tubule-shaped organelles, that were never found in untransfected cells, were found in the region of γA3-GFP expression. B, high magnification images are shown of electron-dense organelles generated by γA3-GFP. The tubules measured ∼60–70 nm across and were in some cases observed to emanate from other round organelles of ∼100–250 nm in diameter, which contained smaller structures inside (arrowhead). C, γB2-YFP also generated tubules at sites of intracellular accumulation and was also sometimes associated with other organelles (arrowhead). D, in contrast, γC4-YFP did not generate tubules at sites of intracellular accumulation but rather produced membrane sheets adjacent to the nucleus. N, nucleus; M, mitochondria.
Intracellular accumulations were also observed for γB2-YFP and γC4-YFP (Fig. 2, C and D, respectively) and were examined by CLEM. Tubules similar to those found in γA3-GFP-transfected cells were found in γB2-YFP-transfected cells (Fig. 2C). In contrast, γC4-YFP did not induce the formation of tubules (Fig. 2D) but instead accumulated in layered membrane sheets similar to the type of accumulations observed for some ER proteins (25, 26). Unlike γA3-GFP-induced tubules, the γC4-YFP-induced sheets were continuous through several serial sections, demonstrating that γC4-YFP induces accumulation of sheets rather than tubules (supplemental Fig. 1). In addition, γC4-YFP sheets were found to be continuous with the nuclear envelope, suggesting an ER origin (supplemental Fig. 1).
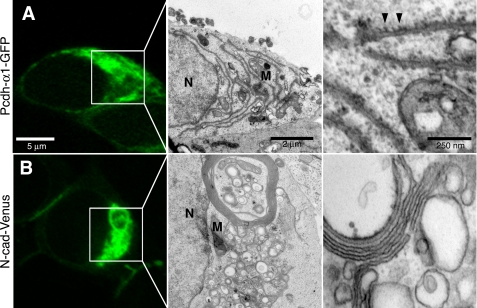
By comparison, α1-GFP was almost exclusively intracellular and absent at cell-cell contacts (Fig. 3A) in agreement with its apparent lack of adhesive activity (9, 10). By CLEM, α1-GFP showed ER-like accumulations that, at higher magnification, appeared to be associated with ribosomes (Fig. 3A, right panel), suggesting an expansion of rough ER. N-cad-Venus was more prominent at cell junctions (not shown) but also could be found intracellularly (Fig. 3B). CLEM of N-cad-Venus intracellular accumulations showed a different ER-like profile with wrapped sheets and vacuoles (Fig. 3B, right panel) that, unlike α1-GFP, did not associate with ribosomes. These whorled membranes have also been shown to be associated with accumulations due to ER expansion (25, 26). Thus, γA3-GFP and γB2-YFP produce a specific type of intracellular tubule versus the ER and sheet-like accumulations observed for the other constructs.
FIGURE 3.
Pcdh-α1-GFP and N-cad-Venus do not produce intracellular tubules. A, α1-GFP, a representative of the α cluster, exhibited almost exclusive intracellular accumulation (left). Low magnification electron micrograph of boxed region (middle). When visualized by electron microscopy, α1-GFP produced elongated rough ER-like organelles that were found associated with ribosomes (right, arrowheads). B, although found more frequently at cell-cell interfaces, N-cad-Venus could be found intracellularly in some cells (box, left). Low magnification electron micrograph of boxed region (middle). Examination of intracellular N-cad-Venus regions revealed membrane whorls and numerous smaller vesicles that lacked ribosomes (right). N, nucleus; M, mitochondria.
γA3-GFP and γB2-YFP Cocluster with LC3
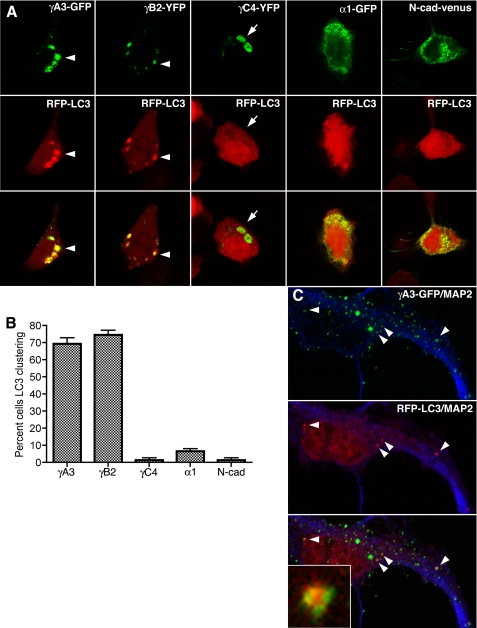
To determine the origin of the tubules within the secretory pathway, we searched for organelle markers that might colocalize with γA3-GFP- and γB2-YFP-induced tubules (see supplemental Fig. 2). The autophagy protein LC3, when fused to either GFP or RFP, has been shown to be a reliable marker for autophagosomes in cultured cells and in vivo (18, 27). We found that both γA3-GFP and γB2-YFP induced the clustering of, and colocalized with, RFP-LC3 (Fig. 4A). In contrast, neither γC4-YFP, α1-GFP, nor N-cad-Venus affected the clustering of RFP-LC3, which remained diffuse in transfected cells (Fig. 4A). Quantification showed that ∼70% of cells transfected with γA3-GFP or γB2-YFP and RFP-LC3 exhibited clustered LC3 (Fig. 4B). All constructs evaluated were expressed at comparable levels in cells (see supplemental Fig. 3). Furthermore, no accumulation of autophagic vesicles or autolysosomes, which have been visualized using the same electron microscopic preparation method (22, 23), could account for the high levels of clustered LC3 in γA3-GFP- or γB2-YFP-transfected cells (see Fig. 2). γA3-GFP, when cotransfected with RFP-LC3 into cultured hippocampal neurons at 12 days in vitro, also produced intracellular clusters, some of which were found to cocluster with RFP-LC3 (Fig. 4C, arrowheads and inset).
FIGURE 4.
Pcdh-γA3 and -γB2 induce clustering of, and colocalize with, LC3. A, the indicated constructs (top panels) were transfected together with RFP-LC3 (middle panels). All Pcdh-γ constructs generated clear intracellular accumulations (arrows and arrowheads). Only γA3-GFP and γB2-YFP induced clustering of, and colocalized with, RFP-LC3 (arrowheads). γC4-YFP did not induce clustering of RFP-LC3 despite its intracellular location (arrows). α1-GFP and N-cad-Venus also did not affect the distribution of RFP-LC3. B, the percentage of cells with punctate RFP-LC3 was determined from three independent experiments. Error bars, S.E. C, colocalization of Pcdh-γA3-GFP and RFP-LC3 in neurons transfected with the two constructs at 12 days in vitro is shown. Blue channel is MAP2 immunostaining. Arrowheads indicate colocalized profiles. Inset shows codistribution of the two constructs in an organelle-like profile.
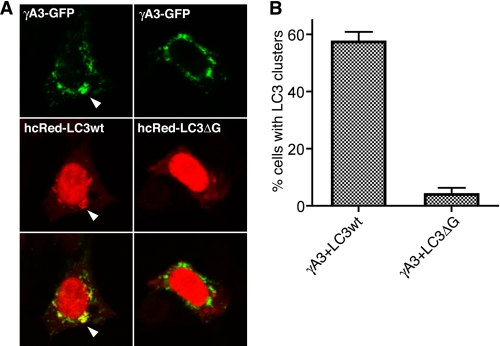
It is known that LC3 clustering artifacts due to protein aggregation can lead to misidentification of LC3 puncta (28, 29). LC3 is lipidated at the glycine residue near its carboxyl terminus via a reaction similar to the ubiquitin ligase pathway, and this lipidation is required for LC3 targeting in autophagy (29). Nonspecific clustering of LC3 at protein aggregates lacks the requirement for lipidation of LC3 (29). We sought to determine whether LC3 lipidation is required for clustering with γA3-GFP. Wild-type hcRed-LC3 or lipidation-defective hcRed-LC3-ΔG (29) was transfected with γA3-GFP, and the extent of LC3 clustering was evaluated quantitatively (Fig. 5). Wild-type hcRed-LC3 was found to cocluster with γA3-GFP (Fig. 5A, left panels) similar to RFP-LC3 (see Fig. 4). In contrast, hcRed-LC3-ΔG did not cocluster with γA3-GFP (Fig. 5A, right panels). Quantitative analysis demonstrated that γA3-GFP induced the formation of wild-type hcRed-LC3 clusters but not that of hcRed-LC3-ΔG. Thus, lipidation of LC3 is required for the colocalization of LC3 with Pcdh-γ-induced intracellular tubules.
FIGURE 5.
Lipidation-defective LC3 does not colocalize with γA3-GFP. A, γA3-GFP was cotransfected with wild-type hcRed tagged LC3 (hcRed-LC3wt) or similarly tagged LC3 truncated at the glycine residue near the carboxyl terminus (hcRed-LC3ΔG), the site for lipid conjugation. hcRed-LC3wt was clustered and colocalized with γA3-GFP, whereas hcRed-LC3ΔG was not. B, results were quantified as the percentage of cells with punctate LC3 from three independent experiments. Error bars, S.E.
LC3 Mediates Pcdh-γ Intracellular Tubule Elongation from Lysosomes
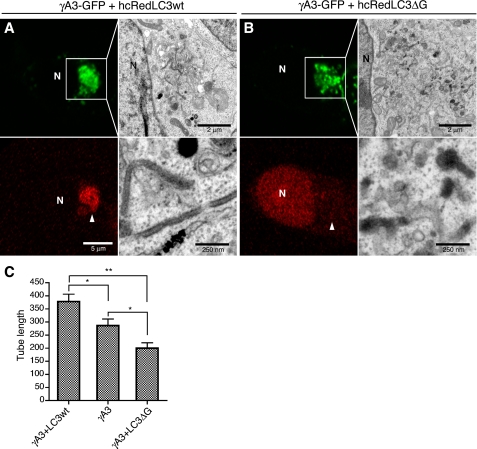
We suspected that wild-type and lipidation-defective LC3 might differentially affect tubule formation or elongation when coexpressed with γA3-GFP. To test this, we compared the tubules produced by γA3-GFP alone or in the presence of wild-type hcRed-LC3 or hcRed-LC3-ΔG. In contrast to the relatively long electron-dense tubules formed by γA3-GFP in the presence of wild-type hcRed-LC3 (Fig. 6A), the electron-dense tubules were significantly shorter in the presence of hcRed-LC3-ΔG (Fig. 6B). Instead of tubules, most of the electron-dense organelles in hcRed-LC3-ΔG/γA3-GFP-cotransfectedcells consisted of vesicles similar to the type that were often connected to the tubules in RFP-LC3 and γA3-GFP-cotransfected cells (see Fig. 2B, right panel). Quantification revealed an almost 50% reduction in tubule length in cells transfected with γA3-GFP plus hcRed-LC3-ΔG relative to γgA3-GFP plus wild-type hcRed-LC3. Cells transfected with γA3-GFP alone had a tubule length intermediate to the two (Fig. 6C). Thus, LC3 promotes γA3-GFP tubule elongation in a lipidation-dependent fashion.
FIGURE 6.
hcRed-LC3ΔG shortens γA3-GFP-induced tubule length. A, γA3-GFP was cotransfected with hcRed-LC3wt and processed for CLEM. B, cotransfection of hcRed-LC3ΔG with γA3-GFP and CLEM. Intracellular accumulation of γA3-GFP (green channel) did not recruit hcRed-LC3ΔG (red channel), and CLEM of this site revealed fewer tubules that were shorter in length than those observed when hcRred-LC3wt was cotransfected. C, quantification of tubule length for γA3-GFP cotransfected with hcRed-LC3wt or ΔG versus γA3-GFP alone. Results were averaged from 20–30 tubules from two different cells/condition. A t test was used to determine significance. **, p < 0.001; *, p < 0.05. Error bars, S.D. N, nucleus.
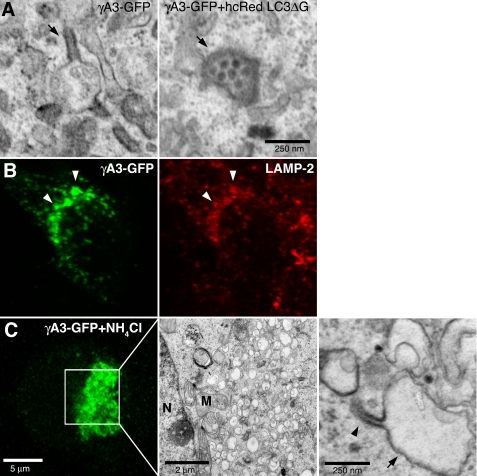
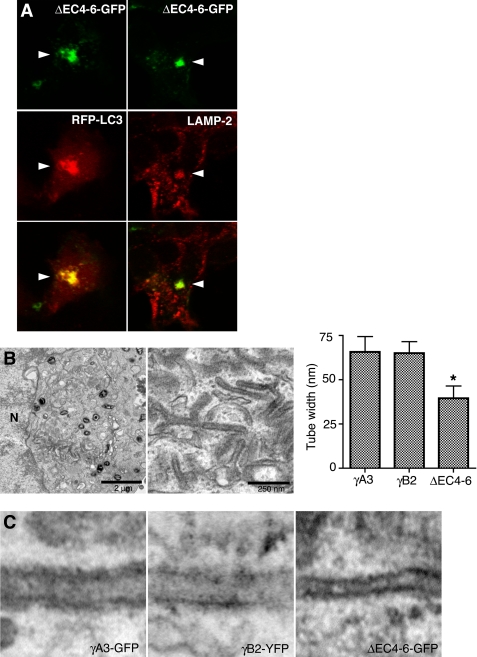
In cells cotransfected with hcRed-LC3-ΔG and γA3-GFP, the predominant electron-dense organelles were 100–250-nm vesicles (Fig. 7A, right panel) rather than tubules (Fig. 7A, left panel), suggesting that the initial substrates for tubule formation may be a subclass of vesicular organelles that sprout tubules when Pcdh-γs accumulate in them. We sought to determine the types of organelles that harbor Pcdh-γs by immunostaining of γA3-GFP-transfected cells with subcellular markers including those for ER, Golgi, early endosomes, and late endosomes/lysosomes (supplemental Fig. 2). We observed a significant colocalization of Pcdh-γs with anti-LAMP-2 immunostaining (Fig. 7B and supplemental Fig. 2). Both γA3-GFP and γB2-YFP, constructs that induced tubules, exhibited significant colocalization with LAMP-2 (supplemental Fig. 4). Thus, in these cells, the combined data suggest that Pcdh-γA and B, but not C, families, together with LC3, induce the sprouting of tubules as they accumulate in late endosomes/lysosomes in a manner dependent on LC3 lipidation.
FIGURE 7.
Lysosomes are the substrates for tubulogenesis. A, electron-dense organelles in γA3-GFP and hcRed-LC3ΔG-cotransfected cells resemble lysosomes (right panel, arrow) versus organelles from γA3-GFP-tranfected cells in which the tubules (left panel, arrow) retained electron-dense properties whereas lysosomal-like organelles were less electron-dense. B, colocalization of γA3-GFP with lysosomal marker LAMP-2 indicates Pcdh-γs target to lysosomes, implicating them as the substrate for tubule growth. C, NH4Cl inhibition of lysosomal acidification reduces tubulogenesis. Enlarged vacuoles (arrow) only occasionally sprouted tubules (arrowhead). N, nucleus; M, mitochondria.
To verify the lysosomal origin of γA3-GFP-induced tubules, we treated γA3-GFP-transfected cells with NH4Cl, an inhibitor of lysosomal acidification. In this case, large vacuoles (Fig. 7C) were induced with a reduction in tubules, indicating that tubulogenesis is tied to lysosomal function. A few tubules (Fig. 7C, arrowhead) were observed, however, to emanate from the enlarged vacuoles (Fig. 7C, arrow).
Differential Effect of Luminal/Extracellular and Cytoplasmic Domain Deletion on Lysosomal Targeting and Intracellular Tubule Biogenesis
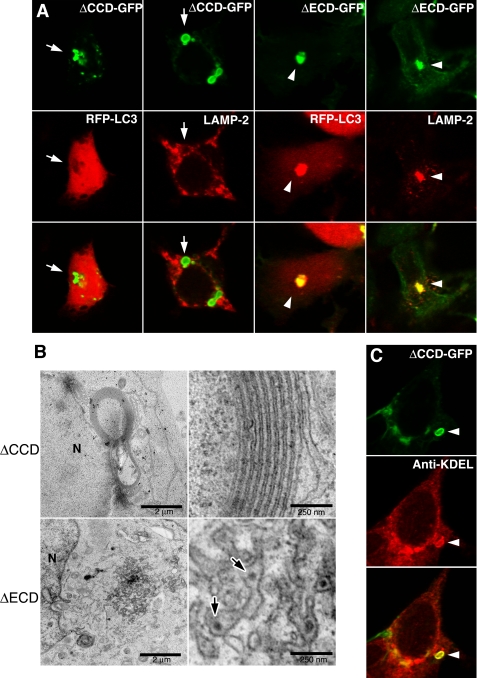
The luminal/extracellular domains of Pcdh-γA3 and Pcdh-γB2, but not Pcdh-γC4, were shown to participate in cell-cell interactions with homophilic properties, whereas the cytoplasmic domains promoted retention (6). We sought to determine whether the luminal/extracellular and/or cytoplasmic domains mediate tubule formation. Cytoplasmic-deleted γA3-GFP (ΔCCD-GFP) was mostly found at the cell surface (6), but intracellular accumulations could be found in a few transfected cells (Fig. 8A, arrows). These never induced clustering of RFP-LC3, nor did they colocalize with LAMP-2 (Fig. 8A, arrows). In contrast, when the luminal/extracellular domain was deleted (ΔECD-GFP; Fig. 8A, arrowheads), both RFP-LC3 clustering and LAMP-2 colocalization were observed. CLEM showed that ΔECD-GFP accumulated in distorted electron-dense organelles that contained smaller profiles similar to those found in lysosomes (Fig. 8B, bottom, arrows). In this case, no tubules were observed. In contrast, ΔCCD-GFP generated membrane whorls (Fig. 8B, top) similar to the type (25, 26) produced upon expression of some ER-targeted proteins. ΔCCD-GFP-induced intracellular membrane whorls were positively immunolabeled with anti-KDEL antibodies (Fig. 8C, arrowheads), confirming their ER origin.
FIGURE 8.
Deletion of the luminal/extracellular and cytoplasmic domains of Pcdh-γA3 differentially affect lysosomal localization, LC3 clustering, and tubule formation. A, intracellular accumulations of ΔCCD-GFP, when found, did not induce clustering of RFP-LC3 and did not colocalize with LAMP-2. In contrast, the luminal/extracellular deletion (ΔECD-GFP) did induce clustering of RFP-LC3 and colocalized with LAMP-2. B, electron micrographs of ΔECD-GFP and ΔCCD-GFP intracellular accumulations are shown. ΔECD-GFP was found in clustered juxtanuclear organelles that contained smaller particles in the lumen (arrows), suggestive of distorted lysosomes whereas ΔCCD-GFP generated larger membrane whorls that resembled ER distortion as has been observed previously (25, 26). C, intracellular ΔCCD-GFP induced membrane whorls labeled positive with anti-KDEL antibodies (arrowhead). N, nucleus.
Shortening the Luminal/Extracellular Domain Decreases Tubule Width
The above results suggested that the cytoplasmic domain promotes lysosomal targeting whereas the luminal/extracellular domain is required for tubulogenesis, potentially by mediating intraluminal interactions. It is known that cadherins mediate their adhesive interactions via the most distal extracellular moiety, the first cadherin repeat (30–32). We deleted the three membrane-proximal cadherin repeats (cadherin repeats 4–6, ΔEC4–6-GFP; see supplemental Fig. 3) to determine whether the distal part of the extracellular domain might be involved in tubulogenesis. We found that this construct induced clustering of RFP-LC3 and colocalized with LAMP-2 (Fig. 9A). CLEM of intracellular ΔEC4–6-GFP revealed that juxtanuclear tubules were generated, but with an apparent narrower width. We quantified the width of tubules generated by γA3-GFP, γB2-YFP, and ΔEC4–6-GFP (Fig. 9B) and found that although both γA3-GFP and γB2-YFP generated tubules of identical width (∼65 nm), ΔEC4–6-GFP-generated tubules were significantly narrower (∼40 nm) (Fig. 9C). Thus, the molecular length of the luminal/extracellular domain is correlated with the diameter of the intracellular tubules, suggesting an intraluminal interaction between Pcdh-γ molecules on apposing membranes stabilizing tubule structure.
FIGURE 9.
Shortening of the extracellular domain decreases tubule width. Cadherin repeats 4–6 were removed, leaving only cadherin repeats 1–3 in the extracellular domain (ΔEC4–6-GFP; see supplemental Fig. 3). A, ΔEC4–6-GFP induces clustering of RFP-LC3 and colocalizes with LAMP-2. B, low (left) and high (center) magnification electron micrographs of intracellular tubules induced by ΔEC4–6-GFP are shown. Right, widths of tubules generated by γA3-GFP, γB2-GFP and ΔEC4–6-GFP constructs were quantified. Decrease in width of ΔEC4–6-GFP tubules was statistically significant (t test, p < 0.0001). Widths of tubules induced by γA3-GFP and γB2-GFP were not different (p > 0.2). Error bars, S.D. C, high magnification of images illustrates widths of γA3-GFP (left), γB2-GFP (center), and ΔEC4–6-GFP induced tubules (right). N, nucleus.
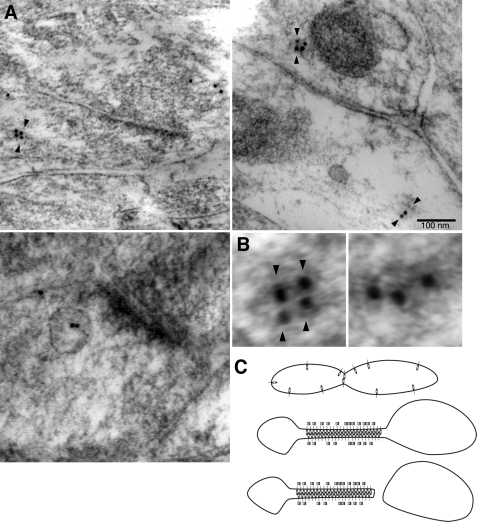
Pcdh-γs Delinate Subdomains of Tubulovesciular Organelles in Vivo
By immunoelectron microscopy, Pcdh-γs were shown to have a prominent intracellular component in vivo (6, 8). In many cases, Pcdh-γ labeled constrictions of, or emanations from, organelle profiles (Fig. 10A, arrowheads), in a paired arrangement on opposing membranes (Fig. 10B, left panel). When quantified, Pcdh-γ-labeled organelles had this arrangement 55% of the time versus 45% of organelles with unpaired gold particles (Fig. 10B, right panel). These results are consistent with a role for Pcdh-γ-mediated intraluminal interactions in organelle dynamics in vivo (Fig. 10C).
FIGURE 10.
Arrangement of Pcdh-γs within organelle profiles. A, immunoelectron microscopy of adult hippocampal CA1 region with antibodies to Pcdh-γ constant intracellular domain. Pcdh-γs have a prominent intracellular distribution (8) and label tubulovesicular organelles in axons and dendrites. Pairing of gold particles (arrowheads) at apparent constriction sites suggests the possibility of an adhesive conformation within organelles. Such paired labeling of gold particles has also been observed at sites of transcellular interactions (8). Pcdh-γs were also observed in lysosomal-like organelles (bottom left). B, example of organelle labeled in paired (left, arrowheads) or unpaired (right) configuration. C, model for intraluminal interactions mediated by Pcdh-γs. Interactions could be stabilized by LC3 (squares). Homophilic intraluminal interactions may facilitate the budding of new organelles containing high concentrations of a single Pcdh-γ isoform.
DISCUSSION
The cellular function of all of the Pcdhs has been poorly defined (33), unlike the well characterized classical cadherins. The Pcdh-γs can mediate transcellular interactions with homophilic properties (5, 6) and, for the Pcdh-αs, less is known about their functions at the cellular level, but these most likely do not mediate transcellular interactions (9, 10, 34). Knock-out phenotypes for Pcdh-γs are consistent with transcellular interactions as one feature but also point to other modes of action (7, 12–14, 24, 35). Pcdh-γs exhibit a prominent intracellular distribution in cultured neurons and in neural tissue (6, 8). Here, we show that Pcdh-γA3 and -γB2 generate, from substrate organelles, intracellular tubules that require the autophagic molecule LC3 for their elongation. Based on the accumulated data, we speculate that Pcdh-γs have the ability to generate their own trafficking organelles at intracellular sites where they accumulate by altering membrane shape or curvature and promoting intraluminal interactions.
Nonconventional Roles for Pcdh-γs
The predominant phenotype when the Pcdh-γ cluster is inactivated in neurons is cell death (7, 12, 13, 24), and this could be either cell-autonomous or non-cell-autonomous (13, 24). Prevention of neuron death in Pcdh-γ-deleted animals by double knock out with Bax revealed synaptic defects in spinal cord (12) presumably due to disrupted Pcdh-γ transcellular interactions. However, completely normal retinal connectivity was restored by rescue from apoptosis in Pcdh-γ retinal knock outs (13), indicating that retinal connectivity has no requirement for Pcdh-γs despite the high expression of the molecules there. The existence of two, apparently separable, phenotypes, neuronal death and synapse loss, indicates the potential for multiple functions for Pcdh-γs in various neuronal systems. Either or both synapse development and cell survival could be dependent on intracellular trafficking of Pcdh-γs.
There is a growing list of molecules that influence intracellular membrane shape, and multiple ER-resident transmembrane proteins that drive the structural dynamics of ER are now beginning to be elaborated (36), including the reticulon family which can transform the ER from sheets into tubules (37). It is possible that Pcdh-γs could define a subdomain of membranes in organelles in which they accumulate and modify these membranes in a manner similar to the reticulons (37, 38). Pcdh-γs lack a cleavable prodomain (39), that for classical cadherins blocks adhesion during intracellular transport (40), and therefore its membrane-membrane adhesive activity is likely to be active inside the lumen of intracellular compartments where it could participate in the stabilization of organelle shape (Fig. 10C). An adhesive-like mechanism for maintenance of ER shape has previously been proposed (36) and could also participate in the stabilization of the tubule morphology. Such Pcdh-γ-generated tubules might emanate from any organelle that harbors them in neurons.
Nonautophagic Role for LC3
LC3 is the mammalian homologue of yeast Atg8 which was identified as a gene required for proper formation of autophagic vesicles or vacuoles in yeast. LC3 was first identified as an ∼18-kDa protein that copurified with microtubules and with MAP1A/1B (41). LC3 was found to translocate to autophagosomes and can be present on the cytosolic and luminal sides (27). In mammals, LC3 has three other homologues, GABARAP, GEC-1, and GATE-16, which also translocate to autophagosomes upon lipidation (18). The exact role of LC3/Atg8 and related proteins in autophagy is still not clear. One study showed that the size of autophagic vacuoles depends on Atg8 levels in yeast (15). Knock outs of different genes of the autophagic cascade, which eliminate LC3 family lipidation, were shown to alter the shape or size of autophagic vesicles, or prevent their closure, but not eliminate them (17). We found that Pcdh-γ-induced intracellular tubules are shorter in the presence of the lipidation-defective LC3. Our results support the idea, previously suggested, that LC3 controls the rate of intracellular membrane expansion (15), which may indicate a generalized role for LC3 in nonautophagic processes, some of which have recently begun to be elaborated (42) and are consistent with a suggested role for LC3 family members in membrane and receptor trafficking (43–48). It is possible that intracellular Pcdh-γs may be involved in regulating LC3 family-mediated organelle trafficking in addition to their roles in cell-cell interactions.
Whether or not the Pcdh-γ-induced tubules represent a precursor to autophagic vesicles or another type of organelle that utilizes LC3 for its biogenesis remains to be conclusively determined, but autophagy is unlikely given that autophagic vesicles were never found in the vicinity of intracellular Pcdh-γs and that the tubules originate from lysosomes. The tubules observed in the present study do bear a resemblance, tubules connected to vesicular profiles, to those found using biochemical purification of LC3-containing membranes (49). Because no increases in autophagic vesicles were observed in Pcdh-γ-transfected cells it is likely that the tubules observed are one of the few examples of nonautophagic organelles that utilize LC3 for its biogenesis. Alternatively, the tubules could be an arrested stage of autophagic organelle biogenesis prior to the generation of the phagophore. Regardless of whether the tubules are autophagy-related or -unrelated, the observation that certain Pcdh-γs can induce organelle formation in an LC3-dependent fashion points to a potential novel intracellular role for the molecules in trafficking in addition to cell-cell interactions.
Mechanism for Trafficking of Recognition Units
The diversity of Pcdh-γs in the nervous system generated by alternative splicing (3) strongly suggests a role in cell surface recognition. It is possible that through intracellular homophilic interactions, Pcdh-γ trafficking might be directed into “packets” (Fig. 10C) that when inserted into the plasma membrane contain a homogeneous population of a single Pcdh-γ isoform. The addition of such recognition packets to the surface may have more immediate consequences on synaptic development versus slow accumulation of individual Pcdh-γ molecules at cell-cell interactions sites in neurons via conventional secretory mechanisms.
Supplementary Material
Acknowledgments
We thank Drs. Qiang Wu, Isei Tanida, Joshua Weiner, Hidekazu Tanaka, Marcus Frank, Ingrid Haas, and Zhenyu Yue for reagents; Drs. Ana Maria Cuervo and Zhenyu Yue for helpful discussions; and William G. Janssen for technical advice.
This work was supported by National Institutes of Health Grant NS051238 and an Irma T. Hirschl award (to G. R. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- Pcdh
- protocadherin
- GFP
- green fluorescent protein
- YFP
- yellow fluorescent protein
- RFP
- red fluorescent protein
- N-cad
- N-cadherin
- LC3
- MAP 1A/1B LC3
- CLEM
- correlative light and electron microscopy
- wt
- wild type
- ER
- endoplasmic reticulum
- CCD
- cytoplasmic domain
- ECD
- extracellular domain.
REFERENCES
- 1.Kohmura N., Senzaki K., Hamada S., Kai N., Yasuda R., Watanabe M., Ishii H., Yasuda M., Mishina M., Yagi T. (1998) Neuron 20, 1137–1151 [DOI] [PubMed] [Google Scholar]
- 2.Wu Q., Maniatis T. (1999) Cell 97, 779–790 [DOI] [PubMed] [Google Scholar]
- 3.Tasic B., Nabholz C. E., Baldwin K. K., Kim Y., Rueckert E. H., Ribich S. A., Cramer P., Wu Q., Axel R., Maniatis T. (2002) Mol. Cell 10, 21–33 [DOI] [PubMed] [Google Scholar]
- 4.Obata S., Sago H., Mori N., Rochelle J. M., Seldin M. F., Davidson M., St John T., Taketani S., Suzuki S. T. (1995) J. Cell Sci. 108, 3765–3773 [DOI] [PubMed] [Google Scholar]
- 5.Frank M., Ebert M., Shan W., Phillips G. R., Arndt K., Colman D. R., Kemler R. (2005) Mol. Cell. Neurosci. 29, 603–616 [DOI] [PubMed] [Google Scholar]
- 6.Fernández-Monreal M., Kang S., Phillips G. R. (2009) Mol. Cell. Neurosci. 40, 344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X., Weiner J. A., Levi S., Craig A. M., Bradley A., Sanes J. R. (2002) Neuron 36, 843–854 [DOI] [PubMed] [Google Scholar]
- 8.Phillips G. R., Tanaka H., Frank M., Elste A., Fidler L., Benson D. L., Colman D. R. (2003) J. Neurosci. 3, 5096–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mutoh T., Hamada S., Senzaki K., Murata Y., Yagi T. (2004) Exp. Cell Res. 94, 494–508 [DOI] [PubMed] [Google Scholar]
- 10.Morishita H., Umitsu M., Murata Y., Shibata N., Udaka K., Higuchi Y., Akutsu H., Yamaguchi T., Yagi T., Ikegami T. (2006) J.Biol.Chem. 281, 33650–33663 [DOI] [PubMed] [Google Scholar]
- 11.Triana-Baltzer G. B., Blank M. (2006) J. Neurobiol. 66, 393–407 [DOI] [PubMed] [Google Scholar]
- 12.Weiner J. A., Wang X., Tapia J. C., Sanes J. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefebvre J. L., Zhang Y., Meister M., Wang X., Sanes J. R. (2008) Development 135, 4141–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrett A. M., Weiner J. A. (2009) J. Neurosci. 29, 11723–11731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Z., Nair U., Klionsky D. J. (2008) Mol. Biol. Cell 19, 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Z., Nair U., Geng J., Szefler M. B., Rothman E. D., Klionsky D. J. (2009) Autophagy, 217–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita N., Hayashi-Nishino M., Fukumoto H., Omori H., Yamamoto A., Noda T., Yoshimori T. (2008) Mol.Biol. Cell 19, 4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanida I., Ueno T., Kominami E. (2004) Int. J. Biochem. Cell Biol. 36, 2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka H., Shan W., Phillips G. R., Arndt K., Bozdagi O., Shapiro L., Huntley G. W., Benson D. L., Colman D. R. (2000) Neuron 25, 93–107 [DOI] [PubMed] [Google Scholar]
- 20.Phillips G. R., Huang J. K., Wang Y., Tanaka H., Shapiro L., Zhang W., Shan W. S., Arndt K., Frank M., Gordon R. E., Gawinowicz M. A., Zhao Y., Colman D. R. (2001) Neuron 32, 63–77 [DOI] [PubMed] [Google Scholar]
- 21.Polishchuk R. S., Polishchuk E. V., Marra P., Alberti S., Buccione R., Luini A., Mironov A. A. (2000) J. Cell Biol. 148, 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Razi M., Chan E. Y., Tooze S. A. (2009) J. Cell Biol. 185, 305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razi M., Tooze S. A. (2009) Methods Enzymol. 452, 261–275 [DOI] [PubMed] [Google Scholar]
- 24.Prasad T., Wang X., Gray P. A., Weiner J. A. (2008) Development 135, 4153–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snapp E. L., Hegde R. S., Francolini M., Lombardo F., Colombo S., Pedrazzini E., Borgese N., Lippincott-Schwartz J. (2003) J.Cell Biol. 63, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingwood D., Schuck S., Ferguson C., Gerl M. J., Simons K. (2009) J. Biol. Chem. 284, 12041–12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuma A., Matsui M., Mizushima N. (2007) Autophagy 3, 323–328 [DOI] [PubMed] [Google Scholar]
- 29.Tanida I., Yamaji T., Ueno T., Ishiura S., Kominami E., Hanada K. (2008) Autophagy 4, 131–134 [DOI] [PubMed] [Google Scholar]
- 30.Shapiro L., Fannon A. M., Kwong P. D., Thompson A., Lehmann M. S., Grübel G., Legrand J. F., Als-Nielsen J., Colman D. R., Hendrickson W. A. (1995) Nature 374, 327–337 [DOI] [PubMed] [Google Scholar]
- 31.Pertz O., Bozic D., Koch A. W., Fauser C., Brancaccio A., Engel J. (1999) EMBO J. 18, 1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel S. D., Ciatto C., Chen C. P., Bahna F., Rajebhosale M., Arkus N., Schieren I., Jessell T. M., Honig B., Price S. R., Shapiro L. (2006) Cell 124, 1255–1268 [DOI] [PubMed] [Google Scholar]
- 33.Jontes J. D., Phillips G. R. (2006) Trends Neurosci. 29, 186–191 [DOI] [PubMed] [Google Scholar]
- 34.Emond M. R., Jontes J. D. (2008) Dev. Biol. 321, 175–187 [DOI] [PubMed] [Google Scholar]
- 35.Su H., Marcheva B., Meng S., Liang F. A., Kohsaka A., Kobayashi Y., Xu A. W., Bass J., Wang X. (2010) Dev. Biol. 339, 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibata Y., Voeltz G. K., Rapoport T. A. (2006) Cell 126, 435–439 [DOI] [PubMed] [Google Scholar]
- 37.Voeltz G. K., Prinz W. A., Shibata Y., Rist J. M., Rapoport T. A. (2006) Cell 124, 573–586 [DOI] [PubMed] [Google Scholar]
- 38.Hu J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M. M., Rapoport T. A., Prinz W. A. (2008) Science 319, 1247–1250 [DOI] [PubMed] [Google Scholar]
- 39.Koch A. W., Farooq A., Shan W., Zeng L., Colman D. R., Zhou M. M. (2004) Structure 12, 793–805 [DOI] [PubMed] [Google Scholar]
- 40.Ozawa M., Kemler R. (1990) J. Cell Biol. 111, 1645–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuznetsov S. A., Gelfand V. I. (1987) FEBS Lett. 212, 145–148 [DOI] [PubMed] [Google Scholar]
- 42.Baisamy L., Cavin S., Jurisch N., Diviani D. (2009) J. Biol. Chem. 284, 28232–28242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elazar Z., Scherz-Shouval R., Shorer H. (2003) Biochim. Biophys. Acta 1641, 145–156 [DOI] [PubMed] [Google Scholar]
- 44.Scherz-Shouval R., Sagiv Y., Shorer H., Elazar Z. (2003) J. Biol. Chem. 278, 14053–14058 [DOI] [PubMed] [Google Scholar]
- 45.Chen Z. W., Chang C. S., Leil T. A., Olsen R. W. (2007) J. Neurosci. 27, 6655–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leil T. A., Chen Z. W., Chang C. S., Olsen R. W. (2004) J. Neurosci. 24, 11429–11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y., Dun S. L., Huang P., Chen C., Chen Y., Unterwald E. M., Dun N. J., Van Bockstaele E. J., Liu-Chen L. Y. (2006) Neuroscience 140, 1265–1276 [DOI] [PubMed] [Google Scholar]
- 48.Marsden K. C., Beattie J. B., Friedenthal J., Carroll R. C. (2007) J. Neurosci. 27, 14326–14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao W., Kang J. H., Liao Y., Ding W. X., Gambotto A. A., Watkins S. C., Liu Y. J., Stolz D. B., Yin X. M. (2010) J. Biol. Chem. 285, 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.