Abstract
The adapter protein 3BP2 (also known as SH3BP2 and Abl SH3-binding protein 2) has been involved in leukocyte signaling and activation downstream immunoreceptors. Genetic studies have further associated 3BP2 mutations to the human disease cherubism and to inflammation and bone dysfunction in mouse. However, how wild type 3BP2 functions in macrophage differentiation remains poorly understood. In this study, using small interfering RNA-mediated silencing of 3BP2 in the RAW264.7 monocytic cell line, we show that 3BP2 was required for receptor activator of NFκB ligand (RANKL)-induced differentiation of RAW264.7 cells into multinucleated mature osteoclasts but not for granulocyte macrophage-colony stimulating factor/interleukin-4-induced differentiation into dendritic cells. 3BP2 silencing was associated with impaired activation of multiple signaling events downstream of RANK, including actin reorganization; Src, ERK, and JNK phosphorylation; and up-regulation of osteoclastogenic factors. In addition, 3BP2 knockdown cells induced to osteoclast by RANKL displayed a reduced increase of Src and nuclear factor of activated T cells (NFATc1) mRNA and protein expression. Importantly, 3BP2 interacted with Src, Syk, Vav, and Cbl in monocytic cells, and the introduction of constitutively active mutants of Src and NFATc1 in 3BP2-deficient cells restored osteoclast differentiation. Finally, the expression of a 3BP2 cherubism mutant was found to promote increased Src activity and NFAT-dependent osteoclast formation. Together, this study demonstrates that wild type 3BP2 is a key regulator of RANK-mediated macrophage differentiation into osteoclast through Src and NFATc1 activation.
Keywords: Adaptor Proteins, Cell Differentiation, Cytokine Action, Cytoskeleton, Macrophage, NFAT Transcription Factor, Signal Transduction, Src, Osteoclast, RANKL
Introduction
Osteoclasts are multinucleated bone-resorbing cells, differentiating from CD11b+ hematopoietic cells of the monocyte/macrophage lineage. The interaction between bone marrow stroma, osteoblasts, hematopoietic cells, and the immune system is determinant for bone homeostasis and osteoclastogenesis (1, 2). Osteoclast differentiation is essentially triggered by two hematopoietic factors, the member of the tumor necrosis factor superfamily of cytokine receptor activator of NFκB ligand (RANKL)4 and macrophage colony-stimulating factor (M-CSF) (1). In response to osteotropic factors, osteoblasts express RANKL, which binds RANK on osteoclast progenitor cell surfaces, leading to osteoclast maturation. Consistently, mice lacking RANKL or RANK exhibit an osteopetrotic phenotype, associated with defects in bone resorption and a lack of osteoclasts (3). Stimulation of mononuclear osteoclast precursors by RANKL and M-CSF regulates growth, differentiation, fusion, and survival, leading to functional multinucleated osteoclasts (1). Mature osteoclasts express typical markers, including tartrate-resistant acidic phosphatase (TRAP) and calcitonin receptor, and adhere to the bone surface through a ring of polymerized actin, adhesion receptors (αvβ3), and cytoskeleton molecules, forming a sealed resorbing compartment in which acidification and secretion of proteolytic enzymes allow the resorption of the bone matrix (1, 4).
Osteoclast stimulation by RANKL and M-CSF triggers the activation of intracellular events involving a large number of signaling molecules, including the adapters TRAF6 (5), Dap12 and FcRγ (6, 7), the Rho GTPases activator Vav3 (8), phosphatidylinositol 3-kinase (9), and MAPK family members ERK, JNK, and p38. The Src (10), Syk (7, 11), and Tec (12) protein-tyrosine kinase families are other key molecules in osteoclastogenesis downstream RANK, ITAM-bearing molecules, integrins, c-Fms, and the M-CSF receptor (1, 12, 13). These signal transduction pathways ultimately converge to the activation of several transcription factors such as NFκB, c-Fos, PU.1, and microphathalmia-associated transcription factor (4). Another crucial transcriptional event during osteoclastogenesis is the up-regulation of NFATc1 downstream RANK and intracellular calcium (Ca2+) signaling (14, 15). NFATc1 is a member of the NFAT family of transcription factors, the transcriptional activities of which are regulated by the Ca2+-dependent serine/threonine phosphatase calcineurin (16). Inhibiting calcineurin activity by cyclosporine A and FK506 suppressed osteoclast differentiation in vitro (17). Conversely, the expression of constitutively activated NFATc1 promotes osteoclast differentiation in the absence of RANKL (14, 18). Therefore, NFATc1 appears to be necessary and sufficient for osteoclastogenesis.
Adapter proteins are key components of leukocyte immunoreceptor signal transduction pathways coupled to protein-tyrosine kinases (19, 20). Molecular scaffolds composed of adapter proteins and enzymes are assembled and activated at the plasma membrane by Src and/or Syk protein-tyrosine kinases. These scaffolds transduce signals to the cytoplasm, cytoskeleton, and nucleus to activate lipid and calcium signaling, gene expression, and metabolic changes involved in leukocyte proliferation, differentiation, and motility. We and others have identified a regulatory role for 3BP2 (c-Abl SH3 domain-binding protein-2, also known as SH3BP2) in immunoreceptor signaling, including T (21, 22), B (23–26), NK (27), and mast (28) cells. Importantly, 3BP2 associates several signaling proteins such as Src/Syk kinases, Vav proteins, and PLCγ, involved in NFAT activation (reviewed in Refs. 29 and 30). Consistently, 3BP2 was found to positively regulate the activity of NFAT in T and B cells (21, 23). In addition, studies in mouse deficient for 3BP2 expression have shown that 3BP2 regulates B cell development and BCR-mediated B cell activation and calcium mobilization (24, 25). Finally, genetic evidence linking 3BP2 to the human genetic bone disease cherubism (31, 32) indicates that 3BP2 also plays a crucial role during inflammation and bone remodeling. Cherubism is an autosomal dominant disorder characterized by the erosion of maxillar and mandibular bone with the resultant dental and facial deformity caused by excessive osteoclast activity and giant cell granuloma formation (33). The signaling alterations of a mutant form of 3BP2 as observed in a mouse model of cherubism include increased tumor necrosis factor α production by hyperactive macrophages associated with systemic inflammation, aberrant osteoclast activities, and osteoporosis (32). Interestingly, genetic inactivation of NFATc1 in mice with cherubism prevented bone loss (34), suggesting that NFAT activation by 3BP2 is a critical step during osteoclastogenesis. However, exactly how wild type 3BP2 functions in RANK-mediated osteoclast differentiation has not yet been elucidated.
In this study, we have investigated the role of 3BP2 during osteoclast differentiation and RANK signaling. Using RNA interference blocking experiments in the RAW264.7 monocyte/macrophage cell line, we show that the absence of 3BP2 in pre-osteoclasts is associated with a severe reduction of osteoclast formation and increased expression of osteoclastogenic factors. The absence of 3BP2 resulted in decreased RANK-mediated actin cytoskeleton remodeling, Src phosphorylation, and activation of multiple signaling pathways involved in RANK signaling, as well as a deregulated expression of NFATc1. 3BP2 interacted with signaling proteins, including Src, Syk, Vav1, and Cbl in resting cells, and the introduction of constitutively active mutants of Src and NFATc1 in 3BP2-deficient cells restored osteoclast differentiation. In addition, the expression of a 3BP2 cherubism mutant was found to promote increased Src activity and NFAT-dependent osteoclast formation. Altogether, this study demonstrates that wild type 3BP2 is a key regulator of RANK-mediated osteoclastogenesis through Src and NFATc1 activation.
EXPERIMENTAL PROCEDURES
Cell Line and Culture
RAW264.7 cells were purchased from American Type Culture Collection (Manassas, VA). The cells were maintained at 37 °C, 5% CO2 in α-minimum Eagle's medium (Lonza, Walkersville, MD) supplemented with 10% fetal bovine serum (Hyclone) and penicillin and streptomycin (Invitrogen). For stimulation, the cells were deprived of serum for 12 h in α-minimum Eagle's medium. 5 × 106 cells/ml were then stimulated at 37 °C for the indicated time with or without the indicated cytokine.
Reagents, Antibodies, and Plasmids
All of the chemicals were from Sigma-Aldrich, except the pharmacological inhibitors PP2 and cyclosporine, which were from Merck. Soluble RANKL was from R & D Systems (Minneapolis, MN), and M-CSF, GM-CSF, and IL-4 were from ImmunoTools (Germany). Anti-3BP2 antibodies were described before (23). Antibodies against phosphorylated forms of Src, Syk, ERK1/2, p38, JNK, MEK1/2, Akt, IKKα/β, total Src, total Akt, and horseradish peroxidase-conjugated secondary antibodies were purchased from Cell Signaling Technology (Beverly, MA). Antibodies against ERK2 and NFATc1 were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-V5 monoclonal antibody was from Invitrogen, and anti-hemagglutinin (HA) was from Roche Applied Science. The wild type and R415P 3BP2 cDNAs bearing a N-terminal V5 epitope tag (23) was cloned into the murine leukemia virus-based LZRS-IRES-GFP retroviral vector. The plasmids encoding the VIVIT peptide, SrcY527F, and constitutively active NFATc1 (plasmids 11106, 13660, and 11102, respectively) were from Addgene (Cambridge, MA).
Generation of 3BP2 Knockdown RAW264.7 Cells
Three short hairpin RNA (shRNA)-expressing plasmids were generated using the Block-iT RNA interference vector kit (Invitrogen). The complementary nucleotide sequences targeting positions 381–401, 488–508, and 507–527 of mouse 3BP2 RNA were the following: 3BP2 381 sense, 5′-CACCGGAAATTGGCCACTTCCATGACGAATCATGGAAGTGGCCAATTTCCTTTT-3′; 3BP2 381 antisense, 5′-AAAAGGAAATTGGCCACTTCCATGATTCGTCATGGAAGTGGACCATTTCC-3′; 3BP2 488 sense, 5′-CACCGCCTGTCTTCATATCCCATGGCGAACCATGGGATATGAAGACAGGCTTTT-3′; 3BP2 488 antisense, 5′-AAAAGCCTGTCTTCATATCCCATGGTTCGCCATGGGATATGAAGACAGGC-3′; 3BP2 507 sense, 5′-CACCGGACAATGAAGATTACGAACACGAATGTTCGTAATCTTCATTGTCCTTTT-3′; and 3BP2 507 antisense, 5′-AAAAGGACAATGAAGATTACGAACATTCGTCTTCGTAATCTTCATTGTCC-3′. Following annealing and ligation into pENTR/H1/TO, the 3BP2 shRNA vectors were transfected into RAW264.7 cells using Lipofectamine (Invitrogen). As a control, RAW264.7 cells were transfected with the same amount of a LacZ shRNA construct. Stably transfected cells were selected using 100 μg/ml of zeocin (Invitrogen), and stable suppression of 3BP2 expression was evaluated by immunoblotting.
In Vitro Differentiation of Osteoclasts and Dendritic Cells
For osteoclastogenesis assays using the RAW264.7 cell line, the cells were plated in a 12-well plate at a cell density of 10,000 cells/well in the presence of 40 ng/ml RANKL, combined or not with 30 ng/ml M-CSF, and cultured for 4–5 days. Fresh culture medium containing RANKL was added on day 3. The cells were stained for TRAP activity with the leukocyte acid phosphatase kit (Sigma-Aldrich). Multinucleated TRAP-positive cells were counted and scored by microscopy. To determine complexity and size differences, osteoclasts were counted by number of nuclei and size. TRAP-positive cells with more than three nuclei and larger than 100 μm in diameter were counted as osteoclasts.
For dendritic cell differentiation, RAW264.7 cells were cultured with recombinant GM-CSF (100 ng/ml) and IL-4 (10 ng/ml) for 6 days. For maturation of dendritic cells, lipopolysaccharide (LPS; 10 μg/ml) was added for 2 additional days. After 8 days, the cells were analyzed for their morphology under a Zeiss Axiovert 40C light microscope or stained with fluorescein isothiocyanate- or phycoerythrin-conjugated antibodies reactive to CD11c, CD80, or CD86 (all purchased from Becton-Dickinson), followed by flow cytometry analysis on a FACScan (Becton Dickinson).
F-actin Fluorescence Staining of Osteoclast
The cells were seeded onto a sterile chamber slide and treated with RANKL (40 ng/ml) for the indicated times. After washing, the cells were fixed with 3.7% formaldehyde in PBS, permeabilized with 0.2% Triton X-100 for 10 min, washed twice in PBS containing 1% bovine serum albumin, and incubated with 100 ng/ml of Texas Red phalloidin (Invitrogen) at 22 °C in a humidified atmosphere for 30 min. After staining, the cells were washed with PBS, rinsed with water, and mounted with Fluoromount (Sigma-Aldrich). Polymerized actin was visualized using a Zeiss Axiovert 200M fluorescence microscope (Zeiss Leica Microsystems) equipped by a Hamamatsu ORCA-ER digital camera (Hamamatsu, Japan). Image analysis was performed with Volocity software (Improvision Inc., Waltham, MA). Quantification of F-actin content in osteoclasts was performed by flow cytometry. The cells were scraped off culture dishes, fixed with 3.7% formaldehyde in PBS, and permeabilized with 0.05% saponin in PBS, 1% bovine serum albumin for 20 min at room temperature. The cells were washed and labeled with 100 ng/ml AlexaFluor 488-conjugated phalloidin (Invitrogen) for 30 min at 22 °C. Following three washes with cold PBS, 1% bovine serum albumin, polymerized actin was measured by flow cytometry. The data are expressed as the mean channel fluorescence intensity for each sample.
Immunoprecipitation and Immunoblotting
The cells were lysed at 1 × 108 cells/ml in ice-cold lysis buffer (1% Triton X-100 in 150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 0.1% SDS, 0.1% sodium deoxycholate, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride) for 30 min on ice. Cleared lysates were then incubated for 3 h at 4 °C with the indicated antibodies and 1 h with protein G-Sepharose beads (Sigma). The pellets were then washed three times with ice-cold lysis buffer and resuspended in SDS sample buffer. Eluted immunoprecipitates or whole cell lysates were separated by SDS-PAGE and analyzed by immunoblotting as described before (23).
Transfection, Luciferase Reporter, and Complementation Assays
The reporter plasmid pTRAP-luc was a kind gift from H. Takayanagi (Tokyo Medical and Dental University, Tokyo, Japan) (14). The luciferase reporter constructs for NFAT and AP-1 activities and the 3BP2 expression constructs were described before (21, 23). Transient transfections into RAW264.7 cells were performed using the Amaxa (Lonza, Germany) nucleofection system. Briefly, 2 × 106 cells were washed and resuspended in 100 μl of Buffer V, mixed with 5 μg of reporter plasmid along with the indicated expression plasmids and a Renilla luciferase plasmid (Promega), and electroporated using program D-032 on the Amaxa nucleofection device. Following incubation at 37 °C for 16 h, the cells were stimulated or not with RANKL (40–100 ng/ml) for 24–48 h at 37 °C. Dual luciferase reporter assays were performed as described (23). Normalized luciferase activities were determined in triplicate and expressed as fold increase relative to the basal activities measured in control vector-transfected cells.
For complementation assays, the cells were transfected with mock retroviral expression vectors, constitutively active NFATc1 expression vector, or a combination of the SrcY527F expression vector and enhanced GFP expression vector, as described above. After 48 h of transfection, the GFP-positive transfected cells were purified on a FACSAria cell sorter (Becton-Dickinson) and subjected to in vitro differentiation assays and immunoblotting analysis.
Real Time Quantitative PCR
Total RNAs from RAW264.7 cells were reverse transcribed using the high capacity cDNA archive random priming kit (Applied Biosystems). Real time quantitative PCR was performed using a 7900HT sequence detector system (Applied Biosystems) and the SYBR Green dye detection protocol as previously described (35). The relative expression level of target genes mRNA between control (X) and sample (Y) was calculated using the formula ΔCTY − DCTX and expressed as fold over control (2ΔΔCT).
Statistical Analysis
The data are expressed as the means ± S.D. The statistical significance of differences between the experimental groups was determined by an unpaired Student's t test. Effects with a p value less than 0.05 were considered statistically significant.
RESULTS
Suppression of Osteoclast Differentiation in 3BP2 Knockdown RAW264.7 Cells
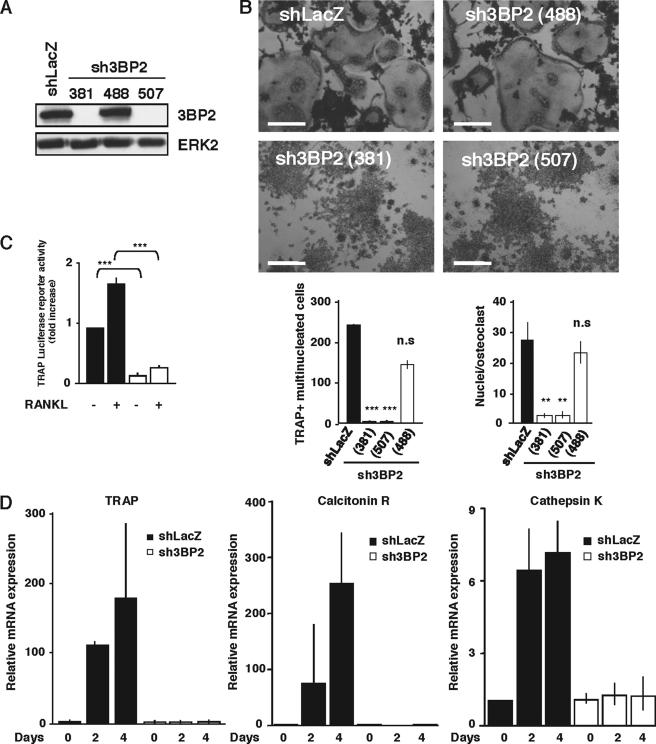
To examine the involvement of 3BP2 in the regulation of osteoclast differentiation, we generated stable and inducible 3BP2 knockdown cell models. Three shRNA-expressing plasmids targeting three different sequences of 3BP2 were generated and were stably transfected into the RAW264.7 cell line. As shown in Fig. 1(A and B), two shRNA constructs designated sh3BP2(381) and sh3BP2(507) completely suppressed the expression of 3BP2 and RANKL-induced formation of TRAP-positive multinucleated cells. In contrast, the shRNA-expressing plasmid sh3BP2(488), which had no effect on 3BP2 expression as compared with the shLacZ control vector, did not interfere with RANKL-induced cell-cell fusion. Quantitative analysis showed that the lack of 3BP2 dramatically reduced the number of TRAP-positive multinuclear osteoclasts and the number of nuclei/TRAP-positive cells induced upon RANKL stimulation (Fig. 1B). We determined that neither spontaneous proliferation of sh3BP2(381) cells was significantly modified compared with shLacZ cells (supplemental Fig. S1), nor was RANK expression different on both cell types (not shown). We also controlled that the combined stimulation of the sh3BP2(381) RAW264.7 cell clone by RANKL in the presence of M-CSF had no effect on the impaired differentiation of 3BP2-silenced cells (supplemental Fig. S2). To further document the role of 3BP2 in osteoclast differentiation, we selected the sh3BP2(381) RAW264.7 cell clone (referred to as sh3BP2 cells for simplicity) to examine the expression of osteoclastogenic factors using real time quantitative PCR analysis and luciferase reporter assays. First, RAW264.7 cells were transiently transfected with a TRAP promoter-luciferase plasmid, and the luciferase activity was measured. As shown in Fig. 1C, 3BP2 knockdown cells (sh3BP2) showed a marked reduction of both basal and RANKL-induced TRAP promoter activities as compared with control cells (shLacZ). Using real time quantitative PCR analysis, we next examined the expression of osteoclastogenic factors in control or 3BP2 knockdown cells treated with sRANKL for different times. We found that sRANKL-induced expression of TRAP, calcitonin receptor, and cathepsin K was severely impaired in 3BP2-deficient cells (Fig. 1D). Thus, expression of 3BP2 is required for RANKL-induced differentiation of RAW264.7 cells into multinucleated TRAP-positive osteoclasts.
FIGURE 1.
Expression of 3BP2 is required for RANKL-stimulated osteoclast formation in RAW264.7 cells. A, generation of stable 3BP2 knockdown in RAW264.7 cells. Three shRNA-expressing plasmids targeting positions 381–401 (381), 488–508 (488), and 507–527 (507) of mouse 3BP2 RNA and a control LacZ shRNA construct were stably transfected into RAW264.7 cells. After selection, stable suppression of 3BP2 expression was evaluated by immunoblotting using antibodies against 3BP2 and ERK2 as loading control. B, 3BP2 knockdown cells (sh3BP2 (381), sh3BP2 (488), and sh3BP2 (507)) and shLacZ control cells were cultured for 4 days with sRANKL (40 ng/ml) and stained for TRAP activity. Morphology (top panels) and multinucleated TRAP-positive cells (bottom panels) were assessed and scored by microscopy. To determine complexity and size difference, TRAP+ multinucleated cell number and nuclei/osteoclast were counted. The data are expressed as the means ± S.D. of three equivalent wells and are representative of three independent experiments. **, p < 0.01 versus shLacZ; ***, p < 0.001 versus shLacZ. Scale bars, 50 μm. C, shLacZ and sh3BP2(381) cells were transfected with a TRAP luciferase reporter construct. 8 h after transfection, the cells were stimulated or not with RANKL (40 ng/ml). Normalized luciferase activity was determined 24 h after stimulation and expressed as the fold increase relative to basal activities measured in control vector-transfected cells. The results are the means ± S.D. of triplicate determinations. ***, p < 0.001 versus shLacZ. D, shLacZ and sh3BP2 cells were stimulated or not with RANKL (40 ng/ml) for the indicated times. The expression of TRAP, calcitonin receptor, and cathepsin K mRNA was determined by real time quantitative PCR. The data are expressed as the means ± S.D. of triplicate determinations and are representative of three independent experiments.
3BP2 Is Not Required for Dendritic Cell Differentiation
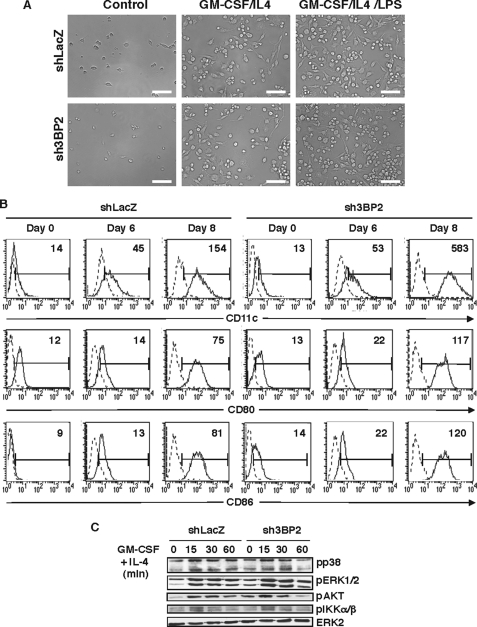
It is known that monocytes/macrophages exposed to GM-CSF/IL-4 can acquire DC phenotype and function (36). To examine whether 3BP2 is involved in DC differentiation, sh3BP2 RAW264.7 cells were cultured with GM-CSF and IL-4 for 6 days. Maturation of dendritic-like cells was further induced by treatment with LPS for 2 additional days. Cell differentiation was then analyzed by phase contrast microscopy and flow cytometry. After 6 days of treatment with GM-CSF/IL-4, a morphological transformation from macrophage-like cells into dendritic-like cells was observed for both control and 3BP2 knockdown cells, a process further increased following incubation with LPS for another 2 days (Fig. 2A). Dendritic morphology was characterized by an increase in cell size and multiple membrane protrusions. Dendritic-like phenotypic change in either control or 3BP2-deficient cells was also evidenced by the increased membrane expression of typical dendritic cell markers such as CD11c, CD80 (B7.1), and CD86 (B7.2) following GM-CSF/IL-4 + LPS treatment (Fig. 2B). Finally, the lack of 3BP2 expression in RAW264.7 cells had no significant effects on the early biochemical events triggered by cell stimulation with GM-CSF/IL-4, including the activation of ERK, p38, Akt, and IKKα/β phosphorylation (Fig. 2C).
FIGURE 2.
Effects of 3BP2 suppression on differentiation of RAW264.7 cells into dendritic cells. A, shLacZ and sh3BP2 cells were cultured with GM-CSF (100 ng/ml) and IL-4 (10 ng/ml) for 6 days and then activated for 2 additional days with LPS (10 ng/ml). Dendritic-like cell morphology was examined by light microscopy. A representative field of each culture condition is shown. Scale bars, 50 μm. B, cells were cultured as described above and stained with fluorescein isothiocyanate- or phycoerythrin-conjugated antibodies against CD11c, CD80, or CD86, followed by flow cytometry analysis. The dotted line histograms represent cells stained with isotype-matched control antibodies. The data shown are representative of three independent experiments. C, cell lines were cultured in the absence of serum for 12 h before stimulation with GM-CSF (100 ng/ml) and IL-4 (10 ng/ml) for 0, 15, 30, and 60 min. The cell lysates were subjected to immunoblot analysis with antibodies against phospho-p38, -IKKα/β, -AKT, and -ERK1/2. The same membrane was stripped and reprobed with anti-ERK2.
3BP2 Is Required for Organization of the Osteoclast Actin Cytoskeleton
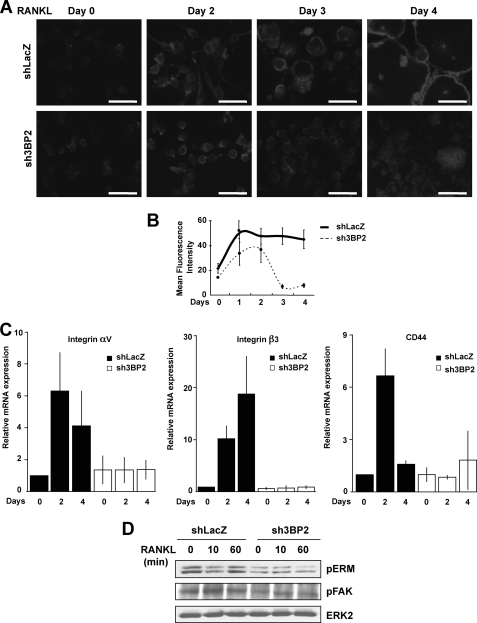
Actin cytoskeleton remodeling plays an essential role during osteoclast differentiation and function (1). To examine the implication of 3BP2 in the osteoclast actin reorganization, sh3BP2 RAW264.7 cells and shLacZ control cells were cultured for 4 days with sRANKL and subjected to immunofluorescence analysis with fluorescent phalloidin to detect actin organization. After 2 days of stimulation, control cells showed increased actin polymerization associated with cell spreading, whereas those lacking 3BP2 exhibited altered F-actin content and spreading. The architectural abnormalities of 3BP2 knockdown osteoclasts were evident after 3 days of RANKL treatment. Whereas control cells showed well formed actin rings and cell fusion at days 3 and 4, no expression of 3BP2 in osteoclasts led to a dramatic reduction of actin polymerization associated with cell-cell fusion (Fig. 3A). By using flow cytometry, we quantified how 3BP2 interferes with actin polymerization in response to RANKL during osteoclast differentiation. Control cells showed a 2–3-fold increase in F-actin content after 1 day of RANKL stimulation, and the total amount of polymerized actin remained stable during the next 3 days of treatment. In contrast, 3BP2 knockdown cells exhibited only a 2-fold increase in F-actin between days 1 and 2 of culture with RANKL, followed by a dramatic fall of actin polymerization associated with impaired cell-cell fusion (Fig. 3B). Functional osteoclasts adhere to the bone surface through adhesion receptors, including αvβ3 integrin and CD44, connected to the actin cytoskeleton network. Lack or failure to up-regulate their expression during osteoclast maturation results in severe cytoskeletal defects and abnormal bone resorption (37, 38). We therefore examined the integrity of integrin αvβ3 and CD44 signaling pathways in the absence of 3BP2. As expected, control osteoclasts treated with RANKL for 2–4 days strongly up-regulated the mRNAs encoding αv and β3 integrin subunits and CD44. In contrast, no significant increase of αvβ3 and CD44 expression was detected in 3BP2 knockdown osteoclasts (Fig. 3C). Consistently, RANKL-induced phosphorylation of FAK and ERM, two cytoplasmic regulators of the αvβ3 and CD44 signaling pathways, respectively, was severely altered in sh3BP2 cells as compared with control cells (Fig. 3D).
FIGURE 3.
Effects of 3BP2 suppression on RANKL-induced organization of the osteoclast actin cytoskeleton. A, shLacZ and sh3BP2 cells were seeded onto sterile chamber slide treated with sRANKL (40 ng/ml) for the indicated times. After washing, the cells were fixed, permeabilized, and incubated with 100 ng/ml of Texas Red phalloidin for 30 min. Polymerized actin was visualized using a fluorescence microscope. The data shown are representative of three independent experiments. Scale bars, 50 μm. B, cells were treated with sRANKL (40 ng/ml) for the indicated times. Following cell fixation and labeling with AlexaFluor 488-conjugated phalloidin (100 ng/ml), actin polymerization was quantified by flow cytometry. The data are expressed as mean channel fluorescence intensity for each sample. The results represent the means ± S.D. of three independent determinations. C, shLacZ and sh3BP2 cells were stimulated or not with RANKL (40 ng/ml) for the indicated times. The expression of integrin αv and β3 subunits and CD44 mRNA was determined by real time quantitative PCR. The data are expressed as the means ± S.D. of triplicate determinations and are representative of three independent experiments. D, cells were cultured without serum for 12 h before stimulation with sRANKL (100 ng/ml) for 0, 10, and 60 min. The cell lysates were subjected to immunoblot analysis with antibodies against phospho-ERM and phospho-FAK. The same membrane was stripped and reprobed with anti-ERK2.
3BP2 Regulates Multiple Signaling Pathways in Response to RANKL
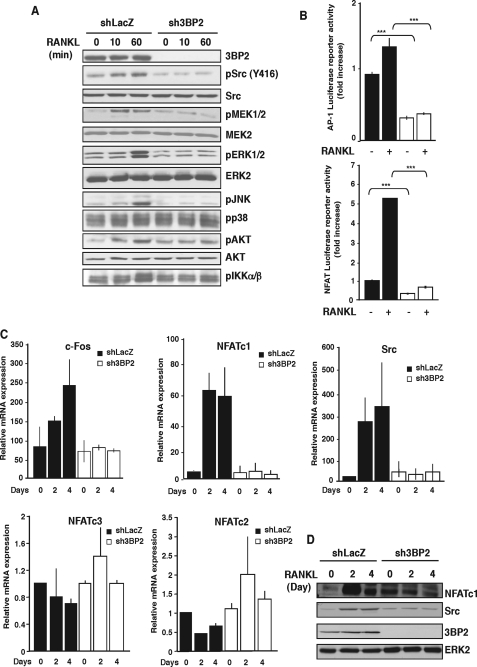
Binding of RANKL to its receptor RANK triggers the recruitment and activation of multiple signaling effectors, leading to transcriptional regulation of osteoclastogenic factors (1). We therefore examined RANKL-induced early signaling pathways in 3BP2 knockdown macrophages. As shown in Fig. 4A, RANKL-stimulated phosphorylation of Src, Akt, and MAPKs ERK1/2 and JNK was dramatically affected in the absence of 3BP2. In contrast, the phosphorylation of p38 was not modified. Upon RANKL stimulation, 3BP2 knockdown cells also displayed reduced phosphorylation of MEK1, an upstream activator of ERK1/2, and reduced phosphorylation of IKKα/β, an upstream activator of the transcription factor NFκB. These early signaling pathways increase the activity of multiple transcription factors playing essential roles during osteoclastogenesis. We therefore examined whether 3BP2 expression can regulate the transcriptional activities of AP-1, NFκB, and NFAT using a luciferase reporter assay in RAW264.7 cells. A marked reduction of basal and RANKL-induced AP-1 and NFAT activities was observed in 3BP2 knockdown cells as compared with control cells (Fig. 4B). In addition, we found reduced levels of the mRNA encoding the transcription factors c-Fos and NFATc1 and the kinase Src (Fig. 4C), whereas the expression of NFATc2 and NFATc3, two transcription factors related to NFATc1, was not significantly modified in the absence of 3BP2 (Fig. 4C). In addition, mRNA expression of PU.1, a transcription factor regulating osteoclastogenesis, was unaffected in the absence of 3BP2 (data not shown). Consistent with the impaired increase of mRNA expression, 3BP2 knockdown cells induced to osteoclast by RANKL for 2–4 days displayed no up-regulation of NFATc1 and c-Src at protein levels, compared with control shLacZ cells (Fig. 4D). Thus, 3BP2 appears to regulate multiple signaling pathways downstream RANKL-RANK interaction, including the major osteoclastogenic factor NFATc1.
FIGURE 4.
3BP2 knockdown affects multiple signaling pathways in response to RANKL. A, shLacZ and sh3BP2 cells were cultured in the absence of serum for 12 h before stimulation with sRANKL (100 ng/ml) for 0, 10, and 60 min. The cell lysates were separated by SDS-PAGE and transferred to nitrocellulose membranes that were subjected to immunoblot analysis with antibodies against phospho-Src, -ERK1/2, -JNK, -p38, -AKT, -IKKα/β, and -MEK1/2. After stripping, the membrane was reprobed with anti-3BP2, Src, AKT, MEK2, and ERK2. B, shLacZ (black bars) and sh3BP2 (white bars) cells were transfected with AP-1 or NFAT luciferase reporter constructs. 8 h after transfection, the cells were stimulated or not with sRANKL (100 ng/ml). Normalized luciferase activity was determined 24 h after stimulation and expressed as the fold increase relative to basal activities measured in control vector-transfected cells. The results are the means ± S.D. of triplicate determinations. ***, p < 0.001 versus shLacZ. C, cells were stimulated with sRANKL for the indicated times. The expression of c-Fos, Src, NFATc1, NFATc2, and NFATc3 mRNA was determined by real time quantitative PCR. The data are expressed as the means ± S.D. of triplicate determinations and are representative of three independent experiments. D, lysates from cells stimulated as indicated above were subjected to immunoblot analysis with antibodies against NFATc1, Src, and 3BP2. Protein loading was controlled by reprobing the membrane with anti-ERK2.
Ectopic Expression of Active Src and NFAT Proteins Rescues Osteoclast Formation by 3BP2 Knockdown Macrophages
Previous studies have implicated 3BP2 in NFAT activation in leukocytes and shown its interaction with members of the Src family of protein-tyrosine kinases (29, 30). As shown in Fig. 5A, 3BP2 co-immunoprecipitated with c-Src in resting monocytic cells. Consistent with other studies in lymphocytes (21, 23), 3BP2 also interacted with Syk, Vav1, and Cbl in RAW264.7 cells. We next determined the contribution of Src and NFATc1 to 3BP2-dependent osteoclast formation. To this end, we introduced in 3BP2 knockdown RAW264.7 cells either control retroviruses or constitutively active forms of c-Src (SrcY527F) (39) (Fig. 5B) or NFATc1 (caNFATc1) (40) (Fig. 6A) and examined osteoclast formation. Expression of SrcY527F in RANKL-stimulated 3BP2 knockdown macrophages partially restored cell-cell fusion and the formation of TRAP-positive multinucleated cells (Fig. 5C). Osteoclast-like cell number (Fig. 5D, left panel) and size (Fig. 5D, right panel) were significantly increased in RANKL-stimulated sh3BP2 cells expressing SrcY527F, as compared with RANKL-stimulated sh3BP2 cells expressing a control construct. Importantly, as shown in Fig. 5B, overexpression of active Src in control and 3BP2 knockdown cells induced the phosphorylation of Syk Tyr352 and ERK1/2, two events critically involved in several aspects of osteoclast formation and function (2). The constitutive phosphorylation of SrcY527F on Tyr416 was confirmed by immunoblot analysis (Fig. 5B).
FIGURE 5.
Ectopic expression of active SrcY527F protein partially rescues RANKL-induced osteoclast formation in the absence of 3BP2 in RAW264.7 cells. A, 3BP2 interacts with c-Src, Syk, Vav1, and Cbl in resting RAW264.7 cells. The cells were lysed at 1 × 108 cells/ml in ice-cold lysis buffer (1% Triton X-100 in 150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 0.1% SDS, 0.1% sodium deoxycholate, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride) for 30 min on ice. Cleared lysates from 1 × 107 cells were incubated with nonrelevant goat antibodies (NR) or sheep anti-3BP2 antibodies for 3 h at 4 °C followed by incubation with protein G-Sepharose beads for 1 h. After three washes, the immunoprecipitates were immunoblotted using antibodies against 3BP2, c-Src, Syk, Vav1, and Cbl. WCL, whole cell lysate. B, shLacZ and sh3BP2 cells were transfected with pLNCX SrcY527F retroviral construct or control vector, together with enhanced GFP expression vector. After 48 h, the GFP-positive cells were purified on a FACSAria cell sorter and subjected to immunoblotting analysis using antibodies against 3BP2, phospho-Src, phospho-Syk, and phospho-ERK1/2. Protein loading was controlled with anti-ERK2 immunoblot. Cells purified as indicated above were cultured for 4 days with sRANKL (40 ng/ml) and stained for TRAP activity. C and D, morphology (C) and multinucleated TRAP-positive cells (D) were assessed and scored by microscopy. Scale bars, 50 μm. TRAP+ multinucleated cell number and nuclei/osteoclast were counted. The data are expressed as the means ± S.D. of three equivalent wells and are representative of three independent experiments. ***, p < 0.001 versus shLacZ.
FIGURE 6.
Complementation of osteoclast formation by ectopic expression of constitutively active NFATc1 in 3BP2 knockdown cells. shLacZ and sh3BP2 cells were transfected with control vector or pRV-HA-caNFAT2-IRES-GFP vector encoding HA-tagged constitutively active caNFATc1 (caNFAT). After 48 h, the GFP-positive cells were purified on a FACSAria cell sorter. A, purified cells were subjected to immunoblotting analysis using antibodies against HA tag. Protein loading was controlled with anti-ERK2 immunoblot. The cells purified as indicated above were then cultured for 4 days in the presence of sRANKL (40 ng/ml) and stained for TRAP activity. B and C, morphology (B) and multinucleated TRAP-positive cells (C) were scored by microscopy. The cells with more than three nuclei were counted multinucleated cells, and the nuclei/osteoclast were counted. The data are expressed as the means ± S.D. of three independent determinations. **, p < 0.01 versus shLacZ; ***, p < 0.001 versus shLacZ.
Next, we assessed the differentiation of cells expressing a retroviral construct encoding a HA-tagged constitutively active form of NFATc1. Expression of caNFATc1 in 3BP2 knockdown cells was found to completely restore the formation of TRAP-positive, multinucleated cells in RANKL-treated cells (Fig. 6B). In addition to morphology, the number (Fig. 6C, left panel) and size (Fig. 6C, right panel) of osteoclast-like cells formed by 3BP2 knockdown macrophages expressing caNFATc1 reached levels comparable with control conditions. The expression of HA-tagged caNFATc1 was confirmed by immunoblot analysis (Fig. 6A). Together, these data suggest that 3BP2 acts upstream of Src and NFATc1 in RANK-induced signaling pathways of osteoclast formation.
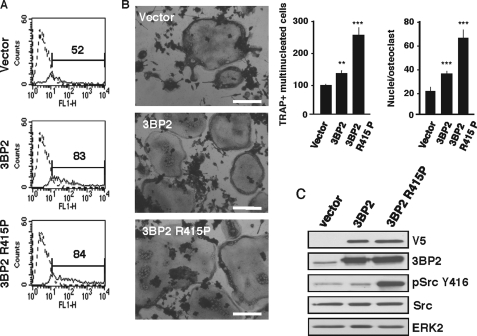
Expression of 3BP2 Cherubism Mutant Increases c-Src Activation and Osteoclast Formation in RAW264.7 Cells
Genetic studies have linked mutations of 3BP2 to the bone disease cherubism in humans and mice (31, 32), and genetic studies have linked 3BP2 mutation to enhanced phosphorylation of Syk in myeloid cells (32). To assess whether 3BP2 cherubism mutant proteins also affect c-Src activity and osteoclast formation in our system, we used retroviral constructs encoding V5-tagged forms of wild type 3BP2 and 3BP2 R415P cherubism mutant, which were expressed in RAW264.7 cells through IRES-GFP bicistronic retroviral constructs (Fig. 7A). After 2 days, GFP+ cells were sorted by fluorescence-activated cell sorter and subjected to RANKL stimulation. As shown in Fig. 7B, expression of the R415P cherubism mutant significantly increased the formation of TRAP+ multinucleated osteoclasts, compared with control and overexpressed wild type 3BP2. Ectopic expression of 3BP2 R415P was also accompanied by a marked increase of Src phosphorylation on the activating site Tyr418 (Fig. 7C). Consistent with studies by Aliprantis et al. (34), we also show that the increased osteoclast formation induced by the 3BP2 R415P mutant required NFAT activity because the observed effect was completely abrogated by expression of the selective NFAT inhibitor VIVIT-GFP (41) (supplemental Fig. S3). Together, these data indicate that 3BP2 mutant proteins promote increased Src activity and NFAT-dependent osteoclast formation in RAW264.7 cells.
FIGURE 7.
Effects of the expression of a 3BP2 cherubism mutant on Src activation and osteoclast differentiation in RAW264.7 cells. RAW264.7 cells were transfected with control vector, LZRS-V5–3BP2-IRES-GFP, or LZRS-V5–3BP2 R415P. A, GFP-positive cells were purified on a FACSAria cell sorter. B, sorted cells were stimulated for 5 days with sRANKL (40 ng/ml) and stained for TRAP activity. Morphology and multinucleated TRAP-positive cells were scored by microscopy as described above. Scale bars, 50 μm. The data are expressed as the means ± S.D. of three independent determinations. **, p < 0.01 versus shLacZ; ***, p < 0.001 versus shLacZ. C, purified cells were subjected to immunoblotting analysis using antibodies against V5 tag, 3BP2, phospho-Src Tyr416. Protein loading was controlled with anti-Src and ERK2.
DISCUSSION
Genetic evidence linking mutant 3BP2 proteins to the bone disease cherubism (31, 32, 34) indicates that 3BP2 may play a crucial role during inflammation and bone remodeling. However, little is known about the molecular mechanisms by which wild type endogenous 3BP2 regulates osteoclastogenesis. Using RNA interference blocking experiments in the RAW264.7 monocyte/macrophage cell line, we show here that the absence of 3BP2 in pre-osteoclasts dramatically impaired their ability to form TRAP+ multinucleated cells following RANKL stimulation. The defects in osteoclastogenesis observed in the absence of 3BP2 resulted from a decreased RANK-mediated actin cytoskeleton remodeling and activation of multiple signaling pathways, including Src phosphorylation and impaired NFATc1 expression. Genetic complementation of 3BP2-deficient cells with constitutively active mutants of Src and NFATc1 restored osteoclast differentiation. In addition, the expression of a 3BP2 cherubism mutant was found to promote increased Src activity and NFAT-dependent osteoclast formation. Our study demonstrates that wild type 3BP2 is required for RANKL-induced osteoclastogenesis through its regulation of signaling pathways involving Src and NFATc1 activation.
In leukocytes, including T and B lymphocytes, NK cells, and mast cells, 3BP2 interacts with intracellular proteins such as Src and Syk protein-tyrosine kinases, Vav proteins, and PLCγ that have been involved in calcium signaling and NFAT activation downstream ITAM-containing immunoreceptors and adaptors (29, 30). Thus, our data indicate that osteoclasts constitute another type of leukocytes in which NFAT activation by wild type 3BP2 plays an important role. Consistent with overexpression experiments (42) and studies in mice genetically engineered to express a gain-of-function mutant of 3BP2 (34), our findings also support the importance of NFATc1 in 3BP2-dependent osteoclast signaling. Interestingly, the absence of 3BP2 expression did not affect the acquisition by monocytic cells of a dendritic-like phenotype, as evidenced by the increased expression of dendritic cell markers such as CD11c, CD80, and CD86, observed following stimulation of 3BP2-deficient cells with GM-CSF and IL-4 or upon further maturation induced by LPS. Early signaling induced by the combination of GM-CSF and IL-4 was also not affected by 3BP2 silencing in RAW264.7 cells. Because myeloid DCs can differentiate upon multiple cytokine signaling pathways (43), a role for 3BP2 in the generation of distinct subsets of DCs cannot be excluded. Recently, a gain-of-function mutant of 3BP2 was involved in increased activity of inflammatory macrophages in a mouse model of cherubism (34). Thus, it would be interesting to determine whether wild type 3BP2 also participates in the differentiation process of inflammatory DCs and/or macrophages. Nevertheless, our data directly implicate wild type 3BP2 in RANK signaling in the differentiation process of monocytic cells toward osteoclasts. Of note, we recently found that bone marrow macrophages from 3BP2-deficient mice failed to differentiate into osteoclasts following RANKL/M-CSF stimulation.5
The process of osteoclast differentiation from monocytic precursors involves the activation downstream RANK of a large number of signaling molecules and transcription factors (4). It is well established that one crucial event during osteoclastogenesis is the up-regulation of NFATc1, a member of the NFAT family of transcription factors, the transcriptional activities of which are regulated by the Ca2+-dependent phosphatase calcineurin (17). During the early phase of osteoclast precursor differentiation, the initial induction of NFATc1 protein following RANK/RANKL interaction is mediated by a TRAF6-dependent pathway and leads to the autoamplification of NFATc1, which then regulates the expression of osteoclastogenic genes involved in cell maturation and bone resorption (1, 4, 17). Differentiation experiments using the RAW264.7 monocyte/macrophage cell line show that the absence of 3BP2 in pre-osteoclasts dramatically impaired early and late signaling events typically associated with RANKL stimulation, including actin cytoskeleton remodeling; activation of Src, ERK1/2, JNK, and Akt; and increased expression and activity of c-Fos and NFATc1. As a consequence, the expression of NFATc1 target genes such as TRAP, calcitonin receptor, cathepsin K, and αVβ3 integrin was severely reduced in the 3BP2-deficient cells following RANK engagement. Interestingly, the expression of the transcription factor PU.1 (data not shown) and of NFATc2 and NFATc3, two other NFAT family members, was not significantly affected by silencing of 3BP2. This suggests that 3BP2 specifically couples RANK signaling to NFATc1 activation, a notion also supported by the rescue experiments with active NFATc1 in 3BP2 knockdown cells. Genetic studies by Aliprantis et al. (34) have established that NFATc1 is downstream of 3BP2 cherubism mutant proteins. Together with our observations, this suggests that wild type and mutant 3BP2 proteins function through overlapping signaling pathways. In 3BP2 knockdown macrophages, RANKL stimulation failed to induce c-Fos mRNA expression and AP-1 activity, suggesting that 3BP2 participates in the initial TRAF6-dependent induction of NFATc1 messenger expression rather than in the NFATc1-dependent amplification of NFATc1 induction. TRAF6 and Src physically interact during osteoclast activation (5), and Src phosphorylation was impaired downstream of RANK in the absence of 3BP2. Conversely, the expression of a gain-of-function cherubism mutant of 3BP2 (3BP2 R415P) in RAW264.7 cells led to enhanced osteoclast formation and increased Src phosphorylation, underlining the importance of Src for 3BP2 signaling in osteoclasts. Importantly, we found that c-Src physically interacted with 3BP2 in monocytic cells. Therefore, it should be interesting to examine whether 3BP2 interacts with TRAF6, directly or indirectly through Src or additional proteins.
Constitutively active Src only partially rescued the impaired osteoclast differentiation observed in 3BP2 knockdown cells. Although osteoclasts rescued by overexpression of active Src differentiated into large multinucleated cells expressing high levels of TRAP, their morphology and size suggested that they are not fully mature. Thus, another question is how does 3BP2 couple RANK to the activation of NFATc1 besides Src? Interestingly, various binding partners of 3BP2, identified in other leukocytes (29, 30), have been involved in bone development, including Abl (44), Src (10), PLCγ (45), Cbl (46), and Vav3 (8). Moreover, the costimulatory signals provided by the ITAM-containing adaptor proteins DAP12 and FcRγ have been shown to play critical roles during osteoclast differentiation through Syk (6, 7). Osteoclasts from DAP12 × FcRγ-null mice exhibit defective NFATc1 expression as a consequence of impaired PLCγ-dependent calcium signaling (6). Of note, B cells from 3BP2−/− mice show impaired PLCγ phosphorylation, calcium mobilization, and NFAT activation (24). Our observations that in myeloid cells 3BP2 forms a signaling complex with c-Src, Syk, Vav, and Cbl provide a mechanistic explanation of how 3BP2 might participate in RANK-mediated osteoclast formation. Also, overexpression of active Src in control and 3BP2 knockdown cells induced the phosphorylation of Syk and ERK1/2, two events critically involved in several aspects of osteoclast function (2). Thus, by regulating Src activity, 3BP2 might impact on other osteoclast signaling pathways. Interestingly, Src-induced phosphorylation of Cbl downstream αvβ3 integrin promotes the recruitment and activation of phosphatidylinositol 3-kinase in a signaling complex composed of Pyk2-Src-Cbl essential for osteoclast bone resorbing activity (51). The interaction of 3BP2 with Cbl in RAW264.7 cells therefore suggests that phosphorylation of Cbl by c-Src represents another possible event of 3BP2 signaling in osteoclast. Src−/− mice are osteopetrotic because of defective formation of ruffled borders and subsequent bone resorption but not because of impaired osteoclast differentiation (47), suggesting the existence of compensatory mechanisms by other Src family members in mice. Whereas a negative regulatory function of Lyn in osteoclastogenesis was reported (48), the expression of Fyn, another Src family member, increased during osteoclast differentiation (12). Our observations that 3BP2 is a partner and substrate of Fyn (21, 23) therefore suggest that Fyn may participate in 3BP2 signaling in addition of Src.
In the absence of 3BP2, osteoclast precursors failed to reorganize their actin cytoskeleton, a process that has been found to be actively regulated by Src and Syk kinases, as well as Rho GTPases and guanine nucleotide exchange factor proteins, such as Vav family members (1, 8, 49). The observed interactions of 3BP2 with Src, Syk, and Vav in myeloid cells again provide some mechanistic insights on how 3BP2 might participate in cytoskeleton reorganization in forming osteoclasts. In genetically engineered mice, a gain-of-function mutant of 3BP2 promoted increased phosphorylation of Syk (32). Interestingly, we observed that a 3BP2 cherubism mutant expressed in macrophages strongly increased Src phosphorylation, compared with wild type 3BP2. It is therefore likely that 3BP2 regulates the localization and/or the activity of some of these proteins through its different protein-binding domains. Further experiments using co-cultured primary bone marrow macrophages and osteoblasts, which bring adhesion receptor ligands, should clarify the mechanism by which endogenous 3BP2 regulates osteoclast actin cytoskeleton remodeling. Interestingly, Src and Syk kinases signaling pathways are involved downstream multiple receptors necessary to osteoclast differentiation, ruffled border formation, and bone resorption, including RANK, costimulatory immunoreceptors coupled to ITAM-containing adapter proteins DAP12/FcRγ, αvβ3 integrin, and c-Fms (11, 13, 50). Thus, 3BP2 may integrate osteoclastogenic signals through its interactions with Src, Syk, Vav, and Cbl proteins.
In conclusion, our study provides evidence that wild type 3BP2 is a crucial regulator of RANK-mediated macrophage differentiation into osteoclasts. Further studies aimed at elucidating the molecular mechanisms by which wild type and pathological 3BP2 proteins regulate leukocyte activation and differentiation should provide a better understanding of inflammatory and bone diseases.
Supplementary Material
Acknowledgments
We thank A. Rao and J. Brugge for VIVIT and CA-NFAT and SrcY527F expression plasmids, respectively; N. Taylor for the LZRS-IRES-GFP retroviral vector; and H. Takayanagi for TRAP promoter-luciferase construct. We also thank N. Gonthier and A. Mallavialle for quantitative PCR analysis and A. Wakkach for helpful discussions.
This work was supported by INSERM, ANR (Agence Nationale pour la Recherche), and l'Association pour la Recherche sur le Cancer.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
R. Rottapel and M. Deckert, unpublished observations.
- RANKL
- receptor activator of NFκB ligand
- GM-CSF
- granulocyte macrophage-colony stimulating factor
- NFκB
- nuclear factor κB
- NFAT
- nuclear factor of activated T cells
- TRAP
- tartrate-resistant alkaline phosphatase
- PLC
- phospholipase C
- IKK
- IκB kinase
- ITAM
- immunoreceptor tyrosine-based activation motif
- sh
- short hairpin
- IL
- interleukin
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- JNK
- c-Jun N-terminal kinase
- MEK
- MAPK/ERK kinase
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- LPS
- lipopolysaccharide
- PBS
- phosphate-buffered saline
- DC
- dendritic cell
- IRES
- internal ribosomal entry site.
REFERENCES
- 1.Boyle W. J., Simonet W. S., Lacey D. L. (2003) Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 2.Walsh M. C., Kim N., Kadono Y., Rho J., Lee S. Y., Lorenzo J., Choi Y. (2006) Annu. Rev. Immunol. 24, 33–63 [DOI] [PubMed] [Google Scholar]
- 3.Kong Y. Y., Yoshida H., Sarosi I., Tan H. L., Timms E., Capparelli C., Morony S., Oliveira-dos-Santos A. J., Van G., Itie A., Khoo W., Wakeham A., Dunstan C. R., Lacey D. L., Mak T. W., Boyle W. J., Penninger J. M. (1999) Nature 397, 315–323 [DOI] [PubMed] [Google Scholar]
- 4.Teitelbaum S. L., Ross F. P. (2003) Nat. Rev. Genet. 4, 638–649 [DOI] [PubMed] [Google Scholar]
- 5.Wong B. R., Besser D., Kim N., Arron J. R., Vologodskaia M., Hanafusa H., Choi Y. (1999) Mol. Cell 4, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 6.Koga T., Inui M., Inoue K., Kim S., Suematsu A., Kobayashi E., Iwata T., Ohnishi H., Matozaki T., Kodama T., Taniguchi T., Takayanagi H., Takai T. (2004) Nature 428, 758–763 [DOI] [PubMed] [Google Scholar]
- 7.Mócsai A., Humphrey M. B., Van Ziffle J. A., Hu Y., Burghardt A., Spusta S. C., Majumdar S., Lanier L. L., Lowell C. A., Nakamura M. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6158–6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faccio R., Teitelbaum S. L., Fujikawa K., Chappel J., Zallone A., Tybulewicz V. L., Ross F. P., Swat W. (2005) Nat. Med. 11, 284–290 [DOI] [PubMed] [Google Scholar]
- 9.Munugalavadla V., Vemula S., Sims E. C., Krishnan S., Chen S., Yan J., Li H., Niziolek P. J., Takemoto C., Robling A. G., Yang F. C., Kapur R. (2008) Mol. Cell. Biol. 28, 7182–7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soriano P., Montgomery C., Geske R., Bradley A. (1991) Cell 64, 693–702 [DOI] [PubMed] [Google Scholar]
- 11.Zou W., Kitaura H., Reeve J., Long F., Tybulewicz V. L., Shattil S. J., Ginsberg M. H., Ross F. P., Teitelbaum S. L. (2007) J. Cell Biol. 176, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinohara M., Koga T., Okamoto K., Sakaguchi S., Arai K., Yasuda H., Takai T., Kodama T., Morio T., Geha R. S., Kitamura D., Kurosaki T., Ellmeier W., Takayanagi H. (2008) Cell 132, 794–806 [DOI] [PubMed] [Google Scholar]
- 13.Zou W., Reeve J. L., Liu Y., Teitelbaum S. L., Ross F. P. (2008) Mol. Cell 31, 422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 15.Asagiri M., Sato K., Usami T., Ochi S., Nishina H., Yoshida H., Morita I., Wagner E. F., Mak T. W., Serfling E., Takayanagi H. (2005) J. Exp. Med. 202, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan P. G., Chen L., Nardone J., Rao A. (2003) Genes Dev. 17, 2205–2232 [DOI] [PubMed] [Google Scholar]
- 17.Takayanagi H. (2007) Ann. N.Y. Acad. Sci. 1116, 227–237 [DOI] [PubMed] [Google Scholar]
- 18.Hirotani H., Tuohy N. A., Woo J. T., Stern P. H., Clipstone N. A. (2004) J. Biol. Chem. 279, 13984–13992 [DOI] [PubMed] [Google Scholar]
- 19.Samelson L. E. (2002) Annu. Rev. Immunol. 20, 371–394 [DOI] [PubMed] [Google Scholar]
- 20.Kurosaki T. (2002) Curr. Opin. Immunol. 14, 341–347 [DOI] [PubMed] [Google Scholar]
- 21.Deckert M., Tartare-Deckert S., Hernandez J., Rottapel R., Altman A. (1998) Immunity 9, 595–605 [DOI] [PubMed] [Google Scholar]
- 22.Le Bras S., Moon C., Foucault I., Breittmayer J. P., Deckert M. (2007) FEBS Lett. 581, 967–974 [DOI] [PubMed] [Google Scholar]
- 23.Foucault I., Le Bras S., Charvet C., Moon C., Altman A., Deckert M. (2005) Blood 105, 1106–1113 [DOI] [PubMed] [Google Scholar]
- 24.de la Fuente M. A., Kumar L., Lu B., Geha R. S. (2006) Mol. Cell. Biol. 26, 5214–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G., Dimitriou I. D., La Rose J., Ilangumaran S., Yeh W. C., Doody G., Turner M., Gommerman J., Rottapel R. (2007) Mol. Cell. Biol. 27, 3109–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla U., Hatani T., Nakashima K., Ogi K., Sada K. (2009) J. Biol. Chem. 284, 33719–33728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jevremovic D., Billadeau D. D., Schoon R. A., Dick C. J., Leibson P. J. (2001) J. Immunol. 166, 7219–7228 [DOI] [PubMed] [Google Scholar]
- 28.Sada K., Miah S. M., Maeno K., Kyo S., Qu X., Yamamura H. (2002) Blood 100, 2138–2144 [DOI] [PubMed] [Google Scholar]
- 29.Deckert M., Rottapel R. (2006) Adv. Exp. Med. Biol. 584, 107–114 [DOI] [PubMed] [Google Scholar]
- 30.Hatani T., Sada K. (2008) Curr. Med. Chem. 15, 549–554 [DOI] [PubMed] [Google Scholar]
- 31.Ueki Y., Tiziani V., Santanna C., Fukai N., Maulik C., Garfinkle J., Ninomiya C., doAmaral C., Peters H., Habal M., Rhee-Morris L., Doss J. B., Kreiborg S., Olsen B. R., Reichenberger E. (2001) Nat. Genet. 28, 125–126 [DOI] [PubMed] [Google Scholar]
- 32.Ueki Y., Lin C. Y., Senoo M., Ebihara T., Agata N., Onji M., Saheki Y., Kawai T., Mukherjee P. M., Reichenberger E., Olsen B. R. (2007) Cell 128, 71–83 [DOI] [PubMed] [Google Scholar]
- 33.Southgate J., Sarma U., Townend J. V., Barron J., Flanagan A. M. (1998) J. Clin. Pathol. 51, 831–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aliprantis A. O., Ueki Y., Sulyanto R., Park A., Sigrist K. S., Sharma S. M., Ostrowski M. C., Olsen B. R., Glimcher L. H. (2008) J. Clin. Invest. 118, 3775–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailet O., Fenouille N., Abbe P., Robert G., Rocchi S., Gonthier N., Denoyelle C., Ticchioni M., Ortonne J. P., Ballotti R., Deckert M., Tartare-Deckert S. (2009) Cancer Res. 69, 2748–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sallusto F., Lanzavecchia A. (1994) J. Exp. Med. 179, 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faccio R., Novack D. V., Zallone A., Ross F. P., Teitelbaum S. L. (2003) J. Cell Biol. 162, 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saltel F., Chabadel A., Bonnelye E., Jurdic P. (2008) Eur. J. Cell Biol. 87, 459–468 [DOI] [PubMed] [Google Scholar]
- 39.Thomas S. M., Brugge J. S. (1997) Annu. Rev. Cell Dev. Biol. 13, 513–609 [DOI] [PubMed] [Google Scholar]
- 40.Monticelli S., Rao A. (2002) Eur. J. Immunol. 32, 2971–2978 [DOI] [PubMed] [Google Scholar]
- 41.Aramburu J., Yaffe M. B., López-Rodríguez C., Cantley L. C., Hogan P. G., Rao A. (1999) Science 285, 2129–2133 [DOI] [PubMed] [Google Scholar]
- 42.Lietman S. A., Yin L., Levine M. A. (2008) Biochem. Biophys. Res. Commun. 371, 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shortman K., Naik S. H. (2007) Nat. Rev. Immunol. 7, 19–30 [DOI] [PubMed] [Google Scholar]
- 44.Li B., Boast S., de los Santos K., Schieren I., Quiroz M., Teitelbaum S. L., Tondravi M. M., Goff S. P. (2000) Nat. Genet. 24, 304–308 [DOI] [PubMed] [Google Scholar]
- 45.Mao D., Epple H., Uthgenannt B., Novack D. V., Faccio R. (2006) J. Clin. Invest. 116, 2869–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka S., Amling M., Neff L., Peyman A., Uhlmann E., Levy J. B., Baron R. (1996) Nature 383, 528–531 [DOI] [PubMed] [Google Scholar]
- 47.Boyce B. F., Yoneda T., Lowe C., Soriano P., Mundy G. R. (1992) J. Clin. Invest. 90, 1622–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim H. J., Zhang K., Zhang L., Ross F. P., Teitelbaum S. L., Faccio R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2325–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ory S., Brazier H., Pawlak G., Blangy A. (2008) Eur. J. Cell Biol. 87, 469–477 [DOI] [PubMed] [Google Scholar]
- 50.Insogna K. L., Sahni M., Grey A. B., Tanaka S., Horne W. C., Neff L., Mitnick M., Levy J. B., Baron R. (1997) J. Clin. Invest. 100, 2476–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyazaki T., Sanjay A., Neff L., Tanaka S., Horne W. C., Baron R. (2004) J. Biol. Chem. 279, 17660–17666 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.