Abstract
Krüppel-like factor 4 (Klf4) is a transcription factor involved in differentiation and proliferation in multiple tissues. We demonstrated previously that tamoxifen-induced deletion of the Klf4 gene in mice accelerated neointimal formation but delayed down-regulation of smooth muscle cell differentiation markers in carotid arteries following injury. To further determine the role of Klf4 in the cardiovascular system, we herein derived mice deficient for the Klf4 gene in smooth and cardiac muscle using the SM22α promoter (SM22α-CreKI+/Klf4loxP/loxP mice). SM22α-CreKI+/Klf4loxP/loxP mice were born at the expected Mendelian ratio, but they gradually died after birth. Although ∼40% of SM22α-CreKI+/Klf4loxP/loxP mice survived beyond postnatal day 28, they exhibited marked growth retardation. In wild-type mice, Klf4 was expressed in the heart from late embryonic development through adulthood, whereas it was not expressed in smooth muscle. No changes were observed in morphology or expression of smooth muscle cell differentiation markers in vessels of SM22α-CreKI+/Klf4loxP/loxP mice. Of interest, cardiac output was significantly decreased in SM22α-CreKI+/Klf4loxP/loxP mice, as determined by magnetic resonance imaging. Moreover, a lack of Klf4 in the heart resulted in the reduction in expression of multiple cardiac genes, including Gata4. In vivo chromatin immunoprecipitation assays on the heart revealed that Klf4 bound to the promoter region of the Gata4 gene. Results provide novel evidence that Klf4 plays a key role in late fetal and/or postnatal cardiac development.
Keywords: Cardiac Muscle, Differentiation, Gene Knock-out, Smooth Muscle, Transcription Factors, Kruppel-like Factors
Introduction
Krüppel-like factor 4 (Klf4)2 (formerly known as gut-enriched Krüppel-like factor or GKLF) is a zinc finger transcription factor involved in both the regulation of differentiation and proliferation in multiple cell types. For example, Klf4 is required for terminal differentiation of the skin (1). Conventional Klf4 knock-out mice are born at the expected Mendelian ratio but die within 15 h after birth due to a failure of normal basement membrane formation. Klf4 knock-out mice also exhibit a 90% decrease in the number of goblet cells in the colon, and the remaining goblet cells are histologically and ultrastructurally abnormal (2). Tissue-specific ablation of Klf4 in mouse stomach results in increased proliferation and altered differentiation of the gastric epithelia (3). In the eye, conditional knock-out of Klf4 results in abnormal corneal epithelium and lack of goblet cells in the conjunctiva (4). As such, Klf4 is implicated in a variety of cellular differentiation and proliferation processes by activating or repressing transcriptional activity of multiple genes. Interest in Klf4 also has increased dramatically over the past year based on observations that it is one of four factors (Oct3/4, Sox2, Klf4, and c-Myc) that, in combination, can induce a variety of somatic cells into an embryonic stem cell-like state or induced pluripotent stem cells (5, 6).
Klf4 also plays a key role in the regulation of gene transcription in the cardiovascular system. We have shown that Klf4 is a potent repressor of multiple smooth muscle cell (SMC) differentiation marker genes, including SM (smooth muscle) α-actin (Acta2), SM-myosin heavy chain (Myh11), and SM22α (7–9). Indeed, Klf4 binds to the promoter-enhancer regions of SMC differentiation marker genes and mediates platelet-derived growth factor BB or oxidized phospholipid-induced suppression of these genes in cultured SMCs (7, 9–11). Although Klf4 normally is not expressed in differentiated SMCs in vivo, it is transiently induced in phenotypically modulated SMCs after vascular injury (9, 12). Of significant importance, we recently demonstrated that conditional deletion of the Klf4 gene in mice results in transient delays in down-regulation of SMC differentiation markers, but subsequent SMC hyperproliferation and enhanced neointimal formation following carotid ligation injury (12). In addition, we presented evidence that enhanced neointimal formation in Klf4-deficient mice was caused by reduced induction of p21WAF1/Cip1, a cell-cycle inhibitor, in SMCs following injury. Klf4 has also been shown to be expressed in vascular endothelial cells. Hamik et al. (13) showed that overexpression of Klf4 increased expression of anti-inflammatory and antithrombotic factors, including eNOS and thrombomodulin, whereas knockdown of Klf4 led to enhancement of tumor necrosis factor α-induced expression of vascular cell adhesion molecule-1 and tissue factor in cultured endothelial cells. As such, results of the preceding studies provide evidence that Klf4 is a critical factor regulating gene transcription in a variety of vascular cells. However, as yet, no studies have examined its function in the heart either in vivo or in vitro.
Recently, mice expressing Cre recombinase under the control of the SM22α promoter (SM22α-CreKI+ mice) have been developed by knocking-in the Cre recombinase-coding sequence into the SM22α gene locus via homologous recombination (14). SM22α is a marker of SMCs, but it also is transiently expressed in the heart between embryonic days (E) 8.0 and 12.5 (15). Therefore, SM22α-CreKI+ mice exhibit Cre recombination in a smooth and cardiac muscle-specific manner but only after these cells have initiated differentiation. To further determine the role of Klf4 in the cardiovascular system, we herein derived smooth and cardiac muscle-specific conditional Klf4-deficient mice by breeding SM22α-CreKI+ mice with mice carrying a loxP allele of Klf4 (Klf4loxP mice) (2) and analyzed their phenotype.
EXPERIMENTAL PROCEDURES
Generation of Smooth and Cardiac Muscle-specific Klf4-deficient Mice
Animal protocols were approved by the University of Virginia Animal Care and Use Committee. Klf4loxP mice were provided by Dr. Klaus H. Kaestner (University of Pennsylvania) (2). The Klf4 gene consists of four exons, (see Fig. 1A) and exon 1 only encodes a first methionine codon. In Klf4loxP mice, recombination of the Klf4loxP allele deletes exons 2 and 3 and causes a frameshift mutation in exon 4, which abolishes Klf4 function completely (2, 12). Klf4loxP mice were bred with transgenic mice expressing Cre recombinase under the control of the SM22α promoter (SM22α-CreKI+ mice) (14) to generate smooth and cardiac muscle-specific Klf4-deficient mice. Indeed, male heterozygous SM22α-CreKI+ mice were bred with female Klf4loxP/loxP mice to generate SM22α-CreKI+/Klf4loxP/wt mice. Male or female SM22α-CreKI+/Klf4loxP/wt mice were then crossed with female or male Klf4loxP/loxP mice to generate SM22α-CreKI+/Klf4loxP/loxP mice (smooth and cardiac-specific Klf4-deficient mice) and SM22α-CreKI−/Klf4loxP/loxP mice (control mice). Both Klf4loxP mice and SM22α-CreKI+ mice were mixed background strains of C57BL/6 and 129, and littermates were used for all comparisons. Genotyping for the Klf4 floxed locus was performed by PCR using three primers, as described previously (2, 12).
FIGURE 1.
Smooth and cardiac muscle-specific deletion of the Klf4 gene was associated with significant postnatal death and growth retardation. A, a schematic representation of smooth and cardiac muscle-specific deletion of the Klf4 gene is shown. The numbers shown represent Klf4 exons. Triangles represent the loxP sites. X, breeding. B, the genotype of 191 offspring from the breeding between SM22α-CreKI+ (CreKI)/Klf4loxP/wt mice and SM22α-CreKI−/Klf4loxP/loxP mice was examined by PCR at the time of birth. C, Kaplan-Meier survival curves for SM22α-CreKI+/Klf4loxP/loxP, SM22α-CreKI+/Klf4loxP/wt, SM22α-CreKI−/Klf4loxP/loxP, and SM22α-CreKI−/Klf4loxP/wt mice are shown (n = 36 per each genotype). A log-rank test for trend yielded. *, p < 0.05. D and E, representative pictures of SM22α-CreKI+/Klf4loxP/loxP and SM22α-CreKI−/Klf4loxP/loxP mice at P1 (D) and P28 (E) are shown. F, changes in the body weight of SM22α-CreKI+/Klf4loxP/loxP, SM22α-CreKI+/Klf4loxP/wt, SM22α-CreKI−/Klf4loxP/loxP, and SM22α-CreKI−/Klf4loxP/wt mice after birth are shown (n = 20∼25 per each genotype). *, p < 0.05 compared with other genotypes. Docta represent the mean ± S.E.
Histology, Immunohistochemistry, and Immunofluorescence
Embryos and multiple tissues were harvested, fixed, and embedded into paraffin as described previously (16). The 5-μm sections were prepared and stained with antibodies for Klf4 (11) or Ki-67 (Santa Cruz Biotechnology, Santa Cruz, CA). Staining was visualized by diaminobenzidine (DAB) and counterstained with hematoxylin. Klf4 staining in the skin was visualized by DAB and counterstained with azure B. TUNEL staining was performed according to the manufacturer's instructions (Roche Diagnostics) (12). Dual immunofluorescence studies were performed with Klf4 antibody raised in rabbit (11) and Gata4 antibody raised in goat (Santa Cruz Biotechnology) (17). Specific staining was detected by Alexa Fluor 555-conjugated anti-rabbit antibody (Invitrogen) and Alexa Fluor 488-conjugated anti-goat antibody (Invitrogen). Sections were counterstained with 4′,6-diamidino-2-phenylindole. For morphological analysis, sections were subjected to Russell-Movat pentachrome staining.
Electron Microscopy
Mice were sacrificed by isoflurane overdose followed by cervical dislocation. The beating hearts were quickly removed and rinsed in oxygenated Krebs buffer with 30 mmol/liter 2,3-butanedione monoxime at 37 °C. The hearts were perfused through the aorta with 2% glutaraldehyde and kept in 2% glutaraldehyde overnight at 4 °C. Small pieces (∼1-mm in diameter) of the lower left ventricle were then postfixed, stained, and embedded following the standard procedure (18, 19): 2% osmium tetroxide for 2 h, saturated aqueous uranyl acetate for 1.5 h, dehydrated in a series of graded alcohols, and embedded in Spurr's resin. Sections were poststained by lead citrate and imaged in a Philips CM12 electron microscope at 120 KeV equipped with a SIA 8C 2,000 × 3,000 CCD camera (Duluth, GA).
Vascular Casting
Pregnant mice 18.5 days post coitus were euthanized by cervical dislocation under isoflurane anesthesia. Embryos were isolated and rinsed in phosphate-buffered saline at 37 °C. The umbilical vein was cut to allow the wash out of blood from the circulation system. Prior to injecting the casting medium, embryos were perfused with 1 ml of warmed 1% lidocaine hydrochloride in 0.9% NaCl containing 1 units/ml heparin via the umbilical artery. Perfusate was injected until the fluid that flowed out from umbilical vein was clear. The vasculature was then fixed for 5 min using 1 ml of warmed 4% paraformaldehyde in phosphate-buffered saline. 1 mm of casting material, Baston's No. 17 (Polysciences) supplemented with jet acrylic liquid (Lang Dental Manufacturing Co., Inc.), was then injected via the umbilical artery. The umbilical cord was tied, and embryos were stored at 4 °C for 12 h to allow polymerization of the casting material. Tissues were then dissolved in 7 mol/liter KOH, and the circulatory cast was cleaned using forceps.
Cardiac Magnetic Resonance Imaging (MRI)
Cardiac MRI was performed in Department of Radiology at the University of Virginia, as previously described (20). Briefly, a 4.7-Tesla MRI system (Varian 200/400 Inova) with Magnex gradients (80 G/cm maximum strength) was used with a custom built Litz radiofrequency coil (Doty Scientific, Columbia, SC). Anesthesia was induced with 3% isoflurane and was maintained with a 1% isoflurane-oxygen mixture throughout the study. Core body temperature was maintained at 37.0 ± 0.1 °C by using an external water bath. Electrocardiogram (ECG) triggering was achieved with a gating/monitoring system (SA Instruments, Stony Brook, NY). Contiguous short-axis bright blood cine images of the left ventricle were obtained with a two-dimensional FLASH gradient echo sequence. Images were analyzed with ARGUS image analysis software (Siemens Medical Solutions, Princeton, NJ).
Microarray and Gene Ontology Analysis
Microarray analysis was performed in quadruplicate by SA Biosciences (Frederick, MD) using Agilent whole mouse genome oligonucleotide microarray kit (Agilent Technologies, Santa Clara, CA). Gene ontology analysis with PANTHER (21) was performed using the database for annotation, visualization, and integrated discovery 2008 (DAVID). Genes considered dysregulated were those with p values < 0.02.
Real-time RT-PCR
Total RNA prepared from the mouse heart, skeletal muscle, brain, and aorta was used for real-time RT-PCR. Primer and probe sequences for Klf4 and 18S rRNA were described previously (9, 17). Primer and probe sequences for Nppa, Nppb, Actc1, Myh7, Serca2, and Gata4 were as follows: Nppa-F, 5′-AGCCGAGACAGCAAACATCAG-3′; Nppa-R, 5′-CAAAAGGCCAGGAAGAGGAAG-3′; Nppb-F, 5′-CAGCTCTTGAAGGACCAAGG-3′; Nppb-R, 5′-AGAGACCCAGGCAGAGTCAG-3′; Actc1-F, 5′-CCCAGAGGAACACCCAACCCTGCT-3′; Actc1-R, 5′-GGCACCATACATTCTACAATGAGC-3′; Actc1-P, 5′-CATCGCCAGAATCCAGAACAATG-3′; Myh7-F, 5′-GATGTTTTTGTGCCCGATGA-3′; Myh7-R, 5′-TGTCGAACTTGGGTGGGTT-3′; Myh7-P, 5′-CAGTCACCGTCTTGCCATTCTCCGT-3′; Serca2-F, 5′-AAATCTCCTTGCCTGTGATCC-3′; Serca2-R, 5′-TGCTAACAACGCACATGCAC-3′; Serca2-P, 5′-ACTACCTGGAACAACCCGCAATACTGGA-3′; Gata4-F, 5′-CAGCAGCAGTGAAGAGATGC-3′; Gata4-R, ATGTCCCCATGACTGTCAGC-3′.
Western Blotting
Western blotting was performed as described previously (11, 12). Antibodies used were as follows: Klf4, cMLC (Abcam, Cambridge, MA), glyceraldehyde-3-phosphate dehydrogenase (6C5, Millipore, Billerica, MA), and Gata4.
Quantitative Chromatin Immunoprecipitation (ChIP) Assays
In vivo quantitative ChIP assays were performed using anti-Klf4 antibody, as described previously (12, 22). Real-time PCR was performed to amplify the promoter region of the Gata4 gene or the c-fos gene (22). Primer sequences for the Gata4 gene promoter were as follows: Gata4-proF, 5′-AGAGCAGCAAACCGCAAG-3′ and Gata4-proR, 5′-AGGACTCTTCCCAAAGCTC-3′.
Statistical Analyses
Data are presented as mean ± S.E. Unless mentioned, statistical analyses were performed by one-way analysis of variance with a post hoc Fisher protected least significant difference test or unpaired t test. p values < 0.05 were considered significant.
RESULTS
Smooth and Cardiac Muscle-specific Deletion of the Klf4 Gene Was Associated with Marked Postnatal Death and Growth Retardation
To generate smooth and cardiac muscle-specific Klf4-deficient mice, we used a two-step breeding protocol. First, heterozygous SM22α-CreKI+ mice were bred with Klf4loxP/loxP mice to generate SM22α-CreKI+/Klf4loxP/wt mice. SM22α-CreKI+/Klf4loxP/wt mice were then crossed with Klf4loxP/loxP mice to generate SM22α-CreKI+/Klf4loxP/loxP mice and SM22α-CreKI−/Klf4loxP/loxP mice. SM22α-CreKI+/Klf4loxP/loxP mice were smooth and cardiac muscle-specific Klf4-knock-out mice, whereas SM22α-CreKI−/Klf4loxP/loxP mice served as controls (Fig. 1A). As shown in Fig. 1B, both SM22α-CreKI+/Klf4loxP/loxP mice and control mice were born at the expected Mendelian ratio and could not be distinguished from one another at birth (Fig. 1D). However, SM22α-CreKI+/Klf4loxP/loxP mice gradually died after birth, and only 40% survived beyond postnatal day (P)28 (Fig. 1C). In addition, the remaining SM22α-CreKI+/Klf4loxP/loxP mice were significantly smaller than their control littermates, a phenotype observed in 100% of surviving SM22α-CreKI+/Klf4loxP/loxP mice (Fig. 1E). At P28, the body weight of SM22α-CreKI+/Klf4loxP/loxP mice was 7.3 ± 0.6 g, whereas control mice weighed 12.0 ± 0.4 g (Fig. 1F). SM22α-CreKI+/Klf4loxP/loxP mice also exhibited shorter tails (2.6 ± 0.7 cm versus 7.0 ± 0.1 cm in control mice at P28, p < 0.05) and partial hair loss on their backs, although it is unclear whether the latter changes are a function of Klf4 loss per se or results from competition with their full-sized littermates. In summary, results clearly indicate that loss of Klf4 in smooth and/or cardiac muscle is associated with a major phenotype during the postnatal period.
Klf4 Was Expressed in the Heart from Late Embryonic Development to Adulthood
To elucidate mechanisms responsible for the postnatal phenotype of conditional Klf4 knock-out mice, we first examined whether Klf4 was expressed in the heart and SMCs during normal development and whether the Klf4loxP allele was recombined in a tissue-specific manner in conditional Klf4 knock-out mice. Klf4 expression patterns were determined in wild-type mice by immunohistochemistry. As shown in Fig. 2, A–C and supplemental Fig. I, A–C, Klf4 was not expressed in the heart at E9.5, E11.5, and E15.5. However, Klf4 was expressed at E18.5 in both the atrium and the ventricle of the heart and was also expressed in P1 and adult hearts (Fig. 2, D–F and supplemental Fig. I, D and E). Of note, Klf4 expression was seen in nearly 50% of the cells in the heart. To determine which cells express Klf4 in the heart, we tested colocalization of Klf4 and Gata4, which is mainly expressed in cardiomyocytes (23). Results of dual immunofluorescence studies showed that Klf4 and Gata4 were coexpressed in the heart, although some of Klf4-positive cells were negative for Gata4 (Fig. 2G). In contrast to the expression of Klf4 in the heart, Klf4 was not detectable in SMCs in any SMC-containing tissues examined during embryonic development or in the adult. For example, it was not expressed in the third, fourth, and sixth branchial arch arteries and the dorsal aorta at E11.5 (Fig. 2B). Moreover, it was not expressed in SMCs of the aorta, the stomach, and the bladder as well as in the skeletal muscle in the adult (Fig. 2, H–K), although it was expressed in aortic endothelial cells, some adventitial cells in the aorta, and epithelial cells in the stomach.
FIGURE 2.
Klf4 was expressed in the heart from late embryonic development to adulthood. A–F, Klf4 expression was determined by immunohistochemistry in wild-type embryos at E9.5 (A), at E11.5 (B), at E15.5 (C), and at E18.5 (D), as well as in the heart of wild-type neonates at P1 (E) and adult (F). Klf4 expression was visualized by DAB, and sections were counterstained with hematoxylin. Bars for A–C, 100 μm; D–F, 50 μm. H, heart; DA, dorsal aorta; III, third branchial arch artery; IV, fourth branchial arch artery; VI, sixth branchial arch artery. G, colocalization of Klf4 and Gata4 in the heart of wild-type mice was examined by dual immunofluorescence staining. Klf4 staining was visualized by Alexa Fluor 555 (red), and Gata4 staining was visualized by Alexa Fluor 488 (green). Sections were counterstained with 4′,6-diamidino-2-phenylindole (blue). Arrowheads indicate cells coexpressing Klf4 and Gata4. Bar, 50 μm. H–K, Klf4 expression was examined in the aorta (H), the skeletal muscle (I), the stomach (J), and the bladder (K) of wild-type mice. Arrowheads indicate aortic endothelial cells (H) and gastric epithelial cells (J), which express Klf4. SM, smooth muscle layer. Bars for H and I, 50 μm; bars for J and K, 100 μm. Representative pictures are shown.
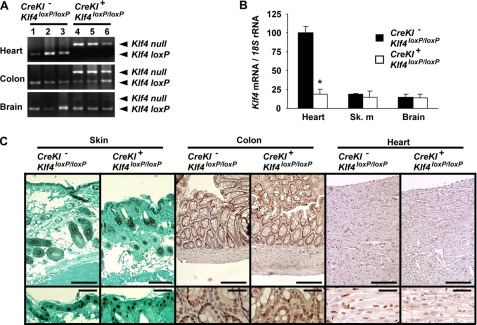
Recombination of the Klf4loxP allele was examined in SM22α-CreKI+/Klf4loxP/loxP mice and control mice. As shown in Fig. 3A, the Klf4loxP allele was efficiently recombined in the heart of SM22α-CreKI+/Klf4loxP/loxP mice. The Klf4loxP allele was also recombined in the colon, which contains a large fraction of SMCs, whereas little recombination was observed in the brain of SM22α-CreKI+/Klf4loxP/loxP mice. Deletion of Klf4 was examined at both the mRNA and protein levels. Klf4 mRNA expression was decreased in the heart of SM22α-CreKI+/Klf4loxP/loxP mice, although it was low and unaffected in the skeletal muscle and the brain (Fig. 3B). Moreover, Klf4 protein expression was dramatically decreased in the heart, whereas it was unaltered in epithelial cells of the skin and colon in SM22α-CreKI+/Klf4loxP/loxP mice (Fig. 3C). Collectively, these results indicate that Klf4 is expressed in the heart but not in SMCs from late embryonic development to adulthood and that loss of Klf4 in the heart, rather than in SMCs, is likely to be responsible for the phenotype of SM22α-CreKI+/Klf4loxP/loxP mice.
FIGURE 3.
SM22α-CreKI+/Klf4loxP/loxP mice exhibited selective loss of Klf4 expression in the heart. A, deletion of the Klf4loxP allele was tested in multiple tissues including the heart, the colon, and the brain of SM22α-CreKI−/Klf4loxP/loxP and SM22α-CreKI+/Klf4loxP/loxP mice at P28, as determined by PCR (n = 3 per each genotype). B, expression of Klf4 mRNA was examined by real-time RT-PCR in the heart, the skeletal muscle (sk. m), and the brain of SM22α-CreKI−/Klf4loxP/loxP and SM22α-CreKI+/Klf4loxP/loxP mice at P28 (n = 6 per each genotype). C, Klf4 expression was examined by immunohistochemistry in the skin, the colon, and the heart of SM22α-CreKI−/Klf4loxP/loxP and SM22α-CreKI+/Klf4loxP/loxP mice at P28. Klf4 was visualized by DAB, and sections were counterstained with azure B for the skin or with hematoxylin for the colon and the heart. Representative pictures are shown (n = 4 per each genotype). Bars, 100 μm for upper panels; 50 μm for lower panels.
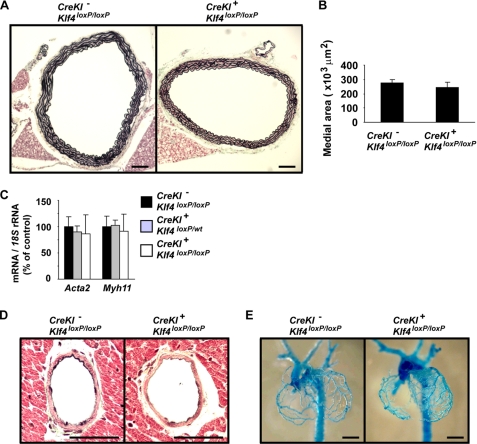
SM22α-CreKI+/Klf4loxP/loxP Mice Had No Discernible Changes in SMC Differentiation or Vascular Morphology
Although Klf4 was not expressed in SMCs during normal development, we analyzed multiple vascular parameters to determine whether defects in SMCs might have contributed to the overall phenotype of SM22α-CreKI+/Klf4loxP/loxP mice. First, we examined the overall morphology of the aorta. The medial area of the thoracic aorta was not different between SM22α-CreKI+/Klf4loxP/loxP and control mice, and no neointima was found in either mice (Fig. 4, A and B). Second, expression of the SMC differentiation markers, Acta2 and Myh11, was unaltered in the aorta of SM22α-CreKI+/Klf4loxP/loxP mice (Fig. 4C). Third, the morphology of smaller arteries including the coronary arteries and the renal arteries was examined, but no differences were found between SM22α-CreKI+/Klf4loxP/loxP and control mice (Fig. 4D and data not shown). Finally, development of coronary arteries was examined in E18.5 embryos using vascular casting (Fig. 4E). No differences in the pattern of formation of coronary arteries were observed between SM22α-CreKI+/Klf4loxP/loxP and control mice. Taken together, these results and the results showing no Klf4 expression in SMCs during normal development (Fig. 2) indicate that the phenotype of SM22α-CreKI+/Klf4loxP/loxP mice was unlikely to be secondary to vascular defects.
FIGURE 4.
SM22α-CreKI+/Klf4loxP/loxP mice had no differences in expression of SMC differentiation marker genes or arterial structure as compared with control mice. A and B, the thoracic aorta from SM22α-CreKI−/Klf4loxP/loxP and SM22α-CreKI+/Klf4loxP/loxP mice at P28 was perfused with 10% formalin under physiological pressure and embedded into paraffin. Cross-sections were stained with Russell-Movat pentachrome method (n = 6 per each genotype). A, representative pictures are shown. Bars, 100 μm. B, medial areas (the average of three sections per each mouse) were measured using ImagePro software. C, expression of SMC differentiation marker genes including Acta2 and Myh11 was examined by real-time RT-PCR in the aorta of SM22α-CreKI−/Klf4loxP/loxP, SM22α-CreKI+/Klf4loxP/wt, and SM22α-CreKI+/Klf4loxP/loxP mice at P14 (n = 5 per each genotype). D, coronary arteries from SM22α-CreKI−/Klf4loxP/loxP and SM22α-CreKI+/Klf4loxP/loxP mice at P28 was sectioned and stained with Russell-Movat pentachrome method (n = 6 per each genotype). Representative pictures of the left anterior descending artery are shown. Bars, 50 μm. E, vascular casting of coronary arteries from E18.5 embryos is shown. Bars, 1 mm.
Cardiac Output Was Decreased in SM22α-CreKI+/Klf4loxP/loxP Mice
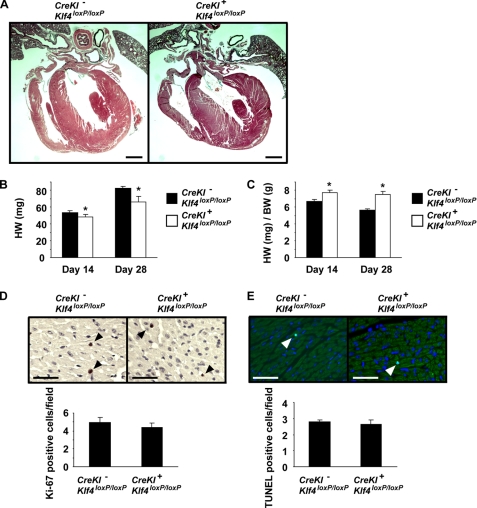
We next ascertained whether there was a cardiac phenotype in SM22α-CreKI+/Klf4loxP/loxP mice. First, serial histological sections of the heart were analyzed in ten SM22α-CreKI+/Klf4loxP/loxP mice and six control mice, including four SM22α-CreKI+/Klf4loxP/loxP mice that died before P28 because they might have more serious cardiac phenotype than the surviving mice (Fig. 5A). No anatomical and morphological abnormalities were found in the heart and the great arteries of SM22α-CreKI+/Klf4loxP/loxP mice. Heart weight was measured at P14 and P28. Although the heart weight itself was slightly decreased in SM22α-CreKI+/Klf4loxP/loxP mice (48 ± 3 mg at P14, 66 ± 7 mg at P28) than control mice (54 ± 2 mg at P14, 82 ± 2 mg at P28), the ratio of heart to body weight was significantly increased in SM22α-CreKI+/Klf4loxP/loxP mice (7.8 ± 0.2 mg/g at P14, 7.5 ± 0.4 mg/g at P28) than control mice (6.7 ± 0.3 mg/g at P14, 5.6 ± 0.1 mg/g at P28) (Fig. 5, B and C). To determine whether these differences were due to altered rates of proliferation or apoptosis, Ki-67 staining and TUNEL staining were performed in the heart at P28. The ratio of Ki-67 positive cells was not different between SM22α-CreKI+/Klf4loxP/loxP (5.0 ± 0.6 per field) and control (4.4 ± 0.5 per field) mice (Fig. 5D). The ratio of TUNEL-positive cells was also not different between SM22α-CreKI+/Klf4loxP/loxP (2.8 ± 0.2 per field) and control (2.6 ± 0.3 per field) mice (Fig. 5E). In addition, the rates of proliferation and apoptosis in the heart were not different between SM22α-CreKI+/Klf4loxP/loxP versus control mouse embryos at E18.5 (supplemental Fig. II). Moreover, we did not detect any substantial differences in the ultrastructure of cardiomyocytes between the two groups, as determined by electron microscopy (supplemental Fig. III). These results suggest that SM22α-CreKI+/Klf4loxP/loxP mice do not exhibit either an altered proliferation rate or apoptotic rate in the heart and that the alteration in the ratio of heart to body weight is mainly due to the reduced body weight of SM22α-CreKI+/Klf4loxP/loxP mice.
FIGURE 5.
The rates of proliferation and apoptosis were unaltered in the hearts of SM22α-CreKI+/Klf4loxP/loxP mice. A, hearts from SM22α-CreKI−/Klf4loxP/loxP and SM22α-CreKI+/Klf4loxP/loxP mice at P28 were sectioned and stained with Russell-Movat pentachrome method. Representative pictures are shown (n = 6 per each genotype). Bars, 1 mm. B and C, heart weight and the ratio of heart to body weight were measured in SM22α-CreKI−/Klf4loxP/loxP and SM22α-CreKI+/Klf4loxP/loxP mice at P14 and P28. *, p < 0.05 compared with SM22α-CreKI−/Klf4loxP/loxP mice. D and E, Ki-67 staining (D) and TUNEL staining (E) were performed in the heart of SM22α-CreKI−/Klf4loxP/loxP and SM22α-CreKI+/Klf4loxP/loxP mice at P28 (n = 5 per each genotype). Ki-67 staining was visualized by DAB, and sections were counterstained with hematoxylin. TUNEL staining was visualized by fluorescein isothiocyanate, and sections were counterstained with 4′,6-diamidino-2-phenylindole. Arrowheads indicate stained cells. Bars, 50 μm.
Cardiac function was analyzed in SM22α-CreKI+/Klf4loxP/loxP and control mice by MRI (Table 1 and supplemental Fig. IV). 5-week-old mice were analyzed because they were the minimum size for cardiac MRI. Consistent with the heart weight measured in Fig. 4C, the left ventricular mass of SM22α-CreKI+/Klf4loxP/loxP mice (40.0 ± 2.3 mg) was smaller than controls (43.0 ± 2.0 mg). Of interest, both the ejection fraction (EF) and the heart rate (HR) were significantly decreased in SM22α-CreKI+/Klf4loxP/loxP mice (EF, 53.8 ± 2.6%; HR, 394 ± 25 beats/min), as compared with control mice (EF, 66.2 ± 1.9%; HR, 441 ± 23 beats/min). No cardiac arrhythmia was found in SM22α-CreKI+/Klf4loxP/loxP mice (supplemental Fig. V). The decreases in these parameters (EF and HR) resulted in a profound reduction in cardiac output (7.5 ± 0.2 μl/min in SM22α-CreKI+/Klf4loxP/loxP mice versus 9.7 ± 0.6 μl/min in control mice). It is well established that decreased cardiac output is associated with abnormal postnatal growth (24, 25). Therefore, results of cardiac MRI suggest that the growth retardation and partial postnatal death in SM22α-CreKI+/Klf4loxP/loxP mice are caused, at least in part, by cardiac insufficiency, although we did not notice obvious signs of heart failure, such as lung edema in SM22α-CreKI+/Klf4loxP/loxP mice.
TABLE 1.
Structural and functional parameters of the heart in 5-week-old SM22α-CreKI+/Klf4loxP/loxP and control mice as measured by cardiac MRI
| SM22α-CreKI−Klf4loxP/loxP | SM22α-CreKI+Klf4loxP/loxP | |
|---|---|---|
| Heart rate (beats/min) | 441 ± 23 | 394 ± 25* |
| Left ventricular end diastolic volume (μl) | 33.5 ± 1.8 | 33.8 ± 3.1 |
| Left ventricular end systolic volume (μl) | 11.5 ± 1.2 | 15.7 ± 1.9* |
| Stroke volume (μl) | 22.0 ± 0.7 | 18.1 ± 1.6* |
| Ejection fraction (%) | 66.2 ± 1.9 | 53.8 ± 2.6* |
| Cardiac output (μl/min) | 9.7 ± 0.6 | 7.5 ± 0.2* |
| Left ventricular mass (mg) | 43.0 ± 2.0 | 40.0 ± 2.3 |
*, p < 0.05 compared with SM22α-CreKI−/Klf4loxP/loxP mice. n = 5 per each genotype.
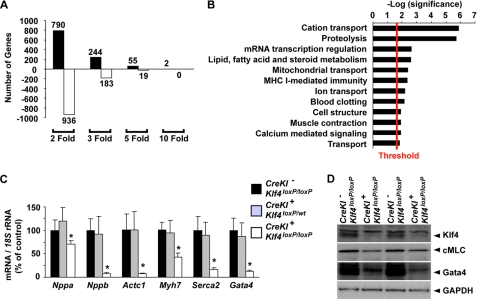
Loss of Klf4 Resulted in Decreased Expression of Multiple Cardiac Genes
To determine the molecular mechanisms for decreased cardiac output in SM22α-CreKI+/Klf4loxP/loxP mice, microarray analyses were performed on hearts of SM22α-CreKI+/Klf4loxP/loxP and control mice at P14. Consistent with results of previous studies showing that Klf4 is both an activator and a repressor of gene transcription (26), results of microarray analyses showed that a large number of genes were increased or decreased by Klf4 deletion (Fig. 6A). Indeed, 244 genes were increased >3-fold, whereas 183 genes were decreased >3-fold in the heart of SM22α-CreKI+/Klf4loxP/loxP mice. A gene ontology analysis of these dysregulated transcripts revealed that the most significantly enriched genes participate in cation transport and that genes related to proteolysis and mRNA transcriptional regulation were also affected (Fig. 6B). Of major interest, loss of Klf4 was associated with coordinate down-regulation of a number of cardiac genes including Myl1 (myosin light polypeptide 1, also known as cardiac myosin light chain), Tnnc1 (cardiac/slow skeletal troponin C), Mybpc2 (myosin binding protein C), Nppb (natriuretic peptide precursor type B, also known as brain natriuretic peptide), and Gata4 (supplemental Table 1). Real-time RT-PCR analysis also showed that multiple cardiac genes including Nppa (natriuretic peptide precursor type A, also known as atrial natriuretic factor), Nppb, Actc1 (cardiac α-actin), Myh7 (myosin heavy chain 7, also known as β-myosin heavy chain), Serca2 (sarcoplasmic/endoplasmic reticulum Ca(2+) pump gene 2), and Gata4 were dramatically decreased in the heart of SM22α-CreKI+/Klf4loxP/loxP mice (Fig. 6C). Expression of these genes was not decreased in the heart of SM22α-CreKI+/Klf4loxP/wt mice, suggesting that loss of one Klf4 allele is insufficient to induce similar changes and that changes are not the consequence of loss of one SM22α allele due to insertion of the Cre recombinase gene. Reduction of expression of cardiac myosin light chain and Gata4 in Klf4-deficient mice was also detectable by Western blotting (Fig. 6D). Taken together, these results suggest that cardiac loss of Klf4 causes a significant decrease in expression of a number of cardiac genes required for contractile function and such a decrease is likely to contribute to the decreased cardiac output in SM22α-CreKI+/Klf4loxP/loxP mice.
FIGURE 6.
Multiple cardiac genes were decreased in the heart of SM22α-CreKI+/Klf4loxP/loxP mice. A and B, microarray analysis was performed in the heart of SM22α-CreKI−/Klf4loxP/loxP and SM22α-CreKI+/Klf4loxP/loxP mice at P14 (n = 4 per each genotype). A, number of genes dysregulated by Klf4 deletion is shown by fold change. B, the 427 genes increased or decreased >3-fold by Klf4 deletion were subjected to gene ontology analysis with PANTHER. Significantly enriched biological processes (p < 0.03) are shown. Plotted is the log (p value) with the threshold set to 1.5 [-log(0.03)]. C, expression of multiple cardiac genes including Nppa, Nppb, Actc1, Myh7, Serca2, and Gata4 was examined by real-time RT-PCR in the heart of SM22α-CreKI−/Klf4loxP/loxP, SM22α-CreKI+/Klf4loxP/wt, and SM22α-CreKI+/Klf4loxP/loxP mice at P14 (n = 5 per each genotype). D, protein expression of Klf4, cardiac myosin light chain (cMLC), Gata4, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was examined in the heart of SM22α-CreKI−/Klf4loxP/loxP and SM22α-CreKI+/Klf4loxP/loxP mice at P14 by Western blotting. Representative pictures are shown (n = 3 for each genotype).
In Vivo ChIP Assays Demonstrated Klf4 Binding to the Gata4 Promoter in Wild-type but Not Conditional Klf4 Knock-out Mouse Hearts
Of the genes decreased by Klf4 deletion, Gata4 is known to be critical for regulating many cardiac genes (23). We performed in vivo ChIP assays to determine whether Klf4 directly binds to the promoter region of the Gata4 gene in the heart. Although little is known regarding the transcriptional control of the Gata4 gene, results of several studies showed that multiple GC boxes and an E box within its promoter (Fig. 7A) were important for expression (27–29). We found a consensus Klf4 binding site at −125/−119 bp, in close proximity to four GC boxes and an E box (Fig. 7A). Indeed, the consensus Klf4 binding sequence is 5′-(G/A)(G/A)GG(C/T)G(C/T)-3′, which is conserved between the mouse, rat, and human Gata4 promoter (30). Results of in vivo ChIP assays showed that Klf4 was associated with the Gata4 promoter region in the heart of control mice, whereas the binding was abolished in SM22α-CreKI+/Klf4loxP/loxP mice (Fig. 7B and supplemental Fig. VI). This binding was selective in that no Klf4 binding was detected at the c-fos promoter. Results provide evidence that Klf4 regulates Gata4 expression by binding to its promoter region and suggest that this is one of the mechanisms whereby knock-out of cardiac Klf4 results in decreased expression of multiple cardiac genes.
FIGURE 7.
Klf4 regulated Gata4 expression by binding to the Gata4 promoter region. A, a schematic diagram of the Gata4 promoter is shown. A consensus Klf4 binding site, 5′-(G/A)(G/A)GG(C/T)G(C/T)-3′, is located within the Gata4 promoter, which contains 4 GC-rich elements and an E box. Numbers represent bp. B, association of Klf4 with the promoter regions of the Gata4 gene and the c-fos gene was determined by in vivo ChIP assays in the heart of SM22α-CreKI−/Klf4loxP/loxP and SM22α-CreKI+/Klf4loxP/loxP mice at P14 (n = 3). *, p < 0.05 compared with control mice. Ab, antibody.
DISCUSSION
Results of the present studies show that selective knock-out of the Klf4 gene in smooth and cardiac muscle is associated with significant postnatal death and severe growth retardation in surviving mice. Moreover, we present several lines of evidence that this phenotype is primarily the result of loss of Klf4 in the heart rather than in SMCs. First, conditional Klf4-deficient mice exhibited a marked decrease in cardiac output and abnormal expression of a larger number of cardiac genes required for its contractile function including Myl1, Tnnc1, Mybpc2, Actc1, Myh7, Serca2, and Gata4. The decrease in cardiac output was seen even in surviving 5-week-old Klf4-deficient mice. Second, surprisingly, Klf4 was highly expressed in the heart of wild-type mice from late embryonic development to adulthood, whereas it was not detectable at all time points examined in either vascular or nonvascular SMCs. Third, results of in vivo ChIP assays showed that Klf4 directly bound to the Gata4 promoter region in the heart of control mice. Fourth, we found no differences in the morphology or the overall structure of arteries including the aorta, the coronary arteries, and renal arteries in conditional Klf4 knock-out mice versus control mice. Finally, there were no differences in the level of expression of SMC differentiation marker genes in the aorta between these mice. Taken together, results of the present studies provide the first evidence that Klf4 is a critical factor in cardiac development and maturation.
In the present studies, we provide evidence that Klf4 regulates Gata4 gene expression by binding to the consensus Klf4 binding site at −125/−119 bp within the Gata4 promoter in the postnatal heart. Gata4 is a zinc finger-containing transcription factor that plays an essential role in promoting cardiac development and differentiation of the myocardium, as well as in regulating hypertrophic growth of the adult heart (23). It is highly expressed in cardiomyocytes throughout embryonic development, postnatal growth, and adulthood, during which it functions as a critical regulator of cardiac-specific gene expression. Despite these pivotal roles, little is known regarding the factors and mechanisms for transcriptional regulation of the Gata4 gene. Indeed, Heicklen-Klein and Evans (27) show that a 14.8-kb fragment upstream of the transcription initiation site of the Gata4 gene is required for its expression in the heart of zebrafish. Guittot et al. (28) show that the first 5-kb of Gata4 promoter-enhancer sequence was sufficient to direct gene expression in the adult mouse heart, although they did not examine the activity of this fragment during embryonic and postnatal development. Recently, two GC boxes and an E box between −124 bp and −36 bp of the mouse Gata4 promoter have been shown to be important for its expression during differentiation of P19.CL6 cells into beating cardiomyocyte-like cells in vitro (29). Results of our present studies showed that Klf4 selectively bound to the Gata4 promoter region in the heart of control mice but not conditional Klf4 knock-out mice, as determined by in vivo ChIP assays. Although complete characterization of the Gata4 promoter, including promoter-reporter assays and electrophoretic mobility shift assays, will be required in further studies, these results at least provide novel evidence indicating that Klf4 plays an important role in regulating transcription of the Gata4 gene. However, factors other than Klf4 likely are responsible for initial induction of Gata4 in the heart because Gata4 has been shown to be expressed within these cells from a very early embryonic time point (23), whereas we found that Klf4 was first induced in the heart at ∼E18.5. In addition, it also should be noted that the phenotype of SM22α-Cre+/Klf4loxP/loxP mice could not be explained solely by the reduction in Gata4 expression because results of previous studies showed that cardiac muscle-specific Gata4 knock-out mice did not die after birth (31). As numerous genes were dysregulated by Klf4 deletion, multiple mechanisms, including the reduction in Gata4 expression, would cooperatively contribute to the phenotype of SM22α-Cre+/Klf4loxP/loxP mice.
Reduction in cardiac output is typically accompanied with compensatory cardiac hypertrophy (24, 25). In the present studies, however, smooth and cardiac muscle-specific Klf4-deficient mice did not exhibit obvious cardiac hypertrophy. Indeed, the heart weight of conditional Klf4 knock-out mice was reduced as compared with that of control mice, whereas the ratio of heart weight to body weight was reciprocally higher in conditional Klf4 knock-out mice. In addition, the rates of proliferation and apoptosis in the heart were not different between the two groups. Lack of cardiac hypertrophy in Klf4-deficient mice is probably caused by reduced expression of Gata4 and other genes affected by Klf4 deletion in these mice. In fact, results of previous studies showed that cardiac Gata4 is required for hypertrophy, compensation, and myocyte viability as determined by analyses of cardiac-specific Gata4 knock-out mice (31). However, it is also possible that we could not detect a transient change in cellular proliferation in our assays, although we measured the fraction of Ki-67 positive cells and phosphohistone H3-positive cells (data not shown) in the hearts of conditional Klf4 knock-out mice and control mice at P28 as well as E18.5. Studies from a number of laboratories (32, 33), including our own (12), have shown that Klf4 can profoundly alter cellular growth by activating the cyclin-dependent kinase inhibitor, p21WAF1/Cip1 in multiple cell types. As such, it is possible that at least some of the effects of Klf4 deletion in the heart may be secondary to alterations in cellular proliferation.
Results of dual immunofluorescence studies in Fig. 2G showed that some of Klf4-positive cells were also positive for Gata4, which is mainly expressed in cardiomyocytes. However, some Klf4-positive cells were negative for Gata4, which might be other cardiac cell types, such as cardiac fibroblasts. To further determine the role of Klf4 in the heart, it is interesting to generate other types of conditional Klf4-deficient mice using multiple cardiomyocyte-specific Cre recombinase expressing mouse lines, including αMHC-Cre mice.
In contrast to the marked effects on cardiac development, smooth and cardiac muscle-specific conditional Klf4 knock-out mice had no discernible phenotype in SMCs within arteries such as the coronary artery versus the aorta where SMCs have distinct embryological origins (34). However, as occurred in the heart, Klf4 deletion in the present studies would have occurred after initial differentiation of SMCs, and we cannot rule out a possible role of Klf4 earlier in SMC development. Consistent with this possibility, Passman et al. (35) recently demonstrated high levels of Klf4 expression in Sca1-positive adventitial cells which can differentiate into vascular SMCs. Indeed, they found that Klf4 expression was markedly down-regulated when these cells differentiated into SMCs in vitro, suggesting a possible role for Klf4 in maintenance of pluripotency and/or self-renewal in these cells. In addition, we have recently shown that tamoxifen-induced deletion of the Klf4 gene in mice exhibited delayed down-regulation of SMC differentiation markers, but exhibited subsequent SMC hyperproliferation and enhanced neointimal formation following vascular injury (12). These results implicate Klf4 as a critical mediator of transitions in SMC phenotype to a more plastic embryonic state in vivo. However, loss of Klf4 in other cell types might also contribute to the phenotype observed in tamoxifen-induced conditional Klf4-deficient mice. Indeed, Klf4 is expressed in aortic endothelial cells and macrophages and has been shown to play anti-inflammatory and proinflammatory roles in each cell type, respectively (13, 36). As such, further studies are needed to clearly elucidate the role of Klf4 in SMC development and phenotypic switching in vivo. However, given the observed defects in cardiac function and gene expression, as well as large reduction in body weight in SM22α-CreKI+/Klf4loxP/loxP mice, these studies will require SMC-specific, rather than SMC-selective, conditional Klf4 knock-out mice.
In summary, we provide novel evidence that Klf4 plays a critical role in late fetal and/or postnatal cardiac development. Indeed, loss of Klf4 in mouse heart caused decreased cardiac output, resulting in severe growth retardation and postnatal death. We also present evidence suggesting that defects in cardiac function within conditional Klf4 knock-out mice are the result of reduced expression of multiple cardiac genes. Further studies are needed to determine the precise mechanisms by which Klf4 regulates cardiac differentiation and maturation during the postnatal period, as well as the role of Klf4 in development of heart disease in experimental animal models and in humans.
Supplementary Material
Acknowledgments
We thank Jack R. Roy and Dominique L. Rose at the University of Virginia for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants P01HL19242, R01HL38854, and R01HL57353 (to G. K. O.). This work was also supported by American Heart Association National Scientist Development Grant 0635253N (to T. Y.)

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. I–VI.
- Klf4
- Krüppel-like factor 4
- E
- embryonic day
- EF
- ejection fraction
- SMC
- smooth muscle cell
- P
- postnatal day
- MRI
- magnetic resonance imaging
- DAB
- diaminobenzidine
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- ChIP
- chromatin immunoprecipitation
- RT
- reverse transcription.
REFERENCES
- 1.Segre J. A., Bauer C., Fuchs E. (1999) Nat. Genet. 22, 356–360 [DOI] [PubMed] [Google Scholar]
- 2.Katz J. P., Perreault N., Goldstein B. G., Lee C. S., Labosky P. A., Yang V. W., Kaestner K. H. (2002) Development 129, 2619–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katz J. P., Perreault N., Goldstein B. G., Actman L., McNally S. R., Silberg D. G., Furth E. E., Kaestner K. H. (2005) Gastroenterology 128, 935–945 [DOI] [PubMed] [Google Scholar]
- 4.Swamynathan S. K., Katz J. P., Kaestner K. H., Ashery-Padan R., Crawford M. A., Piatigorsky J. (2007) Mol. Cell. Biol. 27, 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 6.Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. (2008) Science 322, 949–953 [DOI] [PubMed] [Google Scholar]
- 7.Adam P. J., Regan C. P., Hautmann M. B., Owens G. K. (2000) J. Biol. Chem. 275, 37798–37806 [DOI] [PubMed] [Google Scholar]
- 8.Liu Y., Sinha S., Owens G. (2003) J. Biol. Chem. 278, 48004–48011 [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Sinha S., McDonald O. G., Shang Y., Hoofnagle M. H., Owens G. K. (2005) J. Biol. Chem. 280, 9719–9727 [DOI] [PubMed] [Google Scholar]
- 10.Pidkovka N. A., Cherepanova O. A., Yoshida T., Alexander M. R., Deaton R. A., Thomas J. A., Leitinger N., Owens G. K. (2007) Circ. Res. 101, 792–801 [DOI] [PubMed] [Google Scholar]
- 11.Yoshida T., Gan Q., Owens G. K. (2008) Am. J. Physiol. Cell Physiol. 295, C1175–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida T., Kaestner K. H., Owens G. K. (2008) Circ. Res. 102, 1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamik A., Lin Z., Kumar A., Balcells M., Sinha S., Katz J., Feinberg M. W., Gerzsten R. E., Edelman E. R., Jain M. K. (2007) J. Biol. Chem. 282, 13769–13779 [DOI] [PubMed] [Google Scholar]
- 14.Zhang J., Zhong W., Cui T., Yang M., Hu X., Xu K., Xie C., Xue C., Gibbons G. H., Liu C., Li L., Chen Y. E. (2006) Arterioscler. Thromb. Vasc. Biol. 26, e23–e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L., Miano J. M., Cserjesi P., Olson E. N. (1996) Circ. Res. 78, 188–195 [DOI] [PubMed] [Google Scholar]
- 16.Shang Y., Yoshida T., Amendt B. A., Martin J. F., Owens G. K. (2008) J. Cell Biol. 181, 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida T., Sinha S., Dandré F., Wamhoff B. R., Hoofnagle M. H., Kremer B. E., Wang D. Z., Olson E. N., Owens G. K. (2003) Circ. Res. 92, 856–864 [DOI] [PubMed] [Google Scholar]
- 18.Somlyo A. P., Somlyo A. V., Devine C. E., Peters P. D., Hall T. A. (1974) J. Cell Biol. 61, 723–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones A. W., Somlyo A. P., Somlyo A. V. (1973) J. Physiol. 232, 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isbell D. C., Voros S., Yang Z., DiMaria J. M., Berr S. S., French B. A., Epstein F. H., Bishop S. P., Wang H., Roy R. J., Kemp B. A., Matsubara H., Carey R. M., Kramer C. M. (2007) Am. J. Physiol. Heart Circ. Physiol. 293, H3372–3378 [DOI] [PubMed] [Google Scholar]
- 21.Thomas P. D., Campbell M. J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. (2003) Genome Res. 13, 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrix J. A., Wamhoff B. R., McDonald O. G., Sinha S., Yoshida T., Owens G. K. (2005) J. Clin. Invest. 115, 418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterkin T., Gibson A., Loose M., Patient R. (2005) Semin. Cell Dev. Biol. 16, 83–94 [DOI] [PubMed] [Google Scholar]
- 24.Braunwald E. (1997) in Heart Disease: A Textbook of Cardiovascular Medicine, 5th Ed., W. B. Saunders, Philadelphia, PA [Google Scholar]
- 25.Olson E. N. (2004) Nat. Med. 10, 467–474 [DOI] [PubMed] [Google Scholar]
- 26.Chen X., Whitney E. M., Gao S. Y., Yang V. W. (2003) J. Mol. Biol. 326, 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heicklen-Klein A., Evans T. (2004) Dev. Biol. 267, 490–504 [DOI] [PubMed] [Google Scholar]
- 28.Mazaud-Guittot S., Tétu A., Legault E., Pilon N., Silversides D. W., Viger R. S. (2007) Biol. Reprod. 76, 85–95 [DOI] [PubMed] [Google Scholar]
- 29.Ohara Y., Atarashi T., Ishibashi T., Ohashi-Kobayashi A., Maeda M. (2006) Biol. Pharm. Bull. 29, 410–419 [DOI] [PubMed] [Google Scholar]
- 30.Shields J. M., Yang V. W. (1998) Nucleic Acids Res. 26, 796–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oka T., Maillet M., Watt A. J., Schwartz R. J., Aronow B. J., Duncan S. A., Molkentin J. D. (2006) Circ. Res. 98, 837–845 [DOI] [PubMed] [Google Scholar]
- 32.Zhang W., Geiman D. E., Shields J. M., Dang D. T., Mahatan C. S., Kaestner K. H., Biggs J. R., Kraft A. S., Yang V. W. (2000) J. Biol. Chem. 275, 18391–18398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X., Johns D. C., Geiman D. E., Marban E., Dang D. T., Hamlin G., Sun R., Yang V. W. (2001) J. Biol. Chem. 276, 30423–30428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida T., Owens G. K. (2005) Circ. Res. 96, 280–291 [DOI] [PubMed] [Google Scholar]
- 35.Passman J. N., Dong X. R., Wu S. P., Maguire C. T., Hogan K. A., Bautch V. L., Majesky M. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9349–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feinberg M. W., Cao Z., Wara A. K., Lebedeva M. A., Senbanerjee S., Jain M. K. (2005) J. Biol. Chem. 280, 38247–38258 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.