Abstract
Guanine nucleotide exchange factors (GEFs) catalyze exchange of GDP for GTP by stabilizing the nucleotide-free state of the small GTPases through their Dbl homology/pleckstrin homology (DH·PH) domains. Unconventionally, PDZ-RhoGEF (PRG), a member of the RGS-RhoGEFs, binds tightly to both nucleotide-free and activated RhoA (RhoA·GTP). We have characterized the interaction between PRG and activated RhoA and determined the structure of the PRG-DH·PH-RhoA·GTPγS (guanosine 5′-O-[γ-thio]triphosphate) complex. The interface bears striking similarity to a GTPase-effector interface and involves the switch regions in RhoA and a hydrophobic patch in PRG-PH that is conserved among all Lbc RhoGEFs. The two surfaces that bind activated and nucleotide-free RhoA on PRG-DH·PH do not overlap, and a ternary complex of PRG-DH·PH bound to both forms of RhoA can be isolated by size-exclusion chromatography. This novel interaction between activated RhoA and PH could play a key role in regulation of RhoGEF activity in vivo.
Keywords: G Proteins, Protein Structure, Protein-Protein Interactions, Signal Transduction, X-ray Crystallography
Introduction
The Rho family of monomeric GTPases belongs to the Ras superfamily and is composed of several proteins including the well characterized Rho (A, B, and C), Rac (1 and 2), and Cdc42 proteins. They control a wide range of cellular processes including modulation of cytoskeletal structure, motility, cell division, gene transcription, vesicular transport, and various enzymatic activities (1). Rho GTPases cycle between an inactive GDP-bound state and an active GTP-bound state. This cycling is tightly controlled by sets of protein partners. In the inactive state, Rho proteins are largely sequestered in the cytosol by association with guanine nucleotide dissociation inhibitor proteins. Activation of Rho is mediated by guanine nucleotide exchange factors (GEFs),4 which stimulate the exchange of GDP for GTP (2). When bound to GTP, Rho GTPases engage downstream effectors and influence cellular functions. The regulatory tenure of the Rho proteins is determined by GTPase-activating proteins, which stimulate the hydrolysis of GTP to GDP, thus returning Rho to the inactive state (3). Two structural elements on the GTPases, called switch I and switch II, recognize the nature of the bound nucleotide and change their conformation accordingly. These two switch regions signal the nucleotide status of the GTPase to effectors or regulators by their direct involvement in protein-protein interactions with these binding partners (4).
The large family of GEFs (about 70 in the human genome) that mediate activation of Rho proteins is characterized by tandem Dbl homology (DH) and pleckstrin homology (PH) domains (2, 5). The DH domain provides the site for catalyzing nucleotide exchange on and thus activating Rho proteins. The C-terminal PH domains are diverse with respect to amino acid sequence and biological activity, with functions ranging from facilitation of interaction with Rho to localization of the GEF through association with specific polyphosphoinositides (2). RGS (regulators of G protein signaling)-containing RhoGEFs (RGS-RhoGEFs), including p115RhoGEF, PDZ-RhoGEF (PRG), and leukemia-associated RhoGEF (LARG), are a homologous subfamily of proteins that represent a potential direct regulatory link between G protein-coupled receptors that activate the G12 class of heterotrimeric G proteins and RhoA-mediated pathways that lead to cytokinesis and transformation (6). All RGS-RhoGEFs contain a regulator of G protein signaling (RGS) homology domain (rgRGS or RH), which is situated N-terminal to the DH·PH domains. Some RGS-RhoGEFs function as GTPase-activating proteins for Gα13 and Gα12 subunits through their rgRGS domains, and binding to Gα13 stimulates their guanine nucleotide exchange activity toward RhoA. In addition to the rgRGS domain, PRG and LARG also contain an N-terminal PDZ domain that has been shown to mediate interaction of the RGS-RhoGEFs with regulatory proteins (7–10).
Structures of two RGS-RhoGEF DH·PH domains have been determined, either by themselves or in complexes with nucleotide-free RhoA (11–13). The cores of these DH domains are elongated bundles of six major α helices, similar in structure to those of other DH domains in the broader RhoGEF family. Residues from the DH domains form extensive contacts with switch regions of RhoA. The PH domains of RGS-RhoGEFs form a small interface with nucleotide-free RhoA, suggesting a potential role in catalysis (11). Indeed, both DH and PH domains are required to achieve full GEF activity. It appears that the PH domain increases the exchange activity of the RGS-RhoGEFs, either through stabilization of the DH domain or through direct impact on the substrate, RhoA.
Recently, a highly conserved hydrophobic patch in the PH domain was identified to be important for RhoA activation. Mutation of this hydrophobic patch has no effect on GEF activity in vitro but abolished the ability of LARG to activate RhoA and to induce stress fiber formation in cultured cells (14). The activity of these mutants could be rescued by fusion with exogenous membrane-targeting domains, suggesting that these regions contribute to proper localization of RGS-RhoGEFs by interacting with target(s) at the cell membrane. Attempts to identify partners that interact with the PH domain of RGS-RhoGEFs via this conserved hydrophobic patch, however, have not been successful (14).
In this study, we explore a novel interaction between activated RhoA and the PH domain of PRG. We show by mutagenesis and pulldown assays that the PH domain of PRG can selectively interact with activated RhoA but not Rac1 or Cdc42. We determined the three-dimensional structure of a stable complex between the DH·PH domains of PRG and activated RhoA bound to GTPγS. Contacts of activated RhoA with PRG-DH·PH occur entirely through the PH domain of PRG and are centered at the conserved hydrophobic patch. Conversely, engagement with the PH domain involves largely the switch regions of activated RhoA. A ternary complex of PRG-DH·PH with both activated and nucleotide-free RhoA can be isolated by size-exclusion chromatography. Although activated RhoA does not appear to affect the GEF activity of PRG in vitro, it is possible that it regulates the guanine nucleotide exchange activity by localization of RGS-RhoGEFs to the plasma membrane.
EXPERIMENTAL PROCEDURES
Expression Construct
Coding regions of human PRG were subcloned into a pGEX-KG vector containing the protease recognition site for the tobacco etch virus (pGEX-KG-TEV) for proteolytic cleavage of the expressed domains from glutathione S-transferase. A His6 tag was also inserted at the C termini of the PRG coding sequences. The expression construct for human RhoA was generated by subcloning the coding region of RhoA, either the full length or a C-terminally truncated fragment (ΔC, residues 1–181), into the pGEX-KG-TEV vector.
Expression and Purification of Proteins
PRG or RhoA were expressed in LB medium at 22 °C overnight in Escherichia coli strain BL21(DE3) cells with 100 μm isopropyl-β-d-thiogalactopyranoside. Frozen cells from 1 liter were thawed and suspended with 50 ml of lysis buffer (50 mm NaHEPES, pH 8.0, 200 mm NaCl, 5 mm β-mercaptoethanol, 10 μm GDP, and protease inhibitors). Cells were lysed with the addition of lysozyme, DNase I, and MgCl2 to final concentrations of 2 mg/ml, 50 μg/ml, and 5 mm, respectively. GST-tagged fusion proteins were extracted from the soluble fraction of lysates by affinity chromatography with glutathione-Sepharose 4B (Amersham Biosciences). Resins with GST fusion proteins bound were suspended with lysis buffer and then incubated overnight at 4 °C in the presence of TEV protease to remove the GST tag. Fragments of PRG released from the resin were further purified by IMAC-Ni2+ affinity chromatography (Bio-Rad). Affinity-enriched proteins were subjected to further purification with a Mono Q anion exchange column (Amersham Biosciences) that had been pre-equilibrated with Buffer A (25 mm NaHEPES, pH 8.0, 5 mm β-mercaptoethanol, 2 mm MgCl2, and 10 μm GDP). Elution was accomplished with a linear gradient of 0–0.5 m NaCl in Buffer A.
Activation of GTPases with GTPγS
Purified RhoA, Cdc42, or Rac1 was exchanged into binding buffer (25 mm NaHEPES, pH 8.0, 1 mm EDTA, 1 mm dithiothreitol, 50 mm NaCl, and 10 μm GDP) and concentrated to 200–500 μm. The concentrate was adjusted to 0.5 mm MgSO4 and 1 mm GTPγS and incubated at room temperature for 24 h.
Pulldown Assays
Immobilized GST-tagged RhoA was used to compare the relative ability of purified His6-tagged RhoGEFs to bind RhoA in the presence of different guanine nucleotides. GST-RhoA (80 pmol) was mixed with 10 μl of glutathione-Sepharose 4B resin in 100 μl of incubation buffer (50 mm NaHEPES, pH 7.5, 50 mm NaCl, 1 mm dithiothreitol, 1 mm EDTA, 5 mm MgCl2, and 0.3% (v/v) Triton X-100) and incubated for 30 min at 4 °C. The resin was washed with incubation buffer, and His6-tagged RhoGEF proteins (12 pmol) were added to the immobilized GST-RhoA in incubation buffer (100 μl) containing no additional guanine nucleotide, 10 μm GDP, or 10 μm GTPγS. The mixtures were incubated on a rotating platform for 45–60 min at 4 °C, after which the Sepharose resin was pelleted using a microcentrifuge. Supernatants containing unbound RhoGEF were removed, and the resins were then washed twice with 500 μl of cold incubation buffer. RhoGEF bound to the resin was released by boiling in SDS sample buffer, and respective amounts bound were compared by immunoblot analysis using anti-His6 monoclonal antibody (R&D Systems). Each pulldown assay was repeated at least three times.
Formation of the PRG-DH·PH-RhoA·GTPγS Complex
The DH·PH-RhoA·GTPγS complex was purified by size-exclusion chromatography using Superdex 200/75 tandem gel filtration columns (Amersham Biosciences). Equal moles of RhoA(ΔC)·GTPγS and PRG-DH·PH (residues 712–1085) were mixed and then filtered through the gel filtration columns pre-equilibrated with Buffer B (25 mm Tris-Cl, pH 8.5, 1 mm dithiothreitol, 100 mm NaCl, and 1 mm EDTA) and 2 mm MgCl2. Fractions that contained the DH·PH-RhoA·GTPγS complex (molecular mass of ∼65 kDa as judged by elution volume) were pooled and concentrated using Amicon Ultra-4 (10-kDa) concentrators (Millipore) to a final concentration of 15 mg/ml. Aliquots (50 μl) of the concentrated complex were flash-frozen with liquid nitrogen and stored at −80 °C.
Formation of Ternary Complex of PRG-DH·PH, RhoA·GTPγS, and Nucleotide-free RhoA
EDTA was added to full-length RhoA to a final concentration of 25 mm. The mixture was incubated at room temperature for 15 min to release nucleotide and then concentrated at 4 °C using Amicon Ultra-4 (10-kDa) concentrators (Millipore) to a final concentration of 10 mg/ml. The binary complex between DH·PH and nucleotide-free RhoA (full length) was formed by mixing equal moles of nucleotide-free RhoA (full length) and PRG-DH·PH and then filtered through gel filtration columns at 4 °C pre-equilibrated with Buffer B. Fractions that contained the DH·PH·RhoA complex (molecular mass of ∼67 kDa as judged by elution volume) were pooled, and MgCl2 was added to the pooled fractions to a final concentration of 2 mm. RhoA(ΔC)·GTPγS was then added to the binary complex and incubated for 15 min at room temperature, and the mixture was filtered through gel filtration columns pre-equilibrated with Buffer B and 2 mm MgCl2. Fractions that contained the ternary complex (molecular mass of ∼87 kDa as judged by elution volume) were analyzed by SDS-PAGE.
ITC Assays
Isothermal titration calorimetry (ITC) was performed as described before (15). Briefly, protein samples were dialyzed against Buffer B and 2 mm MgCl2, and a typical titration of DH·PH with activated RhoA involved 20 injections at 3-min intervals of 8 μl of activated RhoA (0.4 mm) into a sample cell containing 1.5 ml of either PRG-DH·PH or PRG-PH (50 μm) at 25 °C (298 K).
Nucleotide Exchange Assay
RhoA was loaded with N-methylanthraniloyl-GDP (mant-GDP, Invitrogen) by incubating 30 μm RhoA with a 10-fold molar excess of mant-GDP in Buffer B and 3 mm MgCl2 at room temperature for 24 h. Loaded RhoA was then purified by gel filtration columns to remove excess nucleotide. Fluorescence assays were performed on a Fluorolog-3 spectrofluorometer at room temperature (λex = 356 nm, λem = 445 nm, slits = 1/1 nm). In each assay, 1 μm mant-GDP-loaded RhoA was incubated with 100 μm GDP in reaction buffer (25 mm NaHEPES, pH 8.0, 100 mm NaCl, 1 mm dithiothreitol, and 5 mm MgCl2) in a 200-μl cuvette. The exchange reaction was started by the addition of 100 nm PRG. Each measurement was repeated at least three times.
Crystallization and Data Collection
The DH·PH-RhoA·GTPγS complex (15 mg/ml) was crystallized by vapor diffusion against a solution of 14–16% polyethylene glycol 10,000, 0.1 m ammonium acetate, and 0.1 m Bis-Tris, pH 5.2–5.8, at 20 °C. Crystals were then cryoprotected with an additional 15% (v/v) ethylene glycol. Native data with an oscillation range of 120° were measured at 100 K at the Structural Biology Center (Beamline 19BM) at Argonne National Laboratory. Diffraction data were reduced using the HKL software package (16).
Structure Determination and Model Refinement
Initial phases were generated by molecular replacement using the coordinates of PRG-DH and PH domains (Protein Data Bank (PDB) entry 1XCG) and RhoA bound to GTPγS (PDB entry 1A2B) as separate search models, using program PHASER (17). Model building was performed using the program Coot (18). The model was refined using Refmac5 from the CCP4 software package (19). Putative water molecules within hydrogen-bonding distance of at least one protein atom or other water oxygen atoms and with refined B-factors <100 Å2 were included in the model. PROCHECK (20) indicates that over 95% of the residues fall in the most favorable regions of ϕ, ψ conformational space (21). Coordinates and structure factors have been deposited in the PDB (22) with accession code 3KZ1. Atomic representations were created using PyMOL (23). Electrostatic potential maps were created with PyMOL and APBS (24).
RESULTS
PDZ-RhoGEF Recognizes GTP-activated RhoA via Its PH Domain
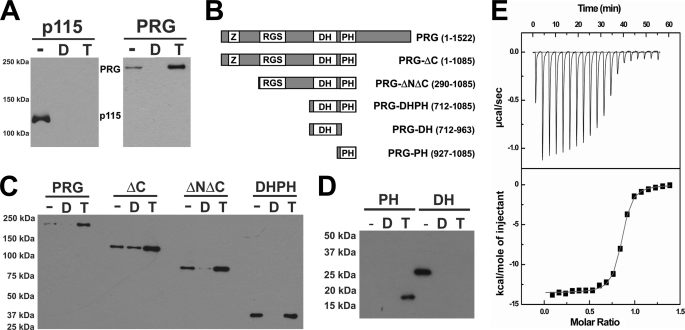
Full-length PRG and p115RhoGEF were each expressed in bacteria with an N-terminal cleavable GST tag and a C-terminal His6 tag for purification. Purified proteins were tested for their ability to bind to immobilized GST-RhoA in the presence or absence of various guanine nucleotides (Fig. 1). Both p115RhoGEF and PDZ-RhoGEF bound well to nucleotide-free RhoA, as expected for binding of the catalytic intermediate of a substrate to its enzyme (Fig. 1A). Although p115RhoGEF bound tightly only to the nucleotide-free form of RhoA, PDZ-RhoGEF unexpectedly showed stronger binding to RhoA in the presence of GTPγS, a non-hydrolyzable analog of GTP (Fig. 1A).
FIGURE 1.
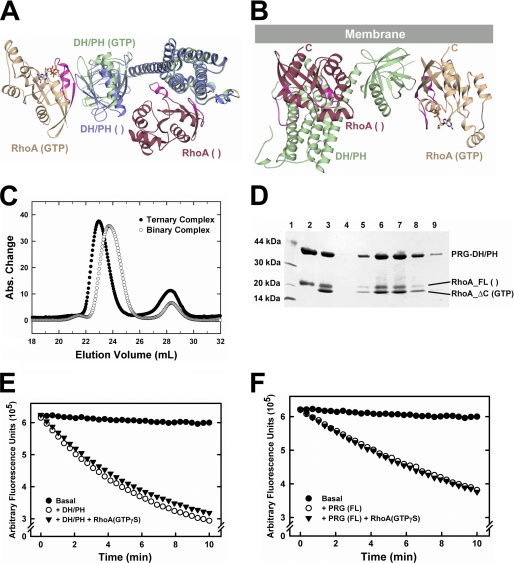
The PH domain from PRG interacts directly with activated RhoA. A, purified p115RhoGEF and PRG (12 pmol) were incubated with 80 pmol of immobilized GST-RhoA with no additional guanine nucleotide (−), 10 μm GDP (D), or 10 μm GTPγS (T), as described under “Experimental Procedures.” RhoGEFs bound to the resin were subjected to SDS-PAGE and visualized by immunoblotting using anti-His6 monoclonal antibody. B, schematic representation of truncated forms of PRG; included amino acids are listed at the right. PDZ domains are designated Z. C, purified PRG (full length or fragments as described in B) were incubated with immobilized GST-RhoA, and RhoGEFs bound to the resin were analyzed by immunoblotting as described in A. D, purified PRG-DH and PRG-PH domains were incubated with immobilized GST-RhoA, with either no additional guanine nucleotide (−) or 10 μm GDP (D) or with immobilized GST-RhoA preloaded with GTPγS (T). DH or PH domains bound to the resin were analyzed by immunoblotting as described in A. E, ITC profile for the binding of PRG-PH to RhoA·GTPγS. Non-linear least squares fit using a “one set of sites” model resulted in the fit shown (solid line) at the bottom.
The domain in PRG that binds to activated RhoA was identified by deletion analysis. Various fragments of PRG (Fig. 1B) were expressed and purified as described under “Experimental Procedures.” The results from pulldown assays (Fig. 1C) reveal that removal of the N-terminal PDZ, the RGS-homology (rgRGS or RH), or the C-terminal domains did not affect binding of PRG to RhoA·GTPγS. Thus, the DH·PH domains of PRG alone are sufficient to bind activated as well as inactive RhoA. It has been shown that the DH·PH domains catalyze the exchange of GDP for GTP on RhoA by interacting directly with the switch regions of RhoA (11, 12). Mutations of key residues of the DH domain, which lead to diminished GEF activity and weak binding to nucleotide-free RhoA, have very little effect on binding of DH·PH to activated RhoA (supplemental Fig. S1). In fact, the DH domain alone of PRG behaves similarly to p115RhoGEF in the pulldown assay in that it only binds tightly to nucleotide-free RhoA. In contrast, the PRG-PH domain is sufficient for binding of RhoA·GTPγS (Fig. 1D). The binding affinity (Kd) between the PRG-DH·PH domains and activated RhoA bound to GTPγS was measured to be about 140 nm by ITC, and similar binding affinity was observed between PRG-PH and activated RhoA (Fig. 1E, Table 1); both reactions are exothermic. PRG-DH·PH shows similar binding affinity toward nucleotide-free RhoA; however, this binding reaction is endothermic. Either DH·PH or PH alone can form a stable complex with activated RhoA bound to GTPγS, and these complexes can be isolated by size-exclusion chromatography. Under the same conditions, the DH domain alone failed to form a stable complex with RhoA·GTPγS (data not shown).
TABLE 1.
Thermodynamic parameters of the binding of PRG to RhoA
Determinations were made using ITC as described under “Experimental Procedures.” Kd, ΔH, and N represent the dissociation constant, enthalpy, and stoichiometry, respectively. RhoA·GTPγS represents activated RhoA preloaded with GTPγS as described under “Experimental Procedures.” RhoA·( ) represents nucleotide-free RhoA. All experimentally derived parameters are an average of two independent experiments done under identical conditions.
| Syringe (400 μm) | Sample cell (50 μm) | N | Kd | ΔH |
|---|---|---|---|---|
| nm | kcal mol−1 | |||
| RhoA·GTPγS | DH·PH | 0.8 | 140 ± 11 | −9.4 ± 1.3 |
| RhoA·GTPγS | PH | 0.8 | 80 ± 6 | −13.6 ± 0.9 |
| RhoA·GTPγS | DH·PH-RhoA·( ) | 0.9 | 110 ± 15 | −10.6 ± 0.9 |
| RhoA·( ) | DH·PH | 0.8 | 110 ± 11 | 3.4 ± 0.3 |
| RhoA·( ) | DH·PH-RhoA·GTPγS | 0.8 | 110 ± 12 | 3.2 ± 0.3 |
Overall Structure of the PRG-DH·PH-RhoA·GTPγS Complex
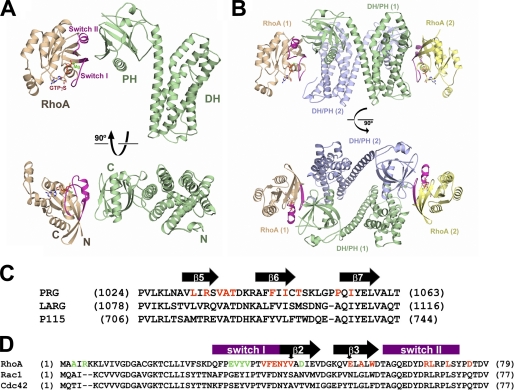
The PRG-DH·PH-RhoA·GTPγS complex was formed by mixing a molar excess of non-prenylated, C-terminally truncated RhoA bound to GTPγS with PRG-DH·PH in the presence of Mg2+ and isolation of the stoichiometric complex by size-exclusion chromatography. Crystals were grown by vapor diffusion. The structure of RhoA·GTPγS bound to the DH·PH domains of PRG was determined at a resolution of 2.7 Å by molecular replacement using separate search models for the PRG-DH domain, PRG-PH domain, and activated RhoA (Fig. 2). The model was refined to conventional and free crystallographic R-factors of 23.4 and 28.3%, respectively. The final atomic model comprises residues 714–1010 and 1020–1082 of the PRG-DH·PH domains and residues 3–181 of RhoA bound to GTPγS and Mg2+. The remaining residues of the DH·PH region are disordered. Data collection and refinement statistics for the complex structure are summarized in Table 2.
FIGURE 2.
Structure of the PRG-DH·PH-RhoA·GTPγS complex. A, ribbon diagrams depicting tertiary structures of PRG-DH·PH in a complex with RhoA·GTPγS. PRG-DH·PH is colored green. RhoA is colored wheat, with switch regions colored purple. GTPγS and magnesium ion are depicted as ball-and-stick models and colored as follows. Oxygen, nitrogen, carbon and phosphorus atoms are colored red, blue, wheat, and yellow, respectively. Magnesium is colored green. B, ribbon diagrams depicting non-crystallographic dimer of the PRG-DH·PH-RhoA·GTPγS complexes (labeled 1 and 2). The first complex (1) is colored as in A. DH·PH and RhoA in the second complex (2) are colored blue and yellow, respectively. C, sequence alignment of the PH domains of RGS-RhoGEFs in the proposed contact region for activated RhoA. Secondary structure (black arrows: β-strands) has been assigned on the basis of the structure of PRG. Residues contacting activated RhoA are colored red. D, partial sequence alignment of Rho family small GTPases. The two conformationally flexible switch elements are indicated by purple blocks on top. Residues in RhoA that are involved in contacts with the PRG-PH domain are colored red. Residues in RhoA contacting the dyad-related PRG-DH domain in the non-crystallographic dimer are colored green. Two residues (Val-43 and Glu-54) shared by both interfaces are marked by arrows on top.
TABLE 2.
Data collection and refinement statistics
| Data collection | |
| Wavelength (Å) | 0.9791 |
| Space group | P212121 |
| Unit cell (Å) | |
| a | 98.17 |
| b | 111.84 |
| c | 138.39 |
| Dmin (Å) | 2.7 |
| Unique reflections | 38,512 |
| Redundancy | 4.8 |
| Completeness (%)a | 90.7 (62.1) |
| <I/σ>a | 16.5 (1.5) |
| Rsyma,b | 0.09 (0.69) |
| Mosaicity (°) | 0.9 |
| Wilson B-factor (Å2) | 62 |
| Refinement | |
| Resolution (Å) | 33.5-2.7 |
| Total reflection used | 36,635 |
| Number of atoms | |
| Protein | 8714 |
| Water | 75 |
| Heterogen | 66 |
| Rwork (%)c | 23.3 |
| Rfree (%)d | 28.3 |
| r.m.s.e deviations | |
| Bond lengths(Å) | 0.014 |
| Bond angles (°) | 1.436 |
| Average B-factor (Å2)f | 71.3 |
a Numbers in parentheses correspond to the last resolution shell (2.75-2.70 Å). Diffraction extended beyond 2.8 Å with I/σ above 2.1 for the resolution shell between 2.86 and 2.80 Å.
b Rsym = ΣhΣi |Ii(h) − <I(h)>|/ΣhΣi Ii(h), where Ii(h) and <I(h)> are the ith and mean measurement of the intensity of reflection h, respectively.
c Rwork = Σh ‖Fo(h)| − |Fc(h)‖/Σh |Fo(h)|, where Fo(h) and Fc(h) are the observed and calculated structure factors, respectively. An I/σ cutoff was not used in the final calculations of R-factors.
d Rfree is the R-factor obtained for a test set of reflections consisting of a randomly selected 5% of the data.
e r.m.s., root mean square.
f Average B-factor calculations include both TLS (translation, libration, screw) and residual B-factors.
The asymmetric unit of the crystal contains two PRG-DH·PH-RhoA·GTPγS complexes related by a two-fold axis of rotation (Fig. 2B). The dimer interface buries about 3200 Å2 of solvent-accessible surface area and is stabilized by interactions between the two DH·PH domains and also between RhoA and the DH domain of the dyad-related molecule (supplemental Fig. S2). However, there is no evidence from size-exclusion chromatography that the DH·PH-RhoA·GTPγS complexes are dimeric in solution (data not shown). Members of the RGS-RhoGEF family have been reported to oligomerize, but interaction involves domains C-terminal to DH·PH (25, 26).
RhoA·GTPγS interacts exclusively with the PH domain of PRG-DH·PH (Fig. 2, A and C). This engagement involves the switch I and switch II regions of RhoA and the β-strands connecting them (Fig. 2D). The structure of the DH·PH domains described here is very similar to that of the same DH·PH domains bound to nucleotide-free RhoA (12). Cα atoms from these two DH·PH structures (residues 714–1081 in PRG) overlap with a root mean square deviation of less than 2 Å. However, instead of having the DH domain engaging nucleotide-free RhoA, the PH domain stabilizes the active conformation of RhoA in this structure. The structure of RhoA in this complex is almost identical to that of RhoA bound to GTPγS alone (27), with a root mean square deviation of 0.5 Å for all Cα atoms (residues 3–181). The electron density is well defined for GTPγS and Mg2+ in the guanine nucleotide binding pocket of RhoA. The structure of the PRG-DH·PH-RhoA·GTPγS complex represents a novel interaction involving activated RhoA.
The PH-RhoA Interface
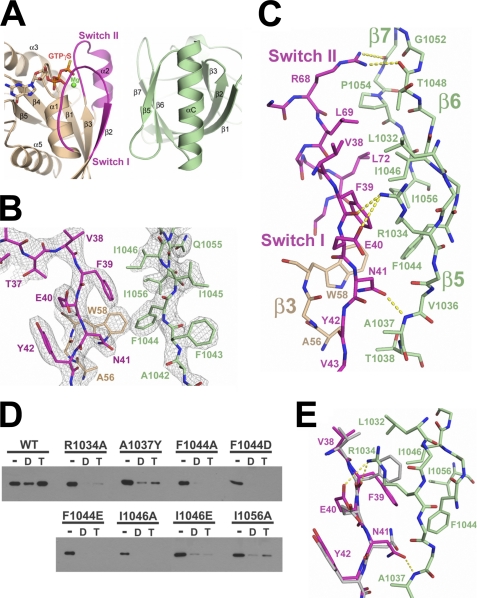
The interface between the PH domain of PRG and RhoA buries a solvent-accessible surface area of about 1300 Å2. This interface is populated by a set of conserved hydrophobic residues from both the PH domain and RhoA. The switch regions of RhoA clamp down on a hydrophobic patch formed by the β5, β6, and β7 strands from the PH domain (Fig. 3A). Phe-39 from switch I of RhoA lies at the center of the interface (Fig. 3, A and C). The side chain of Phe-39 docks into a hydrophobic pocket on the PH domain formed by Leu-1032 from β5, Phe-1044 and Ile-1046 from β6, and Ile-1056 from β7 (Fig. 3C). Point mutations of these hydrophobic residues in PH to either alanine or glutamic acid greatly reduce the binding affinity of PRG-DH·PH for activated RhoA (Fig. 3D). This hydrophobic binding pocket for Phe-39 is flanked on one side by the side chain of Arg-1034 extending from the β5 strand in PH and on the other by the side chain of Trp-58 from the β3 strand in RhoA (Fig. 3C). Arg-1034 forms two hydrogen bonds with the side chain of Glu-40 and the main chain carbonyl group of Phe-39. Mutation of Arg-1034 to alanine significantly reduces binding between activated RhoA and the DH·PH domains of PRG (Fig. 3D). Trp-58 of RhoA forms van der Waals contacts with Phe-1044 of the PH domain.
FIGURE 3.
The PH-RhoA interface. A, ribbon diagram showing the interface between the PRG-PH domain and RhoA·GTPγS using the same color scheme as in Fig. 2A. B, electron density at the PH-RhoA interface. Electron density (cages) from a 2.7 Å σA-weighted 2Fo − Fc difference map (46) is contoured at 1.6 standard deviations above the mean. Residue labels are color-coded as shown in A. C, extensive contacts between switch regions of RhoA and the β5-β6-β7 strands of the PH domain. Hydrogen bonds are drawn as dotted lines and colored yellow. Residues are labeled and color-coded as shown in A. D, purified PRG-DH·PH, wild-type (WT), or mutant were incubated with immobilized GST-RhoA, with no additional guanine nucleotide (−), 10 μm GDP (D), or 10 μm GTPγS (T). DH·PH domains bound to the resin were analyzed by immunoblotting as described in the legend for Fig. 1. Mutations at the predicted RhoA binding site on PH had little effect on binding of PRG-DH·PH to nucleotide-free RhoA but severely impaired the ability of PRG-DH·PH to bind activated RhoA. E, structural comparison of switch I regions in RhoA·GTPγS at the PH-RhoA interface. Elements from RhoA·GTPγS in a complex with DH·PH are colored purple. Elements from RhoA·GTPγS alone are colored gray. PH is colored green. Hydrogen bonds between RhoA·GTPγS and the PH domain are drawn as dotted lines and colored yellow. The side chain conformations of Phe-39, Glu-40, and Asn-41 in switch I of RhoA change significantly upon binding to PH.
At the bottom of the interface (Fig. 3, A and C), the backbone of the β2 strand in RhoA stacks tightly against the loop connecting the β5 and β6 strands in PH. Ala-1037 from the β5-β6 loop forms van der Waals contacts with the main chain atoms of Asn-41, Tyr-42, and Val-43 in β2 of RhoA (Fig. 3C). Mutation of Ala-1037 to a bulky residue reduces binding between DH·PH and activated RhoA (Fig. 3D). The hydrogen bond formed between the main chain amide group of Ala-1037 and the carboxyl group of Asn-41 further stabilizes this interaction.
The rest of the PH-RhoA interface is formed between switch II of RhoA and the β6-β7 loop of the PH domain (Fig. 3, A and C). Two leucine residues (Leu-69 and Leu-72) dock into a hydrophobic pocket on PH formed by side chains of Ile-1046, Thr-1048, Pro-1054, and Ile-1056. Arg-68 from switch II caps this pocket by hydrogen bonding to the main chain carbonyl groups of Thr-1048 and Gly-1052. These interactions appear to alter the conformation of the β6-β7 loop of the PH domain when compared with the PRG-DH·PH domains bound to nucleotide-free RhoA (12). Interaction with switch II of activated RhoA causes the loop to bend toward the DH domain (away from activated RhoA) by 2.0–3.5 Å as indicated by shifts of Cα atom positions within the β6-β7 loop of the PH domain (supplemental Fig. S3).
As noted above, the structure of RhoA·GTPγS in complex with PRG-DH·PH is almost identical to that of RhoA·GTPγS alone. Binding to the PH domain of PRG does not alter the main chain atom positions in the switch regions of RhoA. Rather, the interaction induces substantial changes in side chain conformations of residues in switch I (Phe-39, Glu-40, and Asn-41, Fig. 3E) and switch II (Arg-68) to accommodate the interactions described above. Similar conformational changes of residues in the switch regions are observed when RhoA is bound to other Rho binding domains (28, 29), revealing a general mechanism for interaction of activated RhoA with its effectors (see “Discussion”).
Specificity of PRG-PH for Activated RhoA
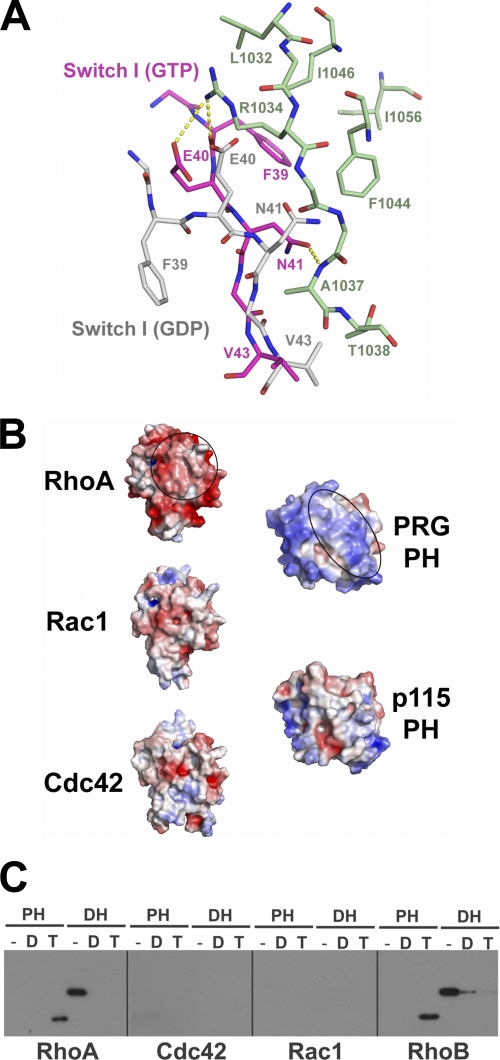
The PH domain of PRG binds tightly to activated RhoA but not to inactive RhoA bound to GDP or in the absence of nucleotide (Fig. 1D). One of the main structural differences between activated and inactive RhoA is the conformation of switch I (30). As discussed above, switch I of RhoA·GTPγS forms extensive interactions with the PH domain of PRG (Figs. 2D and 3). Phe-39 from switch I of activated RhoA docks into a highly conserved hydrophobic pocket on PH. A series of hydrogen bonds provides further support for this interface, including two hydrogen bonds between Arg-1034 of PH and Phe-39/Glu-40 of RhoA and one between Ala-1037 of PH and Asn-41 of RhoA (Fig. 3). In the GDP-bound form, the segment containing Phe-39 of switch I rotates toward the core of RhoA, away from the activated RhoA-PH interface (Fig. 4A). The lateral movement of the Cα atom of Phe-39 from GDP-bound to GTPγS-bound RhoA is about 7 Å, and 11 Å for its side chain. Therefore, Phe-39 in RhoA·GDP is not positioned to interact with the hydrophobic pocket on the PH domain. The change of conformation in switch I also disrupts the hydrogen bond network that supports the RhoA-PH interface. Furthermore, Asn-41, Tyr-42, and Val-43 of switch I in RhoA·GDP would cause steric hindrance with the β5-β6 loop of PH, which contains the key Ala-1037 residue.
FIGURE 4.
Specificity of PRG-DH·PH for activated RhoA. A, structural comparison of RhoA·GDP and RhoA·GTPγS at the PH-RhoA interface. RhoA·GTPγS and PH are color-coded as in Fig. 2A. Elements from RhoA·GDP are colored gray. Hydrogen bonds between RhoA·GTPγS and the PH domain are drawn as dotted lines and colored yellow. The difference between the Cα atom positions of Phe-39 in these two structures of RhoA when superimposed is about 7 Å. B, electrostatic interactions restrict the specificity of PRG-PH for RhoA. On the left, the electrostatic potentials of RhoA, Rac1, and Cdc42 are calculated in the range of −5 kT/e (red) to +5 kT/e (blue) and mapped to the solvent-accessible surface, where k is the Boltzmann's constant, T is temperature (K), and e is the charge of an electron. Molecules are oriented such that the surface on RhoA contacting the PRG-PH domain is shown (indicated by a circle). On the right, similar calculations are carried out with PRG-PH and a modeled p115-PH. The PH domains are oriented such that the surface on PRG-PH contacting the activated RhoA is shown (indicated by an oval). The electrostatic potentials of RhoA and the PH domain of PRG are highly complementary. C, purified PRG-DH and PRG-PH domains were incubated with immobilized GST-RhoA, GST-Cdc42, GST-Rac1, or GST-RhoB, with either no additional guanine nucleotide (−) or 10 μm GDP (D) or with immobilized GST-GTPases preloaded with GTPγS (T). DH and PH bound to the resin were analyzed by immunoblotting as described in the legend for Fig. 1. Only GTPγS-activated RhoA and its close homolog RhoB bound PRG-PH domains.
p115RhoGEF, a homolog of PRG, did not bind activated RhoA in the pulldown assay (Fig. 1A). p115RhoGEF and PRG share high sequence similarity within the PH domain, especially for those residues that contact activated RhoA (Fig. 2C). However, the calculated electrostatic surface potentials of the DH·PH domains of p115RhoGEF and PRG are quite different (Fig. 4B). The β5-β6-β7 segment that contacts activated RhoA is notably electropositive in PRG and complements the electronegative surface of RhoA. The corresponding surface of p115RhoGEF, however, is less electropositive, raising the possibility that electrostatic repulsion disfavors formation of a tight complex between activated RhoA and the PH domain of p115RhoGEF, irrespective of potential differences in specific intermolecular interactions. LARG, the other member of the RGS-RhoGEF family, also possesses a notable electropositive surface in β5-β6-β7 of its PH domain, suggesting that LARG might form a tight complex with activated RhoA.
Most of the residues in RhoA that engage PRG-PH are well conserved in the prototypic Rho family GTPases, Cdc42 and Rac1 (Fig. 2D). Unlike RhoA, activated Cdc42 or Rac1 failed to interact with PRG-PH in the pulldown assays (Fig. 4C). Three amino acids in RhoA that contact PH are less well conserved in Cdc42 and Rac1. These include Glu-40 from switch I and Glu-54 and Ala-56 from the β3 strand (Fig. 2D). Glu-40 forms extensive contacts with the side chain of Arg-1034 in PH (Fig. 3, C and E); the mutation of the latter to an alanine severely impairs binding between activated RhoA and PRG-DH·PH (Fig. 3D). In Cdc42 and Rac1, Glu-40 is replaced by aspartic acid. The shortening of the side chain at this position could eliminate a salt bridge and several van der Waals contacts between Glu-40 and Arg-1034. Glu-54 and Ala-56 from the β3 strand of RhoA interact with Thr-1038 from the β5-β6 loop of PH. Glu-54 is replaced by an asparagine or a threonine in Cdc42 or Rac1, respectively, whereas Ala-56 is replaced with glycine in both. These changes eliminate several van der Waals contacts with Thr-1038 in the PH domain. Therefore, differences in protein sequences among Rho family GTPases could account for the specificity of PRG-DH·PH for activated RhoA, in preference to Cdc42 or Rac1. Furthermore, calculations of the electrostatic surface potentials of these GTPases indicate that RhoA is notably more electronegative than Rac1 or Cdc42 (Fig. 4B). Given that the PH domain of PRG has a highly positive lobe of electrostatic potential, substantial electrostatic attraction favors tight complex formation between RhoA and the PH domain of PRG. Similar differences in electrostatics have been shown to determine the specificity of other GTPase-effector pairs (31, 32).
A Ternary Complex of PRG-DH·PH, RhoA·GTPγS, and Nucleotide-free RhoA
The DH·PH domains of RGS-RhoGEFs promote the exchange of GDP for GTP on RhoA. DH·PH interacts directly with RhoA in its ground state (GDP-bound) and at higher affinity with the intermediate (nucleotide-free state) of the exchange reaction via an interface that largely involves the DH domain (11, 12). Therefore, the PH surface that binds activated RhoA and the DH surface that binds nucleotide-free RhoA do not overlap (Fig. 5A), suggesting that the two forms of RhoA can bind simultaneously to PRG-DH·PH. Comparison of the structure of PRG-DH·PH bound to a nucleotide-free RhoA (12) with that bound to RhoA·GTPγS reveals only minor differences in the structures of the two domains. However, when compared with the DH·PH structure when bound to nucleotide-free RhoA, the DH and PH domains rotate apart when bound to activated RhoA (Fig. 5A). It is not clear whether this rotation is caused by binding to activated RhoA or by interaction between the PH domain and the DH domain from the dyad-related molecule in the crystal (Fig. 2B and supplemental Fig. S2). In the case of LARG, the flexible linker between DH and PH enables considerable relative motion between the two, which upon binding to RhoA, allows PH to form several contacts with RhoA (11). However, such relative motion between DH and PH domains has not been observed in PRG (12, 13).
FIGURE 5.
Ternary complex of PRG-DH·PH, RhoA·GTPγS, and nucleotide-free RhoA (RhoA ( )). A, structural comparison of the DH·PH domains of PRG either bound to activated RhoA (color-coded as in Fig. 2A) or in complex with nucleotide-free RhoA (DH·PH is colored blue, and nucleotide-free RhoA is colored dark red). The two structures are superimposed based on structural alignment of the DH domains. B, a model of the ternary complex docked to the plasma membrane running along the top of the panel. C, chromatograms from size-exclusion chromatography with the binary complex (open circles, DH·PH with nucleotide-free RhoA) and ternary complex (solid circles, binary complex plus activated RhoA) superimposed based on elution volumes. A clear left shift of about 1 ml in elution volumes was observed when comparing the peaks of the two complexes. The peak on the right side with an estimated elution volume of 28 ml represents excess RhoA. D, SDS-PAGE gel showing components of the ternary complex peak. Lane 1, protein standard markers; lane 2, binary complex of DH·PH with nucleotide-free RhoA (full length) (Rho_FL ( )); lane 3, mixture of binary complex with RhoA(ΔC)·GTPγS before size-exclusion chromatography; lanes 4–9, fractions from the ternary complex peak (22–24 ml). This experiment is representative of three trials to show ternary complex formation. E, nucleotide exchange assays with PRG-DH·PH and RhoA. For each time course, 1 μm RhoA loaded with mant-GDP was mixed with 100 μm GDP, and the exchange reaction was started at room temperature by the addition of buffer (Basal, solid circles) or 100 nm PRG-DH·PH alone (open circles) or 100 nm PRG-DH·PH with 1 μm RhoA·GTPγS (solid triangles). The subsequent decrease in fluorescence (λex = 356 nm, λem = 445 nm) was measured for 10 min. F, the same nucleotide exchange assays as described in E, but with full-length PRG and RhoA.
A ternary complex of PRG-DH·PH bound to both nucleotide-free RhoA and activated RhoA can be readily purified by size-exclusion chromatography (Fig. 5C). A binary, nucleotide-free complex between PRG-DH·PH and full-length RhoA was isolated by size-exclusion chromatography first. The binary complex was then mixed with a C-terminally truncated RhoA preloaded with GTPγS and filtered through the same size-exclusion columns. The difference in elution volume of complexes formed in the presence and absence of RhoA·GTPγS translates to a difference of about 20 kDa in apparent molecular mass, consistent with the binding of a RhoA·GTPγS molecule to the binary, nucleotide-free complex, as verified by SDS-PAGE gels (Fig. 5D).
Thermodynamically, the binding reactions between activated RhoA bound to GTPγS and either PRG-DH·PH or a binary, nucleotide-free complex of PRG-DH·PH and RhoA are almost identical (Table 1), with similar ΔH and Kd values. Conversely, binding reactions between nucleotide-free RhoA and either PRG-DH·PH alone or a binary PRG-DH·PH-RhoA·GTPγS complex measured by ITC reveal almost identical ΔH and Kd values (Table 1).
A model of a ternary complex of PRG-DH·PH bound to both nucleotide-free and GTP-bound RhoA predicts the orientation of the Rho proteins with respect to each other (Fig. 5B). In this model, the C termini of both Rho proteins, which are geranylgeranylated in vivo, are oriented to the same surface of the complex and consistent with simultaneous association of both proteins with the plasma membrane bilayer. These results support a putative role for activated RhoA in the regulation of guanine nucleotide exchange on inactive RhoA by PRG.
DISCUSSION
The interaction of activated RhoA with the PH domain of its own regulator, PRG, strongly suggests a coordinate feedback function to either attenuate or enhance function. The structural details of this interaction also enhance our understanding of the specific interaction of RhoA with its effectors.
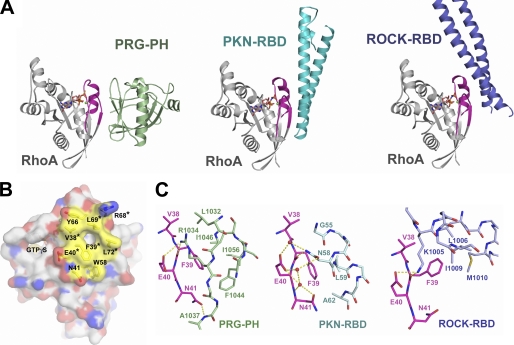
The interface between the PH domain of PRG and activated RhoA bears close similarity to interactions observed in the structures of RhoA complexed to the binding domains of two other RhoA effectors (28, 29). An effector molecule specifically recognizes activated RhoA bound to GTP by direct interaction with the switch regions of RhoA (Fig. 6A). Structural comparison of activated RhoA bound to PRG-PH, Rho kinase (ROCK) (28), or protein kinase N (PKN) (29) reveals that a subset of residues from switch I and switch II form a consensus and mostly hydrophobic binding site for the effectors (Fig. 6B). In all three cases, this site on RhoA complements a hydrophobic patch on the effector. At the center of the consensus binding site are four hydrophobic residues, which include Val-38 and Phe-39 from switch I and Leu-69 and Leu-72 from switch II. Two charged amino acids, Glu-40 from switch I and Arg-68 from switch II, flank the hydrophobic core. Phe-39 from switch I lies at the center of the interfaces. The side chain of Phe-39 adopts different conformations to complement the specific hydrophobic binding pockets of the three effectors (Figs. 3E and 6C). A polar or charged amino acid from the effector (Arg-1034 in PRG-PH, Asn-58 in PKN, or Lys-1005 in ROCK) forms hydrogen bonds with the main chain carbonyl group of Phe-39 and the side chain carboxylate of Glu-40, either directly (PRG-PH and ROCK) or via associated water molecules (PKN). Val-38 from switch I and the two leucine residues from switch II further stabilize the interface by interacting with hydrophobic surfaces on the effectors. Residues outside of this consensus binding site selectively interact with different effectors (Fig. 6). For example, Asn-41 and Trp-58 are involved in the interactions of RhoA with PRG-PH and PKN, but both residues are well exposed in the RhoA-ROCK interface; Tyr-66 from switch II of RhoA does not form direct interactions with PRG-PH but is completely buried when interacting with PKN or ROCK. The docking of Phe-39 into a hydrophobic pocket on effectors and the subsequent reorientation of the side chain of Glu-40 appears to be a key feature for RhoA-effector interactions (Figs. 3E and 6C).
FIGURE 6.
A consensus binding site on RhoA for effector molecules. A, ribbon diagrams depicting the overall structures of RhoA-effector complexes. RhoA is colored gray with switch regions colored purple. GTPγS is depicted as ball-and-stick models and colored as in Fig. 2A. PRG-PH is colored green, the Rho binding domains (RBD) of PKN and ROCK are colored cyan and blue, respectively. B, the solvent-accessible surface of RhoA bound to GTPγS. Nitrogen, oxygen, and carbon atoms are colored blue, red, and gray, respectively. Residues in the consensus binding site for effectors are colored yellow and depicted as ball-and-stick models underneath the surface map. Residues that make contact with all three effectors (PRG-PH, PKN, and ROCK) are marked with asterisks. GTPγS is depicted as ball-and-stick models and colored as in Fig. 2A. C, comparison of the RhoA-effector interface from PH-RhoA (left), PKN-RhoA (middle), and ROCK-RhoA (right) using the same coloring scheme as in panel A.
Residues in the consensus binding site are well conserved among Rho family monomeric GTPases (Fig. 2D) but less well conserved among other members of the Ras superfamily of monomeric GTPases. Phe-39, the key switch I residue that interacts with the hydrophobic patch on effectors of Rho, is replaced by a glutamic acid in Ras and by a valine in Ran. Arg-68 in switch II of Rho is replaced by hydrophobic residues in Ras (alanine), Ran (glycine), and Arf (proline). Also in switch II, the hydrophobic Leu-72 is replaced by glutamine, glycine, or histidine in Ras, Ran, or Arf, respectively. Therefore, this consensus binding site appears to be specific for recognizing effector molecules of the Rho family of small GTPases.
The interaction of activated RhoA with PRG raises the question of its potential physiological impact. It is possible that this association regulates some unknown function of PRG. However, the interaction with the core domains responsible for the nucleotide exchange activity of PRG strongly suggests a modulatory action on this function. The simplest hypothesis is that interaction of activated RhoA with the PRG-PH domain directly influences the guanine nucleotide exchange activity of the molecule, either as a positive or as a negative feedback mechanism. Such a feedback mechanism is observed in Son of Sevenless (SOS), a GEF for Ras (33). Ras-GTP binds to a distinct site on SOS from the site at which RasGEF activity occurs. This binding increases the exchange activity of SOS on Ras by displacing an autoinhibitory domain on SOS. Autoinhibition of PRG has also been reported (34), but the inhibitory element is proposed to be located near the N terminus of the DH domain in PRG, away from the PH-RhoA·GTP interface described here. So far, there is no evidence for such a mechanism as activated RhoA has shown little effect on the exchange activity of PRG-DH·PH domains (Fig. 5E) or full-length PRG (Fig. 5F) for RhoA in solution.
An alternative mechanism for regulation could be enhanced localization of the RhoGEF by binding of the PH domain to RhoA·GTP. There is evidence that targeting of RGS-RhoGEFs to membranes is key to the regulation of the RhoGEF activity (35–37). Translocation of RGS-RhoGEFs could be achieved by the interaction between its rgRGS domain and activated Gα12/13 (15, 38–40). The PDZ domain in LARG and PRG can also contribute to the localization of these RGS-RhoGEFs to membranes by interacting with lysophosphatidic acid receptors (10), plexins (8, 9), or insulin-like growth factors (7). Such translocation is a proposed mechanism for regulation as it brings the enzymes into close proximity with free RhoA (35, 41), which is geranylgeranylated and presumed to be associated with membranes in vivo. Similarly, binding of PRG to activated RhoA in the plasma membrane could lead to further activation of free RhoA, a feed-forward mechanism. This is consistent with the orientation of both activated RhoA and the substrate RhoA, as modeled in Fig. 5 and supported by the isolation of a ternary complex with one DH·PH module simultaneously bound to both nucleotide-free and activated RhoA (Fig. 5, C and D). In this proposed mechanistic paradigm, localization of the RhoGEF by activated RhoA could occur by itself or in conjunction with other localization partners, such as activated G12/13 (via rgRGS domains) or receptors (via PDZ domains). In the first case, activated RhoA could conceivably recruit PRG or other RhoGEFs, regardless of the activating mechanism. In the latter case, activated RhoA would act in concert with a stimulating pathway to enhance or prolong activity; such interactions may be required for full activation of the RGS-RhoGEF in cells.
Other observations support this proposed mechanism. It has been proposed that the PH domain contributes to proper localization of RGS-RhoGEFs by interacting with target(s) at the cell membrane (14). Our study suggests that one of the potential targets interacting with the PH domain of RGS-RhoGEFs at the plasma membrane is activated RhoA. A feature of some PH domains in the Dbl family is to provide a membrane anchor via their interactions with phospholipids; in these proteins, such interaction is required for regulated GEF activity in vivo (42, 43). However, structures of the PH domains from LARG (11) and PRG (12) lack conventional phospholipid binding determinants (44), and the PH domains have not been shown to bind phosphoinositols directly (14) or to localize proteins to membranes by themselves (41, 45). Therefore, the PH domain of RGS-RhoGEFs is likely to be involved in membrane targeting via a novel protein-protein interaction mechanism. The conserved hydrophobic patch on the β5-β6-β7 segment of the PH domain seems to be the key for such interaction, and point mutations of these residues in LARG reduced activation of RhoA by overexpressed LARG in cells (14). Earlier studies ruled out the possibility of binding between the PH domain of RGS-RhoGEFs and Gα12/13, actin, tubulin, or Gβγ (14). Here we show that activated RhoA binds directly to the hydrophobic patch on PH via a RhoA effector-like interface. This hydrophobic patch on the PH domain is highly conserved not only within RGS-RhoGEFs but also among all members of the Lbc subfamily of RhoGEFs. Therefore, the interaction described here between the PRG-PH domain and activated RhoA could be a universal regulatory mechanism employed by all Lbc RhoGEFs for the spatial and temporal regulation of RhoA.
Acknowledgments
We thank Jana Hadas and Stephen Gutowski for superb technical assistance and Diana Tomchick for assistance with data collection. Results shown in this report are derived from work performed at the Argonne National Laboratory, Structural Biology Center, at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grants GM31954 (to P. C. S.) and DK46371 (to S. R. S.). This work was also supported by the Robert A. Welch Foundation Grant I-1262 (to P. C. S.) and a grant from the Alfred and Mabel Gilman Chair in Molecular Pharmacology (to P. C. S.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 3KZ1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- GEF
- guanine nucleotide exchange factor
- DH
- Dbl homology
- PH
- pleckstrin homology
- RGS
- regulators of G protein signaling
- PRG
- PDZ-RhoGEF
- GTPγS
- guanosine 5′-O-[γ-thio]triphosphate
- LARG
- leukemia-associated RhoGEF
- ROCK
- Rho kinase
- PKN
- protein kinase
- TEV
- tobacco etch virus
- GST
- glutathione S-transferase
- ITC
- isothermal titration calorimetry
- mant-GDP
- N-methylanthraniloyl-GDP
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Etienne-Manneville S., Hall A. (2002) Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 2.Rossman K. L., Der C. J., Sondek J. (2005) Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 3.Moon S. Y., Zheng Y. (2003) Trends Cell Biol. 13, 13–22 [DOI] [PubMed] [Google Scholar]
- 4.Vetter I. R., Wittinghofer A. (2001) Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 5.Schmidt A., Hall A. (2002) Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- 6.Sternweis P. C., Carter A. M., Chen Z., Danesh S. M., Hsiung Y. F., Singer W. D. (2007) Adv. Protein Chem. 74, 189–228 [DOI] [PubMed] [Google Scholar]
- 7.Taya S., Inagaki N., Sengiku H., Makino H., Iwamatsu A., Urakawa I., Nagao K., Kataoka S., Kaibuchi K. (2001) J. Cell Biol. 155, 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aurandt J., Vikis H. G., Gutkind J. S., Ahn N., Guan K. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12085–12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swiercz J. M., Kuner R., Behrens J., Offermanns S. (2002) Neuron 35, 51–63 [DOI] [PubMed] [Google Scholar]
- 10.Yamada T., Ohoka Y., Kogo M., Inagaki S. (2005) J. Biol. Chem. 280, 19358–19363 [DOI] [PubMed] [Google Scholar]
- 11.Kristelly R., Gao G., Tesmer J. J. (2004) J. Biol. Chem. 279, 47352–47362 [DOI] [PubMed] [Google Scholar]
- 12.Derewenda U., Oleksy A., Stevenson A. S., Korczynska J., Dauter Z., Somlyo A. P., Otlewski J., Somlyo A. V., Derewenda Z. S. (2004) Structure 12, 1955–1965 [DOI] [PubMed] [Google Scholar]
- 13.Cierpicki T., Bielnicki J., Zheng M., Gruszczyk J., Kasterka M., Petoukhov M., Zhang A., Fernandez E. J., Svergun D. I., Derewenda U., Bushweller J. H., Derewenda Z. S. (2009) Protein Sci 18, 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aittaleb M., Gao G., Evelyn C. R., Neubig R. R., Tesmer J. J. (2009) Cell. Signal. 21, 1569–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z., Singer W. D., Danesh S. M., Sternweis P. C., Sprang S. R. (2008) Structure 16, 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 17.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 19.Collaborative Computational Project Number Four (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 20.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 21.Ramachandran G. N., Sasisekharan V. (1968) Adv. Protein Chem. 23, 283–438 [DOI] [PubMed] [Google Scholar]
- 22.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeLano W. L. (2002) The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC, Portland, OR [Google Scholar]
- 24.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenhaure T. M., Francis S. A., Willison L. D., Coughlin S. R., Lerner D. J. (2003) J. Biol. Chem. 278, 30975–30984 [DOI] [PubMed] [Google Scholar]
- 26.Chikumi H., Barac A., Behbahani B., Gao Y., Teramoto H., Zheng Y., Gutkind J. S. (2004) Oncogene 23, 233–240 [DOI] [PubMed] [Google Scholar]
- 27.Ihara K., Muraguchi S., Kato M., Shimizu T., Shirakawa M., Kuroda S., Kaibuchi K., Hakoshima T. (1998) J. Biol. Chem. 273, 9656–9666 [DOI] [PubMed] [Google Scholar]
- 28.Dvorsky R., Blumenstein L., Vetter I. R., Ahmadian M. R. (2004) J. Biol. Chem. 279, 7098–7104 [DOI] [PubMed] [Google Scholar]
- 29.Maesaki R., Ihara K., Shimizu T., Kuroda S., Kaibuchi K., Hakoshima T. (1999) Mol. Cell 4, 793–803 [DOI] [PubMed] [Google Scholar]
- 30.Wei Y., Zhang Y., Derewenda U., Liu X., Minor W., Nakamoto R. K., Somlyo A. V., Somlyo A. P., Derewenda Z. S. (1997) Nat. Struct. Biol. 4, 699–703 [DOI] [PubMed] [Google Scholar]
- 31.Jezyk M. R., Snyder J. T., Gershberg S., Worthylake D. K., Harden T. K., Sondek J. (2006) Nat. Struct. Mol. Biol. 13, 1135–1140 [DOI] [PubMed] [Google Scholar]
- 32.Hemsath L., Dvorsky R., Fiegen D., Carlier M. F., Ahmadian M. R. (2005) Mol. Cell 20, 313–324 [DOI] [PubMed] [Google Scholar]
- 33.Margarit S. M., Sondermann H., Hall B. E., Nagar B., Hoelz A., Pirruccello M., Bar-Sagi D., Kuriyan J. (2003) Cell 112, 685–695 [DOI] [PubMed] [Google Scholar]
- 34.Zheng M., Cierpicki T., Momotani K., Artamonov M. V., Derewenda U., Bushweller J. H., Somlyo A. V., Derewenda Z. S. (2009) BMC Struct. Biol. 9, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharyya R., Banerjee J., Khalili K., Wedegaertner P. B. (2009) Cell. Signal. 21, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharyya R., Wedegaertner P. B. (2000) J. Biol. Chem. 275, 14992–14999 [DOI] [PubMed] [Google Scholar]
- 37.Jackson M., Song W., Liu M. Y., Jin L., Dykes-Hoberg M., Lin C. I., Bowers W. J., Federoff H. J., Sternweis P. C., Rothstein J. D. (2001) Nature 410, 89–93 [DOI] [PubMed] [Google Scholar]
- 38.Hart M. J., Jiang X., Kozasa T., Roscoe W., Singer W. D., Gilman A. G., Sternweis P. C., Bollag G. (1998) Science 280, 2112–2114 [DOI] [PubMed] [Google Scholar]
- 39.Kozasa T., Jiang X., Hart M. J., Sternweis P. M., Singer W. D., Gilman A. G., Bollag G., Sternweis P. C. (1998) Science 280, 2109–2111 [DOI] [PubMed] [Google Scholar]
- 40.Chen Z., Singer W. D., Sternweis P. C., Sprang S. R. (2005) Nat. Struct. Mol. Biol. 12, 191–197 [DOI] [PubMed] [Google Scholar]
- 41.Wells C. D., Gutowski S., Bollag G., Sternweis P. C. (2001) J. Biol. Chem. 276, 28897–28905 [DOI] [PubMed] [Google Scholar]
- 42.Rossman K. L., Cheng L., Mahon G. M., Rojas R. J., Snyder J. T., Whitehead I. P., Sondek J. (2003) J. Biol. Chem. 278, 18393–18400 [DOI] [PubMed] [Google Scholar]
- 43.Baumeister M. A., Martinu L., Rossman K. L., Sondek J., Lemmon M. A., Chou M. M. (2003) J. Biol. Chem. 278, 11457–11464 [DOI] [PubMed] [Google Scholar]
- 44.Lemmon M. A., Ferguson K. M. (2001) Biochem. Soc. Trans. 29, 377–384 [DOI] [PubMed] [Google Scholar]
- 45.Bhattacharyya R., Wedegaertner P. B. (2003) Biochem. J. 371, 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Read R. J. (1986) Acta Crystallogr. Sect. A 42, 140–149 [Google Scholar]