Abstract
NPR1/GCA (natriuretic peptide receptor 1/guanylyl cyclase A) expression is controlled by several agents, including ANP (atrial natriuretic peptide). After ANP stimulation, NPR1/GCA down-regulates the transcriptional activity of its gene via a cGMP-dependent mechanism. Because we previously identified a cis-acting element responsible for this cGMP sensitivity, we proceed here to explore novel putative protein binding to cGMP-response element (cGMP-RE). Using the yeast one-hybrid technique with a human kidney cDNA library, we identified a strong positive clone able to bind cGMP-RE. The clone was derived from 1083-bp-long cDNA of a gene of yet unknown function localized on human chromosome 1 (1p33.36). We named this new protein GREBP (for cGMP-response element-binding protein). DNA binding assays showed 18-fold higher cGMP-RE binding capacity than the controls, whereas an electromobility shift assay indicated a specific binding for the cGMP-RE, and chromatin immunoprecipitation confirmed the binding of GREBP to the element under physiological conditions. By acting on cGMP-RE, GREBP inhibited the expression of a luciferase-coupled NPR1 promoter construct. In H295R cells, ANP heightened GREBP expression by 60% after just 3 h of treatment while inhibiting NPR1/GCA expression by 30%. Silencing GREBP with specific small interfering RNA increased the activity of the luciferase-coupled NPR1 promoter and GCA/NPR1 mRNA levels. GREBP is a nuclear protein mainly expressed in the heart. We report here the existence of a human-specific gene that acts as a transcriptional repressor of the NPR1/GCA gene.
Keywords: Cyclic GMP (cGMP), DNA-binding Protein, Gene Regulation, Guanylate Cyclase (Guanylyl Cyclase), Peptide Hormones
Introduction
ANP (atrial natriuretic peptide) was discovered 25 years ago by the group of Adolfo J. de Bold (1). Our own team initially demonstrated that cGMP was the signaling pathway of this new hormone (2) acting via particulate guanylyl cyclase (GC)2 (3). Produced in atrial granules, ANP is a small peptide of 28 amino acids that serves as a regulator of blood pressure and blood volume via its natriuretic and vasodilatory actions (1, 4). ANP causes the relaxation of vascular muscles, leading to decreased blood pressure (5, 6). The natriuretic and diuretic effects of ANP on volume homeostasis are achieved through the regulation of water and sodium excretion by the kidneys (7). ANP has also been described as a strong inhibitor of cell growth and hypertrophy (8, 9) as well as a potent antagonist of the renin-angiotensin-aldosterone system via its inhibitory action on aldosterone synthesis by the adrenals (10). Recently, ANP was linked to lipid metabolism because the peptide can stimulate lipolysis through the activation of perilipin and hormone-sensitive lipase (11). In addition to ANP, the natriuretic peptide system includes two other members, brain natriuretic peptide and C-type natriuretic peptide (12, 13) (for a recent review, see Ref. 14).
ANP binds to its receptor and stimulates its intracellular GC domain. The ANP receptor has been named natriuretic peptide receptor 1 or guanylyl cyclase A (NPR1/GCA), a 130-kDa transmembrane protein that converts GTP to the intracellular second messenger cGMP (15, 16). The active NPR1/GCA receptor is a homodimer containing an extracellular ANP-binding domain at its amino-terminal end and an intracellular GC domain at its carboxyl-terminal end (17). The receptor is also able to transduce signals from brain natriuretic peptide but is insensitive to C-type natriuretic peptide stimulation; the latter exerts its effects through a second receptor that shares the same topology as NPR1/GCA, guanylyl cyclase B (GCB or NPR2), which preferentially binds C-type natriuretic peptide (18, 19). Synthesized cGMP molecules then bind to target proteins, including the cGMP-dependent protein kinases (PKG) I and II, the cyclic-nucleotide gated ion channels, and the cyclic nucleotide phosphodiesterases (20), which we initially discovered as cGMP binding activity (21). cGMP also regulates the expression of tumor necrosis factor-α, cyclooxygenase-2, and inducible nitric-oxide synthase (22–27). cGMP is now recognized as a second messenger regulating many cellular functions. Transcriptional regulation by cGMP has been shown to occur both directly and indirectly. Indirect control affects upstream signaling pathways modulating specific targets; e.g. activation of mitogen-activated protein kinases by cGMP controls several transcription factors, such as cAMP-response element-binding protein, ternary complex factor, ATF-2 (activating transcription factor-2), and c-Jun (28–30). Direct control by cGMP involves PKG-dependent phosphorylation of proteins such as cAMP-response element-binding protein, ATF-1, the multifunctional transcription factor TFII-I, and nuclear factor-κB (31–34).
NPR1/GCA expression can be down-regulated by the intracellular accumulation of cGMP under the control of endothelins, glucocorticoids, and angiotensin II and by the natriuretic peptides themselves (15, 35–38). The effect of cGMP on NPR1/GCA consists of a retroinhibition loop that down-regulates the transcriptional activity of the NPR1/GCA gene upon stimulation by ANP or cGMP analogs (39, 40).
Our group has recently reported the existence of a cGMP-response element (cGMP-RE) located in the promoter of NPR1/GCA gene which is responsible for the ANP-NPR1/GCA retroinhibition loop. We have defined a short consensus sequence of 18 bp for this cGMP-RE, AaAtRKaNTTCaAcAKTY, between positions −1372 and −1354 of the NPR1/GCA promoter. The sequence has been initially identified in the rat genome but is also found in mice (95% homology) and humans (75% homology). Deletion of this DNA-responsive element increases the transcriptional activity of the NPR1/GCA promoter by more than 40% (39).
This study identifies the first protein that binds to cGMP-RE and that inhibits the transcriptional activity of the NPR1/GCA gene. We named the novel transcription repressor GREBP (for cGMP-response element-binding protein).
EXPERIMENTAL PROCEDURES
General Methods
All plasmids produced were subcloned in DH5α bacterial strain and purified with a Qiagen maxiprep kit (Mississauga, Ontario, Canada) or Invitrogen's Purelink maxiprep kit (Burlington, Canada). Total RNA was extracted with TRIzol reagent (Invitrogen), followed by DNase I treatment (20 mm Tris, pH 8.3, 50 mm KCl, 2 mm MgCl2, 0.5 unit/ml RNAseOUT, 0.2 unit/μl DNase I), and re-extracted with TRIzol reagent. Semiquantitative reverse transcription-PCR (sqRT-PCR) was performed with a Moloney murine leukemia virus RT kit (Moloney Murine Leukemia Virus Reverse Transcriptase; Invitrogen) and Ambion's Quantum RNA 18 S internal standards (Austin, TX).
Yeast One-hybrid Screening of a Human Kidney cDNA Library
A human kidney cDNA library was obtained from Clontech (Mountain View, CA) as pACT2 plasmids already transformed in the bacterial strain BNN132. The library was titrated, and the colonies were expanded in 30–150-mm Petri dishes to produce maximal amounts of pACT2 plasmids. Plasmids were extracted and purified with the Qiagen Gigaprep kit. One-hybrid screening was conducted with the Matchmaker one-hybrid system from Clontech. Human cGMP-RE (5′-AGGAAATGTACTTCAACATTCTGC-3′) was used as bait. The bait sequence was synthesized in three copies and cloned between the EcoRI and MluI site of pHISi and the EcoRI and SalI site of pLACZi. Plasmids pHISi-hcGMP-RE and pLACZi-hcGMP-RE were introduced in the genome of the yeast strain YM4271 by homologous recombination, and colonies were selected for growth on histidine/uracil-deficient synthetic dextrose medium. The new yeast strain, YM4271pHISipLACZi-hcGMP-RE, was then transformed with human kidney cDNA library plasmids to produce GAL4 activation domain fusion proteins. Colonies, selected for growth at 30 °C on histidine/uracil/leucine-deficient medium, containing 55 mm 3-amino-1,2,4-triazole, were tested for β-galactosidase expression by agarose overlay assays, as described in Ref. 41. Blue colonies were picked up, transferred in his/ura/leu-deficient liquid medium, and allowed to grow at high optical density (∼2). Liquid cultures were then retested for β-galactosidase activity with ortho-nitrophenyl-β-galactoside, a more sensitive assay, as described by the manufacturer; this classified the selected clones by the strength of interaction between the bait and the GALAD fusion protein, thus affecting reporter gene expression. We obtained a strong positive clone expressing high levels of the reporter genes; the pACT2 plasmid was then isolated from crude yeast lysate according to the Clontech protocol and transformed into DH5α Escherichia coli according to standard procedures. pACT2 plasmids were then isolated and sequenced with pACT2-sense (5′-TACCCATACGATGTTCCAGA-3′) and pACT2-antisense (5′-GTGAACTTGCGGGGTTTTTCAGTATCTACGA-3′).
Plasmids
Constructs encoding the GREBP sequence were generated by standard PCR methods with specific primers. pGEX-3X-GREBP was produced with the sense primer 5′-TTGGATCCTTATGAACCATAACCAATAC-3′ and antisense primer 5′-TTGAATTCTGTTAGGGTTGTACGGT-3′; this fragment was cloned in frame with the glutathione S-transferase (GST) gene of pGEX-3X plasmid (GE Healthcare) at the BamHI/EcoRI site. Plasmid peGFP (enhanced green fluorescent protein) (Clontech) was used to produce peGFP-GREBP by introducing, at the KpnI/BamHI site, the GREBP-coding region generated with the sense primer 5′-TTGTCGACGGTACCATGAACCATAACCAATCTACC-3′ and the antisense primer 5′-GTTCTACCGTACAACCCTAAGGATCC-3′. pCDNA1-Neo-GREBP was produced by cloning the coding sequence of the GREBP gene at the BamHI/NotI site of pCDNA1-Neo (Invitrogen). The coding sequence was amplified with the sense primer 5′-CGCCGGATCCGCCACCATGAACCATAACCAATACTACCA-3′ and antisense primer 5′-TTGCGGCCGCTTAGCGTTGTACGGTAGAAC-3′; the sense primer served to introduce the defined Kozak sequence (ccacc) to optimize gene translation, as defined in Ref. 42. For the tagged versions of GREBP, GREBP-StrepTagII, and GREBP-HisTag, antisense primer 5′-AATCTAGATTACTTTTCGAACTGCGGGTGGCTCCAGGCGCCGGCGGGTTGTACGGTAGAACTGCT-3′ was used to add a 3-amino acid linker and a StrepTagII epitope at its carboxyl-terminal end, whereas the antisense primer 5′-AATCTAGATTAGTGGTGGTGATGGTGATGATGGTGGTGATGGGCGCCGGCGGGTTGTACGGTAGAACTGCT-3′ was used to add a 3-amino acid linker and a 10-histidine epitope also at its carboxyl-terminal end. For luciferase experiments, hGCAp-pGL3b was created by amplifying the human NPR1/GCA promoter with the Genome Walker kit from Clontech; the region −2055 to +338 bp of the promoter was then cloned by enzymatic digestion at the KpnI/NcoI site to control the luciferase gene of pGL3b plasmid (Promega, Madison, WI). The cGMP-RE-deleted form of the human NPR1/GCA promoter (hGCAp(ΔcGMP-RE)-pGL3b) was created with hGCAp-pGL3b as template. cGMP-RE was removed by jumping PCR with the sense primer 5′-AGTCTCCTAAAATTCCATATATGTAGTCTGTCTACACAGAATACCT-3′ and antisense primer 5′-GTATTCTGTGTAGACAGACTACATATATGGAATTTTAGGAGACTTG-3′. Jumping PCR allows removal of the cGMP-RE sequence while preserving the structural components of the promoter by cutting down two full turns (20 bp) of the plasmid. Silencing plasmids were generated with pSilencer 2.0-U6 (Ambion) by annealing and cloning inserts for small hairpin RNA (shRNA) at the BamHI/HindIII site. Control plasmid pSilencer 2.0-U6 non-target (NT) contains the upper strand 5′-GATCCCAGTGCTGGTACTTGTACTTCTCTTGAAAGTACAAGTACCAGCACTGTTTTTTTGGAAA-3′ and the lower strand 5′-AGCTTTTCCAAAAAAACAGTGCTGGTACTTGTACTTTCAAGAGAAGTACAAGTACCAGCACTGG-3′. The non-target shRNA sequence was designed for its ability to activate the interference RNA pathways without targeting any gene, allowing rapid screening of gene-specific small interfering RNA, as used successfully by many groups (43–45). The plasmid producing shRNA directed against GREBP was named pSilencer 2.0-U6-shRNA-GREBP and was created with the upper strand 5′- GATCCATGGGCCATTATCGAAGAATTCAAGAGATTCTTCGATAATGGCCCATTTTTTTGGAAA-3′ and the lower strand 5′-AGCTTTTCCAAAAAAATGGGCCATTATCGAAGAATCTCTTGAATTCTTCGATAATGGCCCATG-3′.
Cell Culture, Stimulation, and Transfection
HEK293 (human embryonic kidney) and HeLa (human cervical carcinoma) cells were purchased from ATCC (Manassas, VA) and maintained in Dulbecco's modified Eagle's high glucose medium containing 10% fetal bovine serum, 2% penicillin/streptomycin, 2 g/liter HEPES, and 2.5 g/liter sodium bicarbonate in a 5% CO2 controlled atmosphere. NCI-H295R (human adrenocortical carcinoma, ATCC) cells were maintained in Ham's F-12 medium containing 10% fetal bovine serum, 2% penicillin/streptomycin, 2.0 g/liter HEPES, and 2.5 g/liter sodium bicarbonate. Transfections were performed with Fugene 6 (Roche Applied Science) according to the manufacturer's instructions.
Primer Extension and Full-length mRNA
Primer extension was conducted on HEK293 total RNA. Briefly, 5 μg of HEK293 total RNA was Moloney murine leukemia virus retrotranscribed with 45 fmol of specific radioactive antisense primer 5′-GGAGAGATTTGGTATATG-3′. The length of the extension was then compared with the sequence of pACT2 plasmid containing GREBP cDNA to calculate the number of unknown nucleotides in GREBP. pACT2 plasmid was sequenced by the Thermo sequenase kit (GE Healthcare) and the same radioactive antisense primer. The total length of GREBP mRNA was then confirmed by PCR with the antisense primer 5′-GGTAGGAGTAGCGTGGTAAG-3′ and two sense primers corresponding to regions −5/−25 (5′-GGTTATATCCTTCCCGTACT-3′) or +1/+20 (5′-ATCCCCTGGCCCAACCCGTC-3′) of GREBP cDNA.
GREBP Functional Studies
Tritransfection assays were undertaken to study the effect of GREBP protein on NPR1/GCA promoter activity. The day before transfection, HEK293 cells were seeded in a 12-well plate (7 × 104 cells/well), and plasmids hGCAp-pGL3p or hGCAp(ΔcGMP-RE)-pGL3b, pCDNA1-Neo, and/or pCDNA1-Neo-GREBP (for overexpression experiments), pSilencer 2.0-U6-NT or pSilencer 2.0-U6-shRNA-GREBP (for silencing experiments), and internal control pCMV-βgal (Clontech) plasmids were transfected and allowed to grow for 24 h. The cells were then lysed with Promega reporter buffer. Luciferase activity in the cell extract was quantified by a 20/20n luminometer (Turnerbiosystem, Sunnyvale, CA), and β-galactosidase activity was measured with o-nitrophenyl-d-galactopyranoside as substrate (46). GREBP mRNA overexpression was confirmed by sqRT-PCR with the sense primer 5′-CTTGGTACCGAGCTCGGATC-3′ targeting the 5′-untranslated region (5′-UTR) generated by pCDNA1-Neo-GREBP plasmid and the antisense primer 5′-TTGCGGCCGCTTAGCGTTGTACGGTAGAAC-3′. GREBP-silencing experiments were performed with pSilencer 2.0-U6-shRNA-GREBP and control plasmid pSilencer 2.0-U6-NT. In 6-well plates, HEK293 cells (1.7 × 105 cells/well) were transfected with either 1 μg of NT or shRNA plasmid and allowed to grow for 24 h before lysis. Expression levels were determined by sqRT-PCR with GREBP sense and antisense primers 5′-TCACTTCTGAGTCCCAGAGG-3′ and 5′-GGTAGGAGTAGCGTGGTAAG-3′, whereas NPR1/GCA expression was measured with 5′-ATCCAACTGCGTGGTAGATGGG-3′ and 5′-ATTCGGAAGGAGCGCACAGCAT-3′ as sense and antisense primers.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP was performed as described by Weinmann and Farnham (47). Briefly, 5 × 106 HEK293 cells were seeded in a T-75 flask and transfected with peGFP or peGFP-GREBP and allowed to grow for 12 h. Cross-linking was performed by adding 1% (v/v) formaldehyde in PBS for 10 min at room temperature on a shaker, and the reaction was stopped with 125 mm glycine for 5 min. After washing with ice-cold PBS, cells were allowed to swell for 10 min at 4 °C for nuclei isolation in 5 mm PIPES, pH 8, 85 mm KCl, 0.5% Igepal CA-630 (Sigma), 1 mm phenylmethylsulfonyl fluoride, and 1× complete mini (Roche Applied Science) protease inhibitor. After centrifugation, nuclei were resuspended in nucleus lysis buffer (50 mm Tris-HCl, pH 8, 10 mm EDTA, 1% SDS, 1 mm phenylmethylsulfonyl fluoride, 1× complete mini protease inhibitor) and chromatin was fragmented by sonication using the Misonix sonicator 3000 (4 × 15 s pulse at 90 watts). Debris were removed by centrifugation, and supernatant was cleared with protein A-Sepharose 6MB (GE Healthcare) for 1 h at 4 °C. Beads were pelleted, and their supernatants were diluted in the antibody binding buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, 167 mm NaCl, and protease inhibitors) with 1 μg of normal rabbit IgG (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) catalog no. SC-2027) for the “no antibody” control or with 1 μg of rabbit anti-GFP antibody (Santa Cruz Biotechnology, Inc.; catalog no. SC-8334) and rocked overnight at 4 °C. Before precipitation, one-tenth of the supernatant was kept aside for the PCR of the “input fraction.” Precipitation of the DNA·protein·antibody complexes was done by centrifugation after the addition of protein A-Sepharose for 1 h at 4 °C. The pellets were washed twice with 2 mm EDTA, 50 mm Tris-HCl, pH 8.0, 0.2% N-laurylsarcosyl, and 1 mm phenylmethylsulfonyl fluoride and then four times in 100 mm Tris-HCl, pH 9.0, 500 mm LiCl, 1% deoxycholate, and 1 mm phenylmethylsulfonyl fluoride. Elution of complexes was obtained by the addition of elution buffer (50 mm NaHCO3, 1% SDS), and cross-link was reversed by incubating the samples for 1 h at 37 °C with RNase A, followed by an overnight digestion with proteinase K at 67 °C. The next morning, samples were purified using the Illustra GFX purification column (GE Healthcare) and eluted in 50 μl of Tris buffer. PCR was performed using 1 μl of purified DNA using the sense primer 5′-GGCCTATCGACCACAATT-3′ and antisense primer 5′-TCATTCTGCTTGGATTGGG-3′, respectively, located at −40 and +78 bp of the cGMP-RE. PCR products were analyzed by 2% agarose gels containing ethidium bromide.
In Vitro Binding Assays
pGEX-3X-GREBP or pGEX-3X was transformed into Rosetta bacterial strain (Novagen, Mississauga, Canada) and grown in Terrific Broth (12 g/liter tryptone, 24 g/liter yeast extract, 0.4% glycerol, 2.31 g/liter KH2PO4, 12.54 g/liter K2HPO4) until 600-nm absorbance reached 0.5; proteins were induced with the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside to the liquid culture for 2 h to produce GST or GST-GREBP proteins. Bacterial extracts were prepared as described by Schimtzer et al. (48). 1-ml columns of glutathione-Sepharose 4B (GE Healthcare) were prepared according to the manufacturer's protocols. Bacterial extracts were applied onto the matrix and washed with 5 volumes of PBS to remove unbound proteins. For Western blot experiments, 30 μl of GST or GST-GREBP resin slurry were electrophoresed and transferred onto nitrocellulose according to a standard procedure. The membrane was blocked overnight in 1× TBS and 5% nonfat milk, and the first antibody (anti-GST, dilution 1:8000, catalog no. SC-459, Santa Cruz Biotechnology, Inc.) was added for 1 h in 1× TBS, 5% nonfat milk, and 0.05% Tween 20 at room temperature and followed by three 10-min washes in 1× TBS and 0.05% Tween 20. We used a goat anti-rabbit (dilution 1:15,000, catalog no. SC-2004, 1× TBS, 5% nonfat milk, 0.05% Tween 20) secondary antibody followed again by three 10-min washes (1× TBS, 0.05% Tween 20). Bound protein content was determined by the Bradford method. For the binding assays, resin was poured into the column, and 5 ml of binding buffer (20 mm Hepes, pH 7.9, 35 mm KCl, 1.5 mm MgCl2, 1.5 mm dithiothreitol) containing 10 ng of Klenow 32P-labeled double-stranded rat consensus cGMP-RE (5′-AAAATAGATTTCAACAG-3′) sequence were then added and incubated for 1 h at room temperature. After incubation, the columns were drained and washed twice with 5 ml of binding buffer to remove excess radioactive probes. After washing, a 250-fold excess of cold probe in 5 ml of binding buffer was added and incubated for 1 h. The columns were then drained, and radioactivity was measured at each step.
Electrophoretic Mobility Shift Assays (EMSA)
HEK293 cells were transfected with pCDNA1-Neo-GREBP or control plasmid pCDNA1-Neo. Nuclear extracts were prepared as described previously (39). The 18-bp-long consensus double-stranded human (5′-AAATGTACTTCAACATTC-3′), rat (5′-AAATAGATTTCAACAGTT-3′), or mouse (5′-AAATAGACTTCAACAGTT-3′) cGMP-RE probes were 32P-labeled and purified. For probe conformational studies, we used the 24-bp-long cGMP-RE double-stranded sequence previously defined (39). Human (H24, 5′-AGGAAATGTACTTCAACATTCTGC-3′), rat (R24, 5′-AGAAAATAGATTTCAACAGTTTGC-3′), mouse (M24, 5′-AGAAAATAGACTTCAACAGTTTGC-3′), and two 24-bp-long mutated probes corresponding to the shared sequence between rat and mouse cGMP-RE with nucleotide changes at position 11 (P1, 5′-AGAAAATAGAATTCAACAGTTTGC-3′; P2, 5′-AGAAAATAGAGTTCAACAGTTTGC-3′). 35,000 cpm of probes and 500 ng of poly(dI-dC) were added to 5 μg of nuclear extracts in a total volume of 18 μl of binding buffer (20 mm Hepes, pH 7.9, 35 mm KCl, 1.5 mm MgCl2, 1.5 mm dithiothreitol, 300 μg/ml bovine serum albumin, 12.5% glycerol) and incubated for 30 min at room temperature. Supershift experiments were performed using two tagged constructs of GREBP. 5 μg of nuclear protein extracts of GREBP-StrepTagII-expressing cells were incubated with 1 μg of mouse StreptagII monoclonal antibody (resuspended in PBS) (Novagen (Gibbstown, NJ), catalog no. 71590) or 1 μg of bovine serum albumin/phosphate-buffered saline as control, and, similarly, nuclear extracts of GREBP-HisTag-expressing cells were incubated with 0.5 μg of rabbit His probe antibody (Santa Cruz Biotechnology, Inc., catalog no. SC-803) or with 0.5 μg of normal rabbit IgG (Santa Cruz Biotechnology, Inc., catalog no. SC-2027) as control. Antibodies or controls were added either 10 min before or 20 min after the 32P-labeled probe and 100 ng of poly(dI-dC). Samples were incubated for a total of 30 min at room temperature in 18 μl of supershift binding buffer (10 mm Tris-HCl, pH 7.5, 25 mm NaCl, 0.5 mm MgCl2, 0.5 mm EDTA, 4% glycerol). The samples were resolved by 4.5% non-denaturing PAGE at 4 °C to prevent overheating. The gels were dried and exposed for PhosphorImager scanning.
Fluorescence Microscopy
HEK293 cells were transfected with peGFP or peGFP-GREBP and allowed to grow for 12 h. They were then treated as described previously (49) and viewed with an Olympus IX71 Inverted Microscope at ×40 magnification.
GREBP and NPR1/GCA Expression Analysis
HeLa cells and NCI-H295R cells were seeded in T-25 flasks at 1.0 × 106 and 1.5 × 106 cells, respectively, and grown until near confluence, synchronized by 24-hour serum starvation, and then stimulated with acetic acid (vehicle) or ANP for the times indicated. RNA was extracted to perform sqRT-PCR experiments. GREBP and NPR1/GCA expression was measured by sqRT-PCR with the same previously mentioned primer. The data are expressed as percentages of ANP-stimulated cells compared with vehicle-treated cells.
Multiple Tissue Expression (MTE) Analysis
Human MTE arrays were purchased from Clontech. A 350-bp fragment, corresponding to the coding region of GREBP, was generated by PCR with the sense primer 5′-CGCCGGATCCATGAACCATAACCAATACTA-3′ and antisense primer 5′-TTGCGGCCGCTTAGCGTTGTACGGTAGAAC-3′. This fragment was 32P-labeled with Klenow, hybridized to the membrane according to the manufacturer's instructions, and exposed for 16 h for PhosphorImager scanning.
RESULTS
Yeast One-hybrid Screening of a Human Kidney cDNA Library
With human cGMP-RE as bait, we performed yeast one-hybrid screening of a human kidney cDNA library (Fig. 1A). A total of 60 clones able to grow on minimal media containing 55 mm 3-amino-1,2,4-triazole were tested for β-galactosidase production. One strongly positive clone was found to express high levels of the reporter gene; its β-galactosidase level was much higher than that of other clones (8.19 ± 1.6 versus 0.60 average units of ortho-nitrophenyl-β-galactoside/optical density × min).
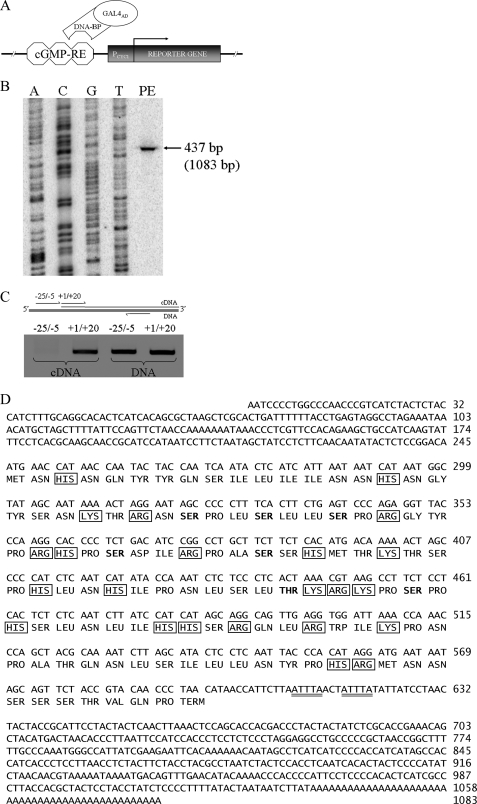
FIGURE 1.
Identification of a cGMP-response element-binding protein. A, yeast one-hybrid screening of a human kidney cDNA library; the putative binding protein interacts with cGMP-RE (present in three copies) to activate the reporter gene via GAL4AD. B, full-length mRNA determination was by primer extension. HEK293 total RNA was retrotranscribed using a radiolabeled primer targeting GREBP mRNA and run beside pACT2-GREBP plasmid sequencing with the same radioactive primer. C, full-length GREBP mRNA was confirmed by RT-PCR with a specific primer targeting upstream and downstream regions of the transcription start site. The positive control was HEK293 DNA, whereas retrotranscribed HEK293 RNA was studied by −25/−5 and +1/+20 RT-PCR. D, nucleotide and amino acid sequences of GREBP. The mRNA destabilization motif is double underlined. Basic residues (framed) constitute 20% of GREBP (23 of 115) and are distributed along the whole sequence. Boldface residues are those with high possibility of serine phosphorylation (>0.800:1.00), and threonine could be strongly phosphorylated (>0.900:1.00) by serine/threonine kinase, including PKG.
Gene, mRNA, and Protein
We then isolated pACT2 plasmid and sequenced the insert. Its sequence (Fig. 1D) revealed a cDNA of 1060 bp containing a 48-bp-long poly(A) tail. Using GenBankTM comparison via BLAST search, we located this gene on human chromosome 1 at position 1p36.33 (as a part of NT_004350.18) near the chromosomal region of the ANP gene (1p36.21). Because the DNA did not appear to belong to any known gene or protein family, we putatively concluded that a novel gene was present, a gene we named GREBP (for cGMP-response element-binding protein).
We performed primer extension assays to determine the total length of mRNA (Fig. 1B); it added an extra 23 bp to our original sequence. We confirmed the sequence of these extra bp by RT-PCR with specific primers for the upstream −25/−5 region and for the +1/+20 region with HEK293 total RNA. No transcript corresponding to the −25/−5 region in RT-PCR was detected, even if we amplified the same region in DNA control experiments (Fig. 1C). These results revealed that the length of endogenous mRNA, including the poly(A) tail, was 1083 bp and that the gene was transcribed as a single exon (Fig. 1D).
Analysis of GREBP cDNA originally fused to the GAL4-AD gene of pACT2 plasmid showed the existence of an open reading frame of 348 bp, coding for a 115-amino acid-long protein with a molecular mass of 13.2 kDa and an isoelectric point of 11.3. The 3′-UTR contained two AU-rich element motifs known to be implicated in rapid mRNA degradation (Fig. 1D, underlined bp) (50). Variation analysis with the National Center for Biotechnology Information single nucleotide polymorphism (SNP) data base revealed 24 SNP distributed within the GREBP gene (Table 1). The nucleotide and amino acid possibilities, in boldface type, are those from our original sequence. Even if population diversity is not provided for given variations, we can assume that SNP located within the coding region can have a strong impact on amino acid content and protein function, whereas those located in the UTR can affect mRNA stability. Protein translation and amino acids analyzed with ExPASy tools indicated that GREBP contains many basic residues (23 of 115) (Fig. 1D, framed residues) characteristic of DNA-binding proteins. The protein sequence contains 6 serine residues that have a high probability of being phosphorylated (>0.800:1) and 1 threonine that could be strongly phosphorylated (>0.900:1) (Fig. 1D, boldface residues) by protein kinases, including PKG (51), reinforcing the idea of the potential role of GREBP in the cGMP signaling cascade.
TABLE 1.
Compilation of SNP located within GREBP gene
SNP analysis of the GREBP transcript was undertaken with the NCBI SNP database. The results are shown as change with associated SNP number, gene position, gene region, and amino acid (AA) variation. Boldface letters are the result of our original GREBP sequence. NA, not applicable.
| Polymorphism | SNP reference number | Gene position | Gene region | AA variation |
|---|---|---|---|---|
| bp | ||||
| A/G | rs41485244 | 321 | Coding | Ser/Gly |
| A/G | rs9701779 | 352 | Coding | Tyr/Cys |
| C/T | rs8179414 | 378 | Coding | Pro/Ser |
| C/T | rs6594029 | 384 | Coding | Pro/Ser |
| C/G | rs55668158 | 397 | Coding | Thr/Arg |
| C/T | rs9701055 | 411 | Coding | His/Tyr |
| C/T | rs7349151 | 432 | Coding | Leu/Phe |
| C/T | rs6594030 | 442 | Coding | Thr/Ile |
| C/T | rs7349153 | 468 | Coding | Leu/Phe |
| A/G | rs9283150 | 486 | Coding | Ser/Gly |
| A/G | rs6594031 | 519 | Coding | Thr/Ala |
| C/T | rs7416152 | 569 | Coding | Silent |
| A/G | rs9782892 | 574 | Coding | Asn/Ser |
| C/G | rs55982362 | 577 | Coding | Ser/Cys |
| C/T | rs56211662 | 594 | 3′-UTR | NA |
| C/T | rs9326619 | 845 | 3′-UTR | NA |
| A/G | rs7411575 | 876 | 3′-UTR | NA |
| C/T | rs55973403 | 910 | 3′-UTR | NA |
| C/T | rs7417504 | 912 | 3′-UTR | NA |
| C/T | rs9283151 | 951 | 3′-UTR | NA |
| A/G | rs6421778 | 996 | 3′-UTR | NA |
| A/G | rs6421779 | 999 | 3′-UTR | NA |
| C/T | rs7340021 | 1016 | 3′-UTR | NA |
| A/G | rs6421780 | 1023 | 3′-UTR | NA |
GREBP Binding to cGMP-RE
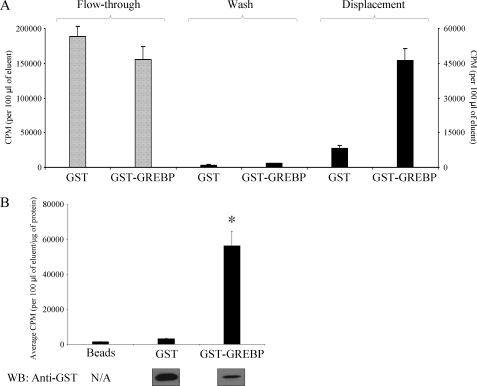
We confirmed the ability of this protein to bind the cGMP-RE by immobilizing a GST-GREBP fusion protein or control GST onto a glutathione-Sepharose 4B matrix. Columns were gravity-drained, and a radioactivity-labeled, double-stranded DNA probe corresponding to the consensus cGMP-RE was applied on the resin. After column washing with binding buffer to remove unbound radioactive probe, bound radioactive probe was eluted from the resin by adding a 250-fold excess of cold probe. GST-GREBP resin retained more radioactivity than the GST column, as can be seen in the flow-through fraction (Fig. 2A, left), whereas elution with the cold probe after washing demonstrated greater specific binding of the probe by the GST-GREBP column (Fig. 2A, right). These experiments were repeated three times with similar results, and, on average, we observed a 5-fold higher binding capacity of GST-GREBP compared with the GST columns (Fig. 2A). We detected 3.5-fold more GST than GST-GREBP protein on the columns, and thus, after correction for the amount of protein present on the columns, we noted an 18-fold higher binding capacity of the cGMP-RE probe for the GST-GREBP protein compared with control GST (Fig. 2B).
FIGURE 2.
DNA binding assays. A, GST or GST-GREBP fusion proteins were immobilized on a glutathione-Sepharose matrix and incubated with 32P-labeled, double-stranded cGMP-RE DNA probe. After draining and washing, a 250-fold excess of cold probe was applied to the matrix to compete for hot probe binding. B, results of three different experiments. Radioactive counts after elution of GST-GREBP, GST, and bead control columns are corrected for protein content (GST and GST-GREBP) and compared. The presence of GST and GST-GREBP proteins on the resin was confirmed by Western blotting (WB) with a specific antibody directed against GST. *, p = 0.012. N/A, not applicable; error bars, S.E.
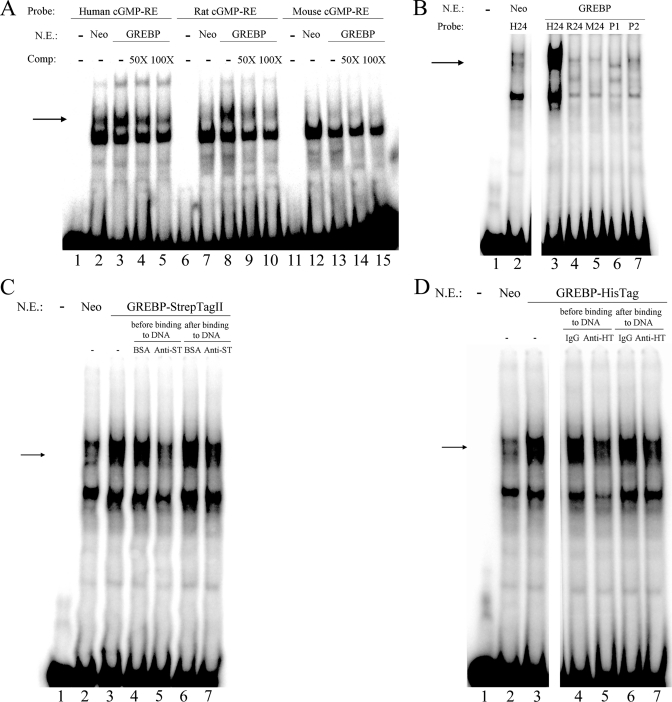
We then proceeded by EMSA to determine the ability of intracellularly expressed GREBP to bind cGMP-RE. HEK293 cells were stably transfected with pCDNA1-Neo-GREBP expressing the full-length coding region of GREBP or the control empty plasmid pCDNA1-Neo. Nuclear proteins were isolated from transfected cells, and EMSA was performed, as described under “Experimental Procedures.” We synthesized double-stranded, 18- and 24-bp-long fragments corresponding to human, rat, and mouse cGMP-RE and tested their potential for interaction with GREBP. Nuclear extracts from HEK293 cells were able to bind the 18-bp-long human and rat but not the mouse probes, as can be seen in Fig. 3A. The arrow in Fig. 3A represents specific interaction between cGMP-RE and overexpressed GREBP protein. Nuclear proteins from cells transfected with control plasmid (Neo) slightly bound the human radioactive probe (Fig. 3A, lane 2), but GREBP overexpression led to an increase in binding capacity (Fig. 3A, lane 3). This augmented binding was specific because it could be competed for by an excess of cold human cGMP-RE; the addition of cold cGMP-RE DNA fragment decreased binding at 50× and returned it to the levels obtained with empty pCDNA1-Neo with a 100-fold excess of cold probe (Fig. 3A, lanes 4 and 5). We saw a similar pattern with the rat probe (Fig. 3A, lanes 6–10). Binding to rat cGMP-RE was increased by GREBP overexpression in HEK293 cells, and the addition of 50- and 100-fold excess of rat cold probe reduced binding of the radioactive probe. We did not observe any band corresponding to GREBP-cGMP-RE interactions with the probe corresponding to the mouse cGMP-RE sequence (Fig. 3A, lanes 11–15). We were intrigued by the different binding capacity of the rat and mouse probes despite only one nucleotide difference and investigated it further. We synthesized longer (24-bp-long) probes corresponding to the human, rat, and mouse sequences and observed that GREBP, although keeping its binding capacity for the human 24-bp-long probe (H24; Fig. 3B, lane 3), was not able to bind the 24-bp-long rat (R24; Fig. 3B, lane 4) and mouse (M24; Fig. 3B, lane 5) probes. Replacing the nucleotide at position 11 with A or G, respectively (P1 and P2), did not restore the binding capacity of the rat or mouse probe (lanes 6 and 7 of Fig. 3B). These results demonstrate differences in binding of GREBP to the cGMP-RE probes.
FIGURE 3.
EMSA of GREBP interacting with cGMP-RE from human, rat, and mouse species. A, 5 μg of nuclear extracts from GREBP or Neo stably transfected HEK293 cells were incubated with 18-bp-long 32P-labeled double-stranded consensus cGMP-RE oligonucleotides corresponding to human, rat, and mouse sequences (39) and were run on non-denaturing PAGE. The figure shows representative EMSA in which the three probes could bind to nuclear proteins (all lanes). The arrow indicates the presence of a protein band visible only with the human and rat RE probes in control (Neo) nuclear extracts (lane 2), which increased in intensity with nuclear extracts from GREBP-transfected cells (lanes 3 and 8). This binding is specific because it could be competed for by 50- and 100-fold excess cold probes (lanes 4, 5, 9, and 10). B, nuclear extracts of GREBP-expressing cells were used to investigate binding to 24-bp-long cGMP-RE oligonucleotides. Binding was visible only with the human 24-bp-long 32P-labeled probe (H24; lane 3) that was more intense after GREBP compared with Neo overexpression (lane 2). The other 24-bp-long probes (rat, R24; mouse, M24; rat/mouse probe with the nucleotide A at position 11, P1; and rat/mouse probe with the nucleotide G at position 11, P2) showed only nonspecific faint bands (lanes 4–7). C, supershift experiments using nuclear extracts of GREBP-StrepTagII-expressing cells. The probe corresponding to the human cGMP-RE sequence was used here. Overexpression of GREBP-StrepTagII protein (lane 3) led to an increased binding over Neo (lane 2). The addition of 1 μg of StrepTagII antibody prior to the 32P-labeled cGMP-RE inhibited DNA-protein interaction (lane 5), whereas the same amount of antibody added after the radiolabeled probe had almost no effect on the binding (lane 7). Bovine serum albumin used as a control added before the probe (lane 4) or after it (lane 6) did not alter the binding compared with GREBP-StrepTagII alone (lane 3). D, overexpression of GREBP-HisTag protein (lane 3) led to an increased binding of the human cGMP-RE probe over Neo (lane 2). The addition of 0.5 μg of HisTag antibody prior to the 32P-labeled probe inhibited DNA-protein interaction (lane 5), whereas the same amount of antibody added after the probe had almost no effect on the binding (lane 7). Normal rabbit IgG added before (lane 4) or after (lane 6) had no effect on probe binding compared with GREBP-HisTag alone (lane 3). N.E., nuclear extracts; Comp., cold competitor; Anti-ST, antibody against StrepTagII; Anti-HT, antibody against polyhistidine tag.
To confirm that GREBP is the protein involved in the DNA·protein complex formation in the nuclear extracts, we performed supershift experiments. For this purpose, we generated two tagged GREBP proteins, each one coupled to a small epitope tag separated by a 3-amino acid linker. Overexpression of these two tagged proteins, GREBP-StrepTagII and GREBP-HisTag, in HEK293 cells led to the same pattern of DNA·protein and DNA·protein·antibody complexes, as shown in Fig. 3, C and D; the addition of antibodies specific to the StrepTagII or the polyhistidine tags inhibited the formation of the cGMP-RE·GREBP complex when added prior to the radiolabeled probe (lanes 5 of Fig. 3, C and D). We also noted that the inhibition of the cGMP-RE·GREBP complex formation was reduced when the antibodies were added after the radiolabeled cGMP-RE probe (lanes 7 of Fig. 3, C and D). By demonstrating that two antibodies directed toward two-tagged GREBP were able to inhibit the DNA·protein complex formation, we confirmed that GREBP is specifically involved in the formation of this complex. Taken together, these experiments demonstrated species-specific binding of GREBP to human cGMP-RE.
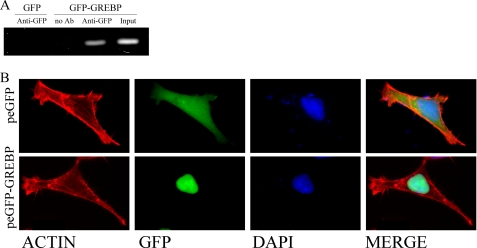
Finally, we investigated the ability of GREBP to bind the cGMP-RE under physiological conditions by performing a ChIP assay. We transfected cells with GFP-GREBP or GFP plasmid, fixed the protein-DNA interaction with formaldehyde, and isolated the nuclear fraction. After immunoprecipitation with IgG or with anti-GFP antibody, we proceeded to PCR amplification of the genomic region spanning the cGMP-RE in the NPR1/GCA promoter. Fig. 4A shows the presence of a band of 178 bp corresponding to the amplified (−1611 to −1433 bp) region of the NPR1/GCA promoter in the input (before immunoprecipitation) as well as in the nuclear fraction of the GFP-GREBP-transfected cells immunoprecipitated with anti-GFP antibody. Controls consisted of rabbit IgG (representing the “no antibody” condition) or nuclear extracts of GFP-overexpressing cells (representing control for GFP binding). No bands could be seen in these latter fractions. These results confirm that GREBP binds to a specific region spanning the cGMP-RE in the genome.
FIGURE 4.
A, ChIP assays for GREBP·cGMP-RE complex. HEK293 cells were transfected with peGFP or peGFP-GREBP for 12 h and fixed with formaldehyde as described under “Experimental Procedures.” GFP-overexpressing cells were used as negative controls. Fixed DNA·protein complexes were immunoprecipitated with an anti-GFP antibody, and normal rabbit IgG served as (no antibody) negative control. Input represented one-tenth of cleared unprecipitated supernatant. B, cellular localization of GREBP in HEK239 cells. HEK293 cells were transfected with peGFP or peGFP-GREBP for 12 h. They were then fixed and stained with phalloidin and 4′,6-diamidino-2-phenylindole. The upper panel shows the whole cell distribution of GFP as control. The lower panels present nuclear localization of the chimeric protein GFP-GREBP. DAPI, 4′,6-diamidino-2-phenylindole.
Cellular Localization of GREBP
We transiently transfected HEK293 cells with peGFP-GREBP to localize GREBP protein. The control experiment, with empty peGFP vector, showed diffuse distribution of GFP (Fig. 4B, upper panels). As expected for a DNA-binding protein, peGFP-GREBP-transfected cells presented a nuclear accumulation of the GFP-GREBP protein overlapping with blue 4′,6-diamidino-2-phenylindole staining (Fig. 4B, lower panel).
GREBP Regulates NPR1/GCA Promoter Activity
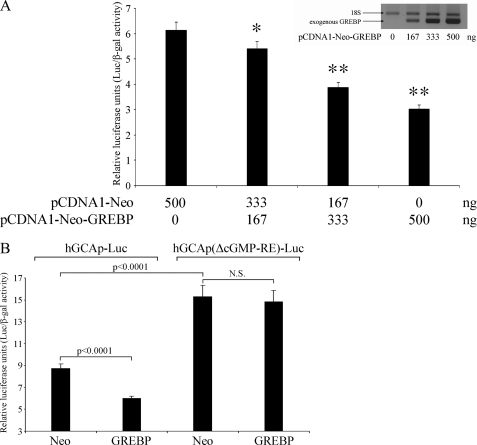
We next examined whether GREBP indeed inhibits the transcriptional activity of the NPR1/GCA promoter by co-transfecting HEK293 cells (7 × 104 cells/well/12 wells) with region −2055 to +338 (cGMP-RE located at −1546 bp) of the human NPR1/GCA promoter, hGCAp-pGL3b, and a plasmid encoding or not encoding GREBP protein, pCDNA1-Neo-GREBP, or pCDNA1-Neo, together with pCMV-βgal as an internal standard plasmid. The cells were transfected with 10 ng of pCMV-βgal, 200 ng of hGCAp-pGL3, and a total amount of 500 ng of pCDNA1 plasmids. Because plasmid quantity is known to affect transfection efficiency, we reduced interexperiment variation by keeping the total amount of pCDNA plasmids constant and varying only the ratio pCDNA1-Neo-GREBP/pCDNA1-Neo, allowing us to capture the specific effect of GREBP. Increasing the ratio of pCDNA1-Neo-GREBP over control plasmid pCDNA1-Neo leads to a dose-dependent increase in the level of GREBP expression and a concomitant decrease in luciferase activity of the NPR1/GCA promoter (Fig. 5A). We next evaluated whether the effect of GREBP on cGMP-RE is specific by deleting cGMP-RE. We transfected HEK293 cells with 10 ng of pCMV-βgal, 500 ng of pCDNA1-Neo plasmids, and 200 ng of hGCAp-pGL3b or the cGMP-RE deletion mutant, hGCAp(ΔcGMP-RE)-pGL3b plasmid. GREBP expression inhibited the intact NPR1/GCA promoter highly significantly (p < 0.0001) by more than 30%, whereas it had no significant effect on the NPR1/GCA promoter without cGMP-RE (Fig. 5B). We also noted that the deletion of cGMP-RE, a negative regulatory element, increased basal NPR1/GCA promoter activity by 175% (Neo versus Neo with and without cGMP-RE, respectively, p < 0.0001). These experiments demonstrated that GREBP expression results in dose-dependent inhibition of luciferase activity of the NPR1/GCA/luciferase-coupled promoter and that GREBP-dependent inhibition of the NPR1/GCA promoter is mediated by cGMP-RE.
FIGURE 5.
NPR1/GCA promoter activity is inhibited by GREBP overexpression, and this inhibition is dependent on cGMP-RE. A, HEK293 cells were co-transfected with 10 ng of pCMV-βgal, 200 ng of hGCAp-pGL3b, and a combination of varying amounts of pCDNA1-Neo and/or pCDNA1-Neo-GREBP for a total plasmid amount of 500 ng/test. GREBP overexpression was confirmed by sqRT-PCR with specific primers targeting GREBP expressed by pCDNA1-Neo-GREBP. B, HEK293 cells were co-transfected with 10 ng of pCMVβ, 400 ng of pCDNA1-Neo or pCDNA1-Neo-GREBP, and 200 ng of hGCAp-pGL3b or the deleted cGMP-RE form of the human NPR1/GCA promoter hGCAp(ΔcGMP-RE)-pGL3b. All values are expressed as means ± S.E. (error bars) of relative luciferase activity from four different experiments performed in triplicate (n = 12). *, p < 0.05; **, p < 0.0001 versus Neo alone.
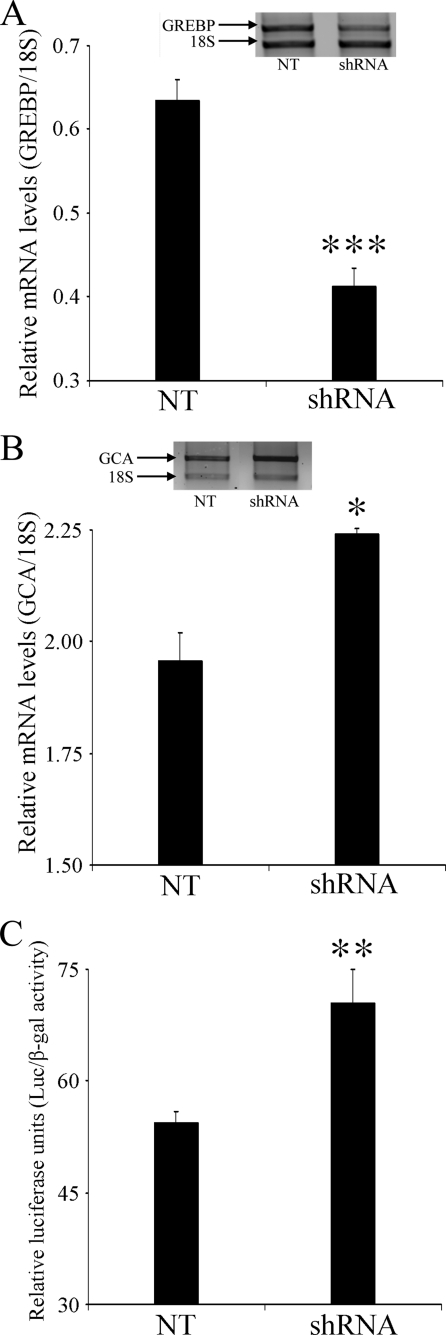
We next looked at the effect of silencing GREBP on NPR1/GCA expression levels. We first transfected HEK293 (1.7 × 105 cells/well/6 wells) with 1 μg of pSilencer 2.0-U6-shRNA-GREBP or control plasmid pSilencer 2.0-U6-NT and let the cells grow for 24 h before RNA extraction and sqRT-PCR. We found that specific shRNA directed to the 781-bp region of the GREBP gene is able to significantly reduce (p < 0.001) (Fig. 6A) its mRNA levels by 35% over the control plasmid. This decrease in GREBP mRNA levels is accompanied by a slight but significant increase (15%) in endogenous NPR1/GCA mRNA levels (Fig. 6B). Moreover, in luciferase experiments, the co-transfection of 10 ng pCMV-βgal with 200 ng of hGCAp-pGL3 and 300 ng of 2.0-U6-shRNA-GREBP or control plasmid pSilencer 2.0-U6-NT elicits a 30% increase in NPR1/GCA promoter activity for GREBP-silenced cells compared with control cells (Fig. 6C). Taken together, these experiments demonstrated that even partial inhibition of GREBP mRNA directly affects the NPR1/GCA promoter by up-regulating its activity, both for the endogenous promoter and for the luciferase-coupled one.
FIGURE 6.
NPR1/GCA transcriptional regulation requires GREBP expression. HEK239 cells were transfected with the pSilencer plasmid producing an NT hairpin sequence (pSilencer 2.0-U6 NT) or shRNA directed against GREBP (pSilencer 2.0-U6-shRNA-GREBP). mRNA levels were analyzed for GREBP expression in A and for NPR1/GCA in B. C, HEK293 cells were co-transfected with 10 ng of pCMVβ, 200 ng of hGCAp-pGL3b, and 300 ng of pSilencer 2.0-U6 NT or of pSilencer 2.0-U6-shRNA-GREBP. All values are expressed as means ± S.E. of relative mRNA levels (A and B) or relative luciferase activity (C) from experiments performed in triplicate. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus NT. Error bars, S.E.
Effects of ANP on NPR1/GCA and GREBP mRNA Expression
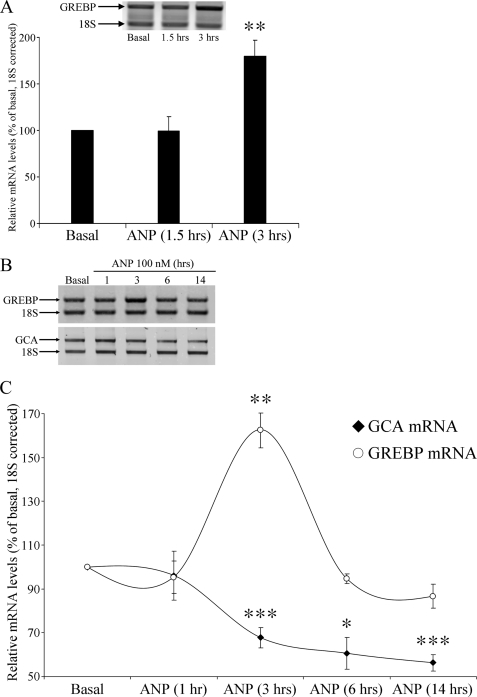
Because ANP is able to regulate the expression of its receptor, we tested the hypothesis that it could also affect GREBP expression levels as a possible intermediate of this action. We incubated HeLa cells with 100 nm ANP and observed a 2-fold increase in GREBP mRNA levels within 3 h of incubation (Fig. 7A). We confirmed this effect in another target cell line for ANP, NCI-H295R cells, which were incubated with a maximal concentration of ANP (100 nm) for the indicated times. After stimulation, they were lysed, and RNA was extracted and subjected to sqRT-PCR assays with expression levels measured as summarized in Fig. 7B. We completed several experiments and observed a 32% decrease in NPR1/GCA mRNA levels at 3 h, which reached 44% at 14 h of ANP incubation (Fig. 7C, circles) concomitantly with a transient but marked increase of GREBP mRNA levels, maximum of 162% at 3 h compared with untreated cells, followed by a return to nearly base-line values by 6 h and to 87% of basal levels at 14 h of incubation with ANP (Fig. 7C, diamonds). These results demonstrate that, in human adrenal cells, ANP binding to its receptor evokes rapid induction of GREBP expression, which, via its binding to cGMP-RE present in the NPR1/GCA promoter, inhibits NPR1/GCA transcription with a concomitant reduction of its mRNA levels.
FIGURE 7.
ANP stimulation influences GREBP and NPR1/GCA expression patterns. A, HeLa cells were treated with 100 nm ANP for the indicated time, and GREBP expression was measured by sqRT-PCR corrected for the 18 S internal standard (from four different experiments). NCI-H295R cells were treated with 100 nm ANP, and gene expression was measured after 1, 3, 6, and 14 h and summarized in B for sqRT-PCR results and compiled in C for graphic representation. All data are from three different experiments performed in duplicate. Values are expressed as means ± S.E. (error bars) relative to the basal state after ANP stimulation. *, p < 0.05; **, p < 0.01; ***, p < 0.005.
GREBP MTE Arrays
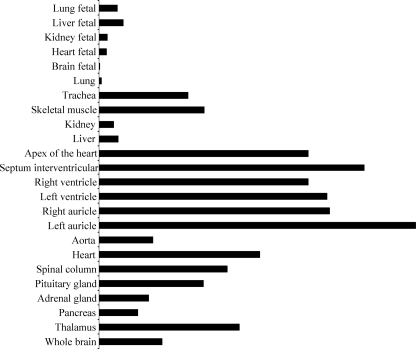
ANP exerts its various biological functions by binding to its receptor, NPR1/GCA. We explored whether the pattern of GREBP expression in humans parallels that of ANP and/or NPR1/GCA in human MTE arrays. Each array contained 75 RNA samples from various adult and fetal tissue sources. Their analysis showed that GREBP expression was lower in fetal than in adult samples. As for ANP, we found that GREBP is predominantly expressed in the atria, ventricles, septum, and apex of the heart (Fig. 8).
FIGURE 8.
GREBP expression profile in human tissue samples. The human MTE array from Clontech is composed of tissues obtained from normal subjects (male/female) from 15 to 66 years old after sudden and/or accidental death. The fetal samples combine 30 embryos, average age 18–30 weeks. GREBP expression is ubiquitous, with the strongest signal in the heart and adult aorta.
DISCUSSION
Because NPR1/GCA plays a key role in transducing various biological functions, including the natriuretic, vasodilatory, and antiproliferative effects of ANP, a better understanding of its mechanism of regulation is essential. Two major processes control NPR1/GCA mRNA and protein levels. GC desensitization involves receptor dephosphorylation after stimulation by its ligand, ANP. This mechanism has been described in several models and is still at the center of many research projects. After short term desensitization, the long term path establishes itself through receptor down-regulation affecting transcriptional regulation of the gene. First described in the mid-1990s, the mechanism involves outcome at the level of the NPR1/GCA gene promoter.
We previously demonstrated the existence of cGMP-RE near the −1500 bp position in mouse, rat, and human NPR1/GCA gene promoters (39). This short element mediates the transcriptional down-regulation of the NPR1/GCA receptor after incubation of the cells with ANP or cGMP analogs. We thus searched for putative binding proteins for this unique cGMP-RE. Here, we report the identification of the first protein that is able to bind cGMP-RE and control NPR1/GCA expression. We present several pieces of evidence in this report to support the hypothesis that GREBP is a transcriptional repressor of NPR1/GCA.
Using the yeast one-hybrid system to screen a human kidney cDNA library, we isolated a clone coding for a protein that binds strongly to cGMP-RE serving as bait. The clone codes for a small protein of previously unknown function that we named GREBP for cGMP-response element-binding protein. The gene is located on human chromosome 1 (1p36.33) close to the ANP gene region (1p36.21). Interestingly, several cross-interacting gene products cluster in the same chromosomal region, leading to the hypothesis that, through evolution, GREBP and ANP genes were kept tightly together because of their strong functional relationship, such as, for example, paraoxonase 1 and 2 genes (52). GREBP mRNA is transcribed as a single exon 1083 bp long and polyadenylated with no splicing process. It has been shown that an elevated concentration of cGMP inhibits the maturation and mRNA splicing process through PKG-dependent phosphorylation of SF1 (splicing factor 1), which prevents prespliceosome assembly (53). Thus, inhibition of the spliceosome machinery should not affect GREBP mRNA, and the protein could reach its target sites, reinforcing the idea that GREBP is involved in a cGMP-signaling pathway. Another interesting fact about this mRNA is the presence of two AU-rich elements at the 3′-UTR, which contributes to rapid mRNA decay. These elements are essential instability determinants for several early response gene mRNAs (54). Tight control of gene expression in response to rapidly changing cellular conditions is important, and, because cGMP concentrations vary rapidly under control of the synthesizing enzymes (GC) and degrading enzymes (phosphodiesterases), retroinhibition loops have to rely on rapid and effective signaling pathways. Another surprising finding about this sequence is the high amount of SNP located within the GREBP gene. In fact, the 1083-bp-long transcript has 24 SNP listed in the National Center for Biotechnology Information SNP data base. However, population diversity is not yet available for these variations. The 5′-UTR does not contain any SNP, probably due to the significant level of regulatory proteins and translation factors targeting this region and making any variation harmful. The coding region contains 14 SNP, which can be as inoffensive as structurally relevant. Indeed, some of the variations can affect amino acid volume (Leu ↔ Phe), side chain charge levels (His ↔ Tyr), global protein structure (Pro ↔ Ser), and phosphate content by destroying or creating phosphorylable residues. SNP located within the 3′-UTR could have an impact on mRNA stability and half-life, but, as for the coding region, further studies will be needed to determine which SNP affect GREBP functions.
Studies of derived amino acid sequence resulting from the translation of GREBP mRNA have led to certain assumptions regarding its possible functions. Indeed, the high number of basic amino acids is representative of DNA-binding proteins (55, 56). ANP-NPR1/GCA signaling results in PKG activation by cGMP and elicits the phosphorylation of downstream proteins. PKG is a serine/threonine kinase. Translational analysis of the gene product by ExPASy tools has revealed putative serine/threonine phosphorylation sites; it is thus possible that cGMP can activate GREBP through PKG-dependent phosphorylation.
The ChIP assays demonstrated that GREBP can bind the cGMP-RE present in the human genome; moreover, this experiment shows that this protein-DNA interaction occurs under physiological conditions and that GREBP is a key protein involved in a (probably) larger complex. Indeed, in our EMSA experiments, the addition of an antibody directed toward the tagged GREBP inhibited its ability to bind the cGMP-RE only when the antibody was added prior to the radiolabeled probe. A similar phenomenon was reported and validated by other groups (57, 58) for other DNA-binding proteins. Interestingly, the addition of antibody after the cGMP-RE probe had almost no effect on the DNA·GREBP complex formation; GREBP may undergo a conformational change upon DNA binding, act within a multiprotein complex, or even homomultimerize around DNA. Further studies are needed to elucidate this mechanism.
Our in vitro DNA binding experiments and EMSA showed that GREBP specifically binds the cGMP-RE. Initially, we observed that GREBP could bind the 18-bp-long human and rat cGMP-RE probes but not the mouse one. This intrigued us because there are several differences in the sequence of the cGMP-RE between humans and rodents but only one nucleotide difference between rat and mouse cGMP-RE, a T/C change, where the C nucleotide is present in both mouse and human cGMP-RE. So, we hypothesized that this lack of interaction between GREBP and the mouse probe was not dependent on the different nucleotide present in the rat/mouse cGMP-RE but on the in vitro conformational structure of the probe. To test this hypothesis, we synthesized longer cGMP-RE (24-bp-long) probes corresponding to the human, rat, and mouse sequences and found that GREBP could bind only the 24-bp-long probe corresponding to the human sequence. Longer probes exhibit different conformations (A-, B-, and Z-conformations, coiled and non-helical) as the number of DNA bp increases, so the difference in binding GREBP between the two rat cGMP-RE probes of different length could be explained by different conformations because nucleotide mutations did not restore the binding capacity of the 24-bp long rat or mouse probes (P1 and P2).
We did not detect any close analog to GREBP in the rat or mouse genome. Several facts can explain the absence of GREBP in the genome from rodent models. Transcriptional controls are different between humans and rodents, being usually more complex in humans; the 5′-UTR of NPR1/GCA is shorter in humans and lacks an Inr element (59). Based on the fact that rat and mouse cells are able to down-regulate NPR1/GCA gene expression, we can assume that a protein with a similar role almost certainly exists in rodents. In fact, GREBP appears to be present in only one other known genome, the chimpanzee. Humans and chimpanzees share a high proportion of their genome; indeed, human ANP mRNA (NM_006172) is 845 bp long and is 98% homologous to 844 bp chimp ANP mRNA (NM_006468). Even if the chimpanzee NPR1/GCA gene has not yet been defined, a region (NW_001229599.1) of its genome is homologous at 99% for 4062 bp of the 4246-bp-long human NPR1/GCA mRNA (NM_000906). These two genes, ANP and NPR1, are co-localized on human and chimpanzee chromosome 1. By comparative genomics, we were able to find a chimpanzee version of human GREBP. This putative gene is also located on chromosome 1 (region NW_001229478.1) and has 89% homology with human GREBP. Translated chimpanzee GREBP protein appears to have the same characteristics as its human counterpart. The hypothetical chimpanzee GREBP is a 113-amino acid long protein, 82% homologous to that of humans, with a molecular mass of 13.2 kDa and an isoelectric point of 11.1. Its protein sequence analysis reveals that 5 serines (>0.750:1) and 1 threonine (>0.603:1) could be phosphorylated by a serine/threonine protein kinase, including PKG. Additional studies are needed to determine if this putative chimp protein is involved in ANP-NPR1/GCA signaling pathways of the chimpanzee. Moreover, we can hypothesize that GREBP is a human-specific gene and may be also present and functional in the chimpanzee ANP-NPR1/GCA pathway. First, it has been proposed that transcription factors/repressors have evolved differently in humans compared with other species (60), and, second, GREBP may belong to a younger age class. These genes, which usually control immunity, perception, and signal transduction, are known to be shorter and intronless and evolved faster than the older classes of gene that are present in every eukaryote cell and run translation, transcription, and replication (61). Evidence that GREBP is really a young gene also came from the fact that only the chimpanzee genome seems to have a homologous GREBP gene, and, in an evolutionary way, the chimpanzee is the human's closest species with 4–5 million years separating the two groups.
Cellular localization studies have shown that GREBP accumulates in the nuclear region of cells. Such compartmentalization is common for proteins associated with transcriptional activity; several studies have revealed that nuclear localization is highly associated with transcription factor function (62, 63).
Our promoter-reporter experiments disclosed that GREBP expression inhibits NPR1/GCA promoter activity in a dose-dependent manner. Moreover, the inhibition caused by GREBP is entirely mediated through cGMP-RE, as demonstrated by the loss of effect of the promoter deletion mutant. In silencing experiments, targeting GREBP with specific shRNA reduced its mRNA levels with consequences on NPR1/GCA regulation. Thus, even a small decrease in GREBP is associated with an increase in endogenous NPR1/GCA transcripts and, concomitantly, augmented activity of the NPR1/GCA luciferase-coupled promoter. Our results also indicate that NPR1/GCA promoter activity can be heightened by either a deletion of cGMP-RE in its promoter (Fig. 5B) and/or by a reduction of GREBP levels (Fig. 6C) with reciprocal regulation and an inverse relationship between GREBP and NPR1/GCA levels.
Endogenous elevations of GREBP mRNA levels are also related to ANP stimulation. Indeed, we were able to show a significant increase of GREBP within 3 h of ANP incubation in two cell lines. This increase correlates with a marked decrease of the NPR1/GCA transcript. NPR1/GCA mRNA still continued to decline after longer ANP incubation but at a much lower pace than with 3-h stimulation. GREBP levels fell after sustained ANP stimulation. Such a decrease in GREBP mRNA is probably under the control of factors, such as decay rate of GREBP mRNA (implication of AU-rich elements), cGMP levels, GREBP partners, trafficking of existing NPR1/GCA receptors, the early responsiveness character of GREBP, and several other factors that remain to be identified. However, we provide strong evidence here for the implication of ANP in the regulation of both GREBP and NPR1/GCA gene expression.
GREBP is widely expressed in human tissues, such as the heart, thymus, brain, and several endocrine glands. A protective role could explain its high level of expression in the heart. Indeed, overexpression in the heart could be protective against overstimulation of NPR1/GCA by ANP, which normally exerts a negative inotropic effect on normal cardiac tissue (64). A constant cardiodepressant effect would be dramatic, so control of GREBP expression through its early and transient response would allow an additional level of fine regulation of NPR1/GCA stimulation.
In conclusion, GREBP emerges as a new transcription repressor acting on unique cGMP-RE present in the NPR1/GCA promoter and controlling the expression of the major receptor of ANP, NPR1/GCA. GREBP (1p33.36) is the third interacting partner in the retroinhibition loop co-located on chromosome 1 with ANP (1p33.21) and NPR1/GCA (1q21–22). Further proteomics and genetics studies are needed to determine the structural domain, protein partners, signaling pathways, and gene mutation that could disable its repressive action.
Acknowledgments
We thank Suzanne Cossette for technical help with the ChIP assays and for the yeast one-hybrid plasmid constructs and Nathalie Bourcier for help with the multiple tissue expression assays.
This work was supported by Canadian Institutes of Health Research Grant MOP-11463 (to J. T.).
- GC
- guanylyl cyclase
- ChIP
- chromatin immunoprecipitation
- GFP
- green fluorescent protein
- eGFP
- enhanced green fluorescent protein
- EMSA
- electromobility shift assay(s)
- cGMP-RE
- cGMP-response element
- GST
- glutathione S-transferase
- MTE
- multiple tissue expression
- PKG
- cGMP-dependent protein kinase(s)
- SNP
- single nucleotide polymorphism(s)
- RT
- reverse transcription
- sqRT
- semiquantitative reverse transcription
- shRNA
- small hairpin RNA
- UTR
- untranslated region
- NT
- non-target
- PIPES
- 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.de Bold A. J., Borenstein H. B., Veress A. T., Sonnenberg H. (1981) Life Sci. 28, 89–94 [DOI] [PubMed] [Google Scholar]
- 2.Hamet P., Tremblay J., Pang S. C., Garcia R., Thibault G., Gutkowska J., Cantin M., Genest J. (1984) Biochem. Biophys. Res. Commun. 123, 515–527 [DOI] [PubMed] [Google Scholar]
- 3.Tremblay J., Gerzer R., Pang S. C., Cantin M., Genest J., Hamet P. (1986) FEBS Lett. 194, 210–214 [DOI] [PubMed] [Google Scholar]
- 4.Inagami T. (1989) J. Biol. Chem. 264, 3043–3046 [PubMed] [Google Scholar]
- 5.Winquist R. J., Faison E. P., Waldman S. A., Schwartz K., Murad F., Rapoport R. M. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 7661–7664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Currie M. G., Geller D. M., Cole B. R., Boylan J. G., YuSheng W., Holmberg S. W., Needleman P. (1983) Science 221, 71–73 [DOI] [PubMed] [Google Scholar]
- 7.Zeidel M. L. (1990) Annu. Rev. Physiol. 52, 747–759 [DOI] [PubMed] [Google Scholar]
- 8.Itoh H., Pratt R. E., Dzau V. J. (1990) J. Clin. Invest. 86, 1690–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh H., Pratt R. E., Ohno M., Dzau V. J. (1992) Hypertension 19, 758–761 [DOI] [PubMed] [Google Scholar]
- 10.Chartier L., Schiffrin E., Thibault G., Garcia R. (1984) Endocrinology 115, 2026–2028 [DOI] [PubMed] [Google Scholar]
- 11.Sengenes C., Bouloumie A., Hauner H., Berlan M., Busse R., Lafontan M., Galitzky J. (2003) J. Biol. Chem. 278, 48617–48626 [DOI] [PubMed] [Google Scholar]
- 12.Sudoh T., Kangawa K., Minamino N., Matsuo H. (1988) Nature 332, 78–81 [DOI] [PubMed] [Google Scholar]
- 13.Sudoh T., Minamino N., Kangawa K., Matsuo H. (1990) Biochem. Biophys. Res. Commun. 168, 863–870 [DOI] [PubMed] [Google Scholar]
- 14.Martel G., Hamet P., Tremblay J. (2010) Mol. Cell Biochem. 334, 53–65 [DOI] [PubMed] [Google Scholar]
- 15.Huo X., Abe T., Misono K. S. (1999) Biochemistry 38, 16941–16951 [DOI] [PubMed] [Google Scholar]
- 16.Potter L. R., Hunter T. (1999) Methods 19, 506–520 [DOI] [PubMed] [Google Scholar]
- 17.Garbers D. L. (1992) Cell 71, 1–4 [DOI] [PubMed] [Google Scholar]
- 18.Garbers D. L., Lowe D. G. (1994) J. Biol. Chem. 269, 30741–30744 [PubMed] [Google Scholar]
- 19.Bennett B. D., Bennett G. L., Vitangcol R. V., Jewett J. R., Burnier J., Henzel W., Lowe D. G. (1991) J. Biol. Chem. 266, 23060–23067 [PubMed] [Google Scholar]
- 20.Beavo J. A., Brunton L. L. (2002) Nat. Rev. Mol. Cell Biol. 3, 710–718 [DOI] [PubMed] [Google Scholar]
- 21.Hamet P., Coquil J. F. (1978) J. Cyclic Nucleotide Res. 4, 281–290 [PubMed] [Google Scholar]
- 22.Cheng H. F., Wang J. L., Zhang M. Z., McKanna J. A., Harris R. C. (2000) Am. J. Physiol. Renal Physiol. 279, F122–F129 [DOI] [PubMed] [Google Scholar]
- 23.Inoue T., Fukuo K., Nakahashi T., Hata S., Morimoto S., Ogihara T. (1995) Hypertension 25, 711–714 [DOI] [PubMed] [Google Scholar]
- 24.Kiemer A. K., Vollmar A. M. (1998) J. Biol. Chem. 273, 13444–13451 [DOI] [PubMed] [Google Scholar]
- 25.Kiemer A. K., Hartung T., Vollmar A. M. (2000) J. Immunol. 165, 175–181 [DOI] [PubMed] [Google Scholar]
- 26.Pérez-Sala D., Cernuda-Morollón E., Díaz-Cazorla M., Rodríguez- Pascual F., Lamas S. (2001) Am. J. Physiol. Renal Physiol. 280, F466–F473 [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto K., Ikeda U., Shimada K. (1997) J. Mol. Cell Cardiol. 29, 2375–2382 [DOI] [PubMed] [Google Scholar]
- 28.Hazzalin C. A., Mahadevan L. C. (2002) Nat. Rev. Mol. Cell Biol. 3, 30–40 [DOI] [PubMed] [Google Scholar]
- 29.Parenti A., Morbidelli L., Cui X. L., Douglas J. G., Hood J. D., Granger H. J., Ledda F., Ziche M. (1998) J. Biol. Chem. 273, 4220–4226 [DOI] [PubMed] [Google Scholar]
- 30.Zaragoza C., Soria E., López E., Browning D., Balbín M., López-Otín C., Lamas S. (2002) Mol. Pharmacol. 62, 927–935 [DOI] [PubMed] [Google Scholar]
- 31.Casteel D. E., Zhuang S., Gudi T., Tang J., Vuica M., Desiderio S., Pilz R. B. (2002) J. Biol. Chem. 277, 32003–32014 [DOI] [PubMed] [Google Scholar]
- 32.Chan S. H., Chang K. F., Ou C. C., Chan J. Y. (2004) Mol. Pharmacol. 65, 319–325 [DOI] [PubMed] [Google Scholar]
- 33.He B., Weber G. F. (2003) Eur. J. Biochem. 270, 2174–2185 [DOI] [PubMed] [Google Scholar]
- 34.Sauzeau V., Rolli-Derkinderen M., Marionneau C., Loirand G., Pacaud P. (2003) J. Biol. Chem. 278, 9472–9480 [DOI] [PubMed] [Google Scholar]
- 35.Garg R., Pandey K. N. (2003) Hypertension 41, 730–736 [DOI] [PubMed] [Google Scholar]
- 36.Katafuchi T., Mizuno T., Hagiwara H., Itakura M., Ito T., Hirose S. (1992) J. Biol. Chem. 267, 7624–7629 [PubMed] [Google Scholar]
- 37.Lanier-Smith K. L., Currie M. G. (1991) Endocrinology 129, 2311–2317 [DOI] [PubMed] [Google Scholar]
- 38.Ye Q., Chen S., Gardner D. G. (2003) Hypertension 41, 675–681 [DOI] [PubMed] [Google Scholar]
- 39.Hum D., Besnard S., Sanchez R., Devost D., Gossard F., Hamet P., Tremblay J. (2004) Hypertension 43, 1270–1278 [DOI] [PubMed] [Google Scholar]
- 40.Cao L., Wu J., Gardner D. G. (1995) J. Biol. Chem. 270, 24891–24897 [DOI] [PubMed] [Google Scholar]
- 41.Duttweiler H. M. (1996) Trends Genet. 12, 340–341 [DOI] [PubMed] [Google Scholar]
- 42.Kozak M. (1987) Nucleic Acids Res. 15, 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Q., Gumireddy K., Schrier M., le Sage C., Nagel R., Nair S., Egan D. A., Li A., Huang G., Klein-Szanto A. J., Gimotty P. A., Katsaros D., Coukos G., Zhang L., Puré E., Agami R. (2008) Nat. Cell Biol. 10, 202–210 [DOI] [PubMed] [Google Scholar]
- 44.Sadat S., Gehmert S., Song Y. H., Yen Y., Bai X., Gaiser S., Klein H., Alt E. (2007) Biochem. Biophys. Res. Commun. 363, 674–679 [DOI] [PubMed] [Google Scholar]
- 45.Lee E. S., Yoon C. H., Kim Y. S., Bae Y. S. (2007) FEBS Lett. 581, 4325–4332 [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J., Fritsch E., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 47.Weinmann A. S., Farnham P. J. (2002) Methods 26, 37–47 [DOI] [PubMed] [Google Scholar]
- 48.Schmitzer A. R., Lépine F., Pelletier J. N. (2004) Protein Eng. Des. Sel. 17, 809–819 [DOI] [PubMed] [Google Scholar]
- 49.El Hader C., Tremblay S., Solban N., Gingras D., Béliveau R., Orlov S. N., Hamet P., Tremblay J. (2005) Am. J. Physiol. Renal Physiol. 289, F1273–F1280 [DOI] [PubMed] [Google Scholar]
- 50.Shaw G., Kamen R. (1986) Cell 46, 659–667 [DOI] [PubMed] [Google Scholar]
- 51.Blom N., Gammeltoft S., Brunak S. (1999) J. Mol. Biol. 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 52.Saeed M., Perwaiz Iqbal M., Yousuf F. A., Perveen S., Shafiq M., Sajid J., Frossard P. M. (2007) Clin. Genet. 71, 238–244 [DOI] [PubMed] [Google Scholar]
- 53.Wang X., Bruderer S., Rafi Z., Xue J., Milburn P. J., Krämer A., Robinson P. J. (1999) EMBO J. 18, 4549–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C. Y., Shyu A. B. (1995) Trends Biochem. Sci. 20, 465–470 [DOI] [PubMed] [Google Scholar]
- 55.Isono K., Yamamoto H., Satoh K., Kobayashi H. (1999) Nucleic Acids Res. 27, 3728–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramji D. P., Foka P. (2002) Biochem. J. 365, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng L. E., Chan F. K., Cado D., Winoto A. (1997) EMBO J. 16, 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu X., Zhu X., Pi W., Ling J., Ko L., Takeda Y., Tuan D. (2005) J. Biol. Chem. 280, 35184–35194 [DOI] [PubMed] [Google Scholar]
- 59.Garg R., Pandey K. N. (2005) Peptides 26, 1009–1023 [DOI] [PubMed] [Google Scholar]
- 60.Gilad Y., Oshlack A., Smyth G. K., Speed T. P., White K. P. (2006) Nature 440, 242–245 [DOI] [PubMed] [Google Scholar]
- 61.Wolf Y. I., Novichkov P. S., Karev G. P., Koonin E. V., Lipman D. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7273–7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stein G. S., van Wijnen A. J., Stein J. L., Lian J. B., Montecino M., Choi J., Zaidi K., Javed A. (2000) J. Cell Sci. 113, 2527–2533 [DOI] [PubMed] [Google Scholar]
- 63.Kopp K., Huang S. (2005) J. Cell Biochem. 95, 217–225 [DOI] [PubMed] [Google Scholar]
- 64.Neyses L., Vetter H. (1989) Biochem. Biophys. Res. Commun. 163, 1435–1443 [DOI] [PubMed] [Google Scholar]