Abstract
Acid sphingomyelinase (ASMase) has been proposed to mediate lipopolysaccharide (LPS) signaling in various cell types. This study shows that ASMase is a negative regulator of LPS-induced tumor necrosis factor α (TNFα) secretion in macrophages. ASMase-deficient (asm−/−) mice and isolated peritoneal macrophages produce severalfold more TNFα than their wild-type (asm+/+) counterparts when stimulated with LPS, whereas the addition of exogenous ceramides or sphingomyelinase reduces the differences. The underlying mechanism for these effects is not transcriptional but post-translational. The TNFα-converting enzyme (TACE) catalyzes the maturation of the 26-kDa precursor (pro-TNFα) to an active 17-kDa form (soluble (s)TNFα). In mouse peritoneal macrophages, the activity of TACE was the rate-limiting factor regulating TNFα production. A substantial portion of the translated pro-TNFα was not processed to sTNFα; instead, it was rapidly internalized and degraded in the lysosomes. TACE activity was 2–3-fold higher in asm−/− macrophages as compared with asm+/+ macrophages and was suppressed when cells were treated with exogenous ceramide and sphingomyelinase. Indirect immunofluorescence analyses revealed distinct TNFα-positive structures in the close vicinity of the plasma membrane in asm−/− but not in asm+/+ macrophages. asm−/− cells also had a higher number of early endosomal antigen 1-positive early endosomes. Experiments that involved inhibitors of TACE, endocytosis, and lysosomal proteolysis suggest that in the asm−/− cells a significant portion of pro-TNFα was sequestered within the early endosomes, and instead of undergoing lysosomal proteolysis, it was recycled to the plasma membrane and processed to sTNFα.
Keywords: Cytokines/Tumor Necrosis Factor, Inflammation, Lipopolysaccharide (LPS), Lysosomes, Secretion, Toll-like Receptors (TLR), TACE, Ceramide, Endosomes, Sphingomyelinase
Introduction
Production of tumor necrosis factor α (TNFα),3 the major mediator of the innate immune response, is tightly regulated by transcriptional, post-transcriptional, and post-translational mechanisms. Dysregulation of TNFα synthesis and/or turnover has been linked to various disease conditions, including rheumatoid arthritis, sepsis, and cancer (1–3). Lipopolysaccharide (LPS), a component of the bacterial cell wall, is a potent inducer of TNFα production and the underlying signaling mechanisms are well understood. LPS binding to its cognitive receptor, MD-2, induces dimerization of the signaling Toll-like receptor-4 (TLR-4) and activation of interleukin-1 receptor-associated kinase-1 (IRAK-1) in a myeloid differentiation factor 88 (MyD88)-dependent manner. Ultimately, the nuclear translocation of nuclear factor κB (4, 5), AP-1, Ets, and Elk-1 (6–8) transcription factors results in multifold induction of TNFα mRNA synthesis. LPS also regulates mRNA stability through AU-rich elements at the 3′-untranslated region of TNFα mRNA (9, 10).
In mice, TNFα mRNA is translated into a 26-kDa precursor protein (pro-TNFα), part of which is immediately N-glycosylated (11). pro-TNFα is selected as a cargo for the Golgin p230-positive vesicles (12) and transported to the plasma membrane via Rab11 recycling vesicles (13, 14). pro-TNFα is integrated in the plasma membrane as a type II membrane protein (15); its ectodomain is cleaved by TNFα-converting enzyme (TACE) (16) and released as a biologically active 17-kDa soluble form (sTNFα). TACE is a member of the a disintegrin and metalloproteinase family of proteases. In addition to TNFα, TACE also processes the two TNFα receptors (p55 and p75), transforming growth factor α, l-selectin, and other secretory proteins (17–20).
The regulation of TNFα mRNA transcription has been extensively studied, and it is well understood. In contrast, evidence for a regulatory role of TACE has emerged only recently. It was reported that a substantial portion of pro-TNFα is not immediately processed by TACE but is rapidly internalized and either degraded in the lysosomes or recycled back to the plasma membrane (21, 22). This observation suggests that TACE activity could be a rate-limiting step in TNFα secretion. Apparently, the activity of TACE is also regulated. Cleavage of an autoinhibitory pro-domain is the first step of TACE activation (23). The active form of TACE is localized primarily at the plasma membrane (24), although activity toward TNFα has also been reported in the cytosol of some cells (25). At the plasma membrane, TACE activity depends upon compartmentalization of the protein in the ordered lipid domains (26–28). It is controversial, however, whether such localization is linked to stimulation or inhibition of activity. Whether TACE localization to the lipid rafts brings the enzyme close to its substrates or induces conformational change is also unknown. In addition, certain stimuli, like phorbol 12-myristate 13-acetate (24) and LPS (29), were shown to induce internalization of TACE, a phenomenon with unknown functional significance.
Acid sphingomyelinase (ASMase) is a lipid hydrolase that converts sphingomyelin to ceramide, a bioactive second messenger molecule. ASMase is activated in response to stimulation with LPS, TNFα, γ-irradiation, CD95, and interleukin-1β (IL-1β) (30–34). ASMase is found mostly in the endosomal/lysosomal compartment, and agonist-dependent translocation of the protein to the plasma membrane is documented in response to CD95 (31), UV irradiation (30), and phorbol 12-myristate 13-acetate (35). A secretory form of the enzyme can be produced through alternative post-translational modification (36) and is activated by inflammatory mediators like LPS, TNFα, and IL-1β (37, 38). Both ASMase and secretory sphingomyelinase are implicated in the formation of ceramide-enriched lipid domains and receptor clustering (39). Mice deficient in ASMase are protected against some of the detrimental effects of CD95 (39), LPS (40), and TNFα (41).
More recent studies in mice have linked ASMase deficiency to increased susceptibility to viral and bacterial infections. ASMase-deficient macrophages exhibit a slow rate of elimination of pathogens like Listeria monocytogenes (42, 43), alphavirus Sindbis (44), and Pseudomonas aeruginosa (45), caused by a protracted phago-lysosomal fusion and membrane budding. A defect in Rab4 recycling pathway has been identified in dermal fibroblasts of patients with Niemann-Pick disease, type A, a condition characterized with low ASMase activity (46). Together, these studies suggest that in addition to a role in signaling, ASMase might be involved in regulation of membrane fusion by regulating membrane fluidity and/or curvature.
This study investigates the role of ASMase in regulation of TNFα production. Our results show that ASMase activity is a novel regulator of post-translational processing of TNFα in macrophages, which impacts TNFα secretion after LPS stimulation. We further found that ASMase-derived ceramide is inhibitor of TACE activity and also affects the intracellular fate of pro-TNFα.
EXPERIMENTAL PROCEDURES
Materials
LPS (Escherichia coli, serotype 026:B6), ammonium chloride, bacterial sphingomyelinase (Staphylococcus aureus), and Brewer thioglycollate broth were purchased from Sigma. The enhanced chemifluorescent kit was from GE Healthcare. Fluorogenic peptide substrate III and ELISA kit came from R & D Systems (Minneapolis, MN). TAPI-1 (N-(R)-[2-(hydroxyaminocarbonyl) methyl]-4-methylpentanoyl-l-naphthyl-alanyl-l-alanine, 2-aminoethyl amide; TNFα protease inhibitor-1) was from Calbiochem. C2-ceramide, C16-ceramide, and d-erythrosphingosine were from Avanti Polar Lipids (Alabaster, AL). Antibodies were from the following manufacturers: anti-phospho-ERK1/2, anti-IRAK-1, anti-EEA1, and anti-TACE were from Santa Cruz Biotechnology (Santa Cruz, CA); anti-TNFα used for Western blotting was from Cell Signaling (Beverly, MA); anti-TNFα used for immunofluorescence was from R & D Systems (Minneapolis, MN); anti-cyclophilin A was from Cell Signaling (Beverly, MA); and anti-β-actin and alkaline phosphatase-conjugated secondary antibodies were from Sigma.
Animals
A colony of ASMase-deficient (asm−/−) mice (47) was maintained in the Association for Assessment and Accreditation of Laboratory Animal Care-approved animal facility of University of Kentucky Medical Center by breeding heterozygous (asm+/−) mice. After weaning, the mice were genotyped and placed on a standard NIH-31 diet and 12-h light/dark cycle in microisolation. Litter-matched asm+/+ and asm−/− mice were injected intraperitoneally with LPS (5.8 mg/kg body weight) or an equivalent volume of 150 mm NaCl. Blood was collected via retro-orbital bleeding, and serum was obtained in serum separator tubes.
Cell Cultures and Treatments
Peritoneal macrophages were elicited from 8-week- old asm+/+ and asm−/− mice. Cells were plated in Dulbecco's modified Eagle's medium, supplemented with 2% fetal bovine serum, and 100 units/ml penicillin/streptomycin mix (Invitrogen) on 6-well plates at density ranging from 0.3 × 106 to 2 × 106 cells/well. Macrophages were allowed to adhere for 3 h in a 37 °C humidified 5% CO2 incubator, and nonadherent cells were removed by aspiration. Treatments with LPS, C2-, C16-ceramide, bacterial SMase, ammonium chloride, and TAPI-1 were done at 19 h after plating. Endocytosis inhibitors (Dynasore, chlorpromazine and monodansylcadaverine) were added 1 h after LPS stimulation.
SDS-PAGE and Western Blotting
Conditioned medium was collected from the wells (2 × 106 cells/well), cleared by centrifugation, and concentrated in Amicon Ultratubes with a cutoff of 10 kDa. Ten microliters from the concentrated medium were subsequently used for Western blotting. Cells were lysed on ice for 30 min in a buffer containing 1 mm EDTA, 1% Triton X-100, 1 mm Na2VO4, 1 mm NaF, and protease inhibitor mixture (1:200) in 10 mm Tris-HCl, pH 7.4. Cell debris was removed by centrifugation at 16,000 × g for 10 min at 4 °C. Proteins were resolved by 10% SDS-PAGE, transferred to polyvinylidene difluoride membranes, and detected using the antibodies described above. Protein-antibody interactions were visualized using ECFTM substrate on a StormTM 860 (GE Healthcare) scanning instrument and analyzed using ImageQuant 5.0 software.
ELISA
The levels of TNFα, IL-1β, and IL-6 were determined by ELISA according to the manufacturer's protocol. The absorbance was measured at 450 nm on a Vmax kinetic microplate reader with Softmax pro software.
TACE Activity Assay
Macrophages were resuspended in 500 μl of 25 mm Tris, pH 8.0, and homogenized by passing through 26-gauge needle several times. A total of 5–10 μg of protein was incubated with fluorogenic substrate III (7-methoxycoumarin-PLAQAV-N-3-(2,4-dinitrophenyl)-l-2,3-diaminopropionyl-RSSSR-NH2) (10 μm) in 25 mm Tris, pH 8.0 (100 μl final volume). The fluorescence emitted from the cleavage product was quantified by spectrofluorometry using excitation and emission wavelength of 320 and 405 nm, respectively. TACE-specific activity was expressed as nanomoles of hydrolyzed substrate per min per mg of protein after calibration for the quantum yield of the fluorogenic substrate.
RNA Isolation, Reverse Transcription, and Quantitative Real Time PCR Analysis
Total RNA was isolated with RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. First-strand cDNA synthesis was performed using random hexamers (Roche Applied Science) and Superscript IITM reverse transcriptase (Invitrogen). TNFα mRNA levels were quantified using TaqManTM mouse TNFα (Mm00443258_m1) and glyceraldehyde-3-phosphate dehydrogenase gene expression assays (Applied Biosystems, Foster City, CA) on ABI PerkinElmer Life Sciences Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA).
Indirect Immunofluorescence
Cells were fixed in cold methanol (permeabilized cells) for 5 min or in 3% formaldehyde (nonpermeabilized cells) for 30 min. After blocking the nonspecific binding with the appropriate serum (2%), proteins of interest were labeled with anti-TACE, anti-TNFα, or anti-EEA1 polyclonal antibodies at a dilution of 1:50, 1:50, and 1:100, respectively. The immune complexes were visualized with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG or TRITC-conjugated bovine anti-goat IgG. In the co-localization studies, the primary antibodies were added sequentially.
Statistical Analysis
Significant changes in multiple comparisons were determined by two-way ANOVA with Bonferroni post test analysis. One-way ANOVA and unpaired t test were also used whenever appropriate. In the figures, an asterisk is used to indicate the significance of main effect (treatment or genotype) and a pound symbol indicates the significance of interaction effect.
RESULTS
Genotype-specific Differences in Cytokine Release in Vivo
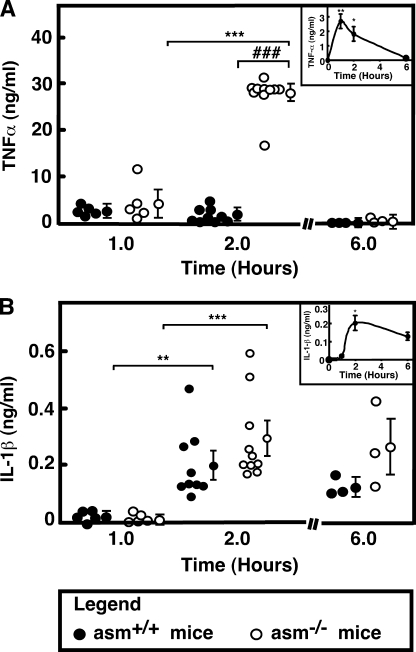
To determine the effect of ASMase deletion on TNFα production, litter-matched asm+/+ and asm−/− mice were injected with saline or LPS (5.8 mg/kg of body weight). The administration of LPS led to a slight elevation in body temperature and lack of tonus within a couple of hours. In all animals, these symptoms disappeared after ∼6 h. TNFα and IL-1β were undetectable in saline-injected mice of both genotypes, although the basal concentration of IL-6 ranged between 2 and 4 ng/ml (data not shown). The serum levels of all measured cytokines increased in response to LPS. In the asm+/+ mice, TNFα levels reached maximum at 1 h after the LPS stimulation (Fig. 1A, inset), although the levels of IL-1β (Fig. 1B, inset) and IL-6 (data not shown) reached maximum at 2 h. In the serum of ASMase-deficient mice, however, TNFα reached concentrations that were 10–15-fold higher than those measured in the serum of asm+/+ mice (Fig. 1A). The levels of IL-1β (Fig. 1B) and IL-6 (data not shown) were similar for both genotypes. These data suggest that ASMase deficiency had a specific effect on the regulation of TNFα concentration in vivo.
FIGURE 1.
Cytokine levels in LPS-injected asm+/+ and asm−/− mice. Mice were injected intraperitoneally with LPS (5.8 mg/kg body weight) or saline. Serum was collected at the indicated times, and the levels of TNFα (A) and IL-1β (B) were measured by ELISA. Data for LPS-injected asm+/+ (filled circles) and asm−/− (open circles) are shown. The levels of the cytokines in saline-injected animals were below the detection levels. Data for individual animals are shown and are the average of identical measurements done in triplicate. The group average ± S.E. is shown on the side. The significance of the main effects of genotype and LPS (***, p < 0.001; **, p < 0.01, *, p < 0.05) and the interaction effect (###, p < 0.001) are based on two-way ANOVA with Bonferroni post-test analysis. The insets are representation of the data from asm+/+ mice on a smaller scale.
Genotype-specific Differences in the Macrophage Response to LPS Stimulation
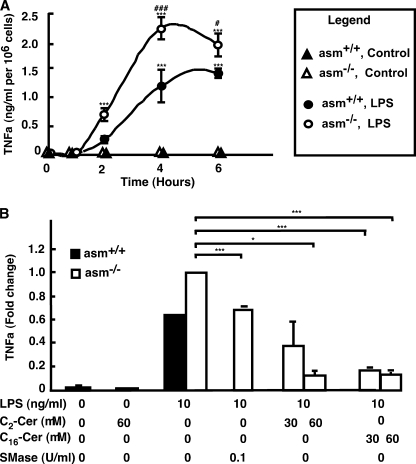
To test whether ASMase deficiency affected the production of TNFα, peritoneal macrophages were isolated from asm+/+ and asm−/− animals, cultured, and stimulated with LPS. The release of TNFα in the medium was monitored by ELISA. As anticipated, treatment with LPS induced TNFα secretion, which reached maximum at 4 h after stimulation (Fig. 2A). As compared with the stimulation observed in vivo, this response was slightly delayed, which might be due to the absence of the LPS-binding proteins and CD14 in vitro. However, TNFα levels were up to 2-fold higher in asm−/− macrophages as compared with asm+/+ cells. No statistically significant differences were detected in respect to the levels of IL-1β or prostaglandin E2 (data not shown). Because the latter is a negative regulator of TNFα secretion (48–50), these results suggest that the differences in TNFα secretion cannot be explained by differences in prostaglandin E2 production. Similarly, the simultaneous treatment with IL-6, which also can suppress TNFα synthesis, had no effect on the genotype-related differences (data not shown). However, supplementation of asm−/− cells with ceramide, the product of ASMase catalytic activity, either by treatment with exogenous SMase or by adding ceramides, inhibited sTNFα secretion in a dose-dependent manner (Fig. 2B). Together, these results indicate that ASMase and ceramide may regulate specific step(s) in TNFα production.
FIGURE 2.
Effect of ASMase, bacterial SMase, and ceramide on TNFα production in primary macrophages. TNFα levels were measured by ELISA in the medium of peritoneal macrophages from asm+/+ (filled symbols) or asm−/− (open symbols). A, cells treated with LPS (100 ng/ml, circles) or phosphate-buffered saline (triangles) for the indicated times. B, cells treated with LPS for 4 h in the presence of bacterial SMase, C2-, C16-ceramide at the indicated concentrations. Control cells were treated with 0.1% ethanol. Data are means ± S.E., n = 3 (A) or n =2 (B). Statistical significance of the main effect (***, p < 0.001) and the interaction effect (###, p < 0.001; #, p < 0.05) was calculated by two-way ANOVA. The statistical significance of bacterial SMase and ceramide effects was calculated by one-way ANOVA (*, p < 0.05; ***, p < 0.001).
ASMase Deficiency Has No Effect on Mitogen-activated Protein Kinase (MAPK) Activation and TNFα mRNA Production
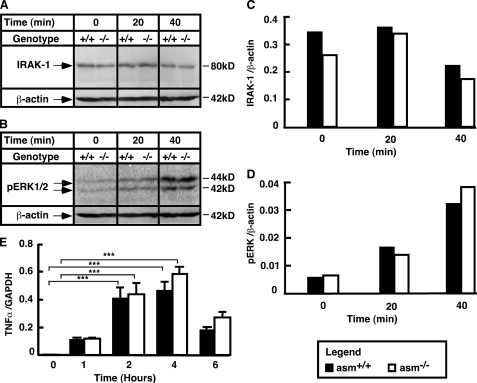
To determine whether the differences in TNFα secretion are related to differences in the LPS-signaling pathway, the activation pattern of proteins proximal to the LPS receptor was first examined. IRAK-1 is a critical mediator of LPS signaling, which becomes phosphorylated and degraded upon stimulation of the receptor. The rate of its degradation determines the magnitude of the LPS response (51, 52). However, the IRAK-1 degradation rates (Fig. 3, A and C), together with the pattern of stimulation of the ERK (Fig. 3, B and D) and c-Jun N-terminal kinase (JNK) phosphorylation (data not shown), were all similar between the two genotypes. Consequently, the magnitude of stimulation of TNFα mRNA transcription was also unaffected by the ASMase deficiency (Fig. 3E).
FIGURE 3.
Stimulation of IRAK-1, ERK, and TNFα mRNA in asm+/+ and asm−/− macrophages. Peritoneal macrophages from asm+/+ and asm−/− mice were treated with LPS (10 ng/ml) for the indicated times. A–D, activation of IRAK-1 (A and C) and ERK1/2 (B and D). Analyses were done using Western blotting and antibodies against IRAK-1 and phosphorylated ERK1/2. Data are representative of three independent experiments. β-Actin levels were used to control for uniform loading. E, stimulation of TNFα mRNA. mRNA levels were determined by real time PCR. A glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA level was used for normalization. Data are presented as mean ± S.E. of three independent experiments. Statistical significance of the treatment effect was calculated (***, p < 0.001) based on two-way ANOVA.
Kinetics of Post-translational Processing of TNFα in Primary Macrophages
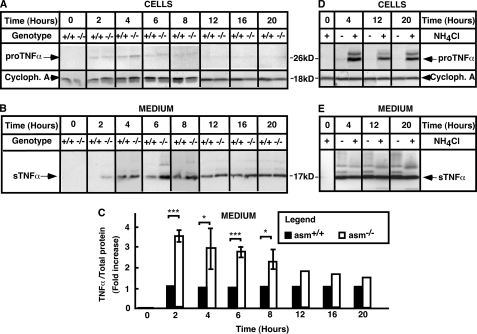
To study the post-translational processing of TNFα, the conversion of pro-TNFα to sTNFα was monitored by Western blotting. As anticipated, neither pro-TNFα nor sTNFα forms were detected in nonstimulated macrophages from both genotypes (Fig. 4, A and B). In asm+/+ macrophages, substantial accumulation of sTNFα in the medium was noticeable at 4 h after stimulation and continued for up to 20 h. However, in asm−/− cells, sTNFα was noticeable as early as 2 h of post-stimulation (Fig. 4B). In an excellent agreement with the differences seen by ELISA, the sTNFa levels were significantly higher in asm−/− cells than in asm+/+ cells, particularly at 2, 6, and 8 h after stimulation (Fig. 4C). The genotype-related differences seemed to decrease after 12 and 16 h.
FIGURE 4.
Post-translational processing of TNFα. Peritoneal macrophages were isolated from asm+/+ and asm−/− mice (A–C) or from asm+/+ mice (D and E) and treated with LPS (10 ng/ml) for the indicated times. Ammonium chloride (10 mm) was added to the culture medium 1 h after stimulation with LPS as indicated. TNFα levels were determined in cell extracts (A and D) or medium (B, C, and E) by Western blotting. Cyclophilin A (Cycloph. A) levels were used to control for uniform loading. The data shown in A and B were compiled from three different experiments, each covering specific time intervals of stimulation with LPS and each repeated at least twice using different macrophage preparation from two to three mice in each genotype. For quantification purposes, the data were normalized for the respective values in asm+/+ macrophages and are means ± S.D. ***, p < 0.001; *, p < 0.05.
Surprisingly, it was difficult to observe pro-TNFα in macrophage cell lysates, even when 100 μg of cellular protein were loaded in a single lane (Fig. 4A). In contrast, pro-TNFα was easily detected in lysates from the mouse macrophage cell line, RAW264.7 cells (supplemental Fig. S1), indicating that the antibody recognized well the intracellular form of TNFα. These differences suggest that the kinetics of TNF post-translational processing in primary macrophages and macrophage-like cell lines may be fundamentally different.
The levels of pro-TNFα in asm+/+ primary macrophages were substantially increased following co-treatment with ammonium chloride, which blocks the activity of the lysosomal hydrolyses (Fig. 4D). These effects are in a good agreement with earlier observations of lysosomal degradation of newly synthesized pro-TNFα (22). Our data further suggest that in macrophages almost half of the newly produced pro-TNFα undergoes such degradation. Because the treatment with ammonium chloride had no effect on the levels of sTNFα in the medium (Fig. 4E), these observations support the conclusion that the activity of TACE is a rate-limiting factor for the processing of pro-TNFα to its mature secretory form, and the nonprocessed pro-TNFα is cleared through lysosomal proteolysis.
Characterization and Regulation of Macrophage TACE Activity
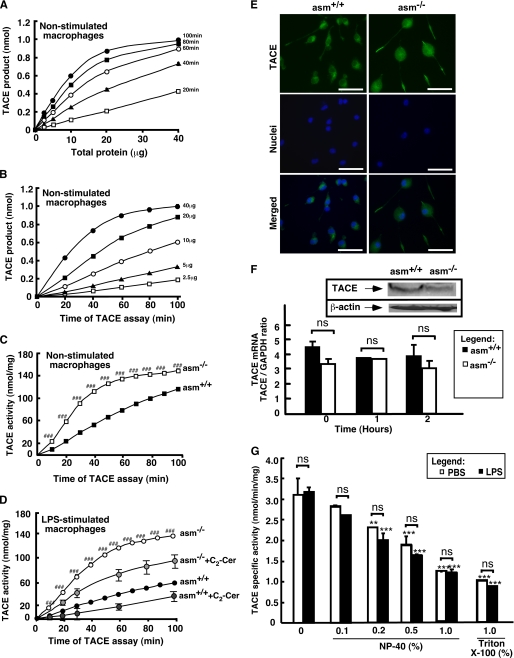
To test directly whether the increased TNFα production in asm−/− was due to higher TACE activity, the latter was measured in vitro using fluorogenic substrate containing the TNFα cleavage site. Preliminary experiments showed that inclusion of detergent in the assay suppressed more than 90% of TACE activity measured in detergent-free cell homogenates (data not shown and Fig. 5G); therefore, all assays were done under detergent-free conditions. After identifying the range of linearity (Fig. 5, A and B), the TACE activity in asm+/+ and asm−/− cells was compared. Both LPS- and control-treated asm−/− macrophages exhibited 2–3-fold higher activity as compared with their respective asm+/+ control cells (Fig. 5, C and D). These differences were not due to differences in TACE mRNA or protein content (Fig. 5F). Real time PCR analyses of TACE mRNA in macrophages from asm+/+ and asm−/− mice revealed no genotype-related differences. Furthermore, LPS treatment also had no effect on TACE mRNA expression. However, TACE protein levels in primary macrophages were too low to be reliably quantified by Western blotting. Instead, two established fibroblast cell lines from a patient with Niemann-Pick disease (a hereditary sphingolipid storage disorder due to loss-of-function mutation in ASMase gene) and a healthy control were used. As anticipated, Niemann-Pick fibroblasts had very low ASMase activity; however, similar to asm−/− macrophages, they exhibited 2–2.5-fold higher TACE activity than cells with functional ASMase (data not shown). Despite these differences in activity, the TACE protein levels were similar in the two fibroblast lines (Fig. 5F, inset), suggesting that the lack of functional ASMase can lead to increased TACE activity without affecting TACE mRNA or protein levels.
FIGURE 5.
TACE activity, expression, and subcellular localization in asm+/+ and asm−/− cells. A and B, test for linearity of TACE activity assay with protein and time. TACE activity was measured in vitro in detergent-free lysates from RAW264.7 cells. C and D, effect of ASMase deficiency on activity of TACE. asm+/+ and asm−/− macrophages were treated with saline (C) or with a combination of C2-ceramide (Cer) (60 μm) or vehicle (0.1% ethanol) (D). Statistical significance (p < 0.001) of the genotype-based difference in TACE activity is designated ###. TACE activity was measured in detergent-free lysates using fluorigenic substrate, and the fluorescence was monitored over time on a microplate reader. The data are means ± S.D. (n = 3). E, subcellular localization of TACE. TACE was visualized by indirect immunofluorescence in permeabilized asm+/+ and asm−/− macrophages using antibodies against mouse TACE and fluorescent microscopy. Hoechst 33258 was used for staining the nuclei. Scale bar represents 20 μm. F, effect of ASMase deficiency on TACE mRNA and protein expression. TACE mRNA levels were determined by real time PCR in asm+/+ and asm−/−macrophages that were treated with phosphate-buffered saline or LPS for the indicated times. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA level was used for normalization. Data are presented as means ± S.E. of three independent experiments. Inset, Western blot analysis in human fibroblasts from healthy individuals or patients with Niemann-Pick disease, type A. β-Actin levels were used to control for uniform loading. G, effect of nonionic detergents on TACE activity. asm+/+ macrophages were treated with LPS (10 ng/ml) (filled bars) or vehicle (open bars) for 4 h. The activity of TACE was measured using fluorigenic substrate and cell lysates (5 μg of protein per assay) in the absence or presence of Nonidet P-40 (NP-40) or Triton X-100 at the indicated concentrations. Data shown are average ± S.D., n = 3. Statistical significance of detergent and LPS effects was determined by two-way ANOVA. Asterisk designates the main effect of detergent (***, p < 0.001; **, p < 0.01), and ns (nonsignificant) refers to the significance of the interaction effect.
Treatment with bacterial SMase to elevate endogenous ceramide content in the macrophages inhibited TACE activity by 80% (data not shown). Treatment of the cells with the cell-permeable ceramide analogue, C2-ceramide (Fig. 5D), had a similar albeit smaller in magnitude effect. Ceramide apparently was not a direct inhibitor of TACE, because it was not effective when added to the assay mixture (data not shown). Ceramide metabolites sphingosine and dihydroceramide also had no effect. Most likely, the inhibition of TACE activity by SMase and ceramide treatment of intact cells was due to indirect effects on TACE. To that extent, we also observed differences in TACE subcellular localization between the two genotypes. Although in asm+/+ macrophages TACE was localized in a Golgi-like compartment in close proximity to the nucleus, in asm−/− cells a uniform and diffused staining was observed throughout the cell body (Fig. 5E).
The TACE activity measured in vitro was not sensitive to stimulation with LPS. Additional optimizations of the enzyme activity assay conditions, including changes in the concentration and type of detergent, failed to reveal any stimulation of TACE catalytic activity in response to LPS (Fig. 5G). This is not surprising because published literature data significantly disagree on this point. Although some groups had provided evidence for stimulation of TACE mRNA transcription, protein content, and catalytic activity by LPS, others had found none. Yet a third group had suggested that LPS can only affect TACE localization within the plasma membrane (53–56).
Effect of ASMase Deficiency on the Subcellular Distribution of TNFα
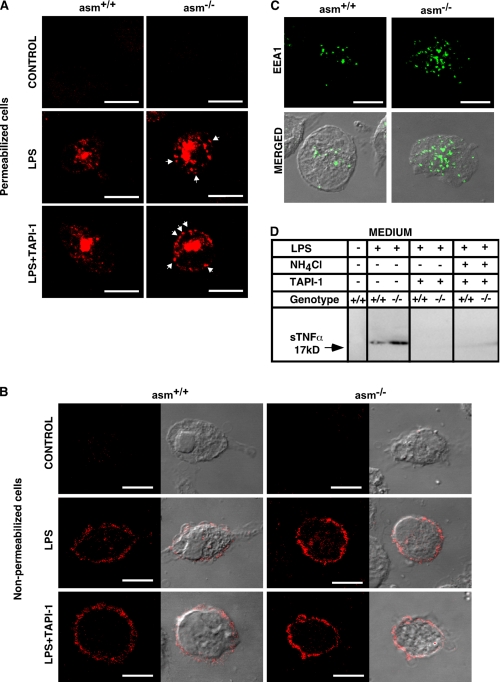
An indirect immunofluorescence approach allowed us to monitor the production of TNFα in the endoplasmic reticulum/Golgi compartment (in permeabilized cells), as well as to observe TNFα at the plasma membrane (in nonpermeabilized cells) (Fig. 6, A and B). Several differences between asm+/+ and asm−/− macrophages emerged. Seemingly, asm−/− cells exhibited more pronounced TNFα staining throughout the cells and specifically in vesicular structures close to the plasma membrane. Co-treatment with TAPI-1, a TACE inhibitor, increased the number of these TNFα positive vesicles in asm−/− but not in asm+/+ cells (Fig. 6A and Fig. 7B) indicating the following: (i) the vesicular structures contained TNFα designated for TACE-mediated cleavage and (ii) TAPI-1 sensitive pool of TNFα was present in asm−/− but not in asm+/+ cells. Keeping in mind that the TACE activity was higher in the asm−/− macrophages, one plausible explanation is that the pro-TNFα molecules that were not processed immediately by TACE were recycled back to the plasma membrane at a greater rate in the asm−/− cells than in asm+/+ cells. To begin testing this possibility, cells were stained for EEA1, a marker for early endosomes. Notably, asm−/− macrophages had a higher number of early endosomes as compared with asm+/+ macrophages (Figs. 6C and 7C).
FIGURE 6.
Subcellular localization of TNFα in asm+/+ and asm−/− macrophages. Macrophages from asm+/+ and asm−/− mice were treated with LPS (10 ng/ml) in the presence or absence of TAPI-1 (20 μm) and NH4Cl (10 mm) for 4 h. A–C, staining for TNFα (A and B) and early endosomes (C). TNFα was visualized in permeabilized (A) and nonpermeabilized (B) cells using antibodies against TNFα and confocal microscopy. The arrows indicate TNFα-positive vesicles proximal to the plasma membrane that are visible in asm−/− cells. Antibodies against the endosomal marker EEA1 were used to visualize early endosomes (C). Transmitted light images (B and C) show cell morphology. The scale bar represents 10 μm. D, suppression of TNFα secretion by TACE inhibitor. Western blot analyses of TNFα in cell culture medium from macrophages stimulated with LPS and indicated inhibitors.
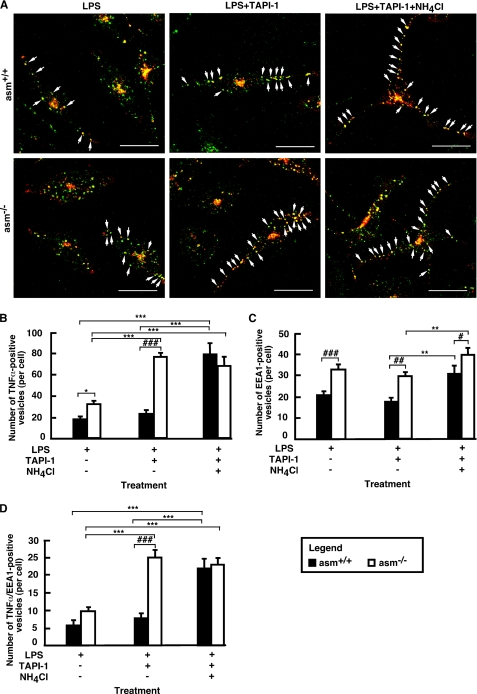
FIGURE 7.
Co-localization of TNFα and EEA1 in asm+/+ and asm−/− macrophages. Peritoneal asm+/+ and asm−/− macrophages were treated with LPS (10 ng/ml) for 4 h. Permeabilized cells were probed sequentially for TNFα (red) and EEA1 (green). Images were collected using confocal microscopy and merged (yellow). Only merged images are shown (A). The TNFα/EEA1 co-localization in a single cell for each panel is depicted by arrows. For quantification purposes, the bulk fluorescence visible in the center of each cell was excluded, and the number of TNFα- positive (B), EEA1-positive (C), and double-positive (D) vesicles (with size bigger than 4 pixels) was counted in 10 cells of each slide. Data shown are average ± S.E. Statistical significance of the main effect (***, p < 0.001; **, p < 0.01; *, p < 0.05) and the interaction effect (###, p <0.001; ##, p < 0.01; and #, p < 0.05) was determined by two-way ANOVA. The scale bar represents 20 μm.
Role of Endosomes in TNFα Processing
To elucidate whether the endosomes are indeed a reservoir of pro-TNFα for plasma membrane recycling, co-localization studies were done (Fig. 7 and supplemental Fig. S2). More TNFα-positive vesicles co-localized with EEA1 in LPS-treated asm−/− cells as compared with asm+/+ cells (Fig. 7, A and B). The number of these double-positive vesicles increased in the presence of TAPI-1 only in asm−/− cells but not in asm+/+ cells (Fig. 7D). In contrast, an increase in TNFα-positive and TNFα/EEA1 double-positive vesicles was observed in asm+/+ macrophages only when cells were co-treated with ammonium chloride, indicating that in asm+/+ cells the majority of intracellular TNFα undergoes lysosomal proteolysis (Fig. 7D). Notably, the addition of ammonium chloride had a negligible effect in asm−/− cells.
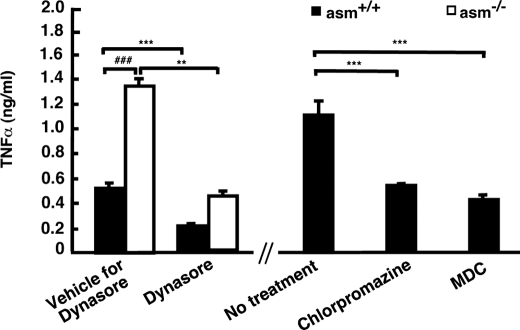
To clarify further the role of endocytosis in TNFα processing, inhibitors of endocytosis were applied 1 h after the LPS treatment to allow for undisturbed stimulation of TNFα mRNA transcription and translation. The inhibitors suppressed TNFα secretion by more than 50%, supporting a role of endocytosis in TNFα secretion (Fig. 8). Two-way ANOVA showed that Dynasore treatment also significantly attenuated the genotype-associated differences. Together, these results suggest that ASMase activity may play a role in partitioning pro-TNFα between two pools as follows: one destined for recycling and TACE-mediated processing and another one that undergoes proteolytic cleavage in the lysosomes.
FIGURE 8.
Effect of inhibition of endocytosis on sTNFα secretion in asm+/+ and asm−/− macrophages. Macrophages from asm+/+ and asm−/− mice were treated for 4 h with LPS (10 ng/ml) in the presence and absence of Dynasore (80 μm), chlorpromazine (2.5 μg/ml), or monodansylcadaverine (MDC) (50 μm) added 1 h after LPS stimulation. sTNFα in the medium was measured by ELISA. Statistical significance of the main effect (***, p < 0.001; **, p < 0.01) and interaction effect (###, p < 0.001) was determined by two-way (for Dynasore treatment) and one-way (for chlorpromazine and monodansylcadaverine treatments) ANOVA.
Effects of Exogenous Sphingomyelinase on TNFα Processing
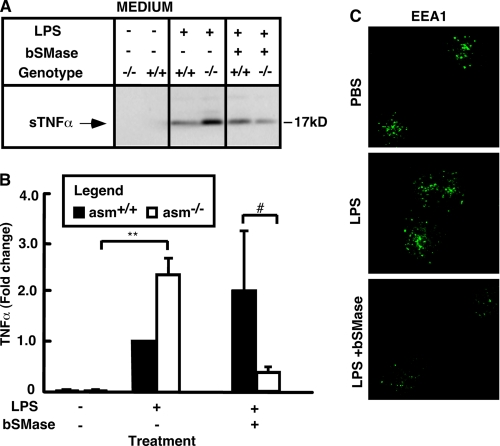
We observed that treatment of asm−/− macrophages with exogenous SMase or ceramides corrects for the differences in TACE activity and TNFα secretion. Similar rescue experiments were done to test whether ASMase and ceramide are involved in regulating TNFα recycling. Treatment of asm−/− cells with exogenous sphingomyelinase was sufficient to bring sTNFα levels close to those measured in asm+/+ cells (Fig. 9, A and B). Furthermore, SMase treatment also significantly decreased the number of early endosomes detected in asm−/− cells (Fig. 9C). These results link SMase and ceramide to early endosomes biogenesis and reveal a correlation between the number of early endosomes and the levels of sTNFα released in response to LPS.
FIGURE 9.
Effect of exogenous sphingomyelinase on sTNFα and EEA1 in asm−/− macrophages. Peritoneal macrophages from asm+/+ and asm−/− mice were treated for 4 h with 10 ng/ml LPS alone or in combination with 0.1 unit/ml bacterial sphingomyelinase (bSMase). A and B, levels of TNFα in medium. TNFα secretion was monitored by Western blotting analyses of cell culture medium. The graph represents the mean values ± S.D. of intensity of TNFα bands from three independent experiments. Statistical significance of the main effect (**, p < 0.01) and the interaction effect of the treatment and the genotype (#, p < 0.05) is shown based on two-way ANOVA. C, visualization of the early endosomes. Immunofluorescence images depicting EEA1-positive early endosomes (green) in permeabilized macrophages. PBS, phosphate-buffered saline.
DISCUSSION
This study provides evidence for the existence of a novel factor, ASMase, that regulates the rate of post-translational processing and secretion of TNFα during LPS stimulation. ASMase is known as a signaling enzyme that mediates several pathways activated by pro-inflammatory stimuli and by cellular stresses. More recent studies also suggest that ASMase participates in membrane biogenesis. The secretory form of ASMase is involved in organization of the signaling domains at the plasma membrane (57), although the endosomal/lysosomal form appears to facilitates intracellular membrane fusion and recycling pathways. Accumulation of sphingomyelin and cholesterol in the endosomal membranes of ASMase-deficient cells seems to impair the establishment of type I cytopathic vacuoles after alphavirus Sindbis virus infection and to interfere with proper maturation of phagosomes to phagolysosomes (44). Evidence to that extent came from experiments indicating that asm−/− macrophages exhibit a slow rate of release of lysosomal hydrolyses to the phagosomes. Defects in the delivery of phagocytosed bacteria to phagolysosomes were also observed, resulting in less efficient pathogen elimination (42, 43). Our results indicate that the recently established role of ASMase in membrane biogenesis may impact the regulation of TNFα processing and secretion.
Two distinct mechanisms seem to be involved. First, ASMase-derived ceramide apparently functions as a negative regulator of TACE activity. This is supported by the inhibition of TACE activity observed after treatment of macrophages with exogenous ceramide or bacterial SMase, both of which increase ceramide content of the plasma membrane. Consistently, the intrinsic activity of TACE was higher in asm−/− macrophages as compared with asm+/+ cells. Most likely, ceramide and ASMase modulate TACE activity by indirect mechanisms because ceramide was effective only when added to cultured cells, but not directly to the TACE assay mixture. As discussed earlier, TACE is a transmembrane protein, and its endogenous activity may depend on its lipid environment. Notably, Tellier et al. (26) found that the mature form of TACE, but not the inactive pro-form, is localized in the sphingomyelin/cholesterol-rich domains and co-purifies with caveolin 1 and flotillin-1. These authors further suggested that sequestration of TACE in the sphingomyelin/cholesterol rafts is rate-limiting for its activity. This is in agreement with our conclusion that ASMase and ceramide control a rate-limiting step in TNFα processing. It is of particular interest that treatment with SMase that not only hydrolyzes sphingomyelin but also depletes the plasma membrane from cholesterol (58) has an inhibitory effect of the endogenous TACE. It is possible that the elevated sphingomyelin (and cholesterol) in asm−/− cells creates a more “favorable” environment for TACE and higher activity.
Second, a seemingly independent mode of regulation was revealed by our indirect immunofluorescent studies. These experiments suggest that ASMase also has an effect on the fate of intracellular pro-TNFα. It has already been proposed that the pro-TNFα, which is not immediately cleaved by TACE, is internalized and either degraded in the lysosomes or recycled back to the plasma membrane and utilized for TACE-mediated cleavage. Our results strongly support such a scenario and provide initial evidence that the partitioning of the nonprocessed TNFα between these two pathways is modulated and may have a profound effect on the levels of TNFα secreted in response to a particular stimulus.
We found that asm−/− macrophages an have increased number of early endosomes suggesting a defect in the endosome maturation to lysosomes. This is consistent with previous studies implicating ASMase in the dynamics of vesicle fusion during endocytosis. Furthermore, the increased co-localization of EEA1 and TNFα in the absence of ASMase suggests that a significant portion of TNFα is sequestered in the early endosomes, recycled back to the plasma membrane, and cleaved by TACE. Further evidence for this came from the observation that inhibition of TACE by TAPI-1 caused significant elevation in intracellular TNFα and TNFα/EEA1 co-localization only in the asm−/− but not in the asm+/+ cells. In contrast, in asm+/+ cells only ammonium chloride treatment had an effect on intracellular TNFα. These observations indicate that a portion of intracellular TNFα, localized in the early endosomes, undergoes TACE-mediated cleavage in asm−/− cells but lysosomal clearance in asm+/+ cells.
Membrane lipids and proteins can be moved from the early endosomes back to the cell surface via two main routes as follows: the Rab4-dependent fast recycling pathway, and the Rab11-mediated slow recycling pathway. Fibroblasts from patients with Niemann-Pick disease, type A, seem to lack the Rab4-mediated recycling but exhibit a more active Rab11-dependent pathway (46). Therefore, the latter may be responsible for the increased TNFα recycling in asm−/− cells.
Negative modulation of TLR signaling pathways has emerged as a critical mechanism of regulating the innate immune response to infection. In the case of TLR-4, several molecules whose expression is induced by LPS have been shown to attenuate and/or block cellular response to LPS, including IRAK-M, the suppressors of cytokine signaling, MyD88s (59, 60). This study provides evidence for a novel putative negative regulator of LPS-induced TNFα secretion, the ASMase. It is unclear which of the two known forms of ASMase, the secretory or the lysosomal, is involved. Because both forms are induced by LPS, one cannot exclude the possibility that the two forms have distinct roles in regulating TNFα secretion. The secretory ASMase might be responsible for regulation of TACE activity at the plasma membrane via its ability to act on sphingomyelin at the plasma membrane, although the lysosomal form might be linked to the defects in endosomal maturation. As discussed in the Introduction, loss-of-function in the lysosomal form is linked to slow phago-lysosomal fusion and membrane budding, as well as increased susceptibility to pathogen infections. These results indicate that cellular ASMase activity and/or ceramide content might have more important role(s) in regulating systemic inflammatory responses than previously thought by affecting the magnitude of TNFα release by the cells of the immune system and perhaps other TNFα-producing cells.
Supplementary Material
Acknowledgments
We sincerely thank Drs. Dayong Wu and Simin Meydani from Tufts University for invaluable help and advice in preparing macrophage cultures and for measuring the levels of prostaglandin E2.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AG 019223 and AG 026711 from NIA (to M. N. N.-K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- TNFα
- tumor necrosis factor α
- ANOVA
- analysis of variance
- ASMase
- acid sphingomyelinase
- SMase
- sphingomyelinase
- EEA1
- early endosomal antigen 1
- ELISA
- enzyme-linked immunosorbent assay
- ERK
- extracellular signal-regulated kinase
- IL
- interleukin
- IRAK-1
- interleukin-1 receptor-associated kinase 1
- LPS
- lipopolysaccharide
- MyD88
- myeloid differentiation factor 88
- TACE
- TNFα-converting enzyme
- TLR4
- Toll-like receptor 4
- sTNFα
- soluble TNFα
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1.Feldmann M., Maini S. R. (2008) Immunol. Rev. 223, 7–19 [DOI] [PubMed] [Google Scholar]
- 2.Sethi G., Sung B., Aggarwal B. B. (2008) Front. Biosci. 13, 5094–5107 [DOI] [PubMed] [Google Scholar]
- 3.Zentella A., Manogue K., Cerami A. (1991) Prog. Clin. Biol. Res. 367, 9–24 [PubMed] [Google Scholar]
- 4.Fujihara M., Muroi M., Tanamoto K., Suzuki T., Azuma H., Ikeda H. (2003) Pharmacol. Ther. 100, 171–194 [DOI] [PubMed] [Google Scholar]
- 5.Pålsson-McDermott E. M., O'Neill L. A. (2004) Immunology 113, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krämer B., Wiegmann K., Krönke M. (1995) J. Biol. Chem. 270, 6577–6583 [DOI] [PubMed] [Google Scholar]
- 7.Tsai E. Y., Falvo J. V., Tsytsykova A. V., Barczak A. K., Reimold A. M., Glimcher L. H., Fenton M. J., Gordon D. C., Dunn I. F., Goldfeld A. E. (2000) Mol. Cell. Biol. 20, 6084–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Donnell P. M., Taffet S. M. (2002) J. Interferon Cytokine Res. 22, 539–548 [DOI] [PubMed] [Google Scholar]
- 9.Zhang T., Kruys V., Huez G., Gueydan C. (2002) Biochem. Soc. Trans. 30, 952–958 [DOI] [PubMed] [Google Scholar]
- 10.MacKenzie S., Fernàndez-Troy N., Espel E. (2002) J. Leukocyte Biol. 71, 1026–1032 [PubMed] [Google Scholar]
- 11.Jue D. M., Sherry B., Luedke C., Manogue K. R., Cerami A. (1990) Biochemistry 29, 8371–8377 [DOI] [PubMed] [Google Scholar]
- 12.Lieu Z. Z., Lock J. G., Hammond L. A., La Gruta N. L., Stow J. L., Gleeson P. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3351–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray R. Z., Kay J. G., Sangermani D. G., Stow J. L. (2005) Science 310, 1492–1495 [DOI] [PubMed] [Google Scholar]
- 14.Kay J. G., Murray R. Z., Pagan J. K., Stow J. L. (2006) J. Biol. Chem. 281, 11949–11954 [DOI] [PubMed] [Google Scholar]
- 15.Idriss H. T., Naismith J. H. (2000) Microsc. Res. Tech. 50, 184–195 [DOI] [PubMed] [Google Scholar]
- 16.Black R. A., Rauch C. T., Kozlosky C. J., Peschon J. J., Slack J. L., Wolfson M. F., Castner B. J., Stocking K. L., Reddy P., Srinivasan S., Nelson N., Boiani N., Schooley K. A., Gerhart M., Davis R., Fitzner J. N., Johnson R. S., Paxton R. J., March C. J., Cerretti D. P. (1997) Nature 385, 729–733 [DOI] [PubMed] [Google Scholar]
- 17.Cook E. B., Stahl J. L., Graziano F. M., Barney N. P. (2008) Invest. Ophthalmol. Vis. Sci. 49, 3992–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solomon K. A., Pesti N., Wu G., Newton R. C. (1999) J. Immunol. 163, 4105–4108 [PubMed] [Google Scholar]
- 19.Peschon J. J., Slack J. L., Reddy P., Stocking K. L., Sunnarborg S. W., Lee D. C., Russell W. E., Castner B. J., Johnson R. S., Fitzner J. N., Boyce R. W., Nelson N., Kozlosky C. J., Wolfson M. F., Rauch C. T., Cerretti D. P., Paxton R. J., March C. J., Black R. A. (1998) Science 282, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 20.Gómez-Gaviro M. V., González-Alvaro I., Domínguez-Jiménez C., Peschon J., Black R. A., Sánchez-Madrid F., Díaz-González F. (2002) J. Biol. Chem. 277, 38212–38221 [DOI] [PubMed] [Google Scholar]
- 21.Solomon K. A., Covington M. B., DeCicco C. P., Newton R. C. (1997) J. Immunol. 159, 4524–4531 [PubMed] [Google Scholar]
- 22.Shurety W., Pagan J. K., Prins J. B., Stow J. L. (2001) Lab. Invest. 81, 107–117 [DOI] [PubMed] [Google Scholar]
- 23.Peiretti F., Canault M., Deprez-Beauclair P., Berthet V., Bonardo B., Juhan-Vague I., Nalbone G. (2003) Exp. Cell Res. 285, 278–285 [DOI] [PubMed] [Google Scholar]
- 24.Doedens J. R., Black R. A. (2000) J. Biol. Chem. 275, 14598–14607 [DOI] [PubMed] [Google Scholar]
- 25.Watanabe N., Nakada K., Kobayashi Y. (1998) Eur. J. Biochem. 253, 576–582 [DOI] [PubMed] [Google Scholar]
- 26.Tellier E., Canault M., Rebsomen L., Bonardo B., Juhan-Vague I., Nalbone G., Peiretti F. (2006) Exp. Cell Res. 312, 3969–3980 [DOI] [PubMed] [Google Scholar]
- 27.Thiel K. W., Carpenter G. (2006) Biochem. Biophys. Res. Commun. 350, 629–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Tresckow B., Kallen K. J., von Strandmann E. P., Borchmann P., Lange H., Engert A., Hansen H. P. (2004) J. Immunol. 172, 4324–4331 [DOI] [PubMed] [Google Scholar]
- 29.Rousseau S., Papoutsopoulou M., Symons A., Cook D., Lucocq J. M., Prescott A. R., O'Garra A., Ley S. C., Cohen P. (2008) J. Cell Sci. 121, 149–154 [DOI] [PubMed] [Google Scholar]
- 30.Zeidan Y. H., Wu B. X., Jenkins R. W., Obeid L. M., Hannun Y. A. (2008) FASEB J. 22, 183–193 [DOI] [PubMed] [Google Scholar]
- 31.Grassme H., Jekle A., Riehle A., Schwarz H., Berger J., Sandhoff K., Kolesnick R., Gulbins E. (2001) J. Biol. Chem. 276, 20589–20596 [DOI] [PubMed] [Google Scholar]
- 32.Falcone S., Perrotta C., De Palma C., Pisconti A., Sciorati C., Capobianco A., Rovere-Querini P., Manfredi A. A., Clementi E. (2004) J. Immunol. 173, 4452–4463 [DOI] [PubMed] [Google Scholar]
- 33.MacKichan M. L., DeFranco A. L. (1999) J. Biol. Chem. 274, 1767–1775 [DOI] [PubMed] [Google Scholar]
- 34.Hofmeister R., Wiegmann K., Korherr C., Bernardo K., Krönke M., Falk W. (1997) J. Biol. Chem. 272, 27730–27736 [DOI] [PubMed] [Google Scholar]
- 35.Zeidan Y. H., Hannun Y. A. (2007) J. Biol. Chem. 282, 11549–11561 [DOI] [PubMed] [Google Scholar]
- 36.Schissel S. L., Jiang X., Tweedie-Hardman J., Jeong T., Camejo E. H., Najib J., Rapp J. H., Williams K. J., Tabas I. (1998) J. Biol. Chem. 273, 2738–2746 [DOI] [PubMed] [Google Scholar]
- 37.Lightle S., Tosheva R., Lee A., Queen-Baker J., Boyanovsky B., Shedlofsky S., Nikolova-Karakashian M. (2003) Arch. Biochem. Biophys. 419, 120–128 [DOI] [PubMed] [Google Scholar]
- 38.Wong M. L., Xie B., Beatini N., Phu P., Marathe S., Johns A., Gold P. W., Hirsch E., Williams K. J., Licinio J., Tabas I. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8681–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grassmé H., Cremesti A., Kolesnick R., Gulbins E. (2003) Oncogene 22, 5457–5470 [DOI] [PubMed] [Google Scholar]
- 40.Haimovitz-Friedman A., Cordon-Cardo C., Bayoumy S., Garzotto M., McLoughlin M., Gallily R., Edwards C. K., 3rd, Schuchman E. H., Fuks Z., Kolesnick R. (1997) J. Exp. Med. 186, 1831–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García-Ruiz C., Colell A., Marí M., Morales A., Calvo M., Enrich C., Fernández-Checa J. C. (2003) J. Clin. Invest. 111, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Utermöhlen O., Herz J., Schramm M., Krönke M. (2008) Immunobiology 213, 307–314 [DOI] [PubMed] [Google Scholar]
- 43.Schramm M., Herz J., Haas A., Kronke M., Utermohlen O. (2008) Cell. Microbiol. 10, 1839–1853 [DOI] [PubMed] [Google Scholar]
- 44.Ng C. G., Coppens I., Govindarajan D., Pisciotta J., Shulaev V., Griffin D. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16326–16331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grassmé H., Jendrossek V., Riehle A., von Kürthy G., Berger J., Schwarz H., Weller M., Kolesnick R., Gulbins E. (2003) Nat. Med. 9, 322–330 [DOI] [PubMed] [Google Scholar]
- 46.Choudhury A., Sharma D. K., Marks D. L., Pagano R. E. (2004) Mol. Biol. Cell 15, 4500–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horinouchi K., Erlich S., Perl D. P., Ferlinz K., Bisgaier C. L., Sandhoff K., Desnick R. J., Stewart C. L., Schuchman E. H. (1995) Nat. Genet. 10, 288–293 [DOI] [PubMed] [Google Scholar]
- 48.Rouzer C. A., Kingsley P. J., Wang H., Zhang H., Morrow J. D., Dey S. K., Marnett L. J. (2004) J. Biol. Chem. 279, 34256–34268 [DOI] [PubMed] [Google Scholar]
- 49.Fieren M. W., van den Bemd G. J., Ben-Efraim S., Bonta I. L. (1992) Immunol. Lett. 31, 85–90 [DOI] [PubMed] [Google Scholar]
- 50.Kim J. G., Hahn Y. S. (2000) Immunol. Invest. 29, 257–269 [DOI] [PubMed] [Google Scholar]
- 51.Cuschieri J., Gourlay D., Garcia I., Jelacic S., Maier R. V. (2004) Cell. Immunol. 227, 140–147 [DOI] [PubMed] [Google Scholar]
- 52.Karakashian A. A., Giltiay N. V., Smith G. M., Nikolova-Karakashian M. N. (2004) FASEB J. 18, 968–970 [DOI] [PubMed] [Google Scholar]
- 53.Ermert M., Pantazis C., Duncker H. R., Grimminger F., Seeger W., Ermert L. (2003) Cytokine 22, 89–100 [DOI] [PubMed] [Google Scholar]
- 54.Doedens J. R., Mahimkar R. M., Black R. A. (2003) Biochem. Biophys. Res. Commun. 308, 331–338 [DOI] [PubMed] [Google Scholar]
- 55.Armstrong L., Godinho S. I., Uppington K. M., Whittington H. A., Millar A. B. (2006) Am. J. Respir. Cell Mol. Biol. 34, 219–225 [DOI] [PubMed] [Google Scholar]
- 56.Robertshaw H. J., Brennan F. M. (2005) Br. J. Anaesth. 94, 222–228 [DOI] [PubMed] [Google Scholar]
- 57.Gulbins E., Kolesnick R. (2003) Oncogene 22, 7070–7077 [DOI] [PubMed] [Google Scholar]
- 58.Slotte J. P., Bierman E. L. (1988) Biochem. J. 250, 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liew F. Y., Xu D., Brint E. K., O'Neill L. A. (2005) Nat. Rev. Immunol. 5, 446–458 [DOI] [PubMed] [Google Scholar]
- 60.O'Neill L. A. (2006) Curr. Opin. Immunol. 18, 3–9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.