Abstract
Cartilage is resistant to tumor invasion. In the present study, we found that the NH2-propeptide of the cartilage-characteristic collagen, type IIB, PIIBNP, is capable of killing tumor cells. The NH2-propeptide is liberated into the extracellular matrix prior to deposition of the collagen fibrils. This peptide adheres to and kills cells from chondrosarcoma and cervical and breast cancer cell lines via the integrins αvβ5 and αvβ3. Adhesion is abrogated by blocking with anti αvβ5 and αvβ3 antibodies. When αv is suppressed by small intefering RNA, adhesion and cell killing are blocked. Normal chondrocytes from developing cartilage do not express αvβ3 and αvβ5 integrins and are thus protected from cell death. Morphological, DNA, and biochemical evidence indicates that the cell death is not by apoptosis but probably by necrosis. In an assay for invasion, PIIBNP reduced the number of cells crossing the membrane. In vivo, in a tumor model for breast cancer, PIIBNP was consistently able to reduce the size of the tumor.
Keywords: Breast Cancer, Cell Adhesion, Cell Death, Collagen, Integrin, Chondrocytes, Chondrosarcoma
Introduction
Cartilage is a unique tissue in being composed of only one cell type, chondrocytes. In addition, cartilage is avascular and resistant to tumor invasion, although the mechanism by which this occurs is unknown. Cartilage ECM2 is predominantly made up of fibrillar type IIB collagen and the large proteoglycan aggrecan. The fibrillar collagens, types I, II, III, V, and XI, are synthesized as procollagens containing NH2- and COOH-terminal extension peptides that are removed prior to deposition of the collagen monomers into fibrils in the extracellular matrix (1). During the chondrogenesis phase of endochondral bone development, large amounts of type IIB collagen are synthesized as the tissue is established. Thereafter, in the cartilaginous growth plate, the cells become hypertrophic, change their predominant collagen synthesis to type X collagen, and eventually die by apoptosis. The hypertrophic cartilage is vascularized and subsequently removed by specialized osteoclasts, and the tissue is replaced by bone synthesized by osteoblasts. By this process, the cartilage provides an anlagen for bone formation (2, 3).
Type II procollagen is unique among the fibrillar collagens in containing vicinal RGD motifs in the NH2-terminal propeptide domain encoded by exon 6 of the COL2A1 gene (supplemental Fig. 1). RGD peptides serve as the primary integrin recognition sites in extracellular matrix proteins and, as such, play an important role in regulating cell/matrix interactions required for proper cell function. Integrins are cell adhesion molecules that consist of two non-covalently associated subunits α and β (4). Integrins are receptors for many ECM matrix proteins, such as for collagens (α1β1, α2β1, α3β1), fibronectin (α3β1, α4β1, α5β1), laminin (α3β1, α6β1), osteopontin (αvβ3, αvβ5, αvβ1), and vitronectin (αvβ3, αvβ1 αvβ5) (4–9). The binding of substrate to integrins on the cell surface stimulates intracellular signaling to affect gene expression (outside-in signaling), and the cell can alter the expression and affinity of integrins (inside-out signaling). This bidirectional signaling controls cellular activity, such as cell-cell and cell-matrix adhesion, internalization, and degradation of matrix molecules, as well as cell migration, proliferation, and apoptosis.
Type II procollagen can be synthesized in two splice forms, type IIA and type IIB. Type IIA contains an additional exon (exon 2) in the NH2-propeptide that encodes a von Willebrand factor C domain and is synthesized by many embryonic tissues, such as basement membrane and prechondrogenic mesenchyme (10–13). Unlike most fibrillar collagens, type IIA procollagen is not processed to remove the NH2-propeptide, and the entire pN-procollagen is deposited into the ECM. It functions to bind growth factors like BMP-2 and transforming growth factor-β via the von Willebrand factor C domain encoded by exon 2 (14). For the type IIB NH2-propeptide made in cartilage, there is no known or even predicted function. Because the RGDRGD sequence, among fibrillar collagens, is unique to type II collagen, conserved throughout mammalian species, and liberated from the procollagen molecule in vivo, we sought to determine the function of this RGD domain of the type IIB procollagen NH2-propeptide. We hypothesized that the function of RGD-encoded regions in type II procollagen NH2-propeptide may be unique to cartilage and required to induce and maintain important biological properties of the tissue. Here we report that the NH2-propeptide of type IIB procollagen (PIIBNP) is capable of inducing death of a chondrosarcoma cell line and other tumor cells, and the interaction with cells is dependent on integrins αVβ3 and αVβ5. Because normal chondrocytes do not express these integrin receptors or express them at low levels, we propose that during development, chondrocytes are resistant to the influence of the type IIB collagen NH2-propeptide that they synthesize.
Because these data suggest that PIIBNP might be a useful therapeutic agent for cancer, we tested the anti-invasive property of PIIBNP and established a breast cancer model in mice to demonstrate that PIIBNP is able to reduce tumor size in vivo.
EXPERIMENTAL PROCEDURES
Histological Analysis
Detection of fibrillar type II collagen, integrin αv, and PIIBNP proteins by immunohistochemistry was performed on 5-μm-thick paraffin-embedded sections of E15.5 mouse embryos using a procedure described previously (15).
Protein Expression, Purification, and Limulus Amebocyte Lysate Test
Total RNA was isolated from 54-day human fetal embryonic tissue obtained from the Central Laboratory for Human Embryology, University of Washington. Reverse transcription-PCR was carried out to obtain a 207-bp cDNA and a 315-bp cDNA encoding type II collagen NH2-propeptide protein exon 2 (16) and exons 3–8 (14), respectively. The cDNAs were cloned into pGEX-4T-2 vector (Clontech), and GST fusion proteins were expressed using BL21 (DE3) host strains. The recombinant proteins were purified by affinity chromatography and filtered with a 0.1-μm low binding filter (Millipore). The purity of proteins was confirmed by SDS-PAGE. The proteins were tested for lipopolysaccharide using a limulus amebocyte lysate kit (Associates of Cape Cod).
Identification of PIIBNP from Developing Human and Mouse Limbs
Human embryonic (days 56 and 57) and mouse E14.5 limbs were used. Samples were frozen, pulverized, mixed with SDS plus dithiothreitol loading buffer, boiled, and then loaded on a 4–20% SDS gel (Pierce). The samples were then blotted to Immobilon FL (Millipore) at 90 V for 1 h at room temperature. The blot was then washed with PBS and blocked with 5% milk (Bio-Rad) in PBS for 1 h at room temperature with shaking. Primary antibody was then added in PBS with 0.1% Tween 20 (Sigma) (PBSt) for 1 h at room temperature with shaking. The blot was washed three times for 10 min each at room temperature with PBSt. Infrared secondary antibody (LICOR) was then incubated for 1 h at room temperature with shaking in 5% milk in PBSt. The blot was then washed six times for 10 min each at room temperature in 1× PBSt with shaking. After a final wash in PBS to remove the Tween 20, the blot was then analyzed on the Odyssey imager (LICOR).
Antibodies
Monoclonal antibodies used for blocking experiments were mouse anti-αvβ3 and -αvβ5 integrin (Chemicon). Polyclonal antibodies for blocking, immunohistochemistry, and Western blotting were rabbit anti human αv, α2, α3, α5, β1, β3, and β5 integrin; goat anti-human actin (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), and chicken anti-PIIBNP (17).
Cell Culture
The human Chondrosarcoma cell line (hCh-1) was developed in our laboratory from a portion of homogenous-appearing tumor (18). HeLa cells were from Dr. William Frazier (Washington University School of Medicine). The hCh-1 cells and HeLa cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum and at 37 °C in a humidified, 5% CO2 atmosphere. Human breast cancer MDA-MB231 cells were provided by Dr. Katherine Weilbaecher (Washington University School of Medicine) and cultured in Dulbecco's modified Eagle's medium (Cellgrows) supplemented with 10% fetal bovine serum (Invitrogen). Normal human chondrocytes were from a 12-year-old and a 6-year-old and provided by Dr. Davis Atkinson (ISTO Technologies, St. Louis, MO). Normal fetal and 2-year-old horse cartilage was provided by Dr. Lisa Fortier (Cornell University), and normal bovine cartilage was from the slaughterhouse at Hansen Packing Meat Company (Jerseyville, IL).
Cell Adhesion Assay
Cell adhesion assays were performed using a spectrophotometric method (19) with some modifications. A 96-well plate was coated with recombinant proteins and blocked with 0.5% bovine serum albumin in PBS. Cells were plated at 5 × 104 cells/well for 1 h. The wells were washed three times with prewarmed PBS to remove non-adherent cells. The adherent cells were fixed with 4% paraformaldehyde in PBS for 10 min and stained with 0.5% toluidine blue for 5 min. The wells were rinsed with water, and attached cells were solubilized with 0.1% SDS. Absorbance at 595 nm was recorded on a Spectra Max plate reader. Synthetic blocking peptides GRGDNP and GRADNP (Biomol International) and antibodies to αvβ3 and αvβ5 integrin were preincubated with cells for 30 min before they were added to the wells.
Site-directed Mutagenesis
Site-directed mutagenesis was performed using a QuikChange kit (Stratagene) to mutate the RGDRGD sequence in GST-PIIBNP. The mutated plasmid was confirmed by DNA sequencing and transformed into BL21 (DE3) host strain to express mutated PIIBNP (mPIIBNP; RADRAD).
siRNA Interference
hCh-1 cells were grown to 60–80% confluence in 6-well plates in Opti-MEM medium (Invitrogen) without antibiotics and serum. The sequence of siRNAs, sense and antisense oligonucleotides, and the method were from Graef et al. (20). The final concentration of oligonucleotides was 0.05 μm. Transfection was performed using a Lipofectamine kit, and cells were stimulated with 200 ng/ml phorbol 12-myristate 13-acetate after a 4-h incubation with oligonucleotides, as described (20). Total RNA was collected and reverse transcribed with Superscript RT II transcriptase. 5 μl of the cDNA were used for semiquantitative [α-32P]dCTP PCR for αv integrin and glyceraldehyde-3-phosphate dehydrogenase; the former were amplified for 26 cycles and the latter for 20 cycles. Samples were run on 6% SDS-PAGE, dried, and exposed to a PhosphorImager screen (Storm, ABI). Bands were quantified with ImageQuant software.
Integrin αv Immunoprecipitation
hCh-1 cells were lysed in buffer containing 1% Triton X-100 and a proteinase inhibitor mixture (Sigma). Cell homogenates were mixed with GST, PIIBNP, or mutant PIIBNP and incubated for 1 h at room temperature. Protein A-agarose (Sigma) previously bound with integrin αv antibody was incubated with cell homogenate mixture for 4 h at 4 °C with gentle shaking. After washing (three times for 5 min each), the precipitated material was analyzed by SDS-PAGE and detected with chicken anti-PIIBNP antibody.
Immunoblotting
Western blotting was performed as described (21). Blots were probed with the following primary antibodies: rabbit anti-αv, -α2, -α3, -α5, -β1, -β3, and -β5 integrin and goat anti-actin (Santa Cruz Biotechnology, Inc.) and chicken anti-PIIBNP antiserum (17).
Cell Surface Labeling and Pull-down Assay
hCh-1 cells were incubated with EZ-link Sulfo-NHS-Biotin reagent for 30 min according to the product instructions (Sigma). After three washes with 0.1 m glycine in PBS, the cells were lysed with buffer containing Triton X-100. The lysate was incubated with glutathione-derivatized agarose beads previously bound with PIIBNP or mutant PIIBNP for 1 h at room temperature. After washing, the bound proteins were separated by SDS-PAGE and transferred to a nylon membrane that was subsequently blocked with 5% nonfat dry milk for 1 h. The membrane was washed and incubated with antibodies and then incubated with a secondary antibody coupled to horseradish peroxidase. The antibody complex was visualized by enhanced chemiluminescence.
Cell Viability Assay
hCh-1, MDA-MB231, or Hela cells were plated, treated with fusion protein, and allowed to incubate for the desired amount of time. When the assay was complete, the medium was removed. The cells were trypsinized, combined with the medium, and centrifuged at 4 °C for 5 min at 1500 rpm. The cell pellet was resuspended in 200 μl of new medium, mixed with an equal volume of trypan blue (Sigma). Both live and dead cells were counted using a hemocytometer.
DNA Laddering
Tissue culture plates were coated with type I collagen. hCH-1 cells were plated at a density of 1 × 106 cells/ml in RPMI (Cellgrow) containing 10% fetal bovine serum (Invitrogen). The cells were treated with staurosporine (Sigma), PIIBNP, or mutant PIIBNP in serum-free medium. The cells were washed with PBS, harvested by trypsinization, centrifuged, resuspended in Hanks' balanced salt solution, and fixed in 70% ethanol for 24 h at −20 °C. The cell pellets were resuspended in phosphate citrate buffer. The supernatant was transferred to a new tube and incubated with Nonidet P-40 and RNase for 30 min at 37 °C. Proteinase K was added for an additional 30 min at 37 °C. Samples were resolved by 1.5% agarose gel electrophoresis.
Cell Morphology Analysis
Cell morphology was visualized under a light microscope (NIKON ECLIPSE E800) with a differential interference contrast objective (×40). Images were digitally photographed using a Q capture Retiga 2000R camera (×400).
Lactate Dehydrogenase (LDH) Release Assay
Lactose dehydrogenase release assays were performed using a CytoTox 96 non-radioactive assay kit (Promega). Briefly, The hCh-1 cells were cultured in a 96-well plate overnight in a complete RPMI 1640 medium. The cells were then treated with 10 μm GST, PIIBNP, mPIIBNP, RGD, or RAD peptide in serum-free medium for 90 h. The plate was centrifuged for 4 min, and 50 μl of the supernatant from each well was transferred to a new flat bottom 96-well plate. 50 μl of reconstituted substrate mix was added to each well. The plate was covered and placed at room temperature for 30 min. After adding 50 μl of stopping solution to each well, the absorbance at 490 nm was recorded in a plate reader.
Transwell Invasion Assay
Transwell invasion assays were adapted from previously described work (22) (MB231 or hCh-1). Briefly, the inserts were washed two times with serum-free medium. Matrigel (20 μl/insert, 1:6 dilution with medium) was polymerized and equilibrated at 37 °C and 5% CO2 for 30 min. After equilibration, inserts were put into a 24-well plate containing 300 μl of serum-free medium in each well. 200 μl of media containing 105 cells with/without 5 μm GST, PIIBNP, or mPIIBNP were added to each insert. The invasion assay was carried out in cell culture incubator for 30 h for MBN231 and 48 h for hCh-1. The cells were fixed with 4% formaldehyde in PBS and stained with 0.2% toluidine blue for 10 min. After washing five times by dipping the chambers in water, the cells at the top of the Matrigel membrane were removed with several Q-tips. The invading cells were visualized (×20 magnification), photographed (at least 12 fields/condition), and counted using Image J software.
Neutral Red Retention Assay
The neutral red retention assay was adapted from previous studies (23). Briefly, hCh-1 cells were seeded in a clear 96-well plate at 1 × 104 cells/well overnight. The medium was removed and replaced with serum-free medium containing 5 μm GST, PIIBNP, or mPIIBNP for 72 h. The medium was removed and replaced with new medium containing 40 μg/ml neutral red. After a 1-h incubation at room temperature, the neutral red taken up by the cells was extracted in 100 μl of 1% acetic acid, 50% ethanol for 15 min with shaking at room temperature, and the absorbance was read at 540 nm. Lysosomal integrity was calculated as a percentage of the absorbance of the untreated control.
Tumor Xenograft Growth in Vivo
All of the experimental animal procedures on nude mice were approved by the University Laboratory Animal Care Committee of Washington University and were performed according to federal regulations for animal research. NOD/SCID mice (a kind gift of Dr. Matthew Ellis, Washington University), at 6–9 weeks of age were injected subcutaneously on the dorsal surface with 10 million MDA-MB231 breast cancer cells. Tumor growth was monitored by palpation, and the onset of when tumors were detectable was noted. Tumor size was measured with calipers, and tumor volume was calculated using a reported method (24). When tumors grew to 0.5 cm in diameter, 6 nmol of PIIBNP or GST were introduced into mice by daily subcutaneous injection for 2 weeks. When tumors grew to sacrifice size (tumor diameter of >2 cm), the tumors were removed and weighed. Representative data were obtained from three mice per experimental group, and the entire experiment was repeated in two independent trials (n = 6 in each experimental group).
RESULTS
PIIBNP Adheres to Cells via Integrins
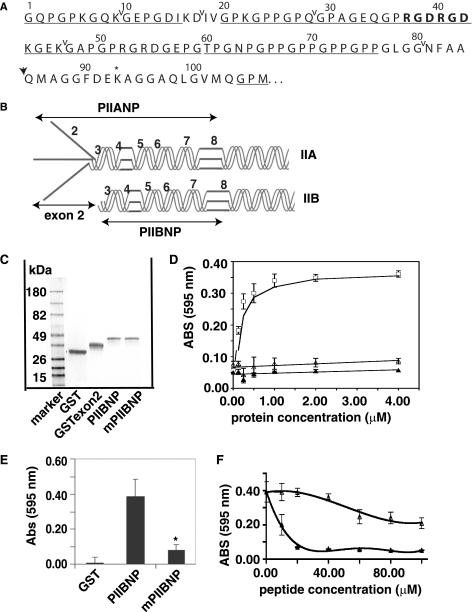
Type II procollagen is unique among the fibrillar collagens in containing vicinal RGD peptides in the NH2-propeptide domain (Fig. 1A). In order to investigate conservation across species, DNA sequence data of the type II NH2-propeptide for human, dog, chicken, horse, rat, Xenopus, and zebrafish were collected from GenBankTM and analyzed using the gene analysis tool DNAstar/MagAlign (supplemental Fig. 1). The RGDRGD sequence in type II NH2-propeptide was conserved across mammalian species; however, only the first RGD was conserved in chicken (RGDRGE), and the second RGD was conserved in Xenopus (RGERGD). In zebrafish, neither RGD is conserved (RGERGA). These differences may reflect the role of cartilage in evolution of endochondral bone formation from zebrafish to xenopus to mammals, where there are significant differences in growth plate formation and bone development.
FIGURE 1.
PIIBNP binds to cell surface in an RGD-dependent manner. A, sequence for type IIB NH2-propeptide. ↓, N-proteinase cleavage site; *, cross-link formation site in N-telopeptide. The RGDRGD motif is shown in boldface type, and the GXY sequence is underlined. Carets delineate exons 3–8. B, the type IIA NH2-propeptide (PIIANP) and protein structure contain the eight exons that are represented by numbers, and type IIB NH2-propeptide (PIIBNP) contains seven exons. C, GST fusion proteins were expressed in bacteria and purified as described under “Experimental Procedures.” Protein purity was assayed using a 4–20% SDS-polyacrylamide gel stained with Coomassie Blue. D, 96-well plates were coated with increasing concentrations of GST (▴), recombinant exon 2 (▵), or GST-PIIBNP (□) at 4 °C overnight and blocked with 0.5% bovine serum albumin, and hCh-1 cells were cultured in the plate for the adhesion assay as described under “Experimental Procedures.” The value of absorbance at 595 nm represents the number of cells bound to the proteins. Absorbance values are represented as mean ± S.D. (n = 8). The smooth curves are fitted linear regression or moving average curves. E, RGDRGD motif in PIIBNP was mutated to RADRAD (mPIIBNP). hCh-1 cell adhesion to GST, PIIBNP, and mutant PIIBNP was analyzed as described above. Mutant PIIBNP significantly reduced cell adhesion (*, p < 0.001 compared with PIIBNP, n = 18). F, hCh-1 cells were incubated with either synthetic RGD or RAD peptide at a concentration of 0–100 μm for 15 min at room temperature and then added into the wells for the attachment assay. Absorbance values are represented as mean ± S.D. (n = 8). The smooth curves are fitted moving average curves.
To undertake these studies, we selected a cell line that was of human origin in order to provide a homogeneous system for testing the function of the human PIIBNP. We chose to use a human cell line developed in our laboratory from the cartilaginous portion of a high grade chondrosarcoma, called hCh-1 (18). This tumor cell line has many properties of chondrocytes in that the morphology of the cells is polygonal, and they express cartilage-characteristic minor collagens and high levels of aggrecan; however, like most chondrosarcoma cell lines, the hCh-1 does not synthesize type II collagen (18). These characteristics are appropriate for our study of the function of type II collagen NH2-propeptide, because there is no endogenous type II collagen to confound the interpretation of the experiments.
To investigate the protein-cell interaction, hCh-1 cell adhesion assays were performed as described under “Experimental Procedures.” GST fusion proteins containing PIIBNP, a region of the COL2A1 gene that does not contain RGD, exon 2, and GST alone were tested in the cell adhesion assay. The recombinant fusion proteins used in this study are diagramed in Fig. 1B in trimeric form (although monomers are produced in the GST fusion protein system), and an example of the purified recombinant protein expression is shown in Fig. 1C. No contaminating proteins were seen on SDS-PAGE, and there was no contamination with lipopolysaccharide, as determined by a limulus amebocyte lysate test. GST-PIIBNP, but not GST alone or GST-exon 2, mediated hCh-1 cell adhesion in a dose-dependent manner (Fig. 1D), suggesting that the RGD motif may play a role in this interaction. Maximal adhesion was seen at a coating concentration of ∼1.5 μm for PIIBNP.
In order to determine whether the cell attachment function was mediated by the RGD sequence, the two RGD triplets were mutated to two RAD triplets by point mutations in the PIIBNP recombinant protein. With the RGDRGD sequence mutated to RADRAD, cell attachment was reduced by 90% (Fig. 1E). As additional evidence for specificity of cell adhesion, competition assays were performed. The synthetic RGD peptide (GRGDNP) inhibited the attachment by 50% at a concentration of 10 μm (Fig. 1F). In contrast, the non-RGD peptide showed no inhibition at a peptide concentration up to 40 μm. At a concentration of more than 40 μm, the non-RGD peptide also inhibited cell adhesion somewhat, but the inhibition was insignificant compared with that by RGD peptide. These data strongly suggest that the RGD motif in PIIBNP mediates the cell attachment.
PIIBNP Adheres to Integrins αvβ5 and αvβ3
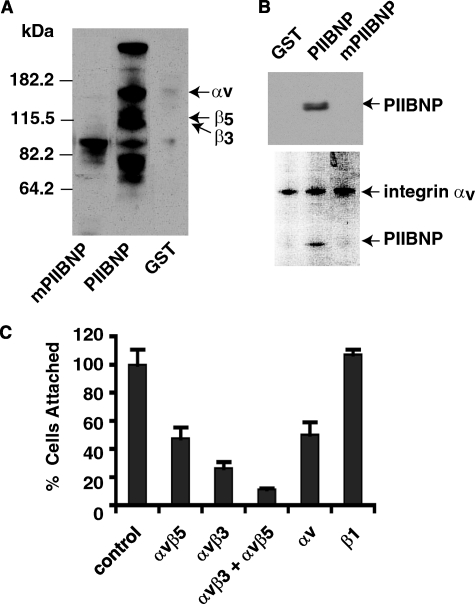
Reverse transcription-PCR analysis was used to determine which integrins were synthesized by the hCh-1 cells. Positive signal was observed for αv, α1, α2, α5, β1, β3, and β5 with no detectable signal for α3 and α4 (date not shown). A weak signal for α10 was detected. To identify the integrins that adhere to the PIIBNP, hCh-1 cell surface proteins were labeled with biotin, and cell membrane proteins were solubilized with lysis buffer containing Triton X-100. The cell membrane proteins were mixed with glutathione-agarose beads that had previously been loaded with recombinant PIIBNP, mutant PIIBNP, or GST. The beads were washed, and proteins associated with the GST fusion proteins were eluted with SDS loading buffer, analyzed by SDS-PAGE, and blotted with horseradish peroxidase-conjugated streptavidin (Fig. 2A). Proteins were identified by Western blotting against various integrin antibodies. As indicated in Fig. 2A, PIIBNP pulled down integrins αv, β5, and β3 but not β1, α3, α4, or α5 (data not shown). No integrins were found in the protein associated with the mutant PIIBNP or GST.
FIGURE 2.
PIIBNP interacts with integrins αvβ5 and αvβ3. A, GST, GST-PIIBNP, and mutant PIIBNP affinity matrix bound to cell surface biotinylated proteins in hCh-1 cells. These proteins were resolved by electrophoresis and identified by horseradish peroxidase-conjugated streptavidin. The blot was then stripped against various integrin antibodies to identify the specific biotinylated integrin. B, Triton X-100-solubilized membrane proteins were incubated with PIIBNP, GST, or mutant PIIBNP. Proteins associated with integrin αV were immunoprecipitated using a polyclonal αV antibody bound to protein A-agarose beads. The immunoprecipitated materials were analyzed using a SDS-PAGE, transferred to a nylon membrane, and incubated with an antiserum to PIIBNP (top panel). The bottom panel shows the Coomassie-stained SDS-PAGE gel. C, integrin antibodies (at equivalent concentration) were incubated with hCh-1 cells for 30 min prior to the cell adhesion assay. Values for cell adhesion to PIIBNP are shown as the percentage of cells attached. They are represented as the means ± S.D. for 4–8 independent measurements.
To further confirm the binding specificity of PIIBNP to αv integrin, immunoprecipitation of integrin αv-binding proteins was performed using a polyclonal integrin αv antibody. Triton X-100-solubilized cell membrane proteins were incubated with PIIBNP, GST, or mutant PIIBNP for 1 h at room temperature. Proteins associated with integrin αv were immunoprecipitated using a polyclonal αv antibody bound to protein A-agarose beads. The proteins bound to antibody were analyzed by SDS-PAGE and Western blotting with PIIBNP antibody. The Western blot results in Fig. 2B (top) show that αv integrin antibody immunoprecipitates PIIBNP, but not mutant PIIBNP or GST, confirming that there is an interaction between PIIBNP and integrin αv. The results from Coomassie Blue staining of SDS-PAGE further show this interaction by demonstrating that only PIIBNP is co-immunoprecipitated with αv (bottom).
To confirm the roles of αvβ5 and αvβ3 as the receptors for PIIBNP, antibody inhibition of cell adhesion was performed. Antibodies to integrins β1, αv, αvβ5, and αvβ3 were incubated with cells prior to cell adhesion assays. As shown in Fig. 2C, cell adhesion to PIIBNP was reduced by 65% (αvβ3), 50% (αvβ5), 90% (αvβ3 plus αvβ5), and 50% (αv) in the presence of equivalent concentrations of integrin αv-containing antibodies. In contrast, antibody to integrin β1 did not inhibit cell adhesion to PIIBNP, confirming that β1 integrin is not involved in the cell adhesion to PIIBNP.
Cell Adhesion to PIIBNP Requires Integrin αv
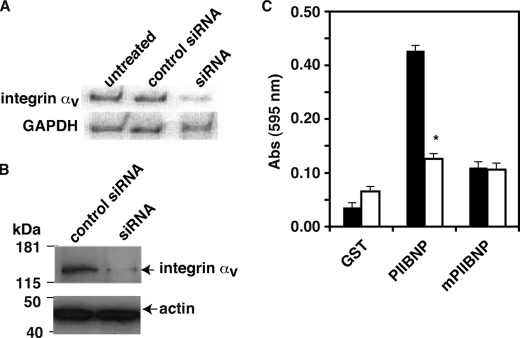
If integrins αvβ3 and αvβ5 are the receptors for PIIBNP, suppression of the common integrin subunit, integrin αv, should reduce cell attachment to PIIBNP. To test this hypothesis, siRNA specific to integrin αv was synthesized, and hCh-1 cells were treated with siRNA prior to cell adhesion assay by the method of Graef et al. (20). As shown in Fig. 3, siRNA suppressed integrin αv expression both at the mRNA (Fig. 3A) and protein (Fig. 3B) levels. Treatment with αv integrin siRNA led to more than 70% reduction of cell adhesion to PIIBNP as compared with unrelated siRNA-treated control (Fig. 3C).
FIGURE 3.
Cell adhesion to PIIBNP is integrin αv-dependent. A, αv integrin siRNA-treated and control siRNA-treated hCh-1 cells were lysed, and mRNA level was detected by a semiquantitative PCR method as described under “Experimental Procedures.” B, siRNA-treated and control siRNA-treated hCh-1 cell membrane proteins were solubilized in cell lysis buffer containing Triton X-100. Cell lysate was resolved by SDS-PAGE and integrin αv expression was analyzed by Western blotting. C, siRNA-treated (white bar) and control siRNA-treated (black bar) hCh-1 cell adhesion to GST, PIIBNP, and mutant PIIBNP were assayed as described above. The value of absorbance at 595 nm represents the number of cells bound to the proteins. Absorbance values for siRNA-treated (white) and control hCh-1 cells (black) are represented as mean ± S.D. (*, p < 0.001 compared with control siRNA-treated cells; n = 8).
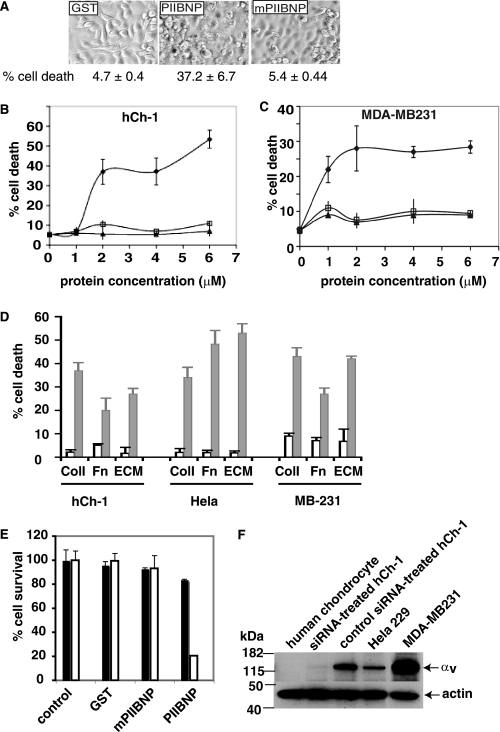
PIIBNP Induces Cell Death in Tumor Cells
In order to begin to define functions for the integrin binding of PIIBNP, recombinant proteins were added to hCh-1 cells. We found that 4 μm GST-PIIBNP, but not mutant PIIBNP or GST, induced 37% cell death after 90 h of incubation (Fig. 4A). In the presence of PIIBNP, some cells were partially detached from the matrix and many had a granular appearance. Cell death was determined by permeability to trypan blue dye. GST and GST-mutant PIIBNP did not cause cell death and the cells remained flattened on the culture dish. PIIBNP-induced hCh-1 cell death was dose-dependent (Fig. 4B). To determine whether PIIBNP can reduce survival of cancer cells other than the chondrosarcoma line, two additional cancer cell lines were used: HeLa cells derived from a cervical carcinoma and MDA-MB231 cells, a breast cancer cell line. PIIBNP induced death of both HeLa and MB231 cells. When PIIBNP was incubated with MDA-MB231 cells for 40 h, the cell numbers were reduced by more than 50% compared with GST control (data not shown); the increase in percentage of cell death was dose-dependent (Fig. 4C) and time-dependent (beginning at 12 h at 3 μm concentration; data not shown). Cell death occurred on a variety of substrate matrices over time, indicating that detachment-mediated cell death (anoikis) did not occur. None of these matrices predominantly uses the αvβ3 and αvβ5 integrins for cell attachment. In other words, whether cultured on fibronectin (which adheres to α3β1, α4β1, and α5β1 integrins), Matrigel ECM (primarily laminin and collagen IV, which adheres to α6β1 and α2β1 integrins), or type I collagen (which adheres to α1β1, α2β1, and α3β1 integrins), PIIBNP was able to induce cell death in the same time period (Fig. 4D).
FIGURE 4.
Induction of tumor cell death by PIIBNP is integrin αv-dependent but substrate-independent. A, 24-well plates were coated with type I collagen. 4 μm GST, PIIBNP, or mutant PIIBNP was incubated with hCh-1 cells in serum-free medium for 3 days. Cells were imaged by light microscopy using a Q Capture Retiga 2000R camera (×20 magnification). The cells were also analyzed by trypan blue exclusion assays as described under “Experimental Procedures,” and the percentage of cell death was calculated and presented. hCh-1 cells (B) and MDA-MB231 cells (C) were incubated with different concentrations of PIIBNP (♦), mutant PIIBNP (□), or GST (▴) for 90 and 41 h. The values represent means ± S.D. for three independent assays. D, hCh-1 cells, HeLa cells, and MB231 cells were plated on type I collagen, fibronectin, or matrigel and incubated with 4 μm GST (white bar) or PIIBNP (black bar) for 90 h (hCh-1), 24 h (HeLa), or 41 h (MB-231). Cell viability was analyzed as described above. The values represent means ± S.D. for three independent assays. E, the control siRNA-treated and anti-integrin αv siRNA-treated hCh-1 cells were plated in 24-well plates previously coated with type I collagen and incubated without or with 4 μm GST, PIIBNP, or mutant PIIBNP in a serum-free medium for 90 h. PIIBNP reduced only control siRNA-treated cell number (white bar) and not the anti-siRNA integrin αv-treated cell number (black bar). F, the integrin αv expression levels were analyzed by Western blotting for hCh-1, anti-integrin αv siRNA-treated hCh-1, control siRNA-treated hCh-1, HeLa, MDA-MB231, and human chondrocytes.
Cell Death Requires Integrin αv
Based on these studies, we hypothesized that PIIBNP adhesion to the cell surface integrins αvβ3 and αvβ5 was a prerequisite for induction of cell death. To test this hypothesis, we treated hCh-1 cells with inhibitor siRNA specific to integrin αv prior to cell viability assay. Cell viability assays indicated that PIIBNP induced death of control siRNA hCh-1 cells but not in anti-αv siRNA-treated hCh-1 cells that no longer expressed integrin αv (Fig. 4E). These results strongly suggested that the PIIBNP-induced cell death requires the RGD sequence and the cellular integrin αvβ3 or αvβ5. To substantiate the correlation of PIIBNP-induced cell death with integrin expression, integrin αv expression in different cell lines was probed by Western blotting (Fig. 4F). Cell viability assays showed that PIIBNP induced cell death of HeLa cells that express primarily αvβ5 integrin (25) and MDA-MB231 breast cancer cells that express both αvβ3 and αvβ5 integrin (25) (Fig. 4D). The normal human chondrocytes did not express αv (Fig. 4F).
Normal Chondrocytes Have Reduced or Undetectable Receptor Integrins for PIIBNP and Thus May Be Protected
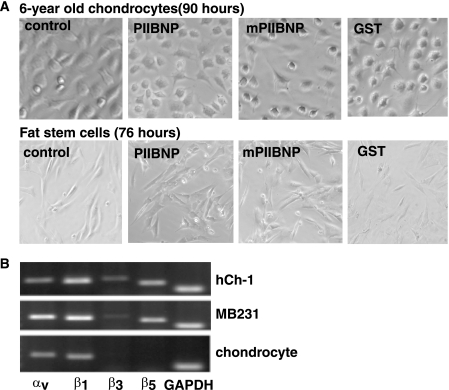
If PIIBNP is able to kill cells, how are chondrocytes protected when they synthesize large amounts of type IIB procollagen? We first demonstrated that normal chondrocytes were not killed by PIIBNP (Fig. 5A). Chondrocytes are shown from a 6-year-old normal human donor (obtained from Dr. Davis Atkisson, ISTO Technologies, St. Louis, MO). Cell viability assays indicated that no trypan blue staining cells were detected in most of the wells when treated with PIIBNP, mPIIBNP, or GST for 90 h. We have also tested chondrocytes from a 12-year-old donor and articular chondrocytes from two young horses (provided by Dr. Lisa Fortier (Cornell University Veterinary School)). Other cell types tested and not killed were human adipose stem cells (Fig. 5A), the murine chondroprogenitor cell line ATDC5, mouse osteoblasts, bone marrow macrophages, the RAW264.7 cell line, and the MC3T3 osteoblast cell line (data not shown). Susceptibility to cell death by PIIBNP correlates with the expression of the integrin receptors, as shown in reverse transcription-PCR analysis of integrin gene expression by some of the cells used in this study (Fig. 5B). In the sample of normal young chondrocytes shown here, there is expression of αv but much less expression of β3 and β5 than in cancer cells. In the young human chondrocytes shown in Fig. 4F, there was no detection of integrin αv.
FIGURE 5.
PIIBNP does not induce death of normal cells with reduced expression of integrin αvβ3 and αvβ5. A, 6-year-old chondrocytes and human fat stem cells were cultured in a 24-well plate and incubated with 4 μm PIIBNP in serum-free medium for 90 h (chondrocytes) and 76 h (fat stem cells). Cells were imaged by light microscopy using a Q Capture Retiga 2000R camera (×20 magnification), and cell viability was confirmed by a trypan blue exclusion assay. B, total RNA was isolated from hCh-1 cells, MDA-MB231 cells, and 6-year-old chondrocytes. Reverse transcription-PCR was performed as described elsewhere (53). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
PIIBNP, but Not Integrin αv, Is Expressed in Developing Cartilage
As mentioned previously and shown in Fig. 5A, normal young human chondrocytes are not killed by PIIBNP. In order to further explore the reason why normal chondrocytes are protected from PIIBNP, we investigated expression of αv in developing cartilage by immunohistochemistry. αv is present in very few tissues in the mouse embryo and is not present in developing cartilage at E14.5 (Fig. 6, A and B) and E17.5 (Fig. 6, C and D). The developing cartilage is easily detected by the high expression of type II collagen (Fig. 6A). The αv expression is confined to the synovial cells lining the developing synovial joint cavity and expressed in the hypertrophic cartilage immediately upon the increase in cell volume (Fig. 6, B and D). Consequently, as cartilage is established, the integrin receptors for PIIBNP, αvβ3, and αvβ5 are not available on chondrocytes for binding. The expression of αv in the hypertrophic cartilage may be related to events involving bone formation, the change in collagen expression from type II to type X, calcification, and invasion by osteoclasts and endothelial cells, and it may be related to the eventual death of the cells. However, this hypothesis is speculative at this time.
FIGURE 6.
PIIBNP, but not integrin αv, is present in developing cartilage. Immunohistochemistry for the type II collagen protein (A and C) and the integrin αv (B and D) in mouse embryonic E14.5 (A and B) humerus and E17.5 (C and D) digits. The type II collagen protein can be detected in the entire humerus or digit, including the developing cartilage (c) and the newly formed hypertrophic regions (h) (the region surrounding the nascent bone (b) in the digits). Detection of the integrin αv is primarily in the synovial cells lining the developing joint cavity (indicated by arrows) and in the hypertrophic region and appears to be excluded from the developing cartilage. Bar (A–G), 200 mm. E, Western blot detection of the PIIBNP protein in normal 56-day embryonic human limb using the chicken anti-exon 3–8 antiserum. Marker molecular weights are indicated. The control lane is recombinant GST-PIIBNP cleaved with thrombin showing the molecular mass of the PIIBNP to be ∼15 kDa. In the last three lanes, increasing concentrations of limb extract are shown with bands reacting with PIIBNP antibody at 16.1 kDa. The band at 16.1 is equivalent to the recombinant PIIBNP that migrates more slowly due to the hydroxylation of proline residues.
In order to reasonably hypothesize a function for the PIIBNP in cartilage, we sought to determine whether the NH2-propeptide, PIIBNP, could be detected in cartilage. Experiments on the biosynthesis of type IIB procollagen strongly suggest that the NH2-propeptide is removed prior to deposition of the fibril in the ECM (1). In extracts from developing cartilage, a protein that reacts with the antibody to PIIBNP was detected at approximately the size of a monomeric NH2-propeptide (Fig. 6E). In this experiment, limbs from E14.5 mice and day 56 and day 57 embryonic human tissues were briefly extracted and subjected to SDS-PAGE. The recombinant PIIBNP cleaved with thrombin was run in the second lane, showing the migration of the PIIBNP produced by bacteria. The third through fifth lanes show extracts from human developing limbs reacted with antibody to PIIBNP (exons 3–8). The band that migrates at 16.1-kDa is the monomer of PIIBNP, migrating slightly slower than the recombinant form due to hydroxylation of proline residues. Taken together, these results demonstrate 1) the presence of cleaved PIIBNP in the tissue, 2) the high expression of type IIB procollagen in developing cartilage, and 3) expression of αv in synovial cells and hypertrophic cartilage but not in chondrocytes of normal developing endochondral bone.
PIIBNP Induces Cell Necrosis in Tumor Cells
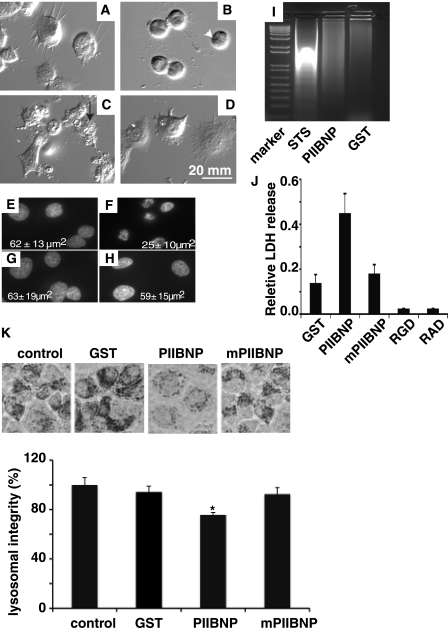
Attempts to demonstrate apoptosis in hCh-1 cells by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling, activated caspase 3 staining, or flow cytometry were unsuccessful. In order to define a mechanism under which the PIIBNP induces hCh-1 cell death, we performed cell morphology analysis using differential interference contrast microscopy. The antitumor agent staurosporine was used to induce apoptosis for comparison. After 12 h of treatment, microscopic examination revealed that GST- and mutant PIIBNP-treated hCh-1 cells have a rounded shape with normal cell characteristics such as lamellipodia extensions and smooth surface (Fig. 7, A and D). In contrast, the treatment with either staurosporine or PIIBNP induced significant morphological alterations of the cellular surface and nucleus. Cells treated with staurosporine for 16 h exhibited rounding, shrinkage, and blebbing of the plasma membrane (Fig. 7B), typical features for cells undergoing an apoptotic process. Cells treated with PIIBNP for 60 h showed features characteristic of necrosis rather than apoptosis, such as a loss of plasma membrane integrity (non-intact plasma membrane), with a jagged discontinuous appearance (Fig. 7C). Nuclear staining with Hoechst dye showed that the nuclear integrity is maintained in PIIBNP-treated cells (Fig. 7E), the same as the GST (Fig. 7G) and mutant PIIBNP (Fig. 7H), but lost in staurosporine-treated cells (Fig. 7F). Quantitation of the average size of the nuclei demonstrated that only the staurosporine-treated cells had smaller nuclei (25 μm2 compared with 61 μm2) (Fig. 7, E–H). DNA laddering assays showed that DNA degradation occurred in staurosporine-treated cells, but not in PIIBNP-treated cells (Fig. 7I). These experiments indicate that apoptosis is not the mechanism of cell death in hCh-1 cells.
FIGURE 7.
PIIBNP induces death of tumor cell lines through a necrosis pathway. hCh-1 cells were plated on coverslips in a 6-well plate previously coated with type I collagen. The cells were incubated with 4 μm GST (A) for 60 h, staurosporine (STS) (B) for 16 h, PIIBNP (C) for 60 h, or mPIIBNP (D) for 60 h. The cells were imaged by a differential interference contrast light microscopy using a Q Capture Retiga 2000R camera (×40 magnification). The cell membrane integrity is lost in PIIBNP-treated cells (arrow, white) but not in staurosporine-treated cells (arrowhead, black). Cell nuclei from cells treated with GST (E), staurosporine (F), PIIBNP (G), and mPIIBNP (H) were stained with Hoechst and photographed by light microscopy using a Q Capture Retiga 2000R camera (×60 magnification). The area of the nucleus was measured using Northern Eclipse software. Nuclear area values in the panels are represented as mean ± S.D. (n = 30). The DNA laddering assays (I) indicated that the DNA integrity was lost only in cells treated with staurosporine (16 h) and not in cells treated with GST, PIIBNP, or mPIIBNP (90 h). Lactate dehydrogenase release assays (J) indicated that the relative LDH release occurred primarily in cells treated with PIIBNP (10 μm, 90 h) and much less in cells treated with GST (10 μm, 90 h), mPIIBNP (10 μm, 90 h), RGD (1 mm, 90 h), or RAD (1 mm, 90 h) synthetic peptides. K, neutral red lysosomal retention assay. The top panel shows neutral red retained in hCh-1 cells. Neutral red retention was quantified by absorption at 540 nm after extraction with 0.5 m HCl, 50% ethanol. The percentage of lysosomal integrity was calculated from the absorption values.
As positive tests for necrosis, we assayed LDH release from the cells and the concentration of lysosomes. Only in the PIIBNP-treated cells was the LDH increased (Fig. 7J). LDH was not released when the apoptosis was induced by treatment with RGD peptides, shown by us and others to induce cell death, probably by a cell detachment-mediated mechanism (26, 27). Programmed necrosis is characterized by a number of cell events caused by calcium permeability, calpain activation leading to lysosomal disruption among other events. We assayed for the number of lysosomes in both hCh-1 cells and MDA-MB231 cells by the method of Kennedy et al. (23). In the presence of 5 μm PIIBNP, lysosomes in MDA-MB231 were reduced by ∼25% (Fig. 7K), and the lysosomes in hCh-1 were reduced by 20% (data not shown). These results suggest that PIIBNP induces cell death via a programmed necrosis pathway rather than an apoptosis pathway.
PIIBNP is Anti-invasive
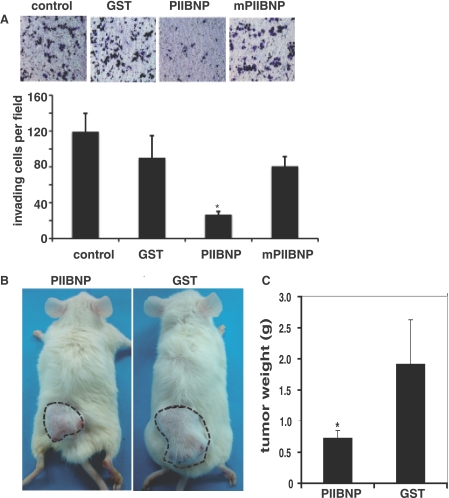
Finally, in order to determine whether there is potential for a therapeutic application of PIIBNP, we tested the effect of PIIBNP on tumor cell invasion using a modified Boyden chamber technique with Matrigel-coated membranes (22). In this assay, tumor cells must overcome a reconstructed basement membrane by a sequential process of proteolytic digestion of the substrate and active migration. The results are shown in Fig. 8A; PIIBNP reduced the number of MDA-MB231 breast cancer cells crossing the membrane by 60%, whereas the mutant PIIBNP was equivalent to GST alone.
FIGURE 8.
PIIBNP inhibits MDA-MB231 breast cancer tumor invasion in vitro and growth in NOD/SCID mice. A, invasion assay. MB231 breast cancer cells were allowed to invade three-dimensional matrigel-coated membranes in the presence of 5 μm GST, PIIBNP, or mPIIBNP for 30 h prior to fixation and quantification of invasion. Toluidine blue-stained cells are shown in the top panel. In the bottom panel, data are expressed as mean numbers of invading cells per high power field (20×) ± S.D. from a minimum of 12 fields/condition. Statistical significance of means was determined using Student's t test (*, p < 0.005 relative to mPIIBNP or GST control). B, in vivo tumor assay. When the tumors grew to 0.5 cm in diameter, the mice were divided into two groups with three mice in each. Mice received 6 nmol of PIIBNP or GST/mouse/day by subcutaneous injection into the tumor for 2 weeks. This experiment has been repeated with identical results. Dashed lines, circumference of the tumor. C, when the largest tumor size in the control group reached to 2 cm in diameter, the mice were sacrificed, and tumors were removed. The tumor weights were represented as mean ± S.D. (*, p < 0.01 as compared with GST control, n = 6).
PIIBNP Suppresses Tumor Growth in Mice
To test the effect of PIIBNP in vivo, a well accepted tumor model was established in nude mice. In this model, MDA-MB231 breast cancer cells are injected into the dorsal surface of NOD/SCID mice, where they form solid tumors (28). In toxicology studies prior to the tumor experiment, we found that within 2 months, 6 nmol of recombinant proteins did not have any toxic effect on NOD/SCID animals (data not shown). Two sets of experiments have been performed with PIIBNP, both with three animals in each of two groups for a total of 12 animals: six treated with GST-PIIBNP and six treated with GST alone. When MDA-MB231 cells were injected into the dorsal surface of the mice, tumors measuring 0.5 cm in diameter were formed within 3 days. Mice were divided into two groups and injected in the same site daily with 6 nmol of GST-PIIBNP or GST for 12 consecutive days as per the established model. As indicated in Fig. 8B, PIIBNP effectively suppressed tumor growth. The mice were sacrificed when tumors on mice in the control group grew to 2 cm in diameter. All tumors were retrieved and tumor weights are presented in Fig. 8C. The tumor size was reduced by greater than 75% in all animals treated with PIIBNP compared with GST control.
DISCUSSION
Type IIB collagen is the predominant collagen in hyaline cartilage and the endochondral bone growth plate, comprising 50% of the protein within the ECM. Type IIB collagen is synthesized and secreted via the endoplasmic reticulum, Golgi, and secretory granules (14). The COOH- and NH2-propeptides are removed prior to deposition of the fibrillar domain in the ECM, but the exact location of their removal is not known. The COOH-propeptides are thought to be removed by the astacin proteinase, BMP-1 (29), whereas the NH2-propeptide is removed by an NH2-proteinase now known as ADAMTS-3 (30). The function of the fibrillar portion of the collagen in the ECM is to provide structural integrity to the tissue; however, there are no known functions for the NH2- or COOH-propeptides. The COOH-propeptide, also named chondrocalcin, has been isolated from cartilaginous growth plates, and it has been suggested that it is involved in mineralization of the tissue (31). Here we demonstrate that type II NH2-propeptide is unique among the fibrillar collagen NH2-propeptides in containing vicinal RGD motifs encoded by exon 6. We show that type IIB procollagen NH2-propeptide, PIIBNP, mediates cell attachment via integrins αvβ3 and αvβ5 via RGD, reduces invasion, and induces cell death in tumor cell lines that express these integrins, human chondrosarcoma (hCh-1), breast cancer (MDA-MB2310), and cervical cancer (HeLa). In vivo, PIIBNP inhibits breast cancer tumor growth in a model of breast cancer. These results strongly suggest that the integrin αvβ3 and αvβ5 are receptors for PIIBNP and that ligation of PIIBNP conducts a death signal.
Prechondrogenic mesenchyme and many other embryonic tissues synthesize the other splice form of type II collagen, IIA. The NH2-propeptide of IIA propeptide is not removed during matrix assembly, being deposited as the pN-procollagen (14). When the recombinant type IIA NH2-propeptide was tested for the integrin binding and cell killing, it was able to adhere to cell surface but did not kill cells.3
Integrins play important physiological roles in endochondral bone development. For example, integrin α2β1 influences proliferation, differentiation, and apoptosis of hypertrophic chondrocytes (32). The natural presence of PIIBNP in developing cartilage and specific binding of the PIIBNP to αvβ3 and αvβ5 integrins suggests specific functions for this integrin signaling. These integrins are characteristic of endothelial cells (33), osteoclasts (34), synovial cells (35), and many tumor cells (36) and have been used to target anti-angiogenic and anti-tumor agents (37, 38). In this light, our initial choice of the human chondrosarcoma cell line, hCh-1, for these investigations proved to be critical; these cells have high levels of expression of αvβ3 and αvβ5 integrins, thus allowing for the identification of the integrins that bind to PIIBNP. Although adult articular chondrocytes have been reported to express αvβ3 and αvβ5 (39), the expression level is very low. In young human growth plate (7–15 years), integrins αvβ3 and αvβ5 are not expressed in developing cartilage. Rather, it has been reported that integrin αvβ3 was found only in osteoclasts and integrin αvβ5 only in the hypertrophic zone (40). These results agree well with our finding that αvβ3 and αvβ5 integrins were not detected in normal young primary chondrocytes used here (Fig. 4F), and integrin αv is undetectable in developing mouse cartilage of long bones (Fig. 3B).
The ability of PIIBNP to induce death of cells with high expression of αVβ3 and αVβ5 integrins may be crucial to protect normal articular chondrocytes and prevent vascular and osteoclast invasion in developing cartilage. Normal young chondrocytes from humans and horses were not killed. Using αv antibody, we show that embryonic chondrocytes do not express αV during chondrogenesis or as mature chondrocytes but do express αV upon hypertrophy at the time of the switch in collagen synthesis from type II to type X collagen. We have made a preliminary report that PIIBNP can inhibit angiogenesis in vivo and endothelial tube formation in vitro (41) and have shown that PIIBNP can also kill osteoclasts via the αvβ3 integrin but not osteoblasts.4 Consequently, the data strongly suggest that the normal function of the PIIBNP is targeted to cells such as endothelial, osteoclast, and synovial cells rather than the chondrocytes. By virtue of the pattern of type II collagen synthesis (highest during cartilage formation) and the ability of PIIBNP to eliminate endothelial cells and osteoclasts from the developing cartilage, we speculate that PIIBNP is an excellent candidate for contributing to the molecular mechanism by which growing cartilage remains avascular and intact.
Other fragments of extracellular matrix proteins have been shown to induce cell death, particularly when released from a larger protein moiety. For example, canstatin, the noncollagenous domain of collagen type IV α-chains, functions as an angiogenic inhibitor. Canstatin triggers tumor cell death through a cross-talk with integrin αvβ3 and αvβ5; cell death is through an apoptotic mechanism (42).
PIIBNP binds to cell surface integrins αvβ3 and αvβ5 but apparently, in hCh-1 cells, does not induce apoptosis. For hCh-1 cells, we did not see positive evidence of apoptosis but did see positive morphological and biochemical evidence of necrosis. In other words, the cells exhibited membrane disruption, the medium was positive for cytosolic enzyme activity as measured by LDH (43), and there was a reduction in lysosomes. Classic necrosis is characterized as a rapid loss of plasmid membrane integrity, organelle swelling, and mitochondrial dysfunctions and the lack of typical apoptotic features such as DNA cleavage and nuclear condensation. The cell death falls into the category of necrosis as judged by LDH leakage, lysosomal integrity loss, and the lack of DNA cleavage and nuclear condensation, but it takes more than 40 h to induce the cell death, indicating that there is a regulated mechanism behind the PIIBNP-induced cell death.
The other fibrillar collagens, types I, III, and V, are also synthesized with NH2- and COOH-propeptides that are removed prior to deposition of fibrils in the ECM. Function has been attributed to the liberated type I NH2-propeptide, which has been suggested to be involved in feedback inhibition of collagen synthesis (44) and, based on the similarity to type IIA procollagen NH2-propeptide, has recently been shown to bind to BMP2 and transforming growth factor-β1 and to act intracellularly (45). This mechanism has nothing to do with integrin binding but functions via the von Willebrand factor C domain of type I and type IIA collagens.
In our study, we have used recombinant GST-PIIBNP to study its function in detail, although a histidine-tagged PIIBNP also acts similarly (data not shown). It is likely that this fusion protein is similar to that found in the tissue in terms of the availability of RGD sequences to bind to the cell surface because we detected monomeric PIIBNP in both human and mouse developing cartilage (Fig. 5E). The NH2-propeptide does not have any glycosylated residues but potentially has four hydroxyproline residues near the COOH terminus. The lysine cross-link is located COOH-terminal to the ADAMTS-3 cleavage site. Our results indicate that monomeric NH2-propeptide can be identified in cartilage. Because there are two RGD sequences next to each other, it is likely that they would be available for binding, particularly because they are near an interruption in the Gly-X-Y sequence and embedded in sequence that would not be tightly wound in a collagen helix (see Fig. 1). It is likely that the PIIBNP would be free in the ECM and available for signaling for a number of reasons. First, there is one NH2-propeptide removed from every procollagen molecule synthesized; consequently, during periods of high collagen synthesis, particularly during development of cartilage and the cartilaginous growth plate, large amounts of NH2-propeptide will be produced. Second, for the type I NH2-propeptide, there is evidence that it is present in the ECM because it has been isolated from bone, where it is synthesized in abundance (46). Last, we show evidence for the presence of PIIBNP in the tissue by biochemical analysis. It appears that the predominant form of PIIBNP in the tissue is the monomeric form. This was true for developing cartilage from both human and mouse embryonic limbs, where a high level of type IIB procollagen synthesis occurs. The presence of monomers in the matrix indicates that our recombinant protein reflects the structure of the NH2-propeptide after cleavage from the type IIB procollagen molecule. There are no disulfide bridges in the type IIB procollagen NH2-propeptide, and the lysine cross-link located in the telopeptide remains on the mature collagen fibril (17). Consequently, this result makes it highly likely that the RGDs of the NH2-propeptide, in vivo, will be available for function in the developing endochondral bone. In support of the function in vivo, new preliminary data from co-culture of hCh-1 cells with embryonic cartilage showed the same tumor cell killing effect as the recombinant PIIBNP (data not shown), further suggesting that recombinant PIIBNP is functionally similar to PIIBNP in vivo.
Certain implications arise from this work. First, cells that are not welcome in cartilage (i.e. tumor cells, endothelial cells, and osteoclasts) express αvβ3 and αvβ5 integrins. Consequently, the type II collagen NH2-propeptide, when liberated from the procollagen, may be one of the protein fragments that establish cartilage anlagen and cartilage growth plates as avascular. Second, because many tumor cells express αvβ3 and αvβ5 integrins and our experiments show that PIIBNP induces chondrosarcoma, HeLa cervical carcinoma, and MDA-MB231 breast cancer cell death, PIIBNP could be a useful therapeutic agent in the treatment of tumors, especially for these that lack effective adjuvant chemotherapy, such as chondrosarcoma and triple negative breast cancers. Chondrosarcoma is the third most common primary malignancy of bone after myeloma and osteosarcoma. The tumors are resistant to commonly used radiation and chemotherapy; therefore, wide surgical excision is currently the best available treatment for intermediate to high grade tumors. Therefore, PIIBNP may be a potential antitumor reagent for chondrosarcoma.
Since the 1970s, investigators have sought the mechanism by which cartilage remains free of tumors and blood vessels (47, 48). Candidates have been identified that have some anti-tumor or anti-angiogenic properties, such as the 25-kDa NC1 domain of chondromodulin-1 (49), TIMP-2 (50), troponin-1 (51), and perlecan (52). Many of these anti-tumor agents are also anti-angiogenic and thus inhibit tumor growth. We propose that PIIBNP would be a good candidate for one of the anti-tumor agents in cartilage for the following reasons: 1) it is naturally produced at high levels during organogenesis of cartilage, and 2) it kills specific cell types that would invade and degrade cartilage. Last, PIIBNP is active in vivo in a breast cancer model system, demonstrating potential for harnessing its activity as a therapeutic agent.
Supplementary Material
Acknowledgments
We thank Dr. Katherine Weilbaecher (Department of Medicine and Co-leader of the Bone Metastasis Working Group) and Dr. William Frazier (Department of Biochemistry) for generous gifts of MDA-MB231 breast cancer cells and HeLa cells and Drs. Matthew Ellis (Co-leader of the Translational and Clinical Research Program of the Siteman Cancer Center) and Katherine Weilbaecher for help with the xenograft tumor model.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 AR R01 036994. This work was also supported by the Siteman Cancer Center Research Development Award.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
L. J. Sandell, Z. Wang, and J. Bryan, unpublished data.
S. Hayashi, Z. Wang, J. Bryan, C. Kobayashi, R. Faccio, and L. Sandell, submitted for publication.
- ECM
- extracellular matrix
- GST
- glutathione S-transferase
- PIIBNP
- NH2-propeptide of type IIB procollagen
- mPIIBNP
- PIIBNP with two RGD motifs mutated to RAD
- siRNA
- small inhibitory RNA
- LDH
- lactate dehydrogenase
- PBS
- phosphate-buffered saline
- En
- embryonic day n.
REFERENCES
- 1.Prockop D. J., Sieron A. L., Li S. W. (1998) Matrix Biol. 16, 399–408 [DOI] [PubMed] [Google Scholar]
- 2.Cancedda R., Descalzi Cancedda F., Castagnola P. (1995) Int. Rev. Cytol. 159, 265–358 [DOI] [PubMed] [Google Scholar]
- 3.Kronenberg H. M. (2003) Nature 423, 332–336 [DOI] [PubMed] [Google Scholar]
- 4.Hynes R. O. (1992) Cell 69, 11–25 [DOI] [PubMed] [Google Scholar]
- 5.Hérard A. L., Pierrot D., Hinnrasky J., Kaplan H., Sheppard D., Puchelle E., Zahm J. M. (1996) Am. J. Physiol. 271, L726–L733 [DOI] [PubMed] [Google Scholar]
- 6.McBrien N. A., Metlapally R., Jobling A. I., Gentle A. (2006) Invest. Ophthalmol. Vis. Sci. 47, 4674–4682 [DOI] [PubMed] [Google Scholar]
- 7.Odrljin T. M., Haidaris C. G., Lerner N. B., Simpson-Haidaris P. J. (2001) Am. J. Respir. Cell Mol. Biol. 24, 12–21 [DOI] [PubMed] [Google Scholar]
- 8.Pande P., Mosleh T. A., Aust A. E. (2006) Toxicol. Appl. Pharmacol. 210, 70–77 [DOI] [PubMed] [Google Scholar]
- 9.Panetti T. S., Wilcox S. A., Horzempa C., McKeown-Longo P. J. (1995) J. Biol. Chem. 270, 18593–18597 [DOI] [PubMed] [Google Scholar]
- 10.Sandell L. J., Morris N., Robbins J. R., Goldring M. B. (1991) J. Cell Biol. 114, 1307–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandell L. J., Nalin A. M., Reife R. A. (1994) Dev. Dyn. 199, 129–140 [DOI] [PubMed] [Google Scholar]
- 12.Nah H. D., Swoboda B., Birk D. E., Kirsch T. (2001) Dev. Dyn. 220, 307–322 [DOI] [PubMed] [Google Scholar]
- 13.Cheah K. S., Lau E. T., Au P. K., Tam P. P. (1991) Development 111, 945–953 [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y., Oganesian A., Keene D. R., Sandell L. J. (1999) J. Cell Biol. 144, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patra D., Xing X., Davies S., Bryan J., Franz C., Hunziker E. B., Sandell L. J. (2007) J. Cell Biol. 179, 687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oganesian A., Zhu Y., Sandell L. J. (1997) J. Histochem. Cytochem. 45, 1469–1480 [DOI] [PubMed] [Google Scholar]
- 17.Fukui N., McAlinden A., Zhu Y., Crouch E., Broekelmann T. J., Mecham R. P., Sandell L. J. (2002) J. Biol. Chem. 277, 2193–2201 [DOI] [PubMed] [Google Scholar]
- 18.Chansky H., Robbins J. R., Cha S., Raskind W. H., Conrad E. U., Sandell L. J. (1998) J. Orthop. Res. 16, 521–530 [DOI] [PubMed] [Google Scholar]
- 19.Leavesley D. I., Stanley J. M., Faull R. J. (1999) Nephrol. Dial Transplant. 14, 1208–1216 [DOI] [PubMed] [Google Scholar]
- 20.Graef T., Steidl U., Nedbal W., Rohr U., Fenk R., Haas R., Kronenwett R. (2005) Angiogenesis 8, 361–372 [DOI] [PubMed] [Google Scholar]
- 21.Wang Z., Ramanadham S., Ma Z. A., Bao S., Mancuso D. J., Gross R. W., Turk J. (2005) J. Biol. Chem. 280, 6840–6849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valster A., Tran N. L., Nakada M., Berens M. E., Chan A. Y., Symons M. (2005) Methods 37, 208–215 [DOI] [PubMed] [Google Scholar]
- 23.Kennedy C. L., Smith D. J., Lyras D., Chakravorty A., Rood J. I. (2009) PLoS Pathog. 5, e1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubo T., Shimose S., Matsuo T., Tanaka K., Yasunaga Y., Sakai A., Ochi M. (2006) J. Orthop. Res. 24, 1138–1144 [DOI] [PubMed] [Google Scholar]
- 25.Borgne-Sanchez A., Dupont S., Langonné A., Baux L., Lecoeur H., Chauvier D., Lassalle M., Déas O., Brière J. J., Brabant M., Roux P., Péchoux C., Briand J. P., Hoebeke J., Deniaud A., Brenner C., Rustin P., Edelman L., Rebouillat D., Jacotot E. (2007) Cell Death Differ. 14, 422–435 [DOI] [PubMed] [Google Scholar]
- 26.Buckley C. D., Pilling D., Henriquez N. V., Parsonage G., Threlfall K., Scheel-Toellner D., Simmons D. L., Akbar A. N., Lord J. M., Salmon M. (1999) Nature 397, 534–539 [DOI] [PubMed] [Google Scholar]
- 27.Kim S. Y., Oh H. K., Ha J. M., Ahn H. Y., Shin J. C., Baek S. H., Lim S. C., Joe Y. A. (2007) Arch. Biochem. Biophys. 459, 40–49 [DOI] [PubMed] [Google Scholar]
- 28.Hirbe A. C., Uluçkan O., Morgan E. A., Eagleton M. C., Prior J. L., Piwnica-Worms D., Trinkaus K., Apicelli A., Weilbaecher K. (2007) Blood 109, 3424–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartigan N., Garrigue-Antar L., Kadler K. E. (2003) J. Biol. Chem. 278, 18045–18049 [DOI] [PubMed] [Google Scholar]
- 30.Fernandes R. J., Hirohata S., Engle J. M., Colige A., Cohn D. H., Eyre D. R., Apte S. S. (2001) J. Biol. Chem. 276, 31502–31509 [DOI] [PubMed] [Google Scholar]
- 31.Van der Rest M., Rosenberg L. C., Olsen B. R., Poole A. R. (1986) Biochem. J. 237, 923–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirsch M. S., Lunsford L. E., Trinkaus-Randall V., Svoboda K. K. (1997) Dev. Dyn. 210, 249–263 [DOI] [PubMed] [Google Scholar]
- 33.Cheresh D. A., Stupack D. G. (2002) Nat. Med. 8, 193–194 [DOI] [PubMed] [Google Scholar]
- 34.Teitelbaum S. L. (2000) J. Bone Miner. Metab. 18, 344–349 [DOI] [PubMed] [Google Scholar]
- 35.Nikkari L., Haapasalmi K., Aho H., Torvinen A., Sheppard D., Larjava H., Heino J. (1995) J. Rheumatol. 22, 16–23 [PubMed] [Google Scholar]
- 36.Mizejewski G. J. (1999) Proc. Soc. Exp. Biol. Med. 222, 124–138 [DOI] [PubMed] [Google Scholar]
- 37.McCarty M. F., Block K. I. (2006) Integr. Cancer Ther. 5, 252–268 [DOI] [PubMed] [Google Scholar]
- 38.McCarty M. F., Block K. I. (2006) Integr. Cancer Ther. 5, 150–171 [DOI] [PubMed] [Google Scholar]
- 39.Salter D. M., Hughes D. E., Simpson R., Gardner D. L. (1992) Br. J. Rheumatol. 31, 231–234 [DOI] [PubMed] [Google Scholar]
- 40.Häusler G., Helmreich M., Marlovits S., Egerbacher M. (2002) Calcif. Tissue Int. 71, 212–218 [DOI] [PubMed] [Google Scholar]
- 41.Sandell L. J., Wang Z., Franz C., Bryan J., Siegel A., Mecham R., Wagensail J., Ell B., Rapraeger A. (2008) FASEB J. 22, 1–418166582 [Google Scholar]
- 42.Magnon C., Galaup A., Mullan B., Rouffiac V., Bouquet C., Bidart J. M., Griscelli F., Opolon P., Perricaudet M. (2005) Cancer Res. 65, 4353–4361 [DOI] [PubMed] [Google Scholar]
- 43.Kroemer G., Galluzzi L., Vandenabeele P., Abrams J., Alnemri E. S., Baehrecke E. H., Blagosklonny M. V., El-Deiry W. S., Golstein P., Green D. R., Hengartner M., Knight R. A., Kumar S., Lipton S. A., Malorni W., Nuñez G., Peter M. E., Tschopp J., Yuan J., Piacentini M., Zhivotovsky B., Melino G. (2009) Cell Death Differ. 16, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bornstein P. (2002) Matrix Biol. 21, 217–226 [DOI] [PubMed] [Google Scholar]
- 45.Oganesian A., Au S., Horst J. A., Holzhausen L. C., Macy A. J., Pace J. M., Bornstein P. (2006) J. Biol. Chem. 281, 38507–38518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisher L. W., Robey P. G., Tuross N., Otsuka A. S., Tepen D. A., Esch F. S., Shimasaki S., Termine J. D. (1987) J. Biol. Chem. 262, 13457–13463 [PubMed] [Google Scholar]
- 47.Brem H., Folkman J. (1975) J. Exp. Med. 141, 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langer R., Brem H., Falterman K., Klein M., Folkman J. (1976) Science 193, 70–72 [DOI] [PubMed] [Google Scholar]
- 49.Kusafuka K., Hiraki Y., Shukunami C., Kayano T., Takemura T. (2002) Acta Histochem. 104, 167–175 [DOI] [PubMed] [Google Scholar]
- 50.Baker E. A., Stephenson T. J., Reed M. W., Brown N. J. (2002) Mol. Pathol. 55, 300–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moses M. A., Wiederschain D., Wu I., Fernandez C. A., Ghazizadeh V., Lane W. S., Flynn E., Sytkowski A., Tao T., Langer R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2645–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mongiat M., Sweeney S. M., San Antonio J. D., Fu J., Iozzo R. V. (2003) J. Biol. Chem. 278, 4238–4249 [DOI] [PubMed] [Google Scholar]
- 53.Choudhuri T., Verma S. C., Lan K., Robertson E. S. (2006) Virology 351, 58–72 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.