Abstract
Alternative cleavage and polyadenylation generate multiple transcript variants of mRNA isoforms with different length of 3′-untranslated region (UTR). Alternative cleavage and polyadenylation enable differential post-transcriptional regulation of transcripts via the availability of different cis-acting elements in 3′-UTRs. Microphthalmia-associated transcription factor (MITF) is a master regulator of melanocyte development and melanogenesis. It has also been implicated in melanoma development. Here we show that melanoma cells favor the expression of MITF mRNA with shorter 3′-UTR. This isoform of mRNA is regulated by microRNA, miR-340. miR-340 interacts with two of its target sites on the 3′-UTR of MITF mRNA, causing mRNA degradation and decreased expression and activity of MITF. On the other hand, the RNA-binding protein coding region determinant-binding protein, shown to be highly expressed in melanoma, directly binds to the 3′-UTR of MITF mRNA and prevents the binding of miR-340 to its target sites, resulting in stabilization of the MITF transcript and elevated expression and transcriptional activity of MITF. This interplay between RNA-binding protein and miRNA describes the important mechanism of regulation of MITF in melanocytes and malignant melanomas.
Keywords: MicroRNA, RNA Abundance, RNA-binding Protein, RNA Turnover, Skin, Alternative Cleavage and Polyadenylation, CRD-BP, IGF2BP1, MITF, Melanoma
Introduction
Microphthalmia-associated transcription factor (MITF)2 is a basic helix-loop-helix leucine zipper dimeric transcription factor belonging to the MYC superfamily of proteins (1). MITF forms dimers and binds to specific sequence motifs present in the promoter of its target genes to activate their transcription. Various studies have documented the role of MITF in the induction of genes required for melanin formation as well as normal melanocyte development. It regulates the transcription of three major pigmentation enzymes, tyrosinase (TYR), tyrosine related protein-1 (TYRP-1), and dopachrome-tautomerase (DCT) (2–4). MITF is also an amplified oncogene in a subset of melanomas and has been shown to regulate a distinct set of target genes that in turn are responsible for neoplasia-related phenotype in these types of cancers. Primarily, this action of MITF in melanoma is mediated by increasing cell proliferation and triggering cell cycle progression. T-Box transcription factor 2 (TBX-2) is one such target that suppresses senescence via down-regulation of p21 (5, 6). CDK2 is another target, the regulation of which modulates cell cycle progression required for melanoma clonogenic growth (7). MITF has also been shown to inhibit apoptosis and thus promote oncogenesis by increasing the expression of its direct target, the anti-apoptotic factor BCL2 (8). Furthermore, MITF contributes to the metastasis of melanoma through transcriptional activation of the c-MET proto-oncogene (9). Therefore, understanding the mechanisms controlling the expression and function of MITF is extremely important, because it will identify key components for melanocyte development, and it may uncover novel targets for the treatment and/or prevention of melanomas.
The regulation of expression of MITF has been shown to occur at multiple levels. The transcriptional regulation of MITF expression has been shown to take place through MITF-M promoter by the Wnt signaling pathway (10–13), the cAMP-cAMP-responsive element-binding protein pathway (14), and also PAX3 (15). There is also some evidence of post-translational regulation of MITF that affects the availability of the functional protein. Mitogen-activated protein kinase signaling pathway components phosphorylate Ser73 in MITF protein, resulting in its ubiquitination and subsequent degradation (16, 17). On the other hand, phosphorylation of the Ser298 residue in MITF by glycogen synthase kinase 3β is believed to increase the DNA binding activity of MITF (18). However, apart from transcriptional and post-translational regulation, another important mode of regulating gene expression is by manipulating the stability of the mature mRNA. Recent reports indicated that MITF is a target of several microRNAs (miRNAs), including the miR-96/183/182 cluster (19) and miR-137 (20), suggesting the importance of a post-transcriptional regulatory step.
miRNAs are small ∼22-nucleotide noncoding RNAs known to act as an important class of gene regulators. They are present abundantly in plant and animal genome, and after being transcribed and processed, the final forms of the mature miRNAs bind to their target sites on the mRNAs present mostly in the 3′-UTR but also were identified in the coding region (21). Several protein factors have been shown to participate in this process toward the formation of the miRNA-induced silencing complex, ultimately leading either to the translational repression or to the degradation of the mRNA.
Alternative cleavage and polyadenylation generate multiple transcript variants of a particular gene with mRNA isoforms having 3′-UTRs of different lengths depending on the position of the alternative cleavage and polyadenylation signals. The exclusion of large parts of the 3′-UTR allows those mRNA isoforms to escape regulation by miRNAs. Recently, it has been observed that in fast proliferating cells, mRNA isoforms with shorter 3′-UTRs are more favored. This preference helps the oncogenes to avoid regulation that would otherwise interfere with the cell cycle progression leading to cellular transformation (22, 23). There are several different isoforms of MITF-M mRNA with varying lengths of 3′-UTR (19, 20). In this study, we investigated the expression pattern of three such mRNA isoforms in melanoma cell lines and normal human melanocytes, finding that MITF mRNAs with short 3′-UTRs are more abundant in melanoma cell lines. We identified that the mRNA isoform with the short 3′-UTR also undergoes miRNA-mediated regulation, and the miR-340 destabilizes the mRNA of MITF in melanoma cell lines. Furthermore, we set out to decipher the role of a RNA-binding protein, coding region determinant-binding protein (CRD-BP, IMP-1, IGF2BP1) in the regulation of MITF. This protein has been shown to attenuate miRNA-dependent degradation of different mRNA (21) and has also been found to be overexpressed in melanoma (24). Our results showed that CRD-BP restricts the action of miR-340 by preventing its access to the mRNA of MITF, thereby proposing a novel mode of regulation of MITF.
EXPERIMENTAL PROCEDURES
Cell Lines, Culture, and Transfection Conditions
Normal human melanocytes (NHM) were maintained in Ham's F-10 medium (Mediatech Inc., Manassas, VA) supplemented with human melanocyte growth supplement (Cascade Biologics Inc., Portland, OR), 5% fetal bovine serum, and PSM solution (1% antibiotic-antimycotic solution containing penicillin, streptomycin, and amphotericin B). 451Lu cell line was maintained in minimal essential medium (Invitrogen) supplemented with 1% sodium pyruvate, 1% nonessential amino acids, 5% fetal bovine serum, and 1% PSM solution. The melanoma cell lines 928, 1011, and 1242 were kindly provided by Dr. Paul Robbins (Center of Cancer Research, NCI, National Institutes of Health, Bethesda, MD). The cells were maintained in Dulbecco's modified Eagle's medium, supplemented with 10% bovine calf serum and 1% PSM solution. The colorectal cancer cell lines HCT116, RKO, and DLD1 and corresponding cell lines with exon 5-disrupted Dicer were kindly provided by Drs. K. Kinzler and B. Vogelstein (Johns Hopkins University School of Medicine) (25). 293T cells were obtained from the ATCC. All of the cell lines were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 1% PSM solution. NHM cells were electroporated using AmaxaTM (Lonza, Switzerland) according to the manufacturer's instructions, and transfections of all other cells were performed using Lipofectamine 2000 reagent, according to manufacturer's recommendations (Invitrogen).
Expression Vectors
Full-length MITF cDNA was purchased from Open Biosystems Inc. (clone identification number 6066096, accession number BC065243, cloned in pCMV-SPORT6) and amplified using Fragment 1 forward and 3′-UTR reverse primers (supplemental Table S1). The PCR product was then cloned in pBIG vector (Clontech) and digested with NotI and SalI and end-filled with Klenow enzyme, and a clone with the right orientation was selected. Three fragments of the MITF mRNA coding region (nucleotides 1–421, 422–841, and 842–1260) as well as full-length cDNA (nucleotides 1–1822) and 3′-UTR (nucleotides 1261–1822) were subcloned into pcDNA3.1. The 3′-UTR of MITF cDNA was PCR-amplified and cloned in pBI-GL (Clontech) just after the stop codon. To obtain the deletion mutants for miR-340 sites based on pBIG or pBI-GL construct, we performed blunt end ligation of PCR products, treated with DpnI as recommended for the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Plasmids for CRD-BP expression or production CRD-BP shRNA were characterized in (26). The pcDNA5-CMV-d2eGFP vector and control sponge-CXCR4 construct were kindly provided by Dr. P. Sharp (MIT, Cambridge, MA). The constructs of sponge-340 and sponge-584c-3p were designed as described (27). Oligonucleotides having seven bulged binding sites for miR-340 and miR-584c-3p, respectively, with 4-nt spacer sequences between them (supplemental Table S1) were annealed, gel-purified, cloned into pcDNA5-CMV-d2eGFP vector, and linearized with XhoI and ApaI.
RNA Isolation and Real Time RT-PCR
Total RNA from cells was isolated using TRI reagent (Molecular Research Center, Cincinnati, OH). For real time RT-PCRs, total RNA was treated with 2 units of DNase I (New England Biolabs). Reverse transcription and quantitative real time PCR were performed using an Advantage RT-for-PCR kit (Clontech) and a Power SYBR Green PCR master mix kit (Applied Biosystems), according to the manufacturer's recommendations. Quantification of the real time PCR was done using an ABI Prism 7000 machine and ABI Prism 7000 SDS software. The sequences of primers used for quantitative real time PCR are presented in supplemental Table S2.
Stem-Loop RT-PCR
The expression of mature miR-183 was detected using a two-step process. Using the total cellular RNA isolated by TRI reagent, first, the stem-loop RT primer for miR-183 (designed according to Ref. 28) was hybridized to the miRNA molecule (incubation at 16 °C for 30 min) and then reverse transcribed for 30 min at 42 °C using an Advantage RT kit (Clontech). After heat inactivating the reverse transcriptase at 95 °C for 5 min, one-tenth of the RT product was used either for end point PCR or for real time PCR with the Power SYBR Green PCR master mix kit (Applied Biosystems) using the miRNA-340 specific forward primer and the universal reverse primer (supplemental Table S2).
mRNA Degradation in Vivo
To investigate the stability of mRNA transcripts, we used the Tet-Off gene expression system (Clontech) as described elsewhere (26). Melanoma or colorectal cell lines or 293T cells were transfected with 4–6 μg of Tet-Off and 2–3 μg of response plasmid pBIG-expressing mRNA transcripts of our interest. Transcription was stopped by adding doxycycline (1 mg/ml) into the media 48 h after co-transfection with other expression vectors. The cells were harvested at different time points after treatment, and the total RNA was extracted as described above. The levels of mRNA were analyzed by real time RT-PCR.
Antibodies and Immunoprecipitation Techniques
Mouse anti-FLAG M2 antibody (Sigma-Aldrich), antibody against β-actin, and MITF (Santa Cruz Biotechnology) as well as secondary antibodies conjugated with horseradish peroxidase (Chemicon) were purchased. The immunoblotting procedures are described elsewhere (29).
Protein Purification
To obtain the whole cell lysate for Western blot analysis, the cells were lysed using denaturing radioimmune precipitation assay buffer containing phosphate-buffered saline, pH 7.4, 0.5% sodium deoxycholate, 0.1% SDS, 1% (v/v) Nonidet P-40, 100 mm sodium orthovanadate, and proteinase inhibitor mixture (Sigma). Protein extracts for UV cross-link reactions were made in nondenaturing lysis buffer (10 mm HEPES, pH 7.6, 3 mm MgCl2, 40 mm KCl, 2 mm dithiothreitol, 5% (v/v) glycerol, 0.5% (v/v) Igepal, and proteinase inhibitor mixture).
Preparation of RNA Substrates for UV Cross-linking Analysis
Plasmids containing full-length MITF mRNA or fragments of its coding region under control of the T7 promoter were linearized with XbaI, purified with phenol-chloroform, and used for in vitro transcription. In vitro transcription was performed with [32P]UTP using a Riboprobe in vitro transcription system (Promega). Radiolabeled RNA substrates were gel-purified before use in UV cross-link reactions.
UV Cross-link Reaction
Analysis of RNA-protein complexes was performed as described before (26). Briefly, protein extracts (20 μg) and internally labeled RNA (1.5–2 × 106 cpm) were incubated in 20 μl of reaction buffer (nondenaturing lysis buffer without protease inhibitors) for 30 min at room temperature. After incubation, RNA-protein complexes were cross-linked by 30 min of exposure to 254-nm UV light, treated with RNase A and RNaseT1, and incubated with anti-FLAG antibodies for 6 h and M2-agarose beads overnight. The immunoprecipitates were washed six times in lysis buffer, boiled, and separated in 10% SDS-PAGE. The gel was dried and exposed to x-ray film for 3–14 days.
Reporter Luciferase Assay
Melanoma cell lines were transfected with the previously described shRNA constructs (26) or miR-Sponge constructs, pSV40 β-galactosidase and pHTRPL4 plasmid (30, 31), or p-BIGL-MITFwt or p-BIGL-MITFδmiR-340 plasmids. 48 h after transfection, luciferase activity was measured using luciferase reporter assay reagent, according to the manufacturer's recommendations (Promega).
Zebrafish Husbandry and Morpholino Knockdown Assays
Adult zebrafish were maintained according to established methods (32). Wild type (AB strain) embryos were obtained from natural mating and staged according to Kimmel et al. (33). The CRD-BP antisense morpholino (MO) designed to block translation of CRD-BP (5′-GAAGTTCAGAAGCAGTCTGTTCATC-3′) was purchased from GeneTools (Philomath, OR). MOs were diluted in 1× Danieau buffer (34) to 10 ng/nl. Standard control MO (GeneTools) was diluted to 2–8 ng/nl. 0.5–1 nl of MO/embryo was injected at the one- to two-cell stage. Embryos at 24 h post-fertilization were lysed for RNA extraction with TRIzol (Invitrogen).
Senescence-associated β-Galactosidase Staining
The cells were fixed with fixative solution (20% formaldehyde, 2% glutaraldehyde) for 20 min, washed with phosphate-buffered saline, and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) solution overnight at 37 °C. The cells stained blue were counted under a microscope (20×), and the percentages of stained cells were estimated.
RESULTS
MITF mRNA with Short 3′-UTR Is Preferentially Expressed in Melanoma Cell Lines
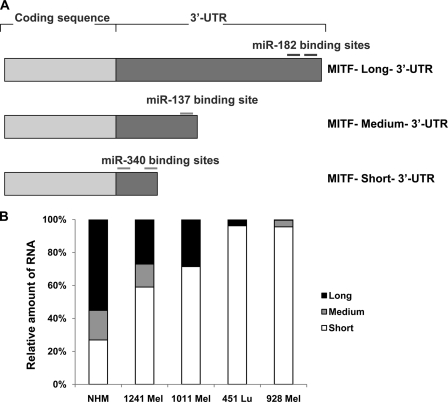
As discussed earlier, several full-length cDNA isoforms of MITF-M were found having the same coding sequence but with 3′-UTRs of varying lengths. Two recent studies that showed regulation of MITF mRNA by two different miRNAs used two different isoforms of MITF mRNA. The study by Bemis et al. (20) showed a functional miR-137-binding site in the 3′-UTR using a full-length cDNA with 1,143 bp of the 3′-UTR sequence (termed hereafter the medium 3′-UTR). Segura et al. (19), on the other hand, used the full-length cDNA with a 3′-UTR length of ∼3 kb (termed the long 3′-UTR) to find a functional miR-182 site (Fig. 1A). We obtained the full-length cDNA clone from Open Biosystems Inc. (clone identification 6066096, accession number BC065243) having a 3′-UTR of ∼0.57 kb in length, and we termed this the mRNA with the short 3′-UTR (Fig. 1A). The phenomenon of the presence of the same mRNA with different 3′-UTRs and also being regulated by different miRNAs led us to investigate the abundance of MITF mRNAs with different 3′-UTRs in different melanoma cell lines. We designed primers unique to the short, medium, and long 3′-UTRs and used them to detect the relative abundance of the individual isoforms by quantitative real time PCR using RNA isolated from four different melanoma cell lines and also from NHMs (Fig. 1B). The results showed that in NHMs the levels of all of the three isoforms were comparable, but in all of the four melanoma cell lines tested, the relative proportion of MITF mRNA with short 3′-UTR was predominating. This suggested that fast proliferating melanoma cell lines preferentially express MITF mRNA with shorter 3′-UTR.
FIGURE 1.
MITF mRNA with short 3′-UTR is more abundant in melanoma cell lines. A, graphical representation of different MITF mRNA isoforms with varying lengths of 3′-UTRs. The approximate positions of the reported miRNAs are shown on long and medium 3′-UTRs. B, real time quantitative PCR with primers specific for long, medium, and short 3′-UTRs (see “Experimental Procedures” for primer sequence) using RNA extracted from NHMs and the indicated melanoma cell lines.
MITF mRNA with Short 3′-UTR Is Also Regulated by miRNA
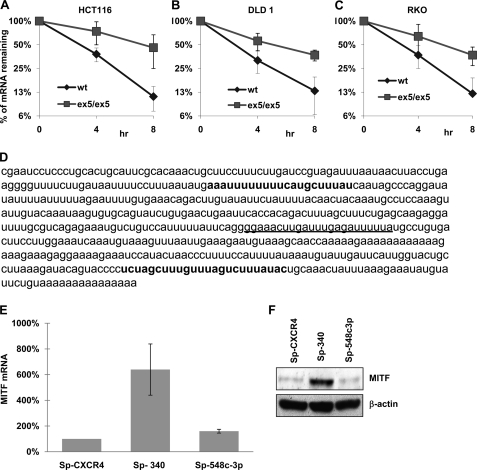
As discussed before, miRNAs have been shown to act on the MITF mRNA with medium and long 3′-UTR. Because we found the mRNA isoform with the short 3′-UTR most prevalent in melanomas, we investigated whether this isoform is also being regulated by miRNAs. We measured the half-life of that mRNA in cells defective in Dicer1 function and thus deficient in miRNA maturation (25). It was observed that the half-life of MITF mRNA with short 3′-UTR was 2–3-fold higher in all three Dicer1Ex5/Ex5 hypomorphic cell lines examined as compared with their normal counterpart where functional miRNAs were present (Fig. 2, A–C). This indicated that MITF with short 3′-UTR is also regulated by miRNAs. We then used a bioinformatic approach (microRNA. org web site) to find out the possible miRNA-binding site in that region. Two miRNAs, miR-340 and miR-548c-3p, were selected for further investigation after short listing of the miRNAs based on their alignment score (Fig. 2D). To inhibit the function of these miRNAs in melanoma cells, we used miR-Sponge constructs as described previously (27) for both of them. Our results showed that the amount of the MITF mRNA isoform with short 3′-UTR increased significantly only when miR-340 function was inhibited (Fig. 2E). Moreover, inhibition of miR-340 function in 451Lu cells also increased the MITF protein level (Fig. 2F). These data demonstrated that MITF mRNA with short 3′-UTR is regulated by miR-340.
FIGURE 2.
The abundant MITF mRNA with short 3′-UTR is also regulated by miRNA. A, Dicerwt and DicerEx5/Ex5 HCT116 cells were co-transfected with Tet-off plasmid and p-BIG-MITF plasmid with short 3′-UTR. Transcription was stopped by treatment with doxycycline for the indicated durations. The stability of MITF transcripts was analyzed by measuring MITF mRNA levels with real time RT-PCR (normalized to GAPDH expression). B, p-BIG-MITF plasmid with short 3′-UTR was expressed in DLD1 cells, Dicerwt (wt), and DicerEx5/Ex5 (ex5/ex5) under the control of the Tet-off system. The stability of MITF mRNA was analyzed as in A. C, stability of MITF transcript with short 3′-UTR expressed in RKOwt and RKO DicerEx5/Ex5 cells was analyzed as in A. All of the results are representative of three separate experiments and are expressed as the mean values ± S.D. (error bars). The average half-lives of mRNAs are presented in supplemental Table S3. D, sequence of the short 3′-UTR of MITF showing binding sites for miR-340 (in bold type) and miR-548c-3p (underlined). E, the levels of endogenous MITF mRNA in 451Lu cells, transfected with the indicated miR-Sponge constructs, were estimated by real time RT-PCR after normalization with respect to GAPDH expression. The results are representative of three separate experiments and are expressed as percentages of control (SP-CXCR4) as the mean values ± S.D. (error bars). F, immunoblot analysis of MITF expression in the 451Lu cells transfected with the indicated miR-Sponge constructs (upper panel). The lower panel shows the expression of β-actin. The levels of endogenous miR-340 are presented in supplemental Fig. S1A.
miR-340 Destabilizes MITF mRNA
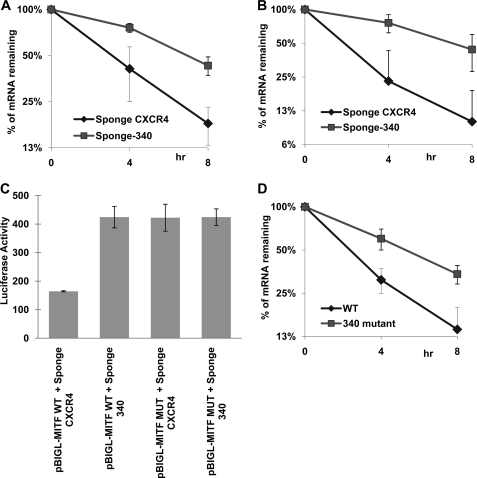
After the initial observations regarding the involvement of miR-340 in the regulation of MITF mRNA, we set out to investigate whether miR-340 regulates the turnover of MITF mRNA. The result showed a 2-fold increase in half-life upon miR-340 inhibition (Fig. 3A), indicating that miR-340 is responsible for destabilization of MITF mRNA. We further tested the efficacy of the miR-340 action using a reporter construct. The short 3′-UTR of MITF mRNA was cloned just after the luciferase coding region in pBI-GL (Clontech). Inhibition of miR-340 function in melanoma cell lines significantly increased the half-life of the chimeric RNA as well as the luciferase enzyme activity (Fig. 3, B and C). This finding suggested that the short 3′-UTR of the MITF mRNA was sufficient for miR-340-mediated inhibition. Next, both of the sites of miR-340 were deleted from the full-length MITF mRNA as well as from the reporter construct to generate the corresponding mutant versions. The half-life of the deleted version of the full-length MITF mRNA was found to be increased significantly (Fig. 3D). The luciferase activity of the mutated chimeric mRNA was also found to be increased ∼2.5-fold, in a manner comparable with the situation when miR-340 function was inhibited. On the other hand, miR-340 inhibition had no additional effect on the luciferase activity of the mutant mRNA (Fig. 3C). Together these data suggested that miR-340 acts through its target sites on 3′-UTR of MITF mRNA, leading to its destabilization.
FIGURE 3.
MITF mRNA is a target of miR-340. A, 451Lu cells were co-transfected with Tet-off plasmid, p-BIG-MITF plasmid with short 3′-UTR, and the indicated miR-Sponge construct. Transcription was stopped by treatment with doxycycline for the indicated durations. The stability of MITF transcripts was analyzed by measuring MITF mRNA levels with real time RT-PCR (normalized to GAPDH expression). B, 451Lu cells were co-transfected with Tet-off plasmid, p-BIGL-MITF-Short MITF 3′-UTR, and the indicated miR-Sponge construct. The turnover of chimeric Luciferase-MITF-3′-UTR transcripts was analyzed as in A. C, 451Lu cells were co-transfected with β-galactosidase-expressing plasmid, Tet-off plasmid, p-BIGL-MITFwt, or p-BIGL-MITFδmiR-340 and the indicated miR-Sponge constructs. After 24 h the luciferase activity was measured. The values represent luciferase activity normalized to β-galactosidase. D, 451Lu cells were co-transfected with Tet-off plasmid, p-BIG-MITFwt, or p-BIG-MITFδmiR-340. The stability of MITF transcripts was analyzed as in A. All of the results are representative of three separate experiments and are expressed as the mean values ± S.D. (error bars). The average half-lives of mRNAs are presented in supplemental Table S3. The levels of endogenous miR-340 are presented in supplemental Fig. S1A.
CRD-BP Stabilizes MITF mRNA
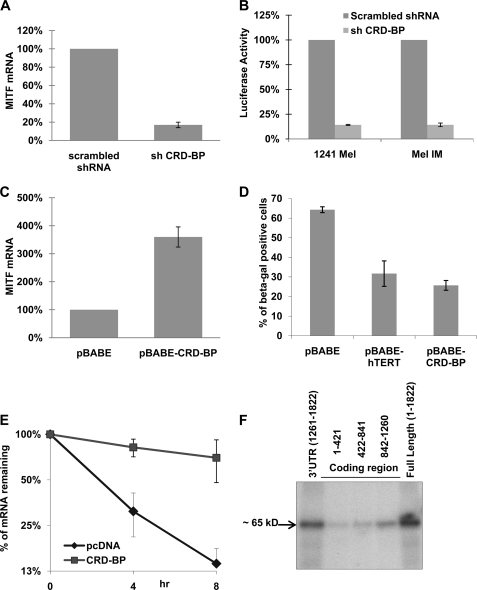
CRD-BP is a multifunctional mRNA-binding protein, modulating the stability, localization, and translation of several RNAs (c-myc, IGF-II, H19, CD4, MDR-1, βTrCP1, etc.) (35–41). As discussed earlier, CRD-BP protects another mRNA from miRNA-mediated destabilization (21). Moreover, CRD-BP has also been shown to be overexpressed in melanoma cell lines and human melanoma samples contributing to oncogenesis (24). Knockdown of CRD-BP in 451Lu cells decreased the level of endogeneous MITF mRNA by more than 5-fold (Fig. 4A). Moreover, MITF-dependent luciferase activity was drastically reduced when CRD-BP was knocked down (Fig. 4B). On the other hand, the levels of endogeneous MITF mRNA were increased 3-fold when CRD-BP was overexpressed in NHMs (Fig. 4C). This overexpression of CRD-BP in NHMs also resulted in significant inhibition of senescence, which is consistent with the function of MITF in the cell cycle progression in melanocytes (Fig. 4D). Interestingly, the effect of CRD-BP on melanocyte senescence was similar to the effect of a powerful senescence inhibitor, hTERT. Together these data indicated that CRD-BP regulates MITF expression as well as its activity. Furthermore, overexpression of CRD-BP dramatically increased the half-life of the MITF mRNA (Fig. 4E). This indicated that MITF expression is regulated post-transcriptionally by CRD-BP. This result corroborates with the nature of how CRD-BP regulates its other target mRNAs. To test whether this regulation is mediated through a direct interaction of CRD-BP protein with MITF mRNA, we performed cross-linking and immunoprecipitation experiments using radiolabeled fragments or full-length MITF mRNA with protein extracts from FLAG-CRD-BP-expressing cells. The result showed that CRD-BP directly interacts with the short 3′-UTR of MITF mRNA (Fig. 4F). These sets of experiments altogether demonstrated that CRD-BP directly interacts with MITF mRNA and stabilizes it. We also measured the expression of the zebrafish ortholog of MITF, mitfa, in embryos when CRD-BP was knocked down using a specific morpholino against CRD-BP. Knockdown of CRD-BP resulted in a dose-dependent decrease in the MITF mRNA level (Fig. 5A), less pigmentation, and a significant reduction in the number of pigment cells (melanophores) in zebrafish embryos (Fig. 5B). These data indicate that the phenomenon of the regulation of MITF expression and activities by CRD-BP is evolutionarily conserved among species.
FIGURE 4.
CRD-BP is a positive regulator of MITF expression. A, 451Lu cells were transfected with either control shRNA or shRNA against CRD-BP. 48 h after transfection, the cells were collected and assayed for levels of MITF mRNA by quantitative RT-PCR, normalized to GAPDH expression, and presented as percentages of control (scrambled shRNA). B, 1241 Mel and Mel IM cells were co-transfected with the indicated shRNA-expressing plasmids, β-galactosidase-expressing plasmid, and pHTRPL4, where the luciferase gene is expressed under the MITF-dependent promoter. The values corresponding to luciferase activity normalized to β-galactosidase expression are presented. C, NHMs were electroporated using AMAXATM with either CRD-BP-overexpressing plasmid or empty vector. 72 h after transfection, the cells were collected, assayed for levels of MITF mRNA by quantitative RT-PCR normalized to GAPDH expression, and presented as percentages of control (pBABE). D, NHM cells were electroporated using AMAXATM with indicated plasmids. 48 h after electroporation, the cells were stained for senescence-associated β-galactosidase, and the percentages of β-galactosidase-positive cells were calculated. E, 451Lu cells were co-transfected with Tet-off plasmid, p-BIG-MITF, and either pcDNA control or CRD-BP-expressing plasmid. The turnover of MITF transcript was analyzed by real time RT-PCR after stopping transcription by doxycycline treatment for the indicated time points. Normalization was done with respect to GAPDH expression. All of the results are representative of three separate experiments and are expressed as the mean values ± S.D. (error bars). The average half-lives of mRNAs are presented in supplemental Table S3. F, FLAG immunoprecipitation of UV cross-linked ribonuclear protein complexes. Protein extracts from 293T cells, transfected with FLAG-CRDBP, were incubated with internally 32P-labeled RNA transcripts of three fragments of the MITF mRNA coding region, MITF full-length mRNA, and the short 3′-UTR. Fragment 1 contains nucleotides 1–421 of the coding region, fragment 2 consists of nucleotides 422–841, and fragment 3 consists of nucleotides 842–1260 of the MITF coding region. Ribonuclear protein complexes were precipitated with anti-FLAG antibodies, analyzed on PAGE, and autoradiographed.
FIGURE 5.
CRD-BP inhibition reduces mitfa expression and pigmentation in zebrafish embryos. A, zebrafish embryos were injected with CRD-BP morpholino (5 or 10 ng) or standard control morpholino. Expression levels of the zebrafish MITF ortholog, mitfa, were assayed by real time quantitative PCR at 24 h post-fertilization and normalized to zebrafish α-tubulin. Morphant expression levels are presented relative to levels of expression in control embryos. B, representative zebrafish embryo at 48 h post-fertilization, injected with control or CRD-BP morpholino. CRD-BP morphants have less pigmentation and reduced numbers of pigment cells (melanophores) in 33 of 33 morphants (from seven independent experiments).
CRD-BP Interferes with the Action of miR-340 Resulting in the Stabilization of MITF mRNA
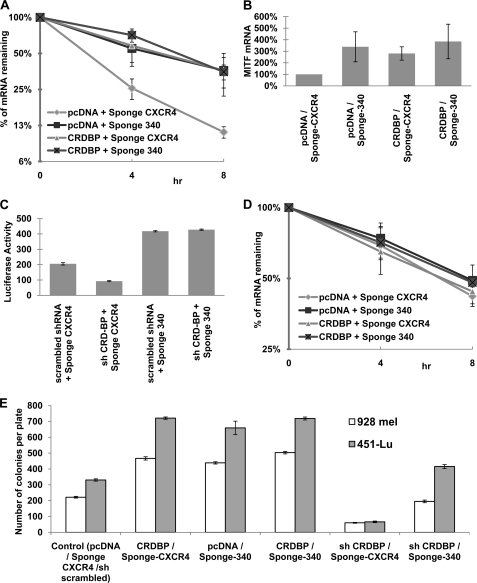
Based on the results that CRD-BP directly interacts with the short 3′-UTR of MITF mRNA, we investigated whether CRD-BP prevents the action of miR-340 on MITF mRNA in a similar fashion as reported earlier for other mRNA (21). Munro et al. (42) have characterized the sequence motif required for the binding of IMP in Drosophila as UUUAY, and we found the presence of that motif either within or juxtaposed to both of the miR-340-binding sites (supplemental Fig. S1B). Moreover, overexpression of CRD-BP increased the half-life of MITF mRNA similar to that of the inhibition of miR-340 function. CRD-BP overexpression, on the other hand, could not increase the half-life further when inhibition of miR-340 function already stabilized the mRNA (Fig. 6A). Similarly, when miR-340 function was inhibited, there was an increase in the endogeneous MITF mRNA level that was not altered further because of CRD-BP overexpression (Fig. 6B). Furthermore, knockdown of CRD-BP decreased MITF-dependent transcription by more than 2-fold, but it had no effect when miR-340 function was already inhibited (Fig. 6C). These data suggested that CRD-BP exerts its effect on MITF mRNA by preventing the access of miR-340 to its sites. When miR-340 action is blocked, the mRNA is stable by itself and becomes unresponsive to the availability of CRD-BP. To further confirm the mechanism of interference of CRD-BP with the miR-340 function, we measured the half-life of the MITF mRNAs with deleted miR-340 sites in the similar experiments. We found that the half-life of the mutated MITF mRNA was increased significantly compared with the wild type MITF mRNA, and overexpression of CRD-BP had no effect on the stability of MITF mRNA that lacks miR-340-binding sites (Fig. 6D). The manifestation that CRD-BP could stabilize MITF mRNA only when miR-340 is functional demonstrates that CRD-BP acts by preventing miR-340 action. To analyze the effect of inhibition of miR-340 on cell proliferation, we have performed colony formation assays and found that inhibition of miR-340 function using specific sponge construct resulted in a significant increase in the number of colonies formed by both 928 mel and 451Lu melanoma cells (Fig. 6E). A similar increase was detected when CRD-BP was overexpressed, but no additive effect on colony formation was observed when CRD-BP and sponge-340 were co-expressed. As expected, knockdown of CRD-BP resulted in drastic inhibition of a number of colonies; however, inhibition of miR-340 rescued the effect of CRD-BP knockdown on the growth of 928 mel and 451Lu melanoma cells (Fig. 6E).
FIGURE 6.
CRD-BP counteracts miR-340 action and stabilizes MITF mRNA. A, wild type MITF-expressing plasmid p-BIG-MITFwt was co-transfected with Tet-off plasmid and the indicated constructs in 451Lu cells. The turnover of MITF transcript was analyzed as Fig. 4E. B, 451Lu cells were transfected with the indicated constructs. 48 h after transfection, the cells were collected and assayed for levels of endogeneous MITF mRNA by quantitative RT-PCR, normalized to GAPDH expression, and presented as percentages of control. C, 451Lu cells were co-transfected with MITF-dependent luciferase-expressing vector pHTRPL4, the indicated shRNA and miR-Sponge-expressing plasmids, and β-galactosidase-expressing plasmid. The values correspond to luciferase activity normalized to β-galactosidase expression. D, construct expressing MITF with both of the miR-340 sites deleted (p-BIG-MITFδmiR-340) was co-transfected with Tet-off plasmid and the indicated constructs in 451Lu cells. The turnover of MITF transcript was analyzed as Fig. 4E. All of the results are representative of three separate experiments and are expressed as the mean values ± S.D. (error bars). The average half-lives of mRNAs are presented in supplemental Table S3. E, 451Lu and 928 Mel cells were co-transfected with indicated constructs and pTk-Puro. The colonies were selected for puromycin resistance, stained with crystal violet, and counted.
DISCUSSION
MITF is widely regarded as the master regulator of melanocyte biology because of its involvement in the regulation of important melanogenic proteins as well as because of its contribution to melanoblast survival, melanocyte lineage commitment, and also melanoma (reviewed in Ref. 43). Therefore, it is the importance of MITF that potentiates the necessity to study its regulatory mechanisms. Post-transcriptional regulation of gene expression involving RNA-binding proteins has been pivotal in the regulation of expression of several genes, and the discovery of miRNAs has added a new dimension to that. MITF is also post-transcriptionally regulated, and two recent studies showed evidence of miR-137 and miR-182 targeting MITF mRNA (19, 20). In the context of this study, we set out to investigate the post-transcriptional regulation of MITF mRNA expression in further detail.
One of the key mechanisms underlying the process of malignant transformation is the activation of oncogenes. The loss of miRNA-binding sites from the 3′-UTRs of the oncogenic mRNAs is considered to be an important mode of oncogene activation (44, 45). This was supported by the recent observations that cancer cells and other proliferating cells tend to express mRNA isoforms with shorter 3′-UTRs to escape from miRNA-mediated regulation (22, 23). MITF mRNA also has several isoforms with different 3′-UTRs, and the most interesting part was that the isoform with short 3′-UTR does not have the binding sites for the previously reported miRNAs (Fig. 1A). We show here that melanoma cells preferentially express the MITF mRNA isoform with short 3′-UTR (Fig. 1B) and comply with other fast proliferating cells in terms of regulation of oncogenic mRNAs. This finding thus designates the MITF mRNA isoform with short 3′-UTR as the most prevalent isoform among melanoma cell lines. Furthermore, our results show that although the MITF mRNA isoform with short 3′-UTR escapes regulation by previously reported miRNAs, it is still being regulated by a different miRNA, miR-340. Interaction of miR-340 with two target sites present on the short 3′-UTR of MITF mRNA results in destabilization of MITF mRNA and inhibition of MITF expression and transcriptional activity (Figs. 2 and 3). Interestingly, miR-340 has been shown to be expressed in primary melanoma cell lines, suggesting its importance in the regulation of melanoma-specific target mRNAs (46). Because the fragment of the MITF mRNA 3′-UTR that contains miR-340-binding sites is a part of all three known 3′-UTRs of MITF, the regulation of MITF by miR-340 appears to be independent of alternative cleavage and polyadenylation and therefore more universal (shared by both melanocytes and melanoma cells).
The results of this study establish CRD-BP as an important positive regulator of MITF expression and function. CRD-BP stabilizes MITF mRNA and increases MITF expression as well as its transcriptional activity (Fig. 4). This effect of CRD-BP is mediated by counteracting the miR-340-mediated degradation of MITF mRNA (Fig. 6). This is not surprising, because we have shown before that CRD-BP also interferes with miR-183 function, resulting in stabilization of βTrCP1 mRNA (21). Our findings here are in line with the recent reports of interplay between RNA-binding proteins and miRNAs, where RNA-binding proteins were shown to interfere with the miRNA function, thereby highlighting a novel mode of post-transcriptional regulation of gene expression (47, 48).
CRD-BP has been shown to be important for the growth and survival of many types of cancer cells (26, 49, 50). It has also been reported to be involved in mammalian development and linked to an effect on cellular adhesion and invasion that takes place during development and malignancy (39, 51). These effects of CRD-BP have been attributed to its regulation of different target mRNAs (i.e. c-myc, βTrCP1, Gli1, etc.). We have also shown previously that CRD-BP is overexpressed in the majority of malignant melanomas (24). However, knockdown of CRD-BP in melanoma cells had a much more robust effect than in other tumor cells, and hence involvement of an additional melanoma specific factor was hypothesized. The data provided in this manuscript suggest that regulation of MITF by CRD-BP may contribute to the observed effects of CRD-BP on melanoma survival and progression and also point to CRD-BP as a potential target for prevention and treatment of this deadly disease.
Supplementary Material
Acknowledgments
We thank Drs. K. Kinzler, P. Robbins, J. Ross, P. Sharp, T. Tuschl, and B. Vogelstein for the generous gifts of reagents and Dr. N. Sanek for help with data collection.
This work was supported, in whole or in part, by National Institutes of Health Grants CA121851 (to V. S. S.), CA12509 (to V. S.), and GM076244 (to Y. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Fig. S1.
- MITF
- microphthalmia-associated transcription factor
- CRD-BP
- coding region determinant-binding protein
- miRNA
- microRNA
- NHM
- normal human melanocyte
- UTR
- untranslated region
- shRNA
- small hairpin RNA
- RT
- reverse transcription
- MO
- morpholino
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Hallsson J. H., Haflidadóttir B. S., Stivers C., Odenwald W., Arnheiter H., Pignoni F., Steingrímsson E. (2004) Genetics 167, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley N. J., Eisen T., Goding C. R. (1994) Mol. Cell Biol. 14, 7996–8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemesath T. J., Steingrímsson E., McGill G., Hansen M. J., Vaught J., Hodgkinson C. A., Arnheiter H., Copeland N. G., Jenkins N. A., Fisher D. E. (1994) Genes Dev. 8, 2770–2780 [DOI] [PubMed] [Google Scholar]
- 4.Yasumoto K., Yokoyama K., Shibata K., Tomita Y., Shibahara S. (1994) Mol. Cell Biol. 14, 8058–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carreira S., Liu B., Goding C. R. (2000) J. Biol. Chem. 275, 21920–21927 [DOI] [PubMed] [Google Scholar]
- 6.Vance K. W., Carreira S., Brosch G., Goding C. R. (2005) Cancer Res. 65, 2260–2268 [DOI] [PubMed] [Google Scholar]
- 7.Du J., Widlund H. R., Horstmann M. A., Ramaswamy S., Ross K., Huber W. E., Nishimura E. K., Golub T. R., Fisher D. E. (2004) Cancer Cell 6, 565–576 [DOI] [PubMed] [Google Scholar]
- 8.McGill G. G., Horstmann M., Widlund H. R., Du J., Motyckova G., Nishimura E. K., Lin Y. L., Ramaswamy S., Avery W., Ding H. F., Jordan S. A., Jackson I. J., Korsmeyer S. J., Golub T. R., Fisher D. E. (2002) Cell 109, 707–718 [DOI] [PubMed] [Google Scholar]
- 9.McGill G. G., Haq R., Nishimura E. K., Fisher D. E. (2006) J. Biol. Chem. 281, 10365–10373 [DOI] [PubMed] [Google Scholar]
- 10.Saito H., Yasumoto K., Takeda K., Takahashi K., Yamamoto H., Shibahara S. (2003) Pigment Cell Res. 16, 261–265 [DOI] [PubMed] [Google Scholar]
- 11.Yasumoto K., Takeda K., Saito H., Watanabe K., Takahashi K., Shibahara S. (2002) EMBO J. 21, 2703–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeda K., Yasumoto K., Takada R., Takada S., Watanabe K., Udono T., Saito H., Takahashi K., Shibahara S. (2000) J. Biol. Chem. 275, 14013–14016 [DOI] [PubMed] [Google Scholar]
- 13.Dorsky R. I., Raible D. W., Moon R. T. (2000) Genes Dev. 14, 158–162 [PMC free article] [PubMed] [Google Scholar]
- 14.Price E. R., Ding H. F., Badalian T., Bhattacharya S., Takemoto C., Yao T. P., Hemesath T. J., Fisher D. E. (1998) J. Biol. Chem. 273, 17983–17986 [DOI] [PubMed] [Google Scholar]
- 15.Watanabe A., Takeda K., Ploplis B., Tachibana M. (1998) Nat. Genet. 18, 283–286 [DOI] [PubMed] [Google Scholar]
- 16.Hemesath T. J., Price E. R., Takemoto C., Badalian T., Fisher D. E. (1998) Nature 391, 298–301 [DOI] [PubMed] [Google Scholar]
- 17.Wu M., Hemesath T. J., Takemoto C. M., Horstmann M. A., Wells A. G., Price E. R., Fisher D. Z., Fisher D. E. (2000) Genes Dev. 14, 301–312 [PMC free article] [PubMed] [Google Scholar]
- 18.Khaled M., Larribere L., Bille K., Aberdam E., Ortonne J. P., Ballotti R., Bertolotto C. (2002) J. Biol. Chem. 277, 33690–33697 [DOI] [PubMed] [Google Scholar]
- 19.Segura M. F., Hanniford D., Menendez S., Reavie L., Zou X., Alvarez-Diaz S., Zakrzewski J., Blochin E., Rose A., Bogunovic D., Polsky D., Wei J., Lee P., Belitskaya-Levy I., Bhardwaj N., Osman I., Hernando E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1814–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bemis L. T., Chen R., Amato C. M., Classen E. H., Robinson S. E., Coffey D. G., Erickson P. F., Shellman Y. G., Robinson W. A. (2008) Cancer Res. 68, 1362–1368 [DOI] [PubMed] [Google Scholar]
- 21.Elcheva I., Goswami S., Noubissi F. K., Spiegelman V. S. (2009) Mol. Cell 35, 240–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayr C., Bartel D. P. (2009) Cell 138, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B. (2008) Science 320, 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elcheva I., Tarapore R. S., Bhatia N., Spiegelman V. S. (2008) Oncogene 27, 5069–5074 [DOI] [PubMed] [Google Scholar]
- 25.Cummins J. M., He Y., Leary R. J., Pagliarini R., Diaz L. A., Jr., Sjoblom T., Barad O., Bentwich Z., Szafranska A. E., Labourier E., Raymond C. K., Roberts B. S., Juhl H., Kinzler K. W., Vogelstein B., Velculescu V. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3687–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noubissi F. K., Elcheva I., Bhatia N., Shakoori A., Ougolkov A., Liu J., Minamoto T., Ross J., Fuchs S. Y., Spiegelman V. S. (2006) Nature 441, 898–901 [DOI] [PubMed] [Google Scholar]
- 27.Ebert M. S., Neilson J. R., Sharp P. A. (2007) Nat. Methods 4, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., Lao K. Q., Livak K. J., Guegler K. J. (2005) Nucleic Acids Res. 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spiegelman V. S., Slaga T. J., Pagano M., Minamoto T., Ronai Z., Fuchs S. Y. (2000) Mol. Cell 5, 877–882 [DOI] [PubMed] [Google Scholar]
- 30.Fang D., Tsuji Y., Setaluri V. (2002) Nucleic Acids Res. 30, 3096–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yasumoto K., Yokoyama K., Takahashi K., Tomita Y., Shibahara S. (1997) J. Biol. Chem. 272, 503–509 [DOI] [PubMed] [Google Scholar]
- 32.Westerfield M. (1995) The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish, University of Oregon Press, Eugene, OR [Google Scholar]
- 33.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995) Dev. Dyn. 203, 253–310 [DOI] [PubMed] [Google Scholar]
- 34.Nasevicius A., Ekker S. C. (2000) Nat. Genet. 26, 216–220 [DOI] [PubMed] [Google Scholar]
- 35.Nielsen J., Christiansen J., Lykke-Andersen J., Johnsen A. H., Wewer U. M., Nielsen F. C. (1999) Mol. Cell Biol. 19, 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen F. C., Nielsen J., Christiansen J. (2001) Scand. J. Clin. Lab. Invest. Suppl. 234, 93–99 [PubMed] [Google Scholar]
- 37.Runge S., Nielsen F. C., Nielsen J., Lykke-Andersen J., Wewer U. M., Christiansen J. (2000) J. Biol. Chem. 275, 29562–29569 [DOI] [PubMed] [Google Scholar]
- 38.Atlas R., Behar L., Elliott E., Ginzburg I. (2004) J. Neurochem. 89, 613–626 [DOI] [PubMed] [Google Scholar]
- 39.Hansen T. V., Hammer N. A., Nielsen J., Madsen M., Dalbaeck C., Wewer U. M., Christiansen J., Nielsen F. C. (2004) Mol. Cell Biol. 24, 4448–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prokipcak R. D., Herrick D. J., Ross J. (1994) J. Biol. Chem. 269, 9261–9269 [PubMed] [Google Scholar]
- 41.Tessier C. R., Doyle G. A., Clark B. A., Pitot H. C., Ross J. (2004) Cancer Res. 64, 209–214 [DOI] [PubMed] [Google Scholar]
- 42.Munro T. P., Kwon S., Schnapp B. J., St Johnston D. (2006) J. Cell Biol. 172, 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levy C., Khaled M., Fisher D. E. (2006) Trends Mol. Med. 12, 406–414 [DOI] [PubMed] [Google Scholar]
- 44.Lee Y. S., Dutta A. (2007) Genes Dev. 21, 1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayr C., Hemann M. T., Bartel D. P. (2007) Science 315, 1576–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mueller D. W., Rehli M., Bosserhoff A. K. (2009) J. Invest. Dermatol. 129, 1740–1751 [DOI] [PubMed] [Google Scholar]
- 47.Kedde M., Agami R. (2008) Cell Cycle 7, 899–903 [DOI] [PubMed] [Google Scholar]
- 48.Kim H. H., Kuwano Y., Srikantan S., Lee E. K., Martindale J. L., Gorospe M. (2009) Genes Dev. 23, 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawakami Y., Kubota N., Ekuni N., Suzuki-Yamamoto T., Kimoto M., Yamashita H., Tsuji H., Yoshimoto T., Jisaka M., Tanaka J., Fujimura H. F., Miwa Y., Takahashi Y. (2009) Biosci. Biotechnol. Biochem. 73, 1811–1817 [DOI] [PubMed] [Google Scholar]
- 50.Noubissi F. K., Goswami S., Sanek N. A., Kawakami K., Minamoto T., Moser A., Grinblat Y., Spiegelman V. S. (2009) Cancer Res. 69, 8572–8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vikesaa J., Hansen T. V., Jønson L., Borup R., Wewer U. M., Christiansen J., Nielsen F. C. (2006) EMBO J. 25, 1456–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.